Background:
Hutchinson-Gilford progeria syndrome (HGPS) is an ultrarare, fatal, premature aging disease caused by a toxic protein called progerin. Circulating progerin has not been previously detected, precluding research using readily available biological samples. This study aimed to develop a plasma progerin assay to evaluate progerin’s quantity, response to progerin-targeted therapy, and relationship to patient survival.
Methods:
Biological samples were collected by The Progeria Research Foundation Cell and Tissue Bank from a non-HGPS cohort cross-sectionally and a HGPS cohort longitudinally. HGPS donations occurred at baseline and intermittently while treated with farnesylation inhibitors lonafarnib±pravastatin and zoledronate, within 3 sequential open-label clinical trials at Boston Children’s Hospital totaling >10 years of treatment. An ultrasensitive single-molecule counting progerin immunoassay was developed with prespecified performance parameters. Intra- and interpatient group statistics were descriptive. The relationship between progerin and survival was assessed by using joint modeling with time-dependent slopes parameterization.
Results:
The assay’s dynamic detection range was 59 to 30 000 pg/mL (R2=0.9987). There was no lamin A cross-reactivity. Mean plasma progerin in non-HGPS participants (n=69; 39 male, 30 female; age, 0.2–71.3 years) was 351±251 pg/mL, and in drug-naive participants with HGPS (n=74; 37 female, 37 male; age, 2.1–17.5 years) was 33 261±12 346 pg/mL, reflecting a 95-fold increase in affected children (P<0.0001). Progerin levels did not differ by sex (P=0.99). Lonafarnib treatment resulted in an average per-visit progerin decrease from baseline of between 35% to 62% (all P<0.005); effects were not augmented by adding pravastatin and zoledronate. Progerin levels fell within 4 months of therapy and remained lower for up to 10 years. The magnitude of progerin decrease positively associated with patient survival (P<0.0001; ie, 15 000 pg/mL decrease yields a 63.9% decreased risk of death). For any given decrease in progerin, life expectancy incrementally increased with longer treatment duration.
Conclusions:
A sensitive, quantitative immunoassay for progerin was developed and used to demonstrate high progerin levels in HGPS plasma that decreased with lonafarnib therapy. The extent of improved survival was associated with both the magnitude of progerin decrease and duration at lower levels. Thus, plasma progerin is a biomarker for HGPS whose reduction enables short- and long-term assessment of progerin-targeted treatment efficacy.
Registration:
URL: https://www.clinicaltrials.gov. Unique identifiers: NCT00879034 and NCT00916747.
Keywords: aging, atherosclerosis, biomarkers, laminopathies, lamins, progeria, progerin
Clinical Perspective.
What Is New?
A quantitative assay was developed to measure plasma progerin, the pathogenic protein in Hutchinson-Gilford progeria syndrome (HGPS).
Average plasma progerin was 95-fold higher in HGPS than in non-HGPS controls.
Treatment with the farnesyltransferase inhibitor lonafarnib, the only Food and Drug Administration–approved therapy for HGPS, resulted in a significant and sustained plasma progerin decrease (average 35%–62%), with evaluations from 4 months to >10 years from the start of therapy.
The extent of improved patient life expectancy was predicted by both magnitude and duration of plasma progerin decrease.
What Are the Clinical Implications?
The quantitative relationship between plasma progerin and HGPS patient survival solidifies the clinical relevance of progerin measurement.
• Plasma progerin has potential for use as a clinical treatment trial outcome measure that is reasonably likely to reflect clinical benefit.
Plasma progerin detection in non-HGPS demonstrates its potential for use in the study of generalized aging and atherosclerosis.
Editorial, see p 1745
Hutchinson-Gilford progeria syndrome (HGPS) is an ultrarare (prevalence of 1 in 18–20 million living individuals1), uniformly fatal premature aging disease.2 There is no sex, ethnic, or geographic bias. Morbidity includes failure to thrive, generalized lipodystrophy, alopecia, bone dysplasia, and progressive atherosclerosis resulting in death predominantly from heart failure at an average age of 14.6 years.3,4
HGPS is a sporadic autosomal dominant disease caused by mutations in the LMNA gene that produce a toxic splice variant of the nuclear membrane protein lamin A called progerin.5,6 Ninety percent of the patients have classic HGPS, caused by a silent mutation (c.1824 C>T) that optimizes the use of an internal splice site within exon 11; the remaining 10% have nonclassic HGPS, caused by single-base mutations that deoptimize the spliceosome recognition sequence of intron 11.3,4 Progerin protein acts in a dominant negative fashion; the presence of progerin, and not the relative decrease in normal lamin A levels, produces disease.7 In support, mice lacking lamin A are phenotypically normal.8
A dose-dependent effect of progerin is supported by murine studies showing graded disease severity and shortened lifespan in heterozygous versus homozygous progerin-expressing mice, and extension of lifespan, as well, in HGPS mice receiving progerin-targeted treatments.9,10 Case reports have demonstrated low and high progerin levels expressed by fibroblast cultures from mildly versus severely affected progeroid patients, respectively.11,12
There is a plethora of downstream cellular and extracellular effects of progerin production,13 many of which are key to cell survival, including impaired nuclear architecture, DNA repair mechanisms, and inflammatory pathways such as the NRF2-based anti-inflammation system,14 mitochondrial dysfunction,15 and premature senescence. Some downstream effects are caused by progerin toxicity, and some are likely rescue mechanisms whose prognostic value is difficult to establish. Direct measurement of the upstream, disease-causing protein averts this issue, and thus could more easily enable early evaluation of therapeutic interventions, both preclinically and clinically.
Unlike lamin A, progerin lacks the cleavage site for a zinc metalloprotease, causing it to remain persistently farnesylated,6 and thereby incorporated as part of the nuclear lamina long-term, where it accumulates and exerts cell damage.16 In vitro, mouse model, and human studies have demonstrated that some phenotypes of HGPS are delayed, reversed by inhibiting farnesylation, or delayed or reversed by inhibiting farnesylation.17 Clinical trials for children with HGPS demonstrated that treatment with the farnesyltransferase inhibitor lonafarnib has cardiovascular, bone, audiologic, and weight benefits,18 and is associated with an average estimated increased survival time of 2.5 years.3,4 This evidence collectively resulted in lonafarnib being the first US Food and Drug Administration–approved drug treatment for HGPS (trade name: Zokinvy).
At present, there is no validated biomarker for HGPS. Because progerin is the disease-causing protein, it is the optimal biomarker candidate. Although progerin protein has been detected in cells and tissues,19–21 these methods are not sufficiently sensitive, quantitative for clinical trial utility, or both, and tissue detection is not feasible for obtaining serial patient samples. The main goal of this study was to detect progerin in a sensitive, specific manner from biological materials that can be obtained longitudinally from children with HGPS. This study reports on the analytical method development and characterization of plasma progerin as a biomarker reasonably likely to predict clinical benefit, filling a major gap in the ability to evaluate treatment efficacy in children with HGPS.
