Abstract
Introduction
Diamond nanoparticles are considered to be one of the most cytocompatible carbon nanomaterials; however, their toxicity varies significantly depending on the analysed cell types. The aim was to investigate the specific sensitivity of endothelial cells to diamond nanoparticles dependent on exposure to nanoparticles.
Methods
Diamond nanoparticles were characterized with Raman spectroscopy, Fourier-transform infrared spectroscopy (FTIR) and dynamic light scattering (DLS). Toxicity of diamond nanoparticles was assessed for endothelial cells (HUVEC), human mammary epithelial cells (HMEC) and HS-5 cell line. The effect of diamond nanoparticles on the level of ROS, NO, NADPH and protein synthesis of angiogenesis-related proteins of endothelial cells was evaluated.
Results and Discussion
Our studies demonstrated severe cell type-specific toxicity of diamond nanoparticles to endothelial cells (HUVEC) depending on nanoparticle surface interaction with cells. Furthermore, we have assessed the effect on cytotoxicity of the bioconjugation of nanoparticles with a peptide containing the RGD motive and a serum protein corona. Our study suggests that the mechanical interaction of diamond nanoparticles with the endothelial cell membranes and the endocytosis of nanoparticles lead to the depletion of NADPH, resulting in an intensive synthesis of ROS and a decrease in the availability of NO. This leads to severe endothelial toxicity and a change in the protein profile, with changes in major angiogenesis-related proteins, including VEGF, bFGF, ANPT2/TIE-2, and MMP, and the production of stress-related proteins, such as IL-6 and IL-8.
Conclusion
We confirmed the presence of a relationship between the toxicity of diamond nanoparticles and the level of cell exposure to nanoparticles and the nanoparticle surface. The results of the study give new insights into the conditioned toxicity of nanomaterials and their use in biomedical applications.
Keywords: diamond nanoparticles, endothelial cells, nanotoxicity, oxidative toxicity
Introduction
Diamond nanoparticles (NDs) are one of the carbon nanomaterials that are increasingly used in industrial products and components as well as in medical products. NDs are used in the biomedical field as biosensors or drug carriers in anti-cancer therapies, regenerative medicine, and imaging diagnostics.1 However, knowledge of ND toxicity is still lacking. The toxic effect of nanoparticles on cells can be assessed at many levels of nanoparticle–cell interactions. The main areas that have been widely analysed over the years are the influence of nanoparticles on the proliferation and viability of various types of cells, the induction of oxidative stress and the production of ROS (reactive oxygen species) in cells, interactions with key organelles (in particular the mitochondria), and the inhibition or stimulation of the secretion of growth factors. The interaction of nanoparticles with cells and tissues is based on direct interaction with the cell membrane and interaction with surface receptors and intracellular proteins after previous endocytosis.2 Therefore, the potential toxicity depends on cell exposure to the nanoparticles, which is higher in standard in vitro 2D cell cultures than in 3D spheroids and in vivo studies.3,4 Moreover, the covalent conjugation of nanoparticles with biomolecules and the often unintentional self-assembly of a protein corona on the surface of nanoparticles decrease the direct contact of nanoparticles with the cell and change the interactions with nanoparticles.5
Interestingly, the toxicity of diamond nanoparticles varies significantly depending on the analysed cell types. Generally, NDs are considered to be one of the most cytocompatible carbon nanomaterials. An analysis of ND toxicity to a variety of cells, including neuroblastomas, macrophages, keratinocytes, and pheochromocytomas, showed no toxicity apart from changes in morphology.6 Our previous reports showed various forms of toxicity of NDs, indicating that NDs reduced proliferation in fast-dividing cells such as glioblastoma cells at longer exposure times but not in fibroblast and hepatocellular cell lines.7–9 There is an increasing body of data showing that ND toxicity is largely dependent on cell type, indicating that NDs can cause severe toxicity to some cell types, including endothelial cells.10,11 Endothelial cells are a unique cell type as they are continuously exposed to the mechanical forces of shear stress, which can lead to a particular sensitivity to nanomaterials that can interact with different cell mechanosensors, including ion channels, the cytoskeleton, and the plasma membrane.12–14 The mechanosensory mechanisms of endothelial cells rapidly react to changes in mechanical force homeostasis, leading to changes in cell physiology.15 Cholesterol-rich membrane microdomains, including caveolae, are important in endothelial cell mechanical stress mechanosensing and mechanotransduction due to activation of various signaling pathways including PI3K-Akt signaling and regulation of endothelial NO synthase (eNOS) and NADPH oxidase (NOX) activity.16
Alterations in mechanical stress lead to various pathologies due to the synthesis of ROS and a decrease in the bioavailability of nitric oxide (NO). Two of the more notable sources of ROS production by mechanical stress are the NOX and eNOS enzymes.16 Endothelial mechanical stress like oscillatory shear stress often leads to an inflammatory response associated with activation of TNFα, IL-8, and IL-6 production. Chronic inflammation of the endothelium leads to dysfunction of the blood vessel through abnormal cell metabolism and increased permeability of the blood vessel.17
A good understanding of ND toxicity to endothelial cells in the context of nanoparticle toxicity is important because endothelial cells are the first barrier that nanoparticles encounter on their way to the target tissue (if the blood vessel itself is not a destination). Here, we hypothesized that endothelial cells show a particular sensitivity to NDs resulting from the NADPH-dependent generation of ROS. Moreover, we assume that endothelial ND toxicity will depend on the exposure of the cells to the surface of the nanomaterials and that this exposure and, consequently, the toxicity differ in different culture methods and after the bioconjugation of the nanoparticles.
Methods
Nanomaterials and Physicochemical Characterization
NDs were purchased from SkySpring Nanomaterials (Houston, USA). The nanopowders were dispersed in ultrapure water to prepare solutions of 1 mg/mL. Immediately after dispersion, the nanopowders were sonicated in a cup horn of a VC 505 sonicator (Sonics & Materials, Newtown, Connecticut, USA) for 2 min, 20% amplitude. NDs were diluted to different concentrations with an appropriate culture medium.
TEM images of nanoparticles were acquired using a JEM-1220 microscope (Jeol, Tokyo, Japan) at 80 kV with a Morada 11-megapixel camera (Olympus Soft Imaging Solutions, Münster, Germany). Samples were prepared by placing droplets of hydrocolloids onto formvar-coated copper grids (Agar Scientific, Stansted, UK) and were air-dried before observations.
Zeta potential measurements were carried out in water and Human Large Vessel Endothelial Cell Basal Medium (Thermo Fisher Scientific, Manassas, USA) with a Nano-ZS90 Zetasizer (Malvern, Worcestershire, UK). After sonication and dilution to a concentration of 20 mg/l in water or cell culture media, NDs were incubated for 24 hours at room temperature. Subsequently, samples were measured after 120 s of stabilization at 25 °C (9 measurements with at least 15 individual replicates) using the Smoluchowski approximation.
The hydrodynamic diameter and polydispersity index of nanoparticles in water and Human Large Vessel Endothelial Cell Basal Medium (Thermo Fisher Scientific) were measured with dynamic light scattering (DLS) using a Nano-ZS90 Zetasizer (Malvern, Worcestershire, UK). As for the zeta potential analysis, after sonication and dilution to a concentration of 20 mg/l in water or cell culture media, NDs were incubated for 24 hours at room temperature. Samples were analysed 9 times with at least 15 individual measurements at 25 °C.
Raman spectra were collected using a Renishaw inVia spectrometer with a 514-nm laser source. The sample was measured as prepared, and the spectra were collected from 5 different spots. Each spot was irradiated for 5 s with a laser power intensity of 0.5 mW. All spectra were very similar, indicating that the sample was homogenous.
FT-IR measurements were performed with a Nicolet iS10 (Thermo Fisher Scientific) spectrometer. Before the sample measurement, the “dry-air” background was recorded, which was subtracted automatically during the registration of the spectra of the investigated samples. The samples were mixed with KBr at a ratio of 1/300 mg and then pressed at 7 MPa cm−2 to form a pellet, and the transmission spectrum was recorded. The spectrum was collected in the range of 400–4000 cm−1.
ND Bioconjugation with an RGD Peptide
ND bioconjugation with a peptide containing the RGD motive was performed with a commonly used method of covalently coupling particles containing the carboxyl group with primary amines in the presence of EDC (1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride) and Sulfo-NHS (N-hydroxysulfosuccinimide).
All reactions were carried out in Protein LoBind tubes (Eppendorf, Hamburg, Germany). ND powder (SkySpring Nanomaterials Inc.) was dispersed in MES buffer (2-[N-morpholino] ethane sulfonic acid) in a concentration of 500 mg/l and sonicated for 45 min. The reaction mixture was prepared by adding 20 µL of 200 mM EDC/MES to 960 µL of ND suspension, followed by mixing and immediately adding 20 µL of 500 mM Sulfo-NHS/MES (Thermo Fisher Scientific). Thereafter, the reaction mixture was incubated for 15 min on an orbital shaker. After the incubation, the unreacted reagent was removed from the sample by centrifuging thrice in an Amicon Ultra-15 3K desalting column (Merck Millipore, Darmstadt, Germany). After the last centrifugation, 900 µL of BupH PBS was transferred to a tube of 1.5 mL.
Subsequently, 100 µL of 2 mg/mL peptide containing the RGD motive (Gly-Arg-Gly-Asp-Ser; Merck Millipore) in BupH PBS was added, and the tube was incubated for 2 h at room temperature on the orbital shaker. The coupling reaction was blocked by the addition of 11 µL of 1 M glycine (Merck Millipore) in PBS, followed by for a further 15 min of incubation. The conjugate was purified by centrifuging thrice in an Amicon Ultra-15 3K column; 4 mL of deionized water was added to the columns between individual spins. After the final centrifugation, the conjugate suspension was filled with water to 960 µL and stored at 4 °C for further use (the final concentration of NDs in the conjugate was 500 mg/l).
