Abstract
The genus Thalassotalea is ubiquitous in marine environments, and up to 20 species have been described so far. A Gram-staining-negative, aerobic bacterium, designated strain PTE2T was isolated from laboratory-reared larvae of the Japanese sea cucumber Apostichopus japonicus. Phylogenetic analysis based on the 16S rRNA gene nucleotide sequences revealed that PTE2T was closely related to Thalassotalea sediminis N211T (= KCTC 42588T = MCCC 1H00116T) with 97.9% sequence similarity. ANI and in silico DDH values against Thalassotalea species were 68.5–77.0% and 19.7–24.6%, respectively, indicating the novelty of PTE2T. Based on genome-based taxonomic approaches, strain PTE2T (= JCM 34608T = KCTC 82592T) is proposed as a new species, Thalassotalea hakodatensis sp. nov.
Introduction
The genus Thalassotalea, a member of the family Colwelliaceae in the order Alteromonadales, was first proposed by Zhang et al. [1] with the description of Thalassotalea piscium. At the same time, four Thalassomonas species, Thalassomonas ganghwensis [2], Thalassomonas loyana [3], Thalassomonas agarivorans [4] and Thalassomonas agariperforans [5], were reclassified into the genus Thalassotalea as Thalassotalea ganghwensis, Thalassotalea loyana, Thalassotalea agarivorans and Thalassotalea agariperforans, respectively. Currently, 20 species have been described in this genus [1, 6–18]. Thalassotalea strains have been isolated from flounder [1], seawater [4, 6, 15, 19], marine sediment [2, 8, 16, 20], marine sand [5, 12], corals [3, 9, 10, 14], aquaculture systems [7], pacific oyster [11], deep-sea seamounts [13], mangrove sediment [17] and red alga [18]. The genus is characterized as being rod-shaped, Gram-negative, aerobic or facultatively anaerobic, catalase and oxidase-positive, and motile with single polar flagellum or non-motile. A previous study showed that one of the members of this genus, Thalassotalea sp. PP2-459 isolated from a carpet-shell clam, possesses quorum-quenching ability against Vibrio anguillarum, which is one of the key pathogens in marine bivalve aquaculture, and thus this strain is proposed as a probiotic candidate [21]. The strain is also known to produce N-acyl dehydrotyrosines, which shows the potential for therapeutic and cosmetic applications [22]. In addition, members of Thalassotalea may contribute to nutrient cycling in marine environments by their polysaccharide degrading abilities [23]. Nevertheless, despite the ubiquity of the Thalassotalea strains and their heterogeneity in phenotypes, no comprehensive genome studies have been performed yet.
During studies on microbiomes associated with the early life stages of the sea cucumber Apostichopus japonicus [24], a new species candidate, strain PTE2, of the genus Thalassotalea was isolated from the pentactula larvae. The authors report that ASVs (ASV0402, ASV0238, and ASV0323), which possess the same V1-V2 region as strain PTE2, increased in abundance in the seawater of late auricularia and pentacutula larvae, suggesting that strain PTE2 may play an important role in host-microbe interaction during sea cucumber development [24]. In this study, modern polyphasic taxonomic studies was employed, including molecular phylogenetic analysis based on 16S rRNA gene nucleotide sequences, phenotypic characterization and genome comparisons, to characterize the newly described species, Thalassotalea sp. PTE2T (= JCM 34608T = KCTC 82592T).
Materials and methods
Bacterial strains and phenotypic characterization
The strain PTE2T was isolated from the pentactula larvae of Apostichopus japonicus [24]. Bacterial colonies were purified using a 1/5 strength ZoBell 2216 agar or broth [24]. T. sediminis KCTC 42588T, T. insulae KCTC 62186T, T. piscium JCM 18590T, T. agarivorans JCM 13379T, and T. atypica JCM 31894T were used as references for genomic and phenotypic comparisons against the strain PTE2T. All strains were cultured on Marine agar 2216 (BD, Franklin Lakes, New Jersey, USA). The phenotypic characteristics were determined according to previously described methods [25–29]. Motility was observed under a microscope using cells suspended in droplets of sterilized 75% artificial seawater (ASW).