Methods
The progerin assay is available from EMD Millipore laboratory (https://www.sigmaaldrich.com), with permission from the Progeria Research Foundation (PRF).
Study Approvals
This study was approved by the Institutional Review Board of Hasbro Children’s Hospital, Providence, RI, and Boston Children’s Hospital (BCH) Committee on Clinical Investigation, Boston, MA. Written informed consent was obtained for all samples collected. Assent was obtained from children old enough to comprehend. When the primary language was not English, consents were performed using translated consent forms and discussions were performed with interpreters. For Hasbro Children’s Hospital consenting, short-form consents were translated; for BCH, full consent forms were translated.
Study Population, Study Samples and Clinical Data, Clinical Trial Drug Dosing and Administration
Patients with HGPS had genetically confirmed progerin-producing mutations in the LMNA gene. Samples from patients with classic HGPS (c.1824 C>T) are presented in the main analyses, whereas nonclassic HGPS comparisons are used in subgroup comparisons and presented in the Supplemental Material. Clinical data were obtained from The PRF International Progeria Registry, Diagnostics Program, and Medical and Research Database (https://www.progeriaresearch.org).
HGPS plasma and serum were obtained from The PRF Cell and Tissue Bank (https://www.progeriaresearch.org). Nontrial samples were not regulated as to the time of day collected. The non-HGPS study cohort consisted of patients who tested negative for suspected HGPS or their relatives. Plasma from patients without HGPS who had stage 2 or 3 kidney disease and congestive heart failure and from healthy controls used for assay development were purchased from Precision for Medicine (Norton, Massachusetts).
Clinical trial study samples were donated to The PRF Cell and Tissue Bank from BCH with informed consent or approval from the BCH Committee on Clinical Investigation. Trial samples were collected at each study visit, in a fasting state, in the morning just before treatment dosing as trough samples. Pharmacokinetics samples were collected at trough and then at specified times after trough. Samples are separated into 3 categories, with acronyms ProLon1,4,18 Triple Therapy,22 and ProLon2,3 that were conducted sequentially as open-label single-center trials of 2 to 4 years duration each, at BCH (Table S1, see previously published study details).
In all clinical trials, patients received oral lonafarnib (Schering-Plough Research Institute, Merck&Co, Inc, or Eiger Biopharmaceuticals). Lonafarnib dosing was 150 mg/m2, except for the first 4 months of ProLon1 when dosing was 115 mg/m2. For the Triple Therapy Trial,22 oral pravastatin (Pravachol, Bristol-Meyers Squibb) and zoledronate (Zometa, Novartis, Inc.) were administered along with lonafarnib. Clinical trial outcome measures included systemwide HGPS-specific phenotype evaluations (see Table S1).
Single Molecule Counting Progerin Immunoassay
A single molecule counting ultrasensitive immunoassay for the detection of plasma progerin was developed. Overall, the assay used a sandwich immunoassay format with an anti–lamin A capture antibody coated on magnetic microparticles, and an anti-progerin–specific detection antibody conjugated to a fluorescent tag (Figure S1) run in a 96-well plate. Progerin-bound detection antibody was eluted and read as single molecules using a confocal laser.
Clinical Sample Processing
For plasma isolation, blood was collected into either sodium heparin or K2EDTA tubes (all BD Biosciences) and centrifuged at 1300g for 10 minutes at room temperature. All samples were aliquoted, stored at –80 °C, and sent to EMD Millipore on dry ice for analysis.
For analysis, samples were diluted in standard diluent, a synthetic serum-based solution containing a proprietary mixture of Tris buffer and carrier protein, before transferring into filter plate wells (MilliporeSigma MSBVN1210). Patient plasma volumes required for analysis were 200 or 100 µL (non-HGPS 1:1 and 1:2) and 8 µL (HGPS 1:25) to generate duplicates. Filter plates with samples were centrifuged for 10 minutes to pull samples through the filters. Filtrate was then assessed as described.
Sample Blinding and Inclusion Criteria
Once the assay development was established, all samples were assessed in a blinded fashion by EMD Millipore technical staff. Repeat samples were assigned differing, random identification numbers. Identification assignments without Hutchinson-Gilford or non–Hutchinson-Gilford prefixes were submitted for analysis and converted to Hutchinson-Gilford and non–Hutchinson-Gilford identifiers by two of the authors (W.N. and L.B.G.) for statistical analysis and publication.
For inclusion in longitudinal analyses, sample sets required a baseline and end-of-trial on-therapy plasma sample. To compare lonafarnib doses, ProLon1 inclusion required a baseline pretherapy, 4-month (dose 115 mg/m2) and end-of-study sample (dose 150 mg/m2). For inclusion in pharmacokinetic analyses, sample sets required at least 1 trough and 1 posttrough plasma sample.
Statistical Analyses
Data summaries are primarily descriptive; comparisons between sample sets used paired Student t tests, 2-sample t tests, and Pearson correlations. Descriptive statistics included sample size, mean, and SD. Statistical significance level was set at a 2-sided P<0.05. There was no adjustment for multiple comparisons.
To assess the relationship between plasma progerin concentration and survival, 2 analyses were performed. The first implemented a joint model with a time-dependent slopes parameterization, which assessed the effect of the current value of progerin on survival, adjusting for the change in progerin. The second implemented a time-dependent Cox model, treating progerin as a time-dependent covariate. Assessment of only baseline progerin on survival outcome was not conducted due to bias, because, immediately after baseline, patients initiated treatment that was anticipated to affect progerin levels.
To quantify increase in life expectancy as it related to both decrease in progerin and duration of time on lonafarnib therapy, the methodology by Yang et al (2021),23 which proposes a dynamic model that uses a series of landmark time points24 on the basis of the conditional restricted mean survival time, was used. Survival and life expectancy analyses were adjusted for sex and age at baseline.
Excel was used for descriptive statistics. SAS version 9.4 was used for the time-dependent Cox model, the JM and dynpred packages in R version 4.0.2 were used for the joint modeling and conditional restricted mean survival time dynamic model, respectively.
Results
Progerin Assay Development and Validation
Assay development and validation details, figures and tables are located in the Supplemental Material (Figures S1 and S2; Tables S2–S5). In brief, the assay had a linear dynamic detection range of 59 to 30 000 pg/mL (R2=0.9987). Performance testing demonstrated average linearity of dilution of 98.2%; average inter- and intra-assay variability coefficients of variation of 7% and 12%, respectively; and average high- and mid–quality control coefficients of variation of 3.8% and 5.8%, respectively. For clinical samples that are frozen before analysis, 4 freeze-thaw cycles did not change progerin detection levels (all P>0.05).