Cell Lines
Human umbilical vein endothelial cells (HUVEC) and human mammary epithelial cells (HMEC) were obtained from Thermo Fisher Scientific, whereas HS-5 cells were obtained from ATCC (LGC Standards, Łomianki, Poland). HUVEC cells were maintained in Human Large Vessel Endothelial Cell Basal Medium or Human Large Vessel Endothelial Cell Basal Medium without phenol red supplemented with low-serum Large Vessel Endothelial Supplement (LVES) (Thermo Fisher Scientific) and antibiotics and antimycotics (Thermo Fisher Scientific), whereas HMEC cells were maintained in HuMEC Basal Serum-Free Medium supplemented with HuMEC Supplement Kit (Thermo Fisher Scientific) and the antibiotic and antimycotic penicillin/streptomycin (Thermo Fisher Scientific). HS-5 cells were maintained in Human Fibroblast Expansion Basal Medium (Medium 106) supplemented with Low-Serum Growth Supplement (LSGS) (Thermo Fisher Scientific) and antibiotics and antimycotics (Thermo Fisher Scientific). Cells were maintained at 37 °C in a humidified atmosphere of 5% CO2 and 95% air.
Cell Proliferation Assay
Cell proliferation was evaluated using the LDH-based CyQUANT Cell Proliferation Assay (Thermo Fisher Scientific). HUVEC, HS-5, and HMEC cells were plated on 96-well plates (5 × 103 cells/well) and incubated for 24 h. New medium containing 10% dH2O (control) or 10% ND hydrocolloid was introduced to the cells at concentrations of 5, 20, 50, and 100 mg/l. Cells were incubated for 48 h and subsequently lysed for 45 min at 37 °C with a lysis buffer. The total amount of LDH (depending on the number of cells) was analysed after incubation with the reaction mixture at room temperature for 30 min. After the addition of a stop solution, absorbance was analysed at 490 nm and 680 nm (reference) using a Tecan Infinite 200 microplate reader (Tecan, Durham, USA). Cell proliferation was expressed as a relative value after subtracting the absorbance from the blank samples. The experiment was performed three times using six replicates for each group.
The experiment in which the effect of the presence of 10% FBS in the culture fluid and the effect of the attachment of the peptide containing the RGD motive to NDs on the reduction of proliferation by nanoparticles was performed in the same manner as described. The study was conducted with NDs with a final concentration of 5 and 50 mg/l. Cell proliferation was expressed as a relative value after subtracting the absorbance from the blank samples. The experiment was performed three times using four replicates for each group.
Membrane Perforation Analysis
Cell membrane perforation was evaluated using the LDH-based CyQUANT Cell Proliferation Assay (Thermo Fisher Scientific). HUVEC, HS-5, and HMEC cells were plated on 96-well plates (5 × 103 cells/well) and incubated for 24 h. New medium containing 10% dH2O (control) or 10% ND hydrocolloid was introduced to the cells at a concentration of 5, 20, 50, and 100 mg/l. After 24 h, the 96-well plates were centrifuged (200 × g; 6 min), and 50 μL of cell medium from each well was transferred to a new 96-well plate. The reaction mixture was subsequently added to each well and incubated for 30 min. The amount of LDH released from the cells was analysed by the addition of a stop solution, and the absorbance was read at 490 nm, with 680 nm used as a reference (Tecan Infinite 200 microplate reader, Tecan). Cell membrane perforation was expressed as a relative value after subtracting the absorbance from the blank samples. The experiment was performed three times using six replicates for each group. Prior to the actual analysis of membrane perforation, the potential interference of NDs with the assay was assessed. The interference test was performed in exactly the same way as the actual analysis but without the presence of cells. The results of the interference tests are presented in Figure 1SA.
HUVEC Metabolic Activity Analysis
Analysis of HUVEC metabolic activity was performed using the Prestoblue assay (Thermo Fisher Scientific). Cells were plated on 96-well plates (5 × 103 cells/well) and incubated for 24 h. New medium containing 10% dH2O (control) or 10% ND hydrocolloid was introduced to the cells at a concentration of 5, 10, 20, 50, and 100 mg/l. After 24 h, fresh medium with 10% Prestoblue reagent was added to each well and incubated for 2 h. Subsequently, the intensity of cell metabolic activity was determined by measuring the fluorescence intensity at an excitation wavelength of 560 nm and an emission wavelength of 590 nm (Tecan Infinite 200 microplate reader, Tecan). Cell metabolic activity was expressed in fluorescence units after subtracting the absorbance from the blank samples. The experiment was performed three times using six replicates for each group. Prior to the actual analysis of cell metabolic activity, the potential interference of NDs with the Prestoblue assay (Thermo Fisher Scientific) was assessed. The interference test was performed in exactly the same way as the actual analysis but without the presence of cells. The results of the interference tests are presented in Figure 1SB.
The analysis of the dependence of ND toxicity on HUVEC cell density was performed similarly to that described above using the Prestoblue test. However, HUVEC cells were seeded on the 96-well plate at a density of 5 × 103, 1 × 104, 1.5 × 104, and 2 × 104 cells/well. After 24-h incubation, the cells were treated with NDs at a concentration of 0.4 ng/cell (the final ND concentration was 20, 40, 60, and 80 mg/L in 100 µL medium, respectively). After 24 h, fresh medium with 10% Prestoblue reagent was added to each well and incubated for 2 h. Subsequently, the fluorescence intensity was measured at an excitation wavelength of 560 nm and an emission wavelength of 590 nm (Tecan Infinite 200 microplate reader, Tecan). Cell metabolic activity was expressed in fluorescence units after subtracting the absorbance from the blank samples. The relative toxicity of HUVEC was expressed as the ratio of the metabolic activity value of ND-treated cells to the metabolic activity of untreated cells. The calculations were performed for each of the HUVEC densities separately. The experiment was performed three times using six replicates for each group.
HUVEC Life/Dead Viability Assay
For the Life/Dead viability assay and the morphology analysis, HUVEC cells were plated on an ibidiTreat µ-Slide VI 0.4 (Ibidi GmbH, Germany) at a density of 1.5 × 104 cells/well. After 24 h, the medium was replaced with fresh medium with 10% ND suspension to obtain a final concentration of 50 mg/l, or 10% dH2O for the control. Cells were incubated with NDs for 24 h. Subsequently, the cells were stained with a mix of NucRed Live 647 reagent (Thermo Fisher Scientific) and NucGreen Dead 488 reagent (Thermo Fisher Scientific). The cells were imaged using an FV1000 confocal microscope (Olympus Corporation, Japan) with a temperature/CO2 incubation system (Solent Scientific). The experiment was performed three times.
HUVEC Spheroid Culture and Analysis
HUVEC spheroids were obtained by plating 1 × 104 cells on a 96-well plate covered with 50 μL of 1% agarose/PBS. After 24 h of incubation, spheroid formation was inspected using an optical microscope. As in previous experiments, the spheroids were treated with NDs at a concentration of 5, 10, 20, 50, and 100 mg/l. After 24 h of incubation, the spheroids were imaged for size analysis and were analysed for cell perforation using the LDH assay described earlier. Spheroid size analysis was performed using FIJI software.18 The experiments were performed three times using six replicates for each group.
HUVEC Time-Lapse
HUVEC cells were seeded at a density of 1 × 104 cells/well in a Nunc Lab-Tek 8-well chamber slide (Thermo Fisher Scientific) and maintained in an incubator for 24 h. Subsequently, the cell medium was changed for fresh Human Large Vessel Endothelial Cell Basal Medium without phenol red supplemented with 25 mM HEPES (Thermo Fisher Scientific). The slide was transferred to a preheated (37 °C) incubation system (Solent Scientific, UK) containing a chamber with 5% CO2/air (Pecon, Germany) on an FV1000 confocal microscope with a motorized stage (Olympus Corporation, Japan). A 10-fold concentrated ND suspension was added to the wells to obtain a final concentration of 20 mg/l, and the time-lapse imaging started immediately thereafter. Imaging lasted for 12 h with 5-min intervals. The experiment was performed four times.
NADPH Level
The intercellular level of NADPH was assessed using the High Sensitivity NADPH Assay Kit (Merck Millipore). For detection, HUVEC cells were cultured until 80% confluency in flasks of 75 cm2. The cells were treated with NDs at a concentration of 20 and 50 mg/l for 4 h. Subsequently, the cells were lysed on ice using a supplied extraction buffer, followed by 1 min of sonication (5-s intervals, 20% amplitude) on ice in Protein LoBind tubes (Eppendorf) using a VC 505 sonicator equipped with a cup horn (Sonics & Materials). Subsequently, the protein extracts were centrifuged for 5 min at 10,000 × g at 4 °C, and the supernatant was transferred to a new tube. Samples were incubated at 60 °C for 30 min to decompose NADP. Fifty microlitres of each experimental protein extract in duplicate and NADPH standard solutions were transferred to a black 96-well plate with a clear bottom (Ibidi GmbH), followed by the addition of the reaction mixture and incubation for 60 min. Subsequently, the fluorescence was measured using a Tecan Infinite 200 microplate reader (Tecan) at an excitation wavelength of 535 nm and an emission wavelength of 587 nm. The values of the blank samples were used to correct the background by subtracting the sample and standard values. A standard curve was prepared by plotting the values obtained from the NADPH standards of 2, 4, 6, and 10 pmol/well, and this was used for the calculation of the NADPH amounts in the samples expressed as fmol per μg of sample total protein. The experiment was performed two times with 4 individual replicates.
Detection of Reactive Oxygen and Nitrogen Species
The evaluation of general reactive oxygen species (ROS) induction after ND treatment was assessed using the general oxidative stress indicator CM-H2DCFDA (Thermo Fisher Scientific), the mitochondrial superoxide indicator MitoSox Red (Thermo Fisher Scientific), and the nitric oxide (NO) synthesis level indicator DAF-FM (Thermo Fisher Scientific). HUVEC cells were seeded at a density of 2.25 × 104 cells/well on an ibiTreat 96-well µ-Plate (a plate with square wells and a flat and clear bottom; Ibidi GmbH) in 250 µL of medium per well. After 24 h, the medium was replaced with fresh medium that was supplemented with 10% ND suspension to obtain a final concentration of 20 mg/l and 50 mg/l, or 10% dH2O for the control. After 4 h of incubation with ND medium, the cells were washed twice with PBS/Ca2+ (Thermo Fisher Scientific). For ROS detection, 300 µL of PBS/Ca2+ with CM-H2DCFDA at a final concentration of 1 µM was added to each well. After 20 min of incubation at 37 °C, the cells were washed, and the nuclei were stained with NucRed Live 647 (Thermo Fisher Scientific) for 20 min in full medium. For mitochondrial superoxide detection, 300 µL of PBS/Ca2+ with MitoSOX Red at a final concentration of 5 μM was added to each well and incubated for 10 min at 37 °C. For NO detection, 300 µL of PBS/Ca2+ with DAF-FM at a final concentration of 2.5 µM was added to each well. After 20 min of incubation at 37 °C, the cells were washed, and the nuclei were stained with NucRed Live 647 (Thermo Fisher Scientific) for 20 min in full medium.