Molecular phylogenetic analysis based on 16S rRNA gene nucleotide sequences
The almost full length 16S rRNA gene sequence (1,424 bp) of strain PTE2T was obtained by direct sequencing of PCR-amplified DNA. 27F and 1509R were used as amplification primers, and four primers: 27F, 800F, 920R and 1509R were used for the Sanger sequencing [29, 30]. Raw sequence reads were assembled to generate a contig using ChromasPro Ver.2.1.10 (Technelysium Pty. Ltd. South Brisbane, Australia). The 16S rRNA gene nucleotide sequences of the type strains of the genus Thalassotalea and other Colwelliaceae species were retrieved from NCBI databases. Sequences were aligned using Silva Incremental Aligner v1.2.11 [29, 31]. A phylogenetic model test and maximum likelihood (ML) tree reconstruction were performed using the MEGAX v.10.1.8 program [29, 32, 33]. ML tree was reconstructed with 1,000 bootstrap replications using Kimura 2-parameter (K2) with gamma distribution (+G) and invariant site (+I) model. In addition, nucleotide similarities among strains were also calculated using “compute pairwise distance” in MEGAX.
Whole genome sequencing
Genomic sequence of PTE2T, T. sediminis KCTC 42588T, T. agarivorans JCM 13379T, T. piscium JCM 18590T T. insulae KCTC 62186T and T. atypica JCM 31894T was obtained using a hybrid assembly method described previously [29]. Genomic sequence of T. loyana LMG 22536T and T. eurytherma JCM 18482T was obtained by using only Illumina sequence reads. Illumina sequence was obtained using a method described previously [29], and assembled using Unicycler 0.4.8 [34]. The whole genome sequences were annotated with DDBJ Fast Annotation and Submission Tool (DFAST) [35]. The complete genome sequences of PTE2T acquired in this study were deposited in GenBank/EMBL/DDBJ under accession number AP027365.
Overall genome relatedness indices (OGRIs)
Overall genome relatedness indices (OGRIs) were calculated to determine the novelty of PTE2T using same methodology described in Yamano et al. [29]. Average nucleotide identities (ANIs) were calculated using the Orthologous Average Nucleotide Identity Tool (OrthoANI) software to calculate OrthoANI with a default setting [36] using genomes of the PTE2T [29]. In silico DDH values were calculated using Genome-to-Genome Distance Calculator (GGDC) 2.1 based on formula 2 being the most robust formula using both complete and incomplete genomes [37]. Average amino acid identities (AAIs) were calculated between PTE2T and other related Colwelliaceae species (Tables 1 and S1) using an enveomics toolbox [38].
Table 1. Genome properties of PTE2T and Thalassotalea species with available genomic sequences.