HGPS-Specific Plasma Progerin Assay Assessments
Details, figures, and tables for HGPS-specific plasma progerin assay performance assessments are located in the Supplemental Material (Figure S3; Tables S6 and S7). In brief, these assessments demonstrate that the assay had no detectable cross-reactivity to lamin A; that both native and recombinant progerin yielded similar progerin recoveries of 96±6% and 120±15% on average, respectively; and that lonafarnib does not interfere with the progerin assay, such that exogenous lonafarnib added to the assay at 3× Cmax in treated patients did not alter progerin detection levels (P>0.05). In addition, there were no significant differences between progerin assessed in NaHeparin plasma versus EDTA plasma (P=0.98); NaHeparin plasma versus serum (P=1.00); or EDTA versus serum (P=0.97).
Plasma Progerin Detection in Untreated Patients
Non-HGPS Plasma
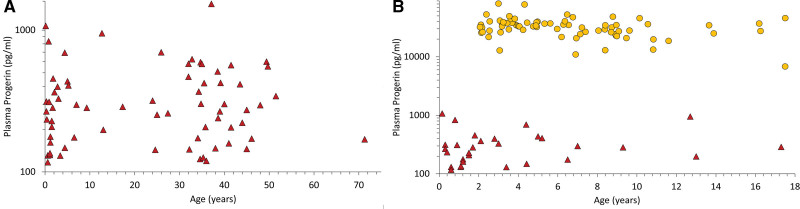
A set of non-HGPS plasma (n=69; 39 male, 30 female) was assessed (Figure 1A). This set of samples consisted of 33 donors evaluated for HGPS or non-HGPS progeroid laminopathies using LMNA ± ZMPSTE24 mutational analysis, 32 healthy relatives of those being evaluated for mutations, and 4 plasma samples from healthy controls purchased commercially. Within the donors being evaluated for HGPS or progeroid laminopathies, none had progerin-producing mutations. Overall, mean progerin was 351±251 pg/mL (median, 284; range, less than lower limit of quantification–1534). Mean donor age was 23.3±18.7 years (range, 0.2–71.3). There was no correlation between donor age and progerin (r=0.3, P=0.81). Progerin levels in a separate group of patients without HGPS with congestive heart failure (n=10; 5 male, 5 female; progerin 325±157 pg/mL) or kidney disease (n=10; 4 male, 6 female; progerin 361±227 pg/mL) were not elevated (P>0.05).
Figure 1.
Plasma progerin levels for patients without HGPS and untreated clinical trial patients. Each symbol represents a unique individual patient level versus age. y axes are log scale. A, Patients without HGPS (n=69; ▲). B, Untreated patients with HGPS (n=74; ● ) displayed along with the same patients without HGPS shown in A that fall within the same age range as the patients with HGPS (▲). HGPS indicates Hutchinson-Gilford progeria syndrome.
Plasma Progerin in Drug-Naive Patients With HGPS
The plasma progerin profile in children with HGPS was assessed at baseline, before trial drug initiation (Figure 1B). Mean donor age was 6.5±3.8 years (range, 2.1–17.5). Mean progerin was 33 261±12 346 pg/mL (median, 33 077) overall (n=74); with females 33 237±11 797 (median, 33 076; n=37) and males 33 285±13 035 (median, 34 214; n=37). There was no association between progerin and sex. Average HGPS progerin was 95-fold increased over non-HGPS (P<0.0001). A subgroup comparison consisting of all age- and sex-matched controls also demonstrated highly significant difference between HGPS (progerin, 34 512±14 432 pg/mL; n=25) and non-HGPS control levels (progerin, 377±226 pg/mL; n=13; P<0.0001).
For individual trials, mean baseline treatment-naive progerin was 27 572±7542 pg/mL (median, 27 669) for ProLon1 (n=26), 31 464±16 958 pg/mL (median, 29 787) for Triple Therapy (n=13), and 38 154±11 546 pg/mL (median=37 225) for ProLon2 (n=35). Although there were no significant differences in progerin in comparison with ProLon1 or ProLon2 with Triple Therapy (P=0.44 and 0.12, respectively), baseline values were higher in ProLon2 versus ProLon1 (P<0.0001).
Plasma Progerin Versus Age
Although most patients donated a single baseline sample just before clinical trial drug administration at the BCH trial site, a subset of patients also donated a blood sample as part of a natural history study conducted before the BCH trial donation, at the National Institutes of Health Clinical Center,2 thus allowing longitudinal progerin assessment free from the influence of drug therapy. These samples were subsequently submitted to the PRF Cell and Tissue Bank. National Institutes of Health study blood samples were processed similarly to the clinical trial samples. The average time between blood draws was 1.6±0.5 years (range, 0.9–2.3). Average initial and follow-up plasma progerin (n=13) was 29 221±7772 versus 33 272±11 959 pg/mL, respectively; there was no significant difference in progerin with time (P=0.14).
Progerin Changes in Patients With HGPS Treated With Farnesylation Inhibitors
Changes in Progerin With Lonafarnib Therapy
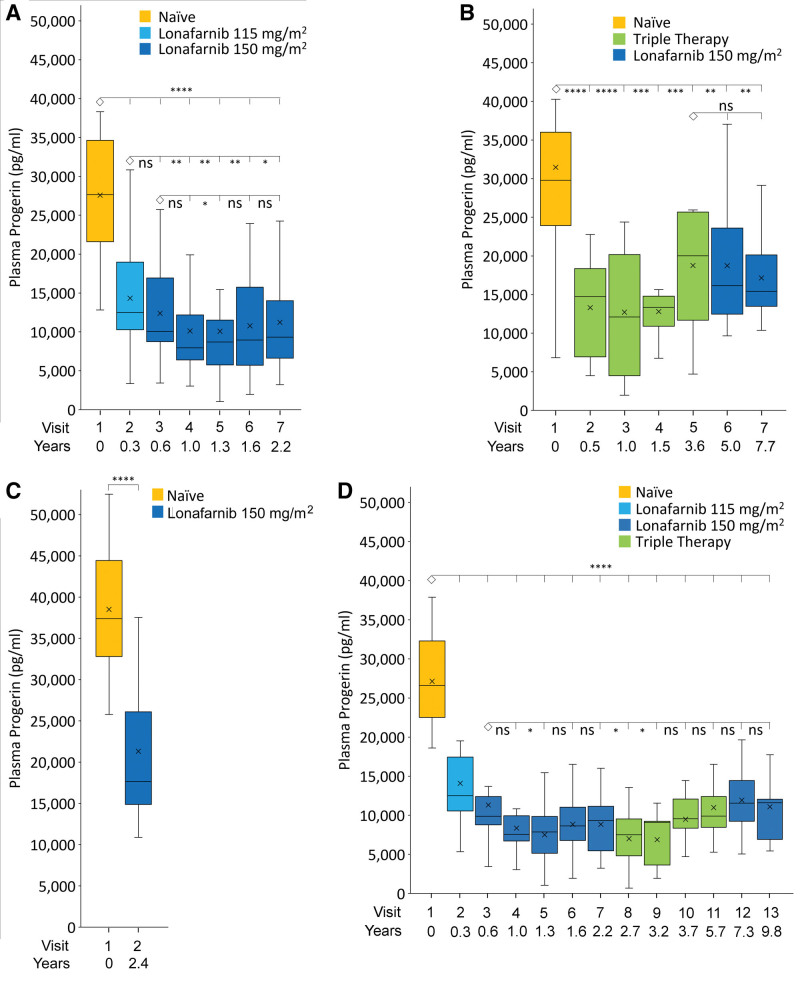
ProLon1 was the first clinical trial administering lonafarnib to children with HGPS and featured several differences from Triple Therapy and ProLon2. Patients visited the trial site and had blood collections every 4 months for a total of 2.2±0.1 years (7 visits), compared with every 6 months for a total of 3.5±0.2 years during Triple Therapy (5 visits) and baseline plus end-of-study at 2.4±0.6 years on therapy for ProLon2 (2 visits). In addition, patients in ProLon1 initiated therapy at 115 mg/m2 for 4 months before elevating to 150 mg/m2. Both Triple Trial and ProLon2 initiated patients at 150 mg/m2. Sample number varied across the individual studies and visits (Table S8); comparisons between study visits included matched plasma sets only. Progerin changes within and between separate but sequential trials were compared (Figure 2 and individual patient data shown in Figure S4). Overall, on-therapy plasma progerin decreased from baseline untreated by 38%, from 32 726±12 659 to 20 211±10 190 pg/mL (P<0.0001).