Subsequently, the cells stained with CM-H2DCFDA, DAF-FM, or MitoSox Red were washed with fresh medium and were analysed with a confocal microscope (FV-1000) with an incubation system at 37° C and a chamber with 5% CO2/air. HUVEC cells were imaged sequentially using a 20× objective at excitation and emission wavelengths of 495/525 nm (CM-H2DCFDA), 510/580 nm (MitoSox Red), 495/515 nm (DAF-FM), or 642/661 nm (NucRed Live 647). All images for one type of stain were taken using exactly the same laser parameters. The experiment was carried out two times with seven replicates, and each well was imaged in four fields of view. The ROS and NO levels were expressed as a sum of the pixel values per cell. Analysis and cell counting were performed using FIJI software.18 The cell number on each image was assessed by the auto-thresholding of the channel with the stained cell nuclei, followed by watershed segmentation and analysis of particle function (>70 px).
Antibody Array Analysis
For antibody array analysis, HUVEC cells were cultured in culture bottles of 75 cm2. At 70% confluency, the cells were treated with NDs at a concentration of 20 mg/l and incubated for 24 h, followed by a phosphate-buffered saline (PBS) wash. Cells not treated with nanoparticles were used as the control. Whole-cell protein extracts were prepared by suspending cells in an ice-cold radioimmunoprecipitation assay buffer (RIPA) containing protease and phosphatase inhibitors (Thermo Fisher Scientific), followed by 1 min of sonication (5-s intervals, 20% amplitude) on ice in Protein LoBind tubes (Eppendorf) using a VC 505 sonicator equipped with a cup horn (Sonics & Materials). Subsequently, the protein extracts were centrifuged for 30 min at 14,000 × g at 4 °C, and the supernatant was transferred to a new tube. The protein extracts from each group obtained from two separate experiments were mixed in equal proportions, and the protein concentration was determined using a Bicinchoninic Acid Kit (Merck Millipore).
Analysis of cytokine synthesis was performed using an antibody array (ab193655; Abcam, Cambridge, UK). The assay was performed in accordance with the manufacturer’s instructions using lysates containing 100 mg/l of total protein per membrane. Membranes were visualized using the ChemiDoc Imaging System (Bio-Rad, Hercules, USA). The raw image of the membranes after visualization is shown in Figure 2S. Protein quantification was performed using FIJI software.18 Before the analysis, the background subtraction function was used. The experiment was performed using cell extracts pooled from two separate experiments. The full array map is available in Tables S1 and S2).
Tube Formation Assay
For the tube formation assay, HUVEC cells were seeded at a density of approximately 2.5 × 104 cells/cm2 on a 24-well plate coated with a Geltrex Matrix (Thermo Fisher Scientific). The cells were incubated for 1 h at 37 °C in a humidified atmosphere of 5% CO2 and 95% air and treated with NDs at a final concentration of 20 mg/l by the gentle addition of 10% final medium (volume) of ND hydrocolloid or dH2O for the control. The cells were further incubated for 12 h in a humidified atmosphere of 5% CO2 and 95% air. Subsequently, the cells were stained for 10 min at 37 °C with 5 µg/mL Wheat Germ Agglutinin, Alexa Fluor 488 Conjugate, in PBS. Tubes were carefully washed with PBS and visualized in full medium without phenol red using a confocal microscope (FV-1000) with an opened pinhole at excitation and emission wavelengths of 495/519 nm. The number of junctions, the total tube length, and the number of meshes were analysed with FIJI software18 and the Angiogenesis Analyzer toolset.19 The experiment was performed two times with four individual replicates.
Results
Physicochemical Characterization of Diamond Nanoparticles
The physicochemical properties of NDs were determined by investigating the nanoparticles’ Raman spectrum, FT-IR spectrum, hydrodynamic size, and zeta potential. The morphology of the nanomaterials was analysed with transmission electron microscopy (TEM).
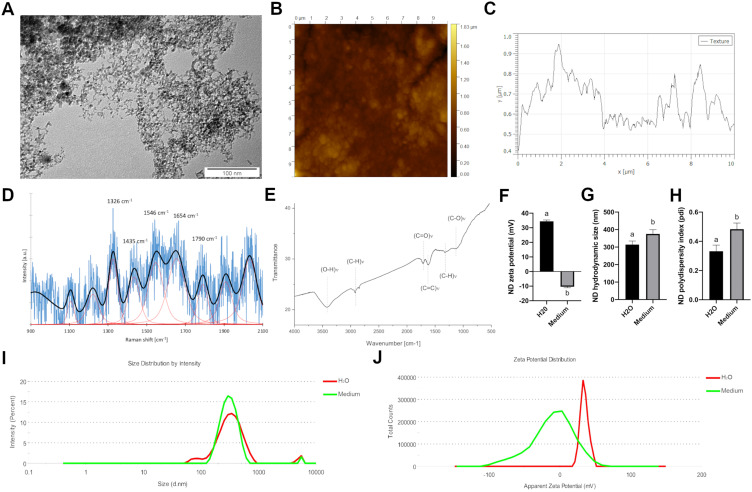
TEM images of NDs (20 mg/l) showed an average diameter of individual nanoparticle ranging from 2 to 10 nm (Figure 1A). AFM analysis of the ND dried hydrocolloid on the silicon wafer showed that ND formed surface with sharp peaks and average roughness of 24.0 nm (Figure 1B and C). Raman spectroscopy (Figure 1D) showed a strong band with a maximum at 1326 cm−1 with a shift towards the lower wavelength. Furthermore, the Raman spectrum showed a broad band of low intensity around 1250 cm−1, a broad band around 1425 cm−1, and the presence of the characteristic G band around 1546 cm−1. The FT-IR spectrum is shown in Figure 1E. The spectrum showed a broad peak between 3000 and 3650 cm−1, a peak around 1630 cm−1, and peaks around 1710 cm−1 and 1115 cm−1.
Figure 1.
Physicochemical characterization of diamond nanoparticles. (A) Transmission electron microscopy image of diamond nanoparticles. (B) Atomic force microscopy image and (C) a topography model of the diamond nanoparticles on a silicon wafer. (D) Raman spectrum of diamond nanoparticles. (E) FTIR spectrum of diamond nanoparticles. (F) Mean zeta potential of diamond nanoparticles after 24 hours incubation in water (“H2O”) and full Human Large Vessel Endothelial Cell Basal Medium (“Medium”). Mean hydrodynamic size (G) and polydispersity index (H) of diamond nanoparticles after 24 hours incubation in H2O and medium. Statistical significance is indicated with different superscripts (P < 0.05; n = 9 with at least 15 individual replicates). Representative hydrodynamic size distribution of diamond nanoparticles after 24 hours incubation in H2O (I) and medium (J). Representative zeta potential distribution of diamond nanoparticles after 24 hours incubation in H2O and medium.
Abbreviations: ND, diamond nanoparticles; a.u., arbitrary unit.
In water, after 24 h of incubation, NDs formed stable agglomerates (mean zeta potential 34.4 mV; Figure 1F and J) with an average hydrodynamic size of 314.5 nm (Figure 1G and I) and average polydispersity index of 0.332 (Figure 1H). Interestingly, after incubation with full Human Large Vessel Endothelial Cell Basal Medium (Thermo Fisher Scientific, Waltham, USA), NDs formed unstable agglomerates (mean zeta potential −10.4 mV; Figure 1F and J) with an average hydrodynamic size of 375.3 nm (Figure 1G and I) and average polydispersity index of 0.475 (Figure 1H).
NDs Show High Toxicity to HUVEC Cells but Not to HMEC or HS-5 Cells
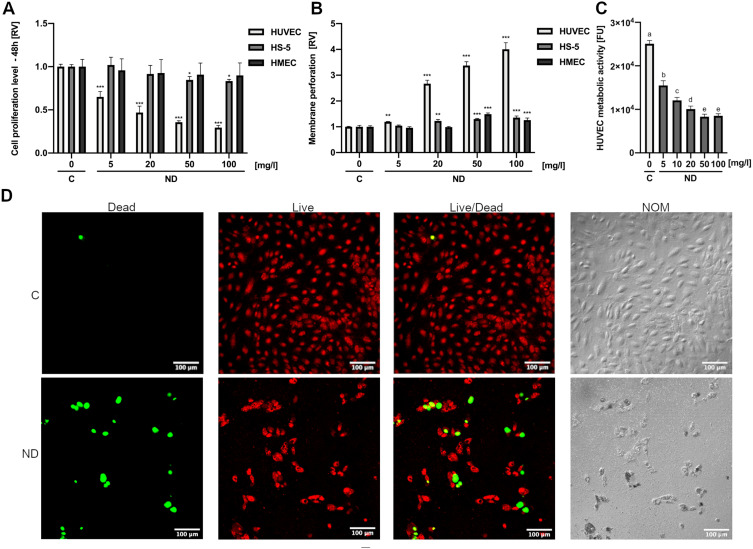
In order to compare the toxicity of NDs to HUVEC cells with the toxicity of NDs to other cell lines cultured under similar conditions, we additionally assessed the proliferation and membrane perforation of ND-treated HMEC and HS-5 cells. All three cell lines were cultured in low-serum media in order to minimize the differences in response to NDs caused by different culture conditions. NDs at concentrations of 5, 20, 50, and 100 mg/l were added to cell cultures of HUVEC, HMEC, and HS-5 24 h after seeding 5 × 103 cells per well on a 96-well plate. For the assessment of membrane perforation, the cells were treated with NDs for 24 h, whereas for the assessment of cell proliferation, they were treated for 48 h. Interestingly, the ND treatment of HMEC and HS-5 cells caused only a minor decrease or no change in cell proliferation (Figure 2A) and membrane perforation, whereas HUVEC cells were affected even at the lowest concentration used (5 mg/l) (Figure 2B). Proliferation after 48 h of incubation with NDs at 50 mg/l was decreased by approximately 65% for HUVEC cells (P < 0.001), whereas for the HMEC and HS-5 cell lines, it was decreased by 6% (P = 0.429, not significant) and 15% (P = 0.031), respectively (Figure 2B).