| Species | Strain | RefSeq accession | Size | Genome assemble status | GC content |
|---|---|---|---|---|---|
| Thalassotalea hakodatensis sp. nov. | PTE2T | AP027365 (in this study) | 4.31 Mb | complete (1 chromosome) | 38.5% |
| Thalassotalea sediminis | KCTC 42588T | AP027361 (in this study) | 3.90 Mb | complete (1 chromosome) | 38.6% |
| Thalassotalea insulae | KCTC 62186T | BSST01000001-BSST01000003 (in this study) | 4.39 Mb | draft (3 contigs) | 40.3% |
| Thalassotalea piscium | JCM 18590T | AP027362 (in this study) | 3.90 Mb | complete (1 chromosome) | 37.5% |
| Thalassotalea agarivorans | JCM 13379T | AP027363 (in this study) | 3.39 Mb | complete (1 chromosome) | 41.9% |
| Thalassotalea loyana | LMG 22536T | BSSV01000001-BSSV01000017 (in this study) | 4.00 Mb | draft (17 contigs) | 40.4% |
| Thalassotalea eurytherma | JCM 18482T | BSSU01000001-BSSU01000035 (in this study) | 3.55 Mb | draft (35 contigs) | 39.7% |
| Thalassotalea atypica | JCM 31894T | AP027364 (in this study) | 4.41 Mb | complete (1 chromosome) | 40.2% |
| Thalassotalea marina | QBLM2T | GCF 014656435.1 | 4.87 Mb | draft (31 contigs) | 39.4% |
| Thalassotalea profundi | YM155T | GCF 014653195.1 | 3.99 Mb | draft (40 contigs) | 36.3% |
| Thalassotalea mangrovi | zs-4T | GCF 005116735.1 | 3.71 Mb | draft (76 contigs) | 45.9% |
| Thalassotalea crassostreae | LPB0090T | GCF 001831495.1 | 3.86 Mb | complete (1 chromosome) | 38.8% |
| Thalassotalea algicola | M1531T | GCF_012932965.1 | 4.06 Mb | draft (19 contigs) | 39.1% |
| Thalassotalea litorea | MCCC IK03283 | GCF 005116735.1 | 3.88 Mb | draft (46 contigs) | 43.9% |
| Thalassotalea euphylliae | H2 | GCF 003390395.1 | 4.36 Mb | complete (1 chromosome) | 43.0% |
Multilocus sequence analysis (MLSA)
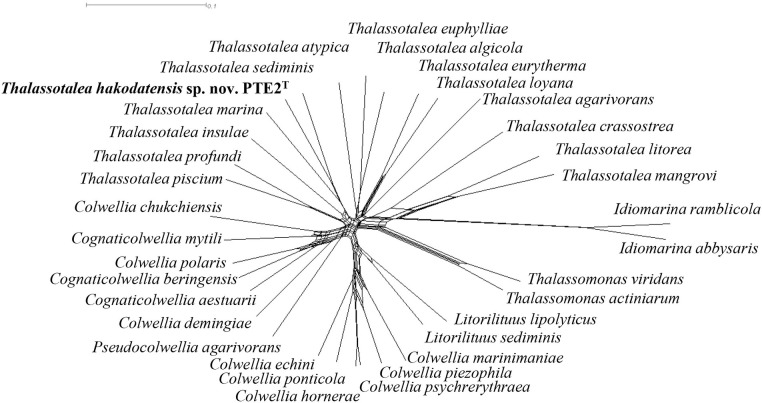
MLSA was performed as previously described [25, 26, 29, 39, 40]. The sequences of four protein-coding genes (ftsZ, mreB, rpoA, and topA), essential single-copy genes in the taxa examined in this study were obtained from the genome sequences of PTE2T, T. sediminis KCTC 42588T, T. insulae KCTC 62186T, T. piscium JCM 18590T, T. agarivorans JCM 13379T, T. loyana LMG 23556T, T. eurytherma JCM 18482T, T. atypica JCM 31894T, T. marina QBLM2T, T. profundi YM155T, T. mangrovi zs-4T, T. crassostreae LPB0090T, T. algicola M1351T, T. litorea MCCC IK03283, T. euphylliae H2 and other related Colwelliaceae and Idiomarinaceae species (Tables 1 and S1) (see genome accession number in the description section below). The sequences of each gene were aligned using ClustalX 2.1 [41]. Concatenation of sequences and phylogenetic NeighborNet reconstruction were performed using SplitsTree 4.16.2 with options of JukesCantor correction, gap-exclusion and 1,000 bootstrap [42]. Regions used for the network reconstruction in Fig 3 were 1–2,549, 1–1,044 1–2,019, and 1–2,549 for ftsZ, mreB, rpoA, and topA, as PTE2 nucleotide sequence positions, respectively.
Fig 3. MLSA network.
Bar, 0.01 substitutions per nucleotide position. Topology of the clade which PTE2 was belonged to was supported by a 100% bootstrap value.