Figure 2.
Plasma progerin levels for patients undergoing clinical trials. On-site Trial center visit numbers and time on therapy versus mean±SEM progerin. Trial visits to Boston Children’s Hospital occurred at various times after visit 1 for different trials. All visit 1 patients were naive to therapy. A, ProLon1, treated with lonafarnib (n=25). B, Triple Trial patients treated with Triple Therapy from baseline to visit 5 (n=13), then switched to lonafarnib monotherapy thereafter (visits 6, 7; n=10). C, ProLon2 lonafarnib monotherapy (n=26). D, Long-term continuous therapy (n=13). Top and bottom box edges represent the 75th and 25th interquartile (IQR) ranges, respectively. Horizontal lines and X within boxes represent medians and means, respectively. Lower and upper whiskers represent Q1 − 1.5 × IQR and Q3 + 1.5 × IQR. *P<0.05; **P<0.01, ***P<0.001, ****P<0.0001; ns, P>0.05. Each visit is compared with the connected ◊ visit.
ProLon1 Progerin Changes With Lonafarnib Therapy
Average progerin decreased from baseline by 48% at month 4, during the dosing period using 115 mg/m2 lonafarnib (n=25, P<0.0001; Figure 2A, Figure S4A). For each of the 5 remaining patient visits where lonafarnib dose was 150 mg/m2, average decreases from baseline ranged between 50% and 62% (n=22–25 patients, all P<0.0001). At the 8-month trial visit, 4 months after transitioning from 115 to 150 mg/m2 dosing, progerin was further decreased to levels close to significance (n=25, P=0.06), and at each of the 4 subsequent trial visits, progerin was significantly decreased while taking 150 mg/m2 of lonafarnib over the 115 mg/m2 dose (n=22–25 patients, all P<0.05). There were no significant differences between average progerin at the 8-month 150 mg/m2 visit and subsequent visits at the same drug dose (n=22–25 patients, all P≥0.05). This dose response demonstrates the assay’s ability to detect an incremental but significant benefit of 150 mg/m2 dosing over 115 mg/m2.
Progerin Changes With Lonafarnib+Pravastatin+Zoledronate Therapy (Triple Therapy)
At the first on-therapy trial visit after 6 months, average progerin decreased from baseline by 41% (n=13, P=0.0018; Figure 2B, Figure S4B). Thereafter, average progerin remained significantly decreased from baseline by 35% to 47% (n=12–13 patients, visits 3–5, P=0.0015–0.0058).
After the formal Triple Therapy period ended, pravastatin and zoledronate were discontinued and 10 patients completed a lonafarnib monotherapy extension for an additional 4.1±0.5 years (range, 3.41–4.65; Figure 2B, Figure S4B). For these 10 patients, there were no significant differences in average progerin between consecutive Triple Therapy and on-treatment lonafarnib monotherapy extension trial visits (P=0.99).
ProLon2 Progerin Changes With Lonafarnib Therapy
Average progerin decreased from baseline by 36.7% during the dosing period (n=26, P<0.0001; Figure 2C, Figure S4C).
Progerin Changes With Long-Term Lonafarnib Therapy
A subgroup of 13 subjects were treated continuously with lonafarnib, without treatment breaks, as part of ProLon1, Triple Therapy Trial, and lonafarnib monotherapy extension of Triple Trial (Figure 2D, Figure S4D). This constitutes an average of 9.8±0.5 (range 9.0–10.3) years of continuous lonafarnib therapy. Average progerin decreased from baseline by 48% at month 4 (P<0.0001) during the dosing period using 115 mg/m2. For all subsequent time points, average decreases from baseline during 150 mg/m2 dosing ranged from 56% to 74% (P<0.0001). Compared with the first time point on 150 mg/m2 (time point 3 at 0.66 year), time points 5, 8, and 9 were significantly lower (P<0.05), and all other time points (4, 6, 7, 10–13) were similar to time point 3 (P≥0.05).
Diurnal Variation in Progerin Using Pharmacokinetics Studies
Plasma was collected at trough, and postdose hours 1, 2, 4, 6, and 8 for pharmacokinetic studies during ProLon1 at lonafarnib doses 115 mg/m2 and 150 mg/m2,18 and during Triple Therapy as well.22 Within each of these sample sets, progerin did not demonstrate significant changes in postdose versus trough levels for ProLon1 at 150 mg/m2 and for Triple Therapy, indicating that progerin does not demonstrate diurnal variation (all P≥0.05). For ProLon1 lonafarnib dose of 115 mg/m2, average progerin trough was similar to 1-hour postdose (P=0.27), but was significantly lower than 2, 4, and 8 hours postdose and differed on average by ≈3000 pg/mL (all P<0.05).