Figure 2.
Diamond nanoparticles are cytotoxic to HUVEC but not to HS-5 and HMEC. HUVEC, HS-5, and HMEC 48-h proliferation (A) and 24-h membrane perforation (B) after treatment with NDs at concentrations of 5, 10, 20, 50, and 100 μg/mL. Statistical significance is indicated with asterisks: *P < 0.033, **P < 0.002, ***P < 0.001 (multifactor ANOVA; P < 0.05; n = 3 with 4 individual replicates for proliferation and 6 individual replicates for membrane perforation). All values are expressed as mean ± standard deviation. (C) HUVEC metabolic activity after treatment with NDs at concentrations of 5, 10, 20, 50, and 100 mg/l for 24 h. Statistical significance is indicated with different superscripts: P<0.001 (a, b, c, d, e) (ANOVA; P < 0.05; n = 3 with 6 individual replicates). (D) Live/Dead assay confocal microscopy images of nontreated HUVEC cells (“C”) and treated cells with 50 mg/l for 24 h (“ND”). The nuclei of dead cells are labelled with a green stain (NucGreen Dead 488), whereas all nuclei are stained with a red stain (NucRed Live 647). Images taken using Nomarski interference contrast (“NOM”) showed light-reflecting nanoparticle agglomerates, suggesting strong endocytosis of NDs.
Abbreviations: RV, relative value; FUs, fluorescent units; C, control; NDs, diamond nanoparticles.
ND toxicity to HUVEC cells was further investigated using the metabolic activity assay Prestoblue and the Live/Dead assay. NDs at concentrations of 5, 10, 20, 50, and 100 mg/l were added to cell cultures of HUVEC 24 h after seeding 5 × 103 cells per well on a 96-well plate. After 24 h of incubation with NDs, the metabolic activity of HUVEC cells was significantly reduced by approximately 40% (P < 0.0001) even at the lowest concentration used (5 mg/l), whereas at the highest concentration (100 mg/l), the metabolic activity was reduced by approximately 70% (P < 0.0001) (Figure 2C).
The Live/Dead assay images were performed similarly, although the cells were treated with one ND concentration (50 mg/l). Apart from an increase in dead cells, there was a severe reduction in cell number in the field of view, suggesting the detachment of dead cells from the culture plate and the destruction of the completely overgrown cell monolayer. Moreover, images taken using Nomarski interference contrast showed light-reflecting nanoparticle agglomerates, suggesting strong endocytosis of NDs (Figure 2D).
ND Bioconjugation Reduces but Not Inhibits HUVEC Toxicity
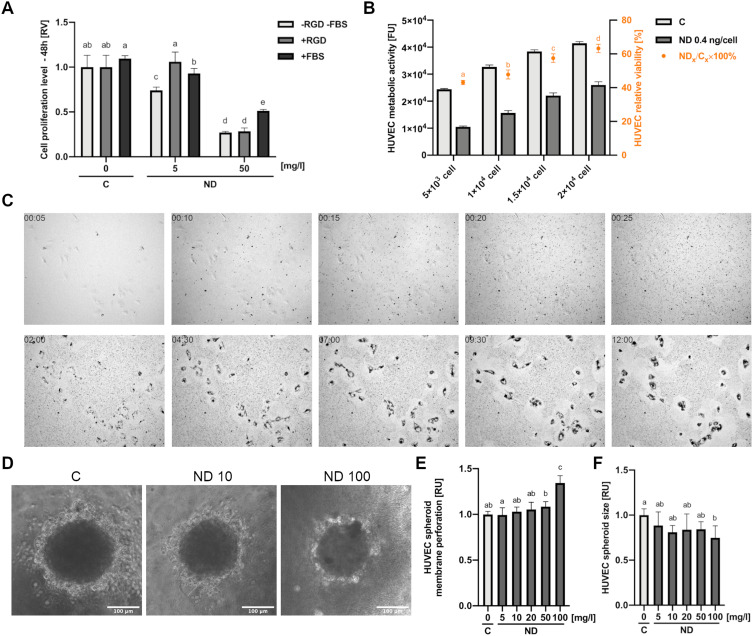
To assess the changes in ND toxicity after bioconjugation, we prepared NDs conventionally bound with a short peptide having an arginine-glycine-aspartic acid (RGD) motive that is popular in biomedical studies using zero-length crosslinker 1-Ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride (EDC). Additionally, we analysed the content of FBS in cell media and thus the influence of the formation of a nonspecific protein corona on the surface of NDs on the toxicity of NDs to HUVEC. For this purpose, we performed a toxicity analysis of NDs in cell medium containing 10% FBS (in contrast to the standard HUVEC medium, which contains no more than 1% FBS). The toxicity of NDs, NDs conjugated with a peptide containing an RGD motive, and NDs in 10% FBS was assessed as in the previous experiments by an analysis of cell proliferation 48 h after treatment with nanomaterials. The analysis of proliferation showed that at a lower concentration (5 mg/l), bioconjugation with a peptide having an RGD motive completely reduced ND toxicity (a reduction by approximately 25%; P < 0.001), whereas a medium with 10% FBS reduced proliferation by approximately 15% (P < 0.001) (Figure 3A). However, at a concentration of 50 mg/l, bioconjugation with a peptide with an RGD motive did not alter the toxicity (P > 0.999). The addition of 10% FBS decreased the ND reduction in proliferation from approximately 27% to 51% (P < 0.001).
Figure 3.
Toxicity of diamond nanoparticles to HUVEC depends on cell density and surface conjugation of nanoparticles. (A) HUVEC 48-h proliferation was analysed for untreated cells (“C”) or cells treated with NDs (“ND”, “-RGD -FBS”) and NDs conjugated with a peptide containing an RGD motive (Gly-Arg-Gly-Asp-Ser; “ND”, “+RGD”) and at concentrations of 5 and 50 mg/l in standard HUVEC culture medium and in medium with the addition of 10% FBS (“+FBS”). Statistical significance is indicated with different superscripts: P<0.001 (a, b, c, d, e) (ANOVA; P < 0.05; n = 3 with 4 individual replicates). (B) Dependence of ND toxicity on HUVEC cell density was assessed in a 96-well plate at densities of 5 x 103, 1 x 104, 1.5 x 104, and 2×104. After 24-h incubation, cells were treated with NDs at a concentration of 0.4 ng/cell (the final ND concentration was 20, 40, 60, and 80 mg/L in 100 µL medium, respectively), and the metabolic activity was assessed. The relative toxicity of HUVEC was expressed as the ratio of the metabolic activity value of the ND-treated cells to the metabolic activity of the untreated cells. Statistical significance is indicated with different superscripts: P<0.001 (a, b, c, d) (ANOVA; P < 0.05; n = 3 with 6 individual replicates). All values are expressed as mean ± standard deviation. (C) HUVEC time-lapse images of a 12-h cell culture with NDs at a final concentration of 20 mg/l. The full video of time-lapse images is available as Supplementary Video. Abbreviations: RV, relative value; FUs, fluorescent units; C, control; NDs, diamond nanoparticles; RGD, Gly-Arg-Gly-Asp-Ser peptide; FBS, fetal bovine serum. (D) HUVEC spheroid morphology after 24-h treatment with NDs at a concentration of 10 and 100 mg/l. HUVEC spheroid membrane perforation (E) and spheroid size (F) for a after 24-h incubation with NDs at a concentration of 5, 10, 20, 50, and 100 mg/l. Statistical significance is indicated with different superscripts:: P<0.001 (a, b, c) (ANOVA; P < 0.05; n = 3 with 6 individual replicates).
ND Toxicity is Decreased at High HUVEC Densities and in Spheroid Cultures
In order to evaluate the impact of cell density on ND toxicity, we seeded HUVEC cells at four different cell densities (5 × 103, 1 × 104, 1.5 × 104, 2 × 104). After 24 h, the HUVEC cells were treated with NDs at a concentration of 0.4 ng per cell (according to the following cell densities: 20, 40, 60, 80 mg/l) or not treated (the control group). As in the previous experiments, after 24 h of treatment with nanomaterials, HUVEC metabolic activity was analysed with the Prestoblue assay. Additionally, the relative viability of the cells was calculated by dividing the metabolic activity of the NP-treated group by the metabolic activity of the control group (Figure 3B). Interestingly, the relative viability of the HUVEC cells was higher at higher cell densities and showed a nearly linear increase. In order to understand this phenomenon, we performed a time-lapse analysis of HUVEC cells at low densities. Images of time-lapse of HUVEC from first 25 minutes of incubation with ND (5 minutes interval) and images of cells after 2 and 12 hours after incubation with ND (2.5-hour interval) are visible in Figure 3C. ND agglomeration was observed during the first 25 min of observation. An intensive uptake of agglomerates by the cells was then observed. Zones free of agglomerates were visible around the cell, suggesting active ND uptake by HUVEC cells, which at lower confluency led to a relatively greater exposure to NDs. The full video of time-lapse images is available as Supplementary Video.
To further evaluate the dependence of endothelial cell cytotoxicity on cell exposure to the nanoparticles, we analysed HUVEC spheroid toxicity after ND treatment. As in the previous experiments, the spheroids were treated with NDs at a concentration of 5, 10, 20, 50, and 100 mg/l. Interestingly, only the highest tested concentration of 100 mg/l led to decreases in spheroid size and a significant increase in membrane perforation (Figure 3D–F).