Pan and core genome analysis
A total of 15 genomes, including eight newly obtained genomes in this study (PTE2T, T. sediminis JCM 42588T, T. insulae KCTC 62186T, T. piscium JCM 18590T, T. agarivorans JCM 13379T, T. loyana LMG 23556T, T. eurytherma JCM 18482T, T. atypica JCM 31894T) and seven retrieved from the NCBI database (T. marina QBLM2T, T. profundi YM155T, T. mangrovi zs-4T, T. crassostreae LPB0090T, T. algicola M1351T, T. litorea MCCC IK03283, T. euphylliae H2), were used for pangenome analysis using the program anvi’o v7 [43] based on previous studies [28, 29, 44–46], with minor modifications. Briefly, contig databases of each genome were constructed by fasta files (anvi-gen-contigs-database) and decorated with hits from HMM models (anvi-run-hmms). Subsequently, functions were annotated for genes in contig databases (anvi-run-ncbi-cogs). KEGG annotation was also performed (anvi-run-kegg-kofams) [29]. The storage database was generated (anvi-gen-genomes-storage) using all contigs databases and pangenome analysis was performed (anvi-pan-genome) [29]. The results were displayed (anvi-display-pan) and adjusted manually [29].
In silico chemical taxonomy: Prediction of fatty acids, polar lipids and isoprenoid quinone using the comparative genomics approach
The genes encoding key enzymes and proteins for the synthesis of fatty acids (FAs), polar lipids and isoprenoid quinones were retrieved from the genome sequences of PTE2T and the related species using in silico MolecularCloning ver. 7 with same methodology by Yamano et al. [29]. Genomic structure and distribution of the genes were compared also using in silico MolecularCloning ver. 7. The 3D structures of genes encoding FA desaturase from some of the strains were predicted using Phyre2 [47].
Results and discussion
Molecular phylogenetic analysis based on 16S rRNA gene nucleotide sequences
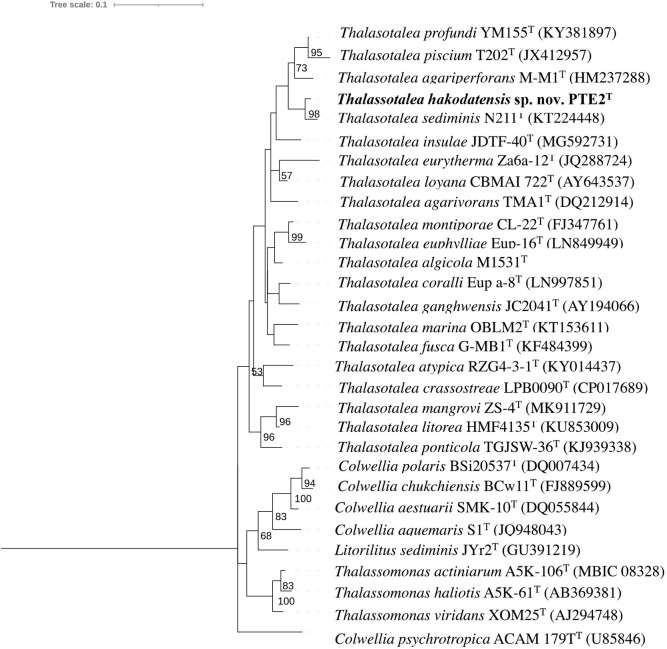
Phylogenetic analysis based on 16S rRNA gene nucleotide sequences showed that strain PTE2T could be affiliated to the members of the genus Thalassotalea (Fig 1). However, the internal node of the genus Thalassotalea was unlikely to be supported by a high bootstrap value (see also MLSA section). The strain showed the highest sequence similarities of 97.9% with T. sediminis, which is below the proposed threshold range of the species boundary, 98.7% [48, 49].
Fig 1. ML tree based on 16S rRNA gene nucleotide sequences of strain PTE2T and related type strains.
The ML tree was reconstructed using the K2+G+I model. Numbers shown on branches are bootstrap values (%) based on 1,000 replicates (>50%). 1,347 bp were compared (69–1,416 position in T. sediminis N2111T, KT224448). Idiomarina ramblicola R22T, of which position was not visible in Fig 1, was used as an outgroup to generate this rooted tree.