Larger Progerin Decrease Is Associated With Improved Patient Survival
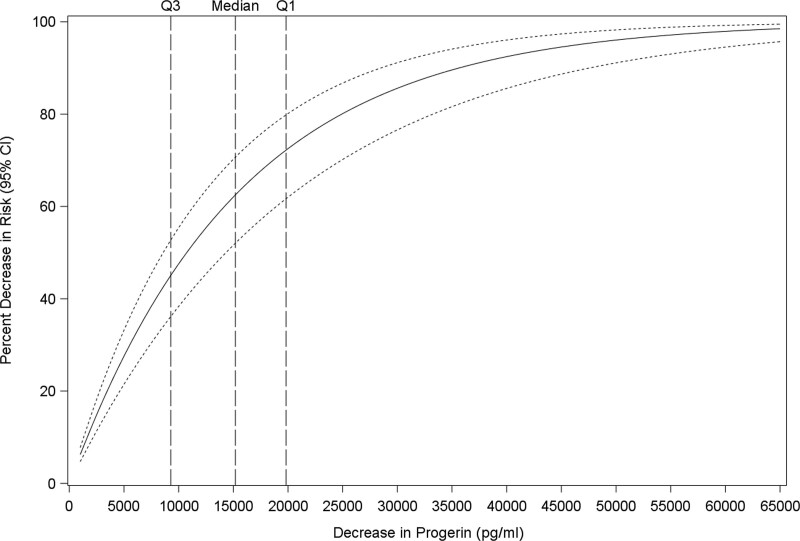
The suitability of plasma progerin to serve as a prognostic biomarker for survival was assessed. First, change in mortality risk per change in measured progerin values was estimated directly from the joint model relating these 2 measures. All patients with plasma samples were included regardless of whether therapy was implemented, length of therapy, or follow-up time (n=74; 26 deceased and 47 living as of April 1, 2022). Progerin levels were significantly related to risk of death (P<0.0001; Figure 3). For example, decreases in plasma progerin of 1000, 10 000, and 15 000 pg/mL (the median decrease on lonafarnib) correspond to 6.6% (95% CI, 5.0%–8.1%), 49.3% (95% CI, 40.0%–57.2%), and 63.9% (95% CI, 53.5%–72.0%) decreases in the risk of death. Progerin was modeled as a function of length of therapy, length of therapy squared, sex, and age at baseline. The joint model adjusted for length of therapy. Additional analyses using standard time-dependent Cox modeling, which does not consider potential measurement error in the longitudinal covariate (progerin), and subgroup analysis, as well, using only those patients with at least 1 on-therapy plasma sample, yielded similar results (Fig. S5, Table S9).
Figure 3.
Progerin level is associated with survival. Risk of death incrementally decreased with lower plasma progerin (P<0.0001) using time-dependent joint modeling. Change in mortality risk (y axis) versus all values in range of observed progerin decrease (x axis) from 0 to 65 000 pg/mL. Solid line is % decrease in risk if death; dashed lines = 95% CI. Median decrease=15 186 pg/mL (Q1;Q3=19 826;9281). n=74 subjects (9 untreated with single samples and 65 treated with untreated baseline plus multiple on-therapy samples).
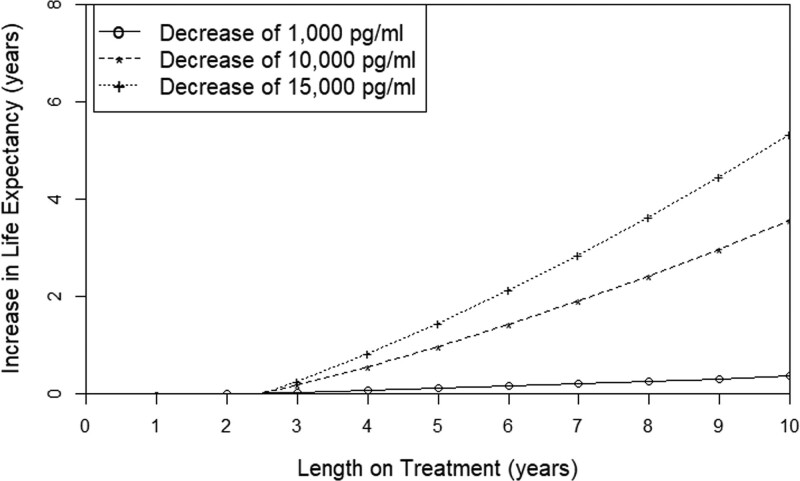
Next, the relationship between lonafarnib treatment duration and change in life expectancy with extent of change in plasma progerin was assessed (Figure 4). This analysis included samples from patients with a baseline pretherapy and at least 1 on-treatment progerin measurement (n=65 patients). Life expectancy incrementally improved with both extent of plasma progerin decrease, and time spent at lower progerin level. Three representative wide-ranging examples of on-therapy progerin decreases (1000, 10 000, and 15 000 pg/mL) spanning 10 years on treatment are graphed in Figure 4. Patient lifespan increased in all 3 examples.
Figure 4.
Extent of increased life expectancy is incrementally associated with lower plasma progerin and longer duration of time on therapy. Increase in life expectancy (y axis) versus length of treatment (x axis) shown for 3 selected changes in plasma progerin (n=65 subjects with baseline and multiple on-therapy samples), using dynamic modeling on the basis of restricted mean survival time.
Discussion
The development of clinically relevant biological markers of disease is critical to identifying and implementing effective treatments for children and young adults with HGPS. Validation of the progerin biomarker in this study that directly quantifies the disease-causing protein has the potential to significantly improve both preclinical and clinical trial assessments of new treatment approaches for this universally fatal disease. Moreover, plasma progerin’s quantitative relationship to patient survival solidifies its clinical relevance.
Progerin has not previously been detected in plasma, serum, or urine, all of which can be sampled longitudinally. Previous investigations of treated and untreated HGPS patient serum and plasma have therefore relied on identifying novel potential biomarkers that lie downstream of the disease-causing protein, progerin,2,18,25–27 and reflect either the direct or indirect effects of this toxic protein. In this study, a sensitive, reproducible immunoassay for detecting progerin was developed and subsequently used to explore progerin’s presence in plasma, filling a major gap in the field of progeria research. The assay did not detect lamin A, progerin’s normal protein counterpart. The sensitivity and low-volume format of the assay allowed analyses of samples of individual patients with HGPS without the need for pooling, which is important for this population of small and fragile children in which blood draw volume must be minimized for safety.
Assay Potential for Investigating Pathobiology of HGPS and Aging
High assay sensitivity also permitted progerin quantification in non-HGPS individuals, which will facilitate future research on aging and aging-related diseases. Progerin levels in plasma from untreated patients with HGPS were on average 95-fold higher than in non-HGPS plasma, although non-HGPS values were sometimes below the assay’s sensitivity (less than lower limit of quantification). This agrees with a previous autopsy study that detected similar ratios of progerin in the vascular wall using immunofluorescence.21 Thus, this assay greatly expands the possibilities for preclinical and clinical research into HGPS and generalized aging.