ND Mediates NADPH-Dependent ROS Formation
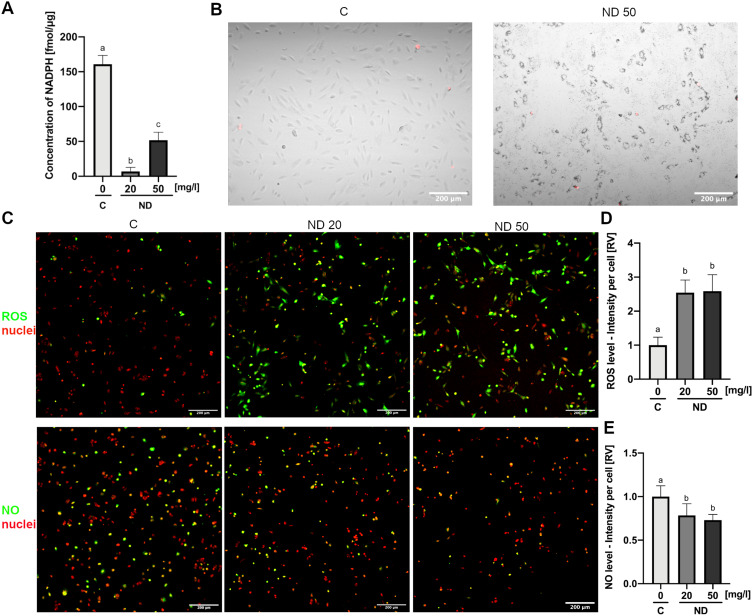
NADPH concentration in cell lysates was assessed after 4 h of incubation of HUVEC cells with NDs at a concentration of 20 or 50 mg/l. Analysis showed a significant decrease, leading to an almost complete depletion of the NADPH pool in cells treated with NDs at a concentration of 20 mg/l (P < 0.001) (Figure 4A). Similarly, NDs at a concentration of 50 mg/l severely decreased the NADPH concentration down to approximately 50 fmol per μg of cell protein lysate, from 160 fmol/μg recorded in the control lysates (P < 0.001) (Figure 4A).
Figure 4.
Diamond nanoparticles led to an NADPH-dependent induction of ROS in HUVEC. (A) Intercellular level of NADPH analysed in lysates of control HUVEC cells (“C”) and HUVEC cells incubated for 4 h with NDs at a concentration of 20 and 50 mg/l (“ND”). Statistical significance is indicated with different superscripts: P<0.001 (a, b, c) (ANOVA; P < 0.05; n = 2 with 4 individual replicates). (B) Merged images of mitochondrial superoxide staining (red channel; MitoSox Red, Thermo Fisher Scientific) and transmitted light with Nomarski interference contrast of control HUVEC cells (“C”) and HUVEC cells incubated for 4 h with NDs at a concentration of 50 mg/l (“ND 50”). (C) Confocal images of ROS and NO in control HUVEC cells (“C”) and HUVEC cells incubated for 4 h with NDs at a concentration of 20 and 50 mg/l (“ND 20”, “ND 50”) detected using the general oxidative stress indicator CM-H2CFDA (green channel; Thermo Fisher Scientific) and the NO synthesis level indicator DAF-FM (green channel; Thermo Fisher Scientific). Cell nuclei were stained with NucRed Live 647 (red channel; Thermo Fisher Scientific). Graphs showing ROS (D) and NO (E) levels expressed as a sum of the pixel values per cell. Statistical significance is indicated with different superscripts: P<0.001 (a, b) (ANOVA; P < 0.05; n = 2 with 7 individual replicates, and each well was imaged in 4 fields of view). All values are expressed as mean ± standard deviation.
Abbreviations: RV, relative value; C, control; NDs, diamond nanoparticles.
Synthesis of the ND-mediated formation of mitochondrial superoxide was analysed using MitoSOX Red (Thermo Fisher Scientific). HUVEC cells were incubated with NDs at a concentration of 50 mg/l for 4 h and analysed using a confocal microscope. There were no significant changes in the formation of mitochondrial superoxide in the control cells and the cells treated with NDs (Figure 4B).
General ROS synthesis was assessed using a general oxidative stress indicator, CM-H2DCFDA (Thermo Fisher Scientific). HUVEC cells were incubated with NDs at a concentration of 20 or 50 mg/l for 4 h and analysed using a confocal microscope. Simultaneous staining of cell nuclei enabled us to convert the total fluorescence signal into the fluorescence signal per cell. The results showed that both concentrations of NDs increased the ROS levels per cell approximately 2.5 times (P < 0.001) (Figure 4C and D).
In order to analyse the ND influence on HUVEC NO synthesis levels, the NO level was assessed using DAF-FM (Thermo Fisher Scientific). HUVEC cells were incubated with NDs at a concentration of 20 or 50 mg/l for 4 h and analysed using a confocal microscope. Simultaneous staining of cell nuclei enabled us to convert the total fluorescence signal into the fluorescence signal per cell. The results showed that NDs at a concentration of 20 and 50 mg/l decreased the NO levels by 22% and 27%, respectively (P = 0.0249 and P = 0.0096) (Figure 4C and E).
NDs Increase the Synthesis Level of Oxidative-Stress-Related Proteins and Reduce NO Synthesis
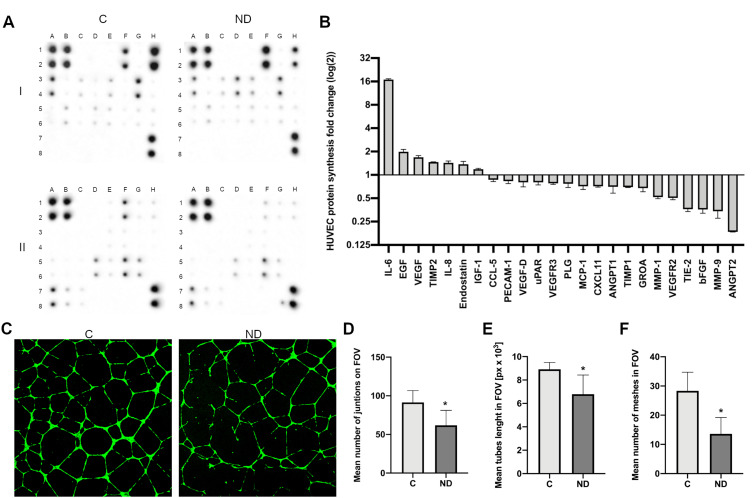
For protein analysis, the HUVEC cells were treated with NDs at a final concentration of 20 mg/l and incubated for 24 h. The protein synthesis level of 43 proteins was analysed using an antibody array (Human Angiogenesis Antibody Array, Abcam), although only 24 proteins were at detectable levels (Figure 5A and B). In comparison with the control, treatment with NDs led to an approximately 16-time increase in the protein level of interleukin-6 (IL-6), whereas epidermal growth factor (EGF), vascular endothelial growth factor (VEGF), and interleukin 8 (IL-8) increased 1.5 to 2 times. Interestingly, the synthesis level of matrix metalloproteinase 1 and 9 (MMP-1 and MMP-9), vascular endothelial growth factor receptor (VEGFR), TIE-2 receptor, basic fibroblast growth factor (bFGF), and angiopoietin 1 and 2 (ANGPT1 and ANGPT2) decreased more than 1.5 times. The strongest decrease was observed for ANGPT2, which decreased approximately 6 times (Figure 5A and B).
Figure 5.
NDs increase the synthesis level of stress-related proteins. (A) Antibody array analysis of cytokine synthesis in control HUVEC cells (“C”) incubated with NDs at a concentration of 20 mg/l for 24 h (“ND”). One assay consists of two membranes, which are labelled “I” and “II”, and the proteins are analysed in duplicate. The localization of the positive controls: I_A1,2; I_B1,2; I_H7,8; II_A1,2; II_B1,2; II_H7,8. The localization of the selected proteins in the order of their fold (log(2)) change shown in the graph (B): IL-6, I_D3,4; EGF, I_F1,2; VEGF, I_G5,6; TIMP2, I_E5,6; IL-8, I_E3,4; endostatin II_H1,2; IGF-1, I_C3,4; CCL-5, I_B5,6; PECAM-1, II_F5,6; VEGF-D, I_H5,6; uPAR, II_A7,8; VEGFR3, II_C7,8; PLG, II_G1,2; MCP-1, I_G3,4; CXCL11, II_A5,6; ANGPT1, II_E1,2; TIMP1, I_D5,6; GRO A, I_A1,2; MMP-1, II_D5,6; VEGFR2, II_B7,8; TIE-2, II_G5,6; bFGF, I_H1,2; MMP-9, II_E5,6; ANGPT2 II_F1,2. The full array map is available in Tables S1 and S2. (C) Images of control HUVEC tube formation (“C”) and tube formation during treatment with NDs at a final concentration of 20 mg/l (“ND”). Graphs show the mean number of junctions of HUVEC tubes in the field of view (D), mean tube length in the field of view (E), and mean number of meshes in the field of view (F). Data were obtained by analysing the images using ImageJ software and the Angiogenesis Analyzer macro. Statistical significance is indicated with asterisks: *P = 0.003, P = 0.002 and P < 0.001, respectively (t-test; P < 0.05; n = 2 with 4 individual replicates). All values are expressed as mean ± standard deviation.
Abbreviations: C, control; NDs, diamond nanoparticles; px, pixels; FOV, field of view.
ND Reduces HUVEC Tube Formation
In order to find out whether the observed toxicity and ROS formation affect the angiogenesis potential of HUVEC, we performed a tube formation assay (Figure 5C). NDs at a final concentration of 20 mg/l were added after 1 h of pre-incubation in order to allow the cells to adhere to the Geltrex Matrix (Thermo Fisher Scientific), followed by a further 12 h of incubation. The number of junctions, the total tube length, and the number of meshes were analysed with FIJI software18 and the Angiogenesis Analyzer toolset.19
ND treatment led to a decrease in the number of junctions by approximately 35% (P = 0.003) (Figure 5D), the tube length by approximately 25% (P = 0.002) (Figure 5E), and the number of meshes created by the HUVEC network by 48% (P < 0.001) (Figure 5F).