Genomic features and overall genome relatedness indices (OGRIs)
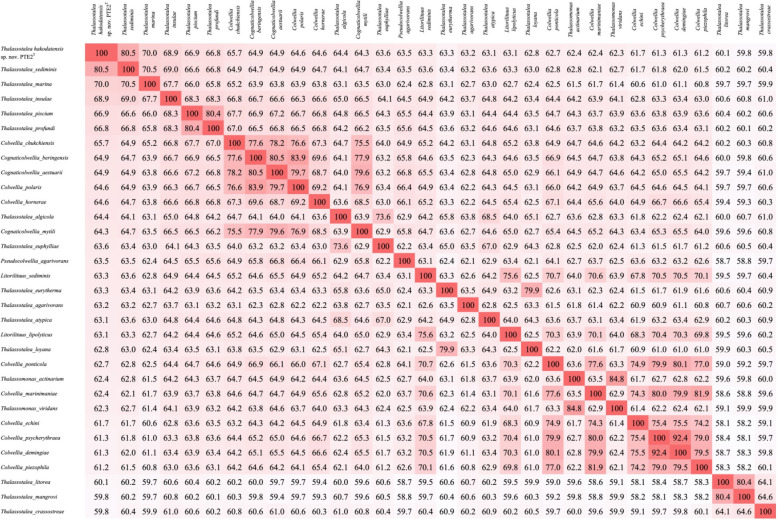
Genomic features of PTE2T and the described Thalassotalea species with available genomic sequences were shown in (Table 1). The complete genomic sequence of PTE2T showed that genome size and G+C content is 4.31 Mb and 38.5%, respectively. In silico DDH and ANI values of the PTE2T against 14 Thalassotalea species were 9.7–24.6% and 68.5–77.0% respectively, which were below the species boundary threshold of 70% and 95% proposed in previous studies (S2 Table) [29]. AAI values were between 59.8–80.5% (Fig 2), which were also below the species delineation boundary of 95–96% [50], confirming that strain PTE2T represents a novel species in the genus Thalassotalea. Interestingly, while five Thalassotalea species (T. sediminis, T. marina, T. insulae, T. piscium and T. profundi) showed relatively high AAI values to strain PTE2T, other Thalassotalea species showed lower AAI values than species in Colwellia, Cognaticolwellia, Pseudocolwellia, Litorilituus or Thalassomonas.
Fig 2. AAI matrix using Thalassotalea and Colwelliaceae genomes.
Multilocus sequence analysis (MLSA)
MLSA network showed that PTE2T is a novel species different from the 14 reference Thalassotalea strains used in this study. This analysis also showed that PTE2T, T. insulae, T. marina T. piscium, T. profundi, and T. sediminis, form a monophyletic clade of the genus Thalassotalea (Fig 3). The MLSA network analysis revealed intriguing findings about the Thalassotalea species and the Colwellia genus. Specifically, the other Thalassotalea species did not form a monophyletic clade, as at least five monophyletic clades were observed. Additionally, the genus Colwellia also did not form a monophyletic clade. Considering these results in combination with AAI values from the last section altogether implies that family Colwelliaceae might be a subject of reclassification in the future.
Phenotypic characterization
PTE2T shared some biochemical features with Thalassotalea species, such as growth at 15, 25 and 30°C, the ability to hydrolyze Tween80, gelatin and DNA. Strain PTE2 T was distinguished from other members with a total of 36 traits (growth at 4, 37 and 40°C, growth in 0% NaCl, oxidase, catalase, indole production, hydrolysis of starch, alginate and agar and 25 carbon assimilation tests) (Table 2).