Although plasma was chosen for primary assay development, other noninvasive biological samples such as urine or saliva from patients and animal models, and cell culture material and disease tissue, as well, might also be evaluable using a similar approach in the future. A comprehensive study of progerin’s relative presence between different organs and tissue types using the assay developed here on either human autopsy specimens or animal models of HGPS would be highly useful to understand how plasma progerin reflects relative tissue contributions to disease.
The limited dataset of samples collected longitudinally from patients naive-to-therapy indicates that progerin deposition into the plasma remains steady during the 2-year period tested. In addition, on average, the plasma levels in the treated patient group were decreased by almost 40% from baseline and stayed lower for up to 10 years. The pathophysiology behind these longitudinal findings deserves further investigation. There is evidence from human autopsy tissue21,28 that progerin can accumulate in tissues with time, implying that progerin production per cell might increase with aging. However, aging in both humans and HGPS mice also results in a relatively acellular vascular media where proteoglycan replaces healthy tissue and vascular stiffening ensues.21,29,30 Other progerin-producing tissues such as liver and kidney do not seem as susceptible to cell loss. A better understanding of the balance between cellular progerin production, cell loss, and the relative contributions of various organs to plasma progerin is needed for insight into the interplay between plasma progerin and tissue pathology, and for a more comprehensive assessment of drug activity.
Advancing Drug Development and Discovery for the Progeria Ultra-rare Disease Community
The only approved drug for HGPS and select progeroid laminopathies is the farnesyltransferase inhibitor lonafarnib (Zokinvy), placing Progeria among the <5% of rare diseases with an approved medication.31 Its clinical trial success was defined by outcomes measured after at least 2 years of treatment, presumably when the sensitivity of clinical testing was adequate to detect change.18,22 The presence of lonafarnib did not interfere with progerin detection. Lonafarnib therapy significantly decreased plasma progerin levels within 4 months, portending a potential shorter-term readout of drug efficacy in future trials with other drugs, depending on the mechanism of drug effect. This would decrease the risk of being exposed to interventions that may not be effective, such as in the Triple Therapy Trial where adding pravastatin and zoledronate did not enhance lonafarnib’s beneficial effects on disease.22 Earlier identification of an effective therapy could be accelerated with regulatory use of progerin as a surrogate end point, in combination with a longer-term clinical outcome measure. In addition, a steady-state on-therapy reduction in progerin for up to 10 years implies that there was no measurable rebound in progerin. Thus, progerin levels in blood are an excellent candidate for both short- and long-term treatment trial outcomes.
A biomarker is a defined characteristic that is measured as an indicator of normal biological or pathogenic processes, or biological responses to an exposure or intervention, including therapeutic interventions.32 This study demonstrates that plasma progerin is an indicator of both pathogenic process and of clinical response to treatment. When untreated patients were included in the survival analysis, plasma progerin levels were associated with mortality risk regardless of whether a treatment had been implemented. Furthermore, plasma progerin not only decreased in response to treatment with lonafarnib, but that decrease was strongly associated with an incrementally conferred survival benefit. For example, because, on average, trial patients’ plasma progerin decreased by >14 000 pg/mL, life expectancy for the 13 patients receiving therapy for >10 years is estimated to increase by almost 5 years. This represents a >35% increase in average lifespan, from 14.5 to almost 20 years of age. Thus, the critical statistical and biological evidence for the use of progerin as a biomarker reasonably likely to predict clinical benefit is present.
Relative Impact Assessment of Progerin Changes Will Depend on Treatment Mechanism of Action
There are a variety of treatment strategies in development for HGPS that may benefit greatly from an assay capable of detecting progerin. These potential treatments are targeted to alter progerin levels using widely varying mechanisms of action (reviewed in Macicior et al17), including genetic editing,33 RNA-based therapies,29,34 and small molecules that target progerin protein’s posttranslational processing, interactions with lamin A or rate of autophagy, or both.
When evaluating the extent to which plasma progerin change reflects disease improvement, the mechanism of drug action must be considered. For example, genetic editing permanently corrects a cell’s mutation, so that its progeny will also be normal. In a HGPS murine study using this approach, a 140% increase in lifespan was achieved with only 20% to 60% mutation correction of cells in tested tissues.33 Part of the efficacy of this correction was presumably engendered by a healthy subpopulation of cells and their creation of a healthier microenvironment, which had ramifications for adjacent cells that were not corrected. Lonafarnib, which, on average, caused a 38% decrease in plasma progerin, penetrates all cell types and would therefore inherently decrease progerin on a large scale, even though the cells it affects are still diseased and continue to produce new progerin molecules. In this context, the gene editor might have less of an effect on plasma progerin than lonafarnib, while still producing greater clinical benefit. Thus, the hypothesis would be that any decrease in plasma progerin reflects a benefit to disease, but the relative magnitude of the decrease between different types of therapies may not be a valid way to assess the extent of benefit.
Study Limitations
There are several study limitations. First, plasma is a “sink” for deposition of progerin from multiple organs and does not differentiate the relative contribution from organs of major disease interest such as the heart and vasculature. However, there is evidence that drug delivery to organs other than vasculature can be beneficial, because extravascular delivery of gene therapy improved both health span and increased lifespan in HGPS mice.35,36 Thus, the natural history of HGPS is not understood well enough to eliminate noncardiovascular organs as pivotal contributors to phenotype.
Conclusions
Treatment with lonafarnib improves some disease features and significantly extends lifespan in progeria, but affected individuals still die of accelerated atherosclerosis and heart failure in their teens and twenties. There is a critical need for improved treatments and a cure. The progerin assay established in this study will enable early identification of new treatments that effectively target progerin, either directly or indirectly.
Article Information
Acknowledgments
The authors are extremely grateful to the children with progeria and their families for participation in this research. The authors thank F. Collins and M. Erdos for supplying HGPS murine plasma for assay controls; M. Meridith, W. Gahl, and W. Introne for National Institutes of Health sample donations to The PRF Cell and Tissue Bank; and S. Shaw for assistance with figures. Dr Gordon had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. Concept and design: Gordon, Massaro, D’Agostino. Acquisition, analysis, or interpretation of data: Gordon, Norris, Hamren, Goodson, Tuminelli, Kleinman, Kieran. Drafting of the manuscript: Gordon, Norris, Massaro, D’Agostino. Critical revision of the manuscript for important intellectual content: Kleinman, Kieran. Statistical analysis: Massaro, D’Agostino, LeClair, Lyass. Obtained funding: Gordon, Kleinman, Kieran. Administrative, technical, or material support: Gordon, Kleinman, Hamren, Goodson. Supervision: Gordon, Kleinman.
Sources of Funding
This study was funded by grants PRF-2012-NH, PRF-2002-CB, from The Progeria Research Foundation, DSF Charitable Foundation, US FDA 1U01FD006886-01, and a Singulex SMC technology innovation award. Schering Plough/Merck/Eiger provided the medication used in the treatment trials at no cost.