Statistical Methods
Data were analysed using one-way, two-way analysis of variance or t-test using GraphPad Prism 8 (GraphPad Software, San Diego, CA, USA). Differences between groups determined via one-way analysis of variance were tested with Tukey’s HSD or Duncan’s post hoc test. Results are shown as means and standard deviations. Differences with P < 0.05 were considered significant.
Discussion
NDs are considered to be one of the most biocompatible carbon nanomaterials; however, their toxicity varies significantly depending on the analysed cell types. The properties of nanoparticles depend not only on their core material but, most importantly, on their surface atoms and functional groups. Generally, the surface of NDs is covered with oxygen-containing functional groups, such as hydroxyl, epoxide, carbonyl, or carboxyl groups, which can function as reducing agents and cause oxidative stress.20 To analyse the structural features of the NDs, we performed Raman spectrum, FT-IR spectrum, and DLS analysis. The morphology of the nanomaterials was analysed with transmission electron microscopy (TEM) and atomic force microscopy (AFM). The analysis of the Raman spectrum showed a strong band with a maximum at 1326 cm−1, which is assigned to the diamond structure.21,22 The shift towards the lower wavelength indicates the presence of diamond crystallites with a diameter below 20 nm.23,24 The appearance of a broad band of low intensity around 1250 cm−1 confirms the presence of fine grains of NDs.24 The bands in the region of 1650 cm−1 and 1800 cm−1 indicate the appearance of surface functional groups (-OH, C=O) or the natural presence of water on the ND surface (the band around 1640 cm−1) in non-functionalized ND samples.21,22 The presence of the characteristic G band around 1546 cm−1 indicates the appearance of stretching vibrations of the graphite plane.21 The broad band around 1425 cm−1 is attributed to the presence of nanocrystalline diamond or ultra-nanocrystalline diamond.25 The broad bands below 1250 cm−1 indicate the presence of sp3 hybridized carbon.21 When NDs are excited with a laser of 355–532 nm, the D band appears only in the background.21 The FT-IR spectrum was performed in order to further analyse the surface functional groups. The broad peak observed between 3000 and 3650 cm−1 is assigned mainly to water and hydroxyl groups. The peak around 1630 cm−1 can be assigned to C=C bonds present in graphitic carbon. The other peaks observed in the FT-IR spectrum show that NDs contain C=O bonds (peaks around 1710 cm−1) and C–O bonds (peak around 1115 cm−1). The smaller features from ~2850 to about 3000 cm−1 can be attributed to the C–H stretch and C–H bending around 1310 cm−1.23 In summary, the NDs used in our experiments have physicochemical parameters characteristic of this type of nanoparticles.
The ND toxicity analysis showed, as expected, a lack of toxicity to HS-5 and HMEC cells. We chose those cell lines because they have a fibroblastic morphology similar to that of HUVEC cells and because they are different cell types (HS-5 is a fibroblast-like stromal cell line, whereas HMEC cells are mammary epithelial cells). Moreover, the HS-5 and HMEC cell lines can be successfully cultured in low-FBS cell media that resemble the medium used for culturing HUVEC cells. Finally, like HUVEC cells, HMEC cells are a primary cell line. In our experiments, HUVEC showed high sensitivity to ND treatment. The metabolic activity assay, the 48-h proliferation assay, the membrane perforation analysis, and the Live/Dead assay showed severe toxicity of NDs. Even the lowest ND concentration tested (5 mg/l) led to a high decrease in cell metabolic activity and proliferation. Additionally, the Live/Dead assay, apart from showing a reduction in cell number and an increase in dead cells, showed changes in cell morphology, including the shortening of cell extensions and the accumulation of ND agglomerates in the cells. Our results are consistent with those obtained for the toxicity of similar NDs tested on immortalized HUVEC cells.11 Moreover, it was observed that ND treatment of HUVEC cells induced apoptosis and/or necrosis, depending on the treatment time and concentration of NDs.10
Chen et al analysed the interactions of human serum albumin with NDs and showed that albumin forms part of the corona on the surface of NDs that inhibits their toxicity,26 which is a well-known and intensively studied phenomenon characteristic of most nanoparticles (for reviews, see 27,28). Similar results were reported by different researchers regarding the toxicity of both NDs and other carbon nanomaterials.29–32 Therefore, we analysed ND toxicity in medium with a standard 10% FBS concentration used for culturing most cell lines, rather than the standard low-serum media used for HUVEC cultures. The increased content of FBS decreased ND toxicity by about 20%; however, at a higher ND concentration of 50 mg/l, the toxicity was still high, decreasing HUVEC proliferation by approximately 50%. The hydrodynamic size of NDs in medium with the addition of 10% FBS was significantly higher, showing stronger agglomeration, probably due to the formation of a more extensive protein corona. Larger agglomerates and a bigger protein corona decrease the direct contact of cellular structures with the active surface of the nanoparticles, but they can be more effectively taken up by cells.33 Furthermore, we analysed the influence of the bioconjugation of NDs with a commonly used short peptide containing the RGD motive. As in the previous experiment with an increased content of FBS, we performed the analysis of the effect of the bioconjugation of NDs with a short peptide. The procedure of bioconjugation was based on the commonly used method of covalently coupling particles containing a carboxyl group with primary amines in the presence of EDC and sulfo-NHS. EDC is a zero-length crosslinker, which enables the direct linking of molecules by covalent bonds. Bioconjugation with a peptide containing the RGD motive decreased the toxicity of NDs, but only at a low concentration of 5 mg/l. At this concentration, functionalization inhibited the toxicity of NDs. Surprisingly, the functionalization of NDs at a concentration of 50 mg/l did not affect the toxicity of NDs, showing an approximately 70% reduction in proliferation (a similar reduction was caused by nonfunctionalized NDs). Our findings show that the conjugation of proteins and peptides decreases toxicity, which is consistent with other studies regarding the toxicity of carbon nanomaterials.26,29–32 However, in the case of cells that are sensitive to NDs, such as endothelial cells, the toxicity of the nanoparticles is still significant despite the presence of a protein crown on the surface of NDs.
In order to fully characterize the toxicity of NDs to HUVEC, we tested the dependence of toxicity on cell density, because if nanomaterial toxicity is related to the direct interaction with the plasma membrane, the toxicity should be lower when less membrane surface is exposed, as in confluent cultures, or when cell migration is hampered due to the presence of accompanying cells; both these conditions mean less interaction of the cell with the nanoparticle agglomerates on the surface of the culture vessel. The obtained results suggest that this is the case with ND toxicity to HUVEC cells. We have shown that in the case of a 2D culture, increasing the number of cells and proportionally increasing the number of nanoparticles (so that the concentration of nanoparticles per cell is constant, which in our study was 0.4 ng NDs per cell) leads to an increase in relative viability. Therefore, toxicity was the highest for the lowest number of cells per well tested (relative viability of about 45%) and the lowest for the highest number of cells tested (relative viability of about 65%).
In addition, we conducted a time-lapse observation for 12 h of HUVEC cells treated with NDs. It can be observed that the nanoparticles begin to agglomerate and fall to the bottom of the culture vessel after about 10 min from the start of the incubation. The cells intensively take up nanoparticles, which leads to a visible deposition of agglomerates in the cell and the number of dead cells over time. It has been reported that nanoparticle uptake is strongly related to their physicochemical characteristics. Positively charged nanoparticles such as NDs were shown to effectively enter the cell through endocytosis and to pass the blood–brain barrier.34 Macropinocytosis is a possible way of entering the cell for nanoparticles with a size of less than 10 nm. However, endocytosis, a more effective and selective way of nanoparticle uptake, is efficient for sizes above 20 nm.35,36 Carboxylated NDs of a size of 100 nm can enter lung cancer cells and embryonic fibroblasts mainly by macropinocytosis and clathrin-mediated endocytosis.37 The NDs used in our research, whose individual size was estimated with TEM to be 2 to 10 nm and whose average hydrodynamic size in medium was 375.3, were probably taken up by the cell after agglomeration. The time-lapse images suggest that cells take up NDs most effectively after their agglomeration on the plate surface, which is also associated with a significant reduction in toxicity in the case of higher cell densities. In addition, the time-lapse observations of the cells suggest that a cell that has a free area for local migration in 2D conditions is more exposed to this type of nanomaterial. Ultimately, these observations were confirmed using HUVEC spheroid models. Spheroid models generally are considered to provide more physiological-like conditions, including the activation of appropriate signalling pathways and structures,38,39 thus providing a smaller free surface of cell membranes that can interact with nanomaterials. Treating the spheroids with NDs, as expected, led to a much lower toxicity than observed in the 2D model.
Endothelial cells generate ROS mainly by the activity of NADPH oxidase (NOX), xanthine oxidase, and the mitochondrial respiratory chain.40 However, the major source of stress-induced ROS is NOX activity. ROS generation by nanoparticles depends on the material and the potential ion release, the functional groups on the surface of nanomaterials, and the size of the nanoparticles.41 In the analysis of total ROS synthesis, we observed a significant increase in synthesis after ND treatment. However, in the analysis of the formation of mitochondrial superoxide oxygen species, we did not observe an increase after treatment with NDs. Therefore, we assume that the ROS spike after ND treatment was not associated with the mitochondrial respiratory chain. Thus, we analysed the changes in the levels of NADPH (without NADP+ levels) in HUVEC cells after treatment with NDs. Interestingly, NDs led to an almost complete depletion of NADPH. The major source of ROS production in endothelial cells is the activity of NADPH oxidases. NADPH oxidases are enzyme complexes that transfer electrons from NADPH to molecular oxygen-generating superoxide and hydrogen peroxide. Seven members of the NOX family have been identified (NOX1-5, DUOX-1,2).42 Endothelial cells constitutively express NOX2 and NOX4 isoforms of NADPH oxidase. NOX4 is a constitutively active isoform that shows vasoprotective activity by increasing nitric oxide bioavailability and suppressing cell death pathways.43 NOX2 is regulated by several signalling pathways, including stress-related induction, and can promote endothelial dysfunction, inflammation, and apoptosis.44 In our studies, NDs probably induced NOX2 activation due to mechanical induction, but the exact mechanism should be further evaluated. Interestingly, NDs of similar size and concentration showed low or no toxicity and did not induce the formation of ROS in neuroblastoma macrophages, keratinocytes, and PC-12 cells;6 therefore, this is another argument for the fact that endothelial cells were more sensitive than other cells to NDs. However, inhalation exposure of sub-acute exposure to NDs in mice led to hematological and biochemical changes and increased ROS formation,45 suggesting that the method of exposure is critical.