Table 2. Phenotypic characteristics of PTE2T and Thalassotalea reference strains.
| Characteristics | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
|---|---|---|---|---|---|---|---|---|
| Growth at | ||||||||
| 4°C | + | - | + | + | - | + | - | - |
| 37°C | - | + | - | + | + | - | + | + |
| 40°C | - | NT | - | - | + | - | - | NT |
| Growth in NaCl (broth) | ||||||||
| 0% | - | NT | + | - | + | - | + | NT |
| OF test | O | O | N | O | O | O | O | O |
| Oxidase | + | - | + | + | + | + | + | + |
| Catalase | + | + | + | w | + | w | w | + |
| Indole production | - | NT | - | - | - | w | - | NT |
| Hydrolysis of | ||||||||
| Starch | + | - | - | + | + | + | + | - |
| Alginate | + | - | - | - | w | - | - | - |
| Agar | - | - | - | - | + | - | - | - |
| Antibiotic susceptibility | ||||||||
| GM120(Gentamicin 120) | + | NT | +*1 | + | +*1 | NT | + | NT |
| GAT5(Gemifloxacin 5) | + | NT | NT | + | NT | NT | + | NT |
| GTX30(Cefotaxime 30) | + | NT | NT | + | NT | NT | + | NT |
| CB100(Carbenicillin 100) | + | NT | NT | + | +*1 | +*3 | + | NT |
| CLR15(Clarithromycin 15) | + | NT | -* 2 | + | NT | -*3 | + | NT |
| SXT(Sulfamethoxazole/Trimethoprim) | + | NT | NT | + | NT | NT | + | NT |
| AM10(Ampicillin 10) | w | NT | +*1 | + | +*1 | +*3 | + | NT |
| Utilization of | ||||||||
| D-Mannose | w | - | - | - | - | - | - | - |
| D-Galactose | + | + | - | - | - | - | + | - |
| D-Fructose | + | - | - | - | - | - | - | - |
| Maltose | - | + | - | - | - | - | - | - |
| N-Acetylglucosamine | - | + | - | - | - | - | + | - |
| Succinate | + | - | - | - | - | - | + | - |
| γ-Aminobutyrate | - | - | - | - | - | + | - | - |
| Xylose | + | - | - | - | - | - | - | - |
| D-Glucose | + | + | - | - | - | - | w | - |
| Acetate | - | + | - | - | - | - | + | - |
| D-Glucosamine | - | + | w | - | - | - | + | - |
| Pyruvate | + | + | - | - | - | - | - | - |
| Cellobiose | - | + | - | - | - | - | - | - |
| L-Proline | + | + | - | - | - | + | + | - |
| L-Glutamate | + | + | - | - | - | + | + | - |
| Putrescine | - | - | - | - | - | + | - | - |
| Salicine | - | + | - | - | - | - | - | - |
| DL-Lactate | - | + | - | - | - | - | + | - |
| L-Arginine | + | + | - | - | - | + | + | - |
| L-Asparagine | + | - | - | - | - | + | + | - |
| L-Citrulline | - | - | - | - | - | + | - | - |
| Glycine | + | - | - | - | - | - | - | - |
| L-Histidine | - | + | - | - | - | - | - | - |
| L-Ornithine | + | - | - | - | - | - | - | - |
| L-Serine | + | - | - | - | - | - | - | - |
All strains showed growth at 15°C, 25°C and 30°C and NaCl concentration of 1%, 3%, 6%, 8%, and 10%. All strains were able to hydrolyze Tween80, gelatin, and DNA. All strains tested positive for nitrate reduction. All strains tested negative for utilization of sucrose, melibiose, lactose, D-gluconate, N-acetylglucosamine, fumarate, citrate, aconitate, meso-erythritol, D-mannitol, glycerol, L-tyrosine, D-sorbitol, α-ketoglutarate, xylose, trehalose, glucuronate, δ-aminovalerate, cellobiose, putrescine, propionate, amygdalin, arabinose, D-galacturonate, glycerate, D-raffinose, L-rhamnose, D-ribose, salicine, DL-lactate, L-alanine and histidine.