Disclosures
Dr Gordon is the mother of a subject that was included in this study.
Supplemental Material
Supplemental Methods
Supplemental Results
Tables S1–S9
Figures S1–S5Reference 37
Supplementary Material
Nonstandard Abbreviations and Acronyms
- BCH
- Boston Children’s Hospital
- HGPS
- Hutchinson-Gilford progeria syndrome
- PRF
- The Progeria Research Foundation
Supplemental Material, the podcast, and transcript are available with this article at https://www.ahajournals.org/doi/suppl/10.1161/CIRCULATIONAHA.122.060002.
For Sources of Funding and Disclosures, see page 1743.
Circulation is available at www.ahajournals.org/journal/circ
Contributor Information
Wendy Norris, Email: wnorris@lifespan.org.
Sarah Hamren, Email: sarahhamren@gmail.com.
Robert Goodson, Email: robert1_goodson@yahoo.com.
Jessica LeClair, Email: jleclai2@bu.edu.
Joseph Massaro, Email: jmm@bu.edu.
Asya Lyass, Email: asya@bu.edu.
Ralph B. D’Agostino, Sr, Email: leilanie722@gmail.com.
Kelsey Tuminelli, Email: ktuminelli@progeriaresearch.org.
Mark W. Kieran, Email: mwkieran@gmail.com.
Monica E. Kleinman, Email: monica.kleinman@childrens.harvard.edu.
References
- 1.Gordon LB. PRF by the numbers. The Progeria Research Foundation. 2013. Accessed February 1, 2023. https://www.progeriaresearch.org/prf-by-the-numbers/?hilite=PRF+numbers [Google Scholar]
- 2.Merideth MA, Gordon L, Clauss S, Sachdev V, Smith A, Perry M, Brewer C, Zalewski C, Kim H, Solomon B, et al. Phenotype and course of Hutchinson-Gilford progeria syndrome. N Engl J Med. 2008;358:592–604. doi: 10.1056/NEJMoa0706898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gordon LB, Shappell H, Massaro J, D’Agostino RB, Sr, Brazier J, Campbell SE, Kleinman ME, Kieran MW. Association of lonafarnib treatment vs no treatment with mortality rate in patients with Hutchinson-Gilford progeria syndrome. JAMA. 2018;319:1687–1695. doi: 10.1001/jama.2018.3264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gordon LB, Massaro J, D’Agostino RB, Sr, Campbell SE, Brazier J, Brown WT, Kleinman ME, Kieran MW; Progeria Clinical Trials Collaborative. Impact of farnesylation inhibitors on survival in Hutchinson-Gilford progeria syndrome. Circulation. 2014;130:27–34. doi: 10.1161/CIRCULATIONAHA.113.008285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Sandre-Giovannoli A, Levy N. Altered splicing in prelamin A-associated premature aging phenotypes. Prog Mol Subcell Biol. 2006;44:199–232. doi: 10.1007/978-3-540-34449-0_9 [DOI] [PubMed] [Google Scholar]
- 6.Eriksson M, Brown WT, Gordon L, Glynn MW, Singer J, Scott L, Erdos MR, Robbins CM, Moses TY, Berglund P, et al. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature. 2003;423:293–297. doi: 10.1038/nature01629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scaffidi P, Misteli T. Reversal of the cellular phenotype in the premature aging disease Hutchinson-Gilford progeria syndrome. Nat Med. 2005;11:440–445. doi: 10.1038/nm1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fong LG, Ng JK, Lammerding J, Vickers TA, Meta M, Cote N, Gavino B, Qiao X, Chang SY, Young SR, et al. Prelamin A and lamin A appear to be dispensable in the nuclear lamina. J Clin Invest. 2006;116:743–752. doi: 10.1172/JCI27125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benedicto I, Dorado B, Andrés V. Molecular and cellular mechanisms driving cardiovascular disease in Hutchinson-Gilford progeria syndrome: lessons learned from animal models. Cells. 2021;10:1157. doi: 10.3390/cells10051157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cabral WA, Tavarez UL, Beeram I, Yeritsyan D, Boku YD, Eckhaus MA, Nazarian A, Erdos MR, Collins FS. Genetic reduction of mTOR extends lifespan in a mouse model of Hutchinson-Gilford Progeria syndrome. Aging Cell. 2021;20:e13457. doi: 10.1111/acel.13457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bar DZ, Arlt MF, Brazier JF, Norris WE, Campbell SE, Chines P, Larrieu D, Jackson SP, Collins FS, Glover TW, et al. A novel somatic mutation achieves partial rescue in a child with Hutchinson-Gilford progeria syndrome. J Med Genet. 2017;54:212–216. doi: 10.1136/jmedgenet-2016-104295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hisama FM, Lessel D, Leistritz D, Friedrich K, McBride KL, Pastore MT, Gottesman GS, Saha B, Martin GM, Kubisch C, et al. Coronary artery disease in a Werner syndrome-like form of progeria characterized by low levels of progerin, a splice variant of lamin A. Am J Med Genet A. 2011;155:3002–3006. doi: 10.1002/ajmg.a.34336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilbert HTJ, Swift J. The consequences of ageing, progeroid syndromes and cellular senescence on mechanotransduction and the nucleus. Exp Cell Res. 2019;378:98–103. doi: 10.1016/j.yexcr.2019.03.002 [DOI] [PubMed] [Google Scholar]
- 14.Kubben N, Zhang W, Wang L, Voss Ty C, Yang J, Qu J, Liu G-H, Misteli T. Repression of the antioxidant NRF2 pathway in premature aging. Cell. 2016;165:1361–1374. doi: 10.1016/j.cell.2016.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fang EF, Scheibye-Knudsen M, Chua KF, Mattson MP, Croteau DL, Bohr VA. Nuclear DNA damage signalling to mitochondria in ageing. Nat Rev Mol Cell Biol. 2016;17:308–321. doi: 10.1038/nrm.2016.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sola-Carvajal A, Revêchon G, Helgadottir HT, Whisenant D, Hagblom R, Döhla J, Katajisto P, Brodin D, Fagerström-Billai F, Viceconte N, et al. Accumulation of progerin affects the symmetry of cell division and is associated with impaired Wnt signaling and the mislocalization of nuclear envelope proteins. J Investig Dermatol. 2019;139:2272–2280. doi: 10.1016/j.jid.2019.05.005 [DOI] [PubMed] [Google Scholar]