ND treatment of HUVEC cells also led to a decrease in nitric oxide (NO) levels. NO signalling in endothelial cells is mediated by the activation of nitric oxide synthetase (NOS), which induces anti-inflammatory signalling and modulates basic endothelial processes, such as vascular contraction, through the activation of soluble guanylate cyclase (sGC), which produces cyclic guanosine monophosphate (cGMP), regulating cGMP-dependent protein kinases and ion channels.46 Superoxide may react with NO, leading to the generation of peroxynitrite (ONOO−), which can provide additional ROS generation by the uncoupling of endothelial nitric oxide synthase (eNOS), causing decreased NO levels.47,48 The relation between NO levels and ROS production by NOX was confirmed by the inhibition of NOX with gp91ds-tat or apocynin, which led to a significant restoration of NO production.49 Therefore, the decrease in NO levels indirectly confirms the increase in ROS production by NOX after ND treatment.
Treatment with NDs changed the protein synthesis level of several of the 43 proteins tested. The most notable changes were shown for the IL-6 protein, which is an important pro-inflammatory cytokine synthesized during tissue injury50,51 but also during mechanical stress, including shear stress.52 Moreover, NDs led to decreases in the synthesis level of angiopoietins. Whereas ANGPT1 decreased only approximately 1.5 times, APGPT2 and TIE-2 showed an approximately 3-and 6-fold decrease, respectively. Angiopoietins are angiogenic growth factors that regulate the angiopoietin/Tie signalling pathway through the phosphorylation of Tie receptors for the regulation of angiogenesis. ANGPT2 acts as an antagonist inhibiting ANGPT1 TIE-2 phosphorylation.53 Interestingly, it has been shown that ANPT2 expression was downregulated during shear stress, whereas ANGPT1 expression was not changed.54 Therefore, treatment with NDs led to effects seen in endothelial cells subjected to mechanical stress, such as shear stress. Similarly, increases in EGF, VEGF, and IL-8 and decreases in MMP-1, MMP-9, and TIE-2 synthesis levels were observed after ND treatment of endothelial cells under high shear stress.55–57 Indeed, other research groups showed effects of nanoparticle treatment similar to those of mechanical stress, with nanoparticles of TiO2 and SiO2 in the size range of 20 to 30 nm. However, nanoparticle treatment did not cause a decrease in proliferation but led to increased endothelial monolayer permeability, elevated intracellular calcium, and cytoskeleton rearrangement.58 Other reports show endothelial cell dysfunction induced by oxidative stress caused by silica nanoparticles59,60 and silver nanoparticles.61 Our last experiment, showing a decrease in tube formation in an angiogenesis tube formation assay, confirms earlier results showing the toxicity of NDs, most likely resulting from combined oxidative and mechanical stress.
Conclusion
NDs are considered to be one of the most biocompatible carbon nanomaterials; however, their toxicity varies significantly depending on the analysed cell types. Our studies demonstrated the sensitivity of endothelial cells to NDs in particular at high exposure to unconjugated nanoparticles. We have confirmed the presence of a relationship between the toxicity of NDs and the level of cell exposure to nanoparticles and the nanoparticle surface. Our study suggests that the mechanical interaction with cell membranes and the endocytosis of nanoparticles lead to severe endothelial toxicity due to NADPH-dependent ROS synthesis, a decrease in the bioavailability of NO, and the synthesis of stress-related proteins such as IL-6 and IL-8. The results yielded new information about the conditioned toxicity of NDs, which may provide new insights into the safe and effective use of nanomaterials in biomedical applications.
Funding Statement
This research was funded by the National Science Centre, Poland, project number 2020/37/B/NZ7/03532.
Data Sharing Statement
The datasets analyzed during the current study are available from the corresponding author on reasonable request.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Chauhan S, Jain N, Nagaich U. Nanodiamonds with powerful ability for drug delivery and biomedical applications: recent updates on in vivo study and patents. J Pharm Anal. 2020;10(1):1–12. doi: 10.1016/j.jpha.2019.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang YW, Cambre M, Lee HJ. The toxicity of nanoparticles depends on multiple molecular and physicochemical mechanisms. Int J Mol Sci. 2017;18(12):2702. doi: 10.3390/ijms18122702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kumar V, Sharma N, Maitra SS. In vitro and in vivo toxicity assessment of nanoparticles. Int Nano Lett. 2017;7(4):243–256. doi: 10.1007/s40089-017-0221-3 [DOI] [Google Scholar]
- 4.Rahimi Kalateh Shah Mohammad G, Seyedi SMR, Karimi E, Homayouni-Tabrizi M. The cytotoxic properties of zinc oxide nanoparticles on the rat liver and spleen, and its anticancer impacts on human liver cancer cell lines. J Biochem Mol Toxicol. 2019;33(7):e22324. doi: 10.1002/jbt.22324 [DOI] [PubMed] [Google Scholar]
- 5.Kopac T. Protein Corona, understanding the nanoparticle-protein interactions and future perspectives: a critical review. Int J Biol Macromol. 2021;169:290–301. doi: 10.1016/j.ijbiomac.2020.12.108 [DOI] [PubMed] [Google Scholar]
- 6.Schrand AM, Huang H, Carlson C, et al. Are diamond nanoparticles cytotoxic? J Phys Chem B. 2007;111(1):2–7. doi: 10.1021/jp066387v [DOI] [PubMed] [Google Scholar]
- 7.Strojny B, Grodzik M, Sawosz E, et al. Diamond nanoparticles modify curcumin activity: in vitro studies on cancer and normal cells and in ovo studies on chicken embryo model. PLoS One. 2016;11(10):e0164637. doi: 10.1371/journal.pone.0164637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zakrzewska KE, Samluk A, Wierzbicki M, et al. Analysis of the cytotoxicity of carbon-based nanoparticles, diamond and graphite, in human glioblastoma and hepatoma cell lines. PLoS One. 2015;10(3):e0122579. doi: 10.1371/journal.pone.0122579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grodzik M, Szczepaniak J, Strojny-Cieslak B, et al. Diamond nanoparticles downregulate expression of CycD and CycE in glioma cells. Molecules. 2019;24(8):1549. doi: 10.3390/molecules24081549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Solarska K, Gajewska A, Bartosz G, Mitura K. Induction of apoptosis in human endothelial cells by nanodiamond particles. J Nanosci Nanotechnol. 2012;12(6):5117–5121. doi: 10.1166/jnn.2012.4952 [DOI] [PubMed] [Google Scholar]
- 11.Solarska K, Gajewska A, Kaczorowski W, Bartosz G, Mitura K. Effect of nanodiamond powders on the viability and production of reactive oxygen and nitrogen species by human endothelial cells. Diam Relat Mater. 2012;21:107–113. doi: 10.1016/j.diamond.2011.10.020 [DOI] [Google Scholar]
- 12.Grady ME, Parrish E, Caporizzo MA, Seeger SC, Composto RJ, Eckmann DM. Intracellular nanoparticle dynamics affected by cytoskeletal integrity. Soft Matter. 2017;13(9):1873–1880. doi: 10.1039/c6sm02464e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodríguez-Hernández AG, Vazquez-Duhalt R, Huerta-Saquero A. Nanoparticle-plasma membrane interactions: thermodynamics, toxicity and cellular response. Curr Med Chem. 2020;27(20):3330–3345. doi: 10.2174/0929867325666181112090648 [DOI] [PubMed] [Google Scholar]
- 14.Yin S, Liu J, Kang Y, Lin Y, Li D, Shao L. Interactions of nanomaterials with ion channels and related mechanisms. Br J Pharmacol. 2019;176(19):3754–3774. doi: 10.1111/bph.14792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gray KM, Stroka KM. Vascular endothelial cell mechanosensing: new insights gained from biomimetic microfluidic models. Semin Cell Dev Biol. 2017;71:106–117. doi: 10.1016/j.semcdb.2017.06.002 [DOI] [PubMed] [Google Scholar]
- 16.Fang Y, Wu D, Birukov KG. Mechanosensing and mechanoregulation of endothelial cell functions. Compr Physiol. 2019;9(2):873–904. doi: 10.1002/cphy.c180020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng H, Zhong W, Wang L, et al. Effects of shear stress on vascular endothelial functions in atherosclerosis and potential therapeutic approaches. Biomed Pharmacother. 2023;158:114198. doi: 10.1016/j.biopha.2022.114198 [DOI] [PubMed] [Google Scholar]
- 18.Schindelin J, Arganda-Carreras I, Frise E, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carpentier G, Berndt S, Ferratge S, et al. Angiogenesis Analyzer for ImageJ — a comparative morphometric analysis of “Endothelial Tube Formation Assay” and “Fibrin Bead Assay. Sci Rep. 2020;10(1):11568. doi: 10.1038/s41598-020-67289-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holt KB. Undoped diamond nanoparticles: origins of surface redox chemistry. Phys Chem Chem Phys. 2010;12(9):2048–2058. doi: 10.1039/B920075D [DOI] [PubMed] [Google Scholar]
- 21.Korepanov VI, Hamaguchi HO, Osawa E, et al. Carbon structure in nanodiamonds elucidated from Raman spectroscopy. Carbon. 2017;121:322–329. doi: 10.1016/j.carbon.2017.06.012 [DOI] [Google Scholar]
- 22.Mochalin VN, Shenderova O, Ho D, Gogotsi Y. The properties and applications of nanodiamonds. Nat Nanotechnol. 2012;7(1):11–23. doi: 10.1038/nnano.2011.209 [DOI] [PubMed] [Google Scholar]