Pangenomic analysis
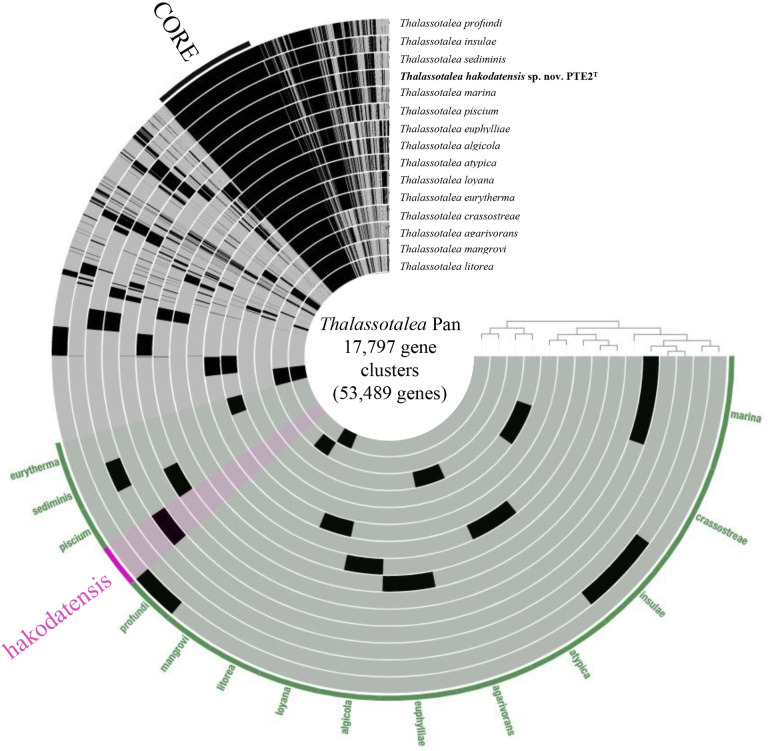
The pangenome of Thalassotalea species consists of 17,797 gene clusters (53,489 genes) (Fig 4). Genes were classified into Core for the genes present in all strains, and species-specific bins for the genes that are unique for each species. Core consisted of 1,175 gene clusters (17,979 genes).
Fig 4. Anvi’o representation of the pangenome of the Thalassotalea species.
Layers represent each genome, and the bars represent the occurrence of gene clusters. The darker colored areas of the bars belong to Core or PTE2T specific bin.
N-acyl homoserine lactone hydrolase gene was present in PTE2T, T. sediminis and T. insulae. The enzyme shows the quorum-quenching activity of hydrolyzing N-acyl homoserine lactones (AHLs), a signal molecule used by many Gram-negative bacteria in quorum-sensing pathways [51]. This activity of quorum quenching has been shown to strengthen host animal resistance to pathogenic bacteria by disrupting their quorum-sensing activities [52].
Strain PTE2T had two genes for with alginate degradation enzyme, namely, poly-(β-D-mannuronate) lyase and oligo-alginate lyase. The phenotype test also show that this strain has alginate hydrolysis ability (Table 2). The presence of these genes implies that strain PTE2T may aid digestion of alginate in sea cucumber gut. In previous study, it was observed that bacteria associated with the algal polysaccharide degradation were abundant in fast-growing Apostichopus japonicus compared to slow-growing individuals, and such bacteria are believed to assist host in obtaining energy [53].
In silico chemical taxonomy
The cellular fatty acid (FA) profile of Thalassotalea species is reported to consist mainly of even-numbered linear mono-unsaturated or saturated chains (i.e. C14:0, C16:0, C16:1 and C18:1), with small amount of odd-numbered linear mono-unsaturated or saturated chains (i.e. C13:0, C15:1 and C17:0), 3-hydroxy FAs (i.e. C11:0 3-OH, C12:0 3-OH) and branched-chain FAs (i.e. iso-C14:0, iso-C16:0) (S3 Table) [1, 3, 4, 8, 11, 13, 15–18]. Pangenomic analysis among described Thalassotalea species reconstructed the basic type II fatty acid biosynthesis (FAS II) pathway driven by FabABFDGVYZ and AccABCD, which is very similar to that of E. coli [54] (S4 Table and S1 Fig). The FAS II pathway could contribute to even-numbered linear mono-unsaturated or saturated chains (C14:0, C16:0, C16:1 ω7c of summed features 3 and C18:1 ω7c), which occupied approximately 20–60% of the total fatty acid (S3 Table). C16:0 is one of the major products produced from the FAS II pathway, which means PTE2T could produce C16:0 (S1 Fig). ω7c mono-unsaturated fatty acids (C16:1 ω7c and C18:1 ω7c) are also major in this group of bacteria and can be produced through ω7 mono-unsaturated fatty acid synthesis initiated by isomerization of trans-2-decenoyl-ACP into cis-3-decenoyl-ACP by FabA (S1 Fig). After elongation by FabB, the acyl chain is returned to the FASII pathway and goes through further elongation, producing C16:1ω7c and C18:1ω7c [55]. All strains, including PTE2T have fabA and fabB, thus it is suggesting that PTE2T is also capable of producing C16:1ω7c and C18:1ω7c.
Odd-numbered saturated fatty acids can also be produced by the FAS II pathway by using propanoyl-CoA as a primer molecule for the initial condensation, instead of acetyl CoA used for even-numbered saturated fatty acids [56]. Amongst multiple pathways for the synthesis of propanoyl-CoA, pangenome analysis in this study revealed that all 15 strains possess bkdAAB genes which are responsible for producing propanoyl-CoA from 2-oxobutanoate via 1-hydroxy propyl-Thpp and S-propanyl-dihydrolipoamide-E. Moreover, 10 strains including PTE2T had genes responsible to another propanoyl-CoA production pathway, accABCD, fadB, fadJ and acuI, which uses acetyl-CoA (S4 Table). These results provide evidence that strain PTE2T has the capacity to synthesize propanoyl-CoA, a precursor for the synthesis of odd-numbered saturated fatty acids. C13:0 and C17:0 derived from this pathway consist approximately 0–18% of the Thalassotalea fatty acid profile (S3 Table). Notably, C17:0 plays a vital role in the production of heptadecenoyl-CoA (C17:1 ω8c), which is described in further detail below.
3-hydroxylated FAs such as C11:0 3-OH and C12:0 3-OH, which are the primary fatty acids in lipid A as well as in ornithine-containing lipids, could be supplied by the FAS II pathway, since 3-hydroxy-acyl-ACP is known to be normally intermediated in the FAS II elongation cycle [55]. Thalassotalea core genes also included lpxA and lpxD, a gene responsible for the incorporation of 3-hydroxy-acyl chains to UDP-N-acetyl-alpha-D-glucosamine and UDP-3-O-(3-hydroxytetradecanoyl)-D-glucosamine, respectively, which are essential for the biosynthesis of lipid A [57, 58].
Fatty acid desaturase (Des) catalyzes to produce unsaturated fatty acid by introducing double bonds to the fatty acid under aerobic conditions [59]. Fatty acid desaturase, des1 was found in the core gene. 3D-structure prediction using Phyre2 program shows that these enzymes are likely to be stearoyl-CoA desaturase (SCD) (S5 Table). SCD introduces cis double bond at the Δ9 position of palmitoyl-CoA (C16:0), heptadecanoyl-CoA (C17:0) or stearoyl-CoA (C18:0), producing palmitoleoyl-CoA (C16:1 ω7c), heptadecenoyl-CoA (C17:1 ω8c) or oleoyl-CoA (C18:1 ω9c) [60]. Using this enzyme, Thalassotalea species could produce C17:1 ω8c, a major fatty acid consisting approximately 10–25% of the total fatty acid profile (S3 Table).
The biosynthesis pathway responsible for producing linear fatty acids also produces branched-chain fatty acids, but with distinct primer molecules. Iso-fatty acids with even-numbered chains found in Thalassotalea species are produced using 3‑methylbutyryl-CoA as a primer molecule instead of acetyl-CoA, which is used for production of linear fatty acids. All strains in this study contain the ilvE and dkdAAB genes that are involved in the degradation of L-leucine to 3‑methylbutyryl-