- 17.Macicior J, Marcos-Ramiro B, Ortega-Gutiérrez S. Small-molecule therapeutic perspectives for the treatment of progeria. Int J Mol Sci . 2021;22:7190. doi: 10.3390/ijms22137190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gordon LB, Kleinman ME, Miller DT, Neuberg DS, Giobbie-Hurder A, Gerhard-Herman M, Smoot LB, Gordon CM, Cleveland R, Snyder BD, et al. Clinical trial of a farnesyltransferase inhibitor in children with Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci U S A. 2012;109:16666–16671. doi: 10.1073/pnas.1202529109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Camafeita E, Jorge I, Rivera-Torres J, Andrés V, Vázquez J. Quantification of farnesylated progerin in Hutchinson-Gilford progeria patient cells by mass spectrometry. Int J Mol Sci . 2022;23:11733. doi: 10.3390/ijms231911733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McClintock D, Ratner D, Lokuge M, Owens DM, Gordon L, Collins FS, Djabali K. The mutant form of lamin A that causes Hutchinson-Gilford progeria is a biomarker of cellular aging in human skin. PLoS One. 2007;2:1–9. doi: 10.1371/journal.pone.0001269.t001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olive M, Harten I, Mitchell R, Beers JK, Djabali K, Cao K, Erdos MR, Blair C, Funke B, Smoot L, et al. Cardiovascular pathology in Hutchinson-Gilford progeria: correlation with the vascular pathology of aging. Arterioscler Thromb Vasc Biol. 2010;30:2301–2309. doi: 10.1161/ATVBAHA.110.209460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gordon LB, Kleinman ME, Massaro J, D’Agostino RB, Shappell H, Gerhard-Herman M, Smoot LB, Gordon CM, Cleveland RH, Nazarian A, et al. Clinical trial of the protein farnesylation inhibitors lonafarnib, pravastatin, and zoledronic acid in children with Hutchinson-Gilford progeria syndrome. Circulation. 2016;134:114–125. doi: 10.1161/CIRCULATIONAHA.116.022188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang Z, Wu H, Hou Y, Yuan H, Chen Z. Dynamic prediction and analysis based on restricted mean survival time in survival analysis with nonproportional hazards. Comput Methods Programs Biomed. 2021;207:106155. doi: 10.1016/j.cmpb.2021.106155 [DOI] [PubMed] [Google Scholar]
- 24.Dafni U. Landmark analysis at the 25-year landmark point. Circ Cardiovasc Qual Outcomes. 2011;4:363–371. doi: 10.1161/CIRCOUTCOMES.110.957951 [DOI] [PubMed] [Google Scholar]
- 25.Gordon LB, Harten IA, Patti ME, Lichtenstein AH. Reduced adiponectin and HDL cholesterol without elevated C-reactive protein: clues to the biology of premature atherosclerosis in Hutchinson-Gilford progeria syndrome. J Pediatr. 2005;146:336–341. doi: 10.1016/j.jpeds.2004.10.064 [DOI] [PubMed] [Google Scholar]
- 26.Harten IA, Zahr RS, Lemire JM, Machan JT, Moses MA, Doiron RJ, Curatolo AS, Rothman FG, Wight TN, Toole BP, et al. Age-dependent loss of MMP-3 in Hutchinson-Gilford progeria syndrome. J Gerontol A Biol Sci Med Sci. 2011;66:1201–1207. doi: 10.1093/gerona/glr137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gordon LB, Campbell SE, Massaro JM, D’Agostino RB, Sr, Kleinman ME, Kieran MW, Moses MA. Survey of plasma proteins in children with progeria pre-therapy and on-therapy with lonafarnib. Pediatr Res. 2018;83:982–992. doi: 10.1038/pr.2018.9 [DOI] [PubMed] [Google Scholar]
- 28.McClintock D, Gordon LB, Djabali K. Hutchinson-Gilford progeria mutant lamin A primarily targets human vascular cells as detected by an anti-Lamin A G608G antibody. Proc Natl Acad Sci U S A. 2006;103:2154–2159. doi: 10.1073/pnas.0511133103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Erdos MR, Cabral WA, Tavarez UL, Cao K, Gvozdenovic-Jeremic J, Narisu N, Zerfas PM, Crumley S, Boku Y, Hanson G, et al. A targeted antisense therapeutic approach for Hutchinson-Gilford progeria syndrome. Nat Med. 2021;27:536–545 doi: 10.1038/s41591-021-01274-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murtada SI, Kawamura Y, Caulk AW, Ahmadzadeh H, Mikush N, Zimmerman K, Kavanagh D, Weiss D, Latorre M, Zhuang ZW, et al. Paradoxical aortic stiffening and subsequent cardiac dysfunction in Hutchinson-Gilford progeria syndrome. J R Soc Interface. 2020;17:20200066. doi: 10.1098/rsif.2020.0066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tambuyzer E, Vandendriessche B, Austin CP, Brooks PJ, Larsson K, Miller Needleman KI, Valentine J, Davies K, Groft SC, Preti R, et al. Therapies for rare diseases: therapeutic modalities, progress and challenges ahead. Nat Rev Drug Discovery. 2020;19:93–111. doi: 10.1038/s41573-019-0049-9 [DOI] [PubMed] [Google Scholar]
- 32.FDA-NIH Biomarker Working Group. BEST (Biomarkers, EndpointS, and other Tools) Resource. Silver Spring, MD: Food and Drug Administration, Bethesda, MD: National Institutes of Health; 2016. [PubMed] [Google Scholar]
- 33.Koblan LW, Erdos MR, Wilson C, Cabral WA, Levy JM, Xiong ZM, Tavarez UL, Davison LM, Gete YG, Mao X, et al. In vivo base editing rescues Hutchinson-Gilford progeria syndrome in mice. Nature. 2021;589:608–614. doi: 10.1038/s41586-020-03086-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Puttaraju M, Jackson M, Klein S, Shilo A, Bennett CF, Gordon L, Rigo F, Misteli T. Systematic screening identifies therapeutic antisense oligonucleotides for Hutchinson-Gilford progeria syndrome. Nat Med. 2021;27:526–535. doi: 10.1038/s41591-021-01262-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Santiago-Fernandez O, Osorio FG, Quesada V, Rodriguez F, Basso S, Maeso D, Rolas L, Barkaway A, Nourshargh S, Folgueras A, et al. Development of a CRISPR/Cas9-based therapy for Hutchinson-Gilford progeria syndrome. Nat Med. 2019;25:423–426. doi: 10.1038/s41591-018-0338-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beyret E. Single-dose CRISPR-Cas9 therapy extends lifespan of mice with Hutchinson-Gilford progeria syndrome. Nat Med. 2019;25:419–422. doi: 10.1038/s41591-019-0343-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Todd J, Freese B, Lu A, Held D, Morey J, Livingston R, Goix P. Ultrasensitive flow-based immunoassays using single-molecule counting. Clin Chem. 2007;53:1990–1995. doi: 10.1373/clinchem.2007.091181 [DOI] [PubMed] [Google Scholar]