- 23.Afandi A, Howkins A, Boyd IW, Jackman RB. Nanodiamonds for device applications: an investigation of the properties of boron-doped detonation nanodiamonds. Sci Rep. 2018;8(1):17–19. doi: 10.1038/s41598-018-21670-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Popov M, Churkin V, Kirichenko A, et al. Raman spectra and bulk modulus of nanodiamond in a size interval of 2–5 nm. Nanoscale Res Lett. 2017;12:4–9. doi: 10.1186/s11671-017-2333-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gottlieb S, Wöhrl N, Schulz S, Buck V. Simultaneous synthesis of nanodiamonds and graphene via plasma enhanced chemical vapor deposition (MW PE-CVD) on copper. SpringerPlus. 2016;5:1. doi: 10.1186/s40064-016-2201-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen M, Zuo X, Xu Q, Wang R, Fan S, Wu H. Investigating the interaction of nanodiamonds with human serum albumin and induced cytotoxicity. J Spectrosc. 2019;2019:e4503137. doi: 10.1155/2019/4503137 [DOI] [Google Scholar]
- 27.Tomak A, Cesmeli S, Hanoglu BD, Winkler D, Oksel Karakus C. Nanoparticle-protein Corona complex: understanding multiple interactions between environmental factors, Corona formation, and biological activity. Nanotoxicology. 2021;15(10):1331–1357. doi: 10.1080/17435390.2022.2025467 [DOI] [PubMed] [Google Scholar]
- 28.Park SJ. Protein–nanoparticle interaction: corona formation and conformational changes in proteins on nanoparticles. Int J Nanomedicine. 2020;15:5783–5802. doi: 10.2147/IJN.S254808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu W, Peng C, Lv M, et al. Protein corona-mediated mitigation of cytotoxicity of graphene oxide. ACS Nano. 2011;5(5):3693–3700. doi: 10.1021/nn200021j [DOI] [PubMed] [Google Scholar]
- 30.Garriga R, Herrero-Continente T, Palos M, et al. Toxicity of carbon nanomaterials and their potential application as drug delivery systems: in vitro studies in Caco-2 and MCF-7 cell lines. Nanomaterials. 2020;10(8):1617. doi: 10.3390/nano10081617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu Y, Li W, Li Q, et al. Effects of serum proteins on intracellular uptake and cytotoxicity of carbon nanoparticles. Carbon. 2009;47(5):1351–1358. doi: 10.1016/j.carbon.2009.01.026 [DOI] [Google Scholar]
- 32.Li J, Zhu Y, Li W, Zhang X, Peng Y, Huang Q. Nanodiamonds as intracellular transporters of chemotherapeutic drug. Biomaterials. 2010;31(32):8410–8418. doi: 10.1016/j.biomaterials.2010.07.058 [DOI] [PubMed] [Google Scholar]
- 33.Chithrani BD, Chan WCW. Elucidating the mechanism of cellular uptake and removal of protein-coated gold nanoparticles of different sizes and shapes. Nano Lett. 2007;7(6):1542–1550. doi: 10.1021/nl070363y [DOI] [PubMed] [Google Scholar]
- 34.Fröhlich E. The role of surface charge in cellular uptake and cytotoxicity of medical nanoparticles. Int J Nanomedicine. 2012;7:5577–5591. doi: 10.2147/IJN.S36111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang S, Li J, Lykotrafitis G, Bao G, Suresh S. Size-Dependent Endocytosis of Nanoparticles. Adv Mater Deerfield Beach Fla. 2009;21:419–424. doi: 10.1002/adma.200801393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao H, Shi W, Freund LB. Mechanics of receptor-mediated endocytosis. Proc Natl Acad Sci U S A. 2005;102(27):9469–9474. doi: 10.1073/pnas.0503879102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu KK, Wang CC, Cheng CL, Chao JI. Endocytic carboxylated nanodiamond for the labeling and tracking of cell division and differentiation in cancer and stem cells. Biomaterials. 2009;30(26):4249–4259. doi: 10.1016/j.biomaterials.2009.04.056 [DOI] [PubMed] [Google Scholar]
- 38.Fang Y, Eglen RM. Three-dimensional cell cultures in drug discovery and development. Slas Discov. 2017;22(5):456–472. doi: 10.1177/1087057117696795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang H, Brown PC, Chow ECY, et al. 3D cell culture models: drug pharmacokinetics, safety assessment, and regulatory consideration. Clin Transl Sci. 2021;14(5):1659–1680. doi: 10.1111/cts.13066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Armitage ME, Wingler K, Schmidt HH, La M. Translating the oxidative stress hypothesis into the clinic: NOX versus NOS. J Mol Med. 2009;87(11):1071. doi: 10.1007/s00109-009-0544-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fu PP, Xia Q, Hwang HM, Ray PC, Yu H. Mechanisms of nanotoxicity: generation of reactive oxygen species. J Food Drug Anal. 2014;22(1):64–75. doi: 10.1016/j.jfda.2014.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lassègue B, San Martín A, Griendling KK. Biochemistry, physiology, and pathophysiology of NADPH oxidases in the cardiovascular system. Circ Res. 2012;110(10):1364–1390. doi: 10.1161/CIRCRESAHA.111.243972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burtenshaw D, Hakimjavadi R, Redmond EM, Cahill PA. Nox, reactive oxygen species and regulation of vascular cell fate. Antioxidants. 2017;6(4):90. doi: 10.3390/antiox6040090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Drummond GR, Sobey CG. Endothelial NADPH oxidases: which NOX to target in vascular disease? Trends Endocrinol Metab TEM. 2014;25(9):452–463. doi: 10.1016/j.tem.2014.06.012 [DOI] [PubMed] [Google Scholar]
- 45.Khosravi Y, Salimi A, Pourahmad J, Naserzadeh P, Seydi E. Inhalation exposure of nano diamond induced oxidative stress in lung, heart and brain. Xenobiotica. 2018;48(8):860–866. doi: 10.1080/00498254.2017.1367974 [DOI] [PubMed] [Google Scholar]
- 46.Moncada S, Higgs A, Higgs A. The L-arginine-nitric oxide pathway. N Engl J Med. 1993;329(27):2002–2012. doi: 10.1056/NEJM199312303292706 [DOI] [PubMed] [Google Scholar]
- 47.Mollnau H, Wendt M, Szöcs K, et al. Effects of angiotensin II infusion on the expression and function of NAD(P)H oxidase and components of nitric oxide/cGMP signaling. Circ Res. 2002;90(4):e58–e65. doi: 10.1161/01.RES.0000012569.55432.02 [DOI] [PubMed] [Google Scholar]
- 48.Chen F, Kumar S, Yu Y, et al. PKC-dependent phosphorylation of eNOS at T495 regulates eNOS coupling and endothelial barrier function in response to G+ -toxins. PLoS One. 2014;9(7):e99823. doi: 10.1371/journal.pone.0099823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Godbole AS, Lu X, Guo X, Kassab GS. NADPH oxidase has a directional response to shear stress. Am J Physiol Heart Circ Physiol. 2009;296(1):H152–H158. doi: 10.1152/ajpheart.01251.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kishimoto T. IL-6: from its discovery to clinical applications. Int Immunol. 2010;22(5):347–352. doi: 10.1093/intimm/dxq030 [DOI] [PubMed] [Google Scholar]
- 51.Kang S, Kishimoto T. Interplay between interleukin-6 signaling and the vascular endothelium in cytokine storms. Exp Mol Med. 2021;53(7):1116–1123. doi: 10.1038/s12276-021-00649-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liton PB, Luna C, Bodman M, Hong A, Epstein DL, Gonzalez P. Induction of IL-6 expression by mechanical stress in the trabecular meshwork. Biochem Biophys Res Commun. 2005;337(4):1229–1236. doi: 10.1016/j.bbrc.2005.09.182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Akwii RG, Sajib MS, Zahra FT, Mikelis CM. Role of angiopoietin-2 in vascular physiology and pathophysiology. Cells. 2019;8:5. doi: 10.3390/cells8050471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chlench S, Mecha Disassa N, Hohberg M, et al. Regulation of Foxo-1 and the angiopoietin-2/Tie2 system by shear stress. FEBS Lett. 2007;581(4):673–680. doi: 10.1016/j.febslet.2007.01.028 [DOI] [PubMed] [Google Scholar]
- 55.Cheng M, Liu X, Li Y, et al. IL-8 gene induction by low shear stress: pharmacological evaluation of the role of signaling molecules. Biorheology. 2007;44(5–6):349–360. [PubMed] [Google Scholar]
- 56.Yun S, Dardik A, Haga M, et al. Transcription factor Sp1 phosphorylation induced by shear stress inhibits membrane type 1-matrix metalloproteinase expression in endothelium. J Biol Chem. 2002;277(38):34808–34814. doi: 10.1074/jbc.M205417200 [DOI] [PubMed] [Google Scholar]
- 57.Dela paz NG, Walshe TE, Leach LL, Saint-Geniez M, D’Amore PA. Role of shear-stress-induced VEGF expression in endothelial cell survival. J Cell Sci. 2012;125(4):831–843. doi: 10.1242/jcs.084301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu Y, Yoo E, Mahler GJ, Doiron AL, Doiron AL. Endothelial barrier dysfunction induced by nanoparticle exposure through actin remodeling via caveolae/raft-regulated calcium signalling. NanoImpact. 2018;11:82–91. doi: 10.1016/j.impact.2018.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guo C, Xia Y, Niu P, et al. Silica nanoparticles induce oxidative stress, inflammation, and endothelial dysfunction in vitro via activation of the MAPK/Nrf2 pathway and nuclear factor-κB signaling. Int J Nanomedicine. 2015;10(1):1463–1477. doi: 10.2147/IJN.S76114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu X, Sun J. Endothelial cells dysfunction induced by silica nanoparticles through oxidative stress via JNK/P53 and NF-κB pathways. Biomaterials. 2010;31(32):8198–8209. doi: 10.1016/j.biomaterials.2010.07.069 [DOI] [PubMed] [Google Scholar]
- 61.Shi J, Sun X, Lin Y, et al. Endothelial cell injury and dysfunction induced by silver nanoparticles through oxidative stress via IKK/NF-κB pathways. Biomaterials. 2014;35(24):6657–6666. doi: 10.1016/j.biomaterials.2014.04.093 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets analyzed during the current study are available from the corresponding author on reasonable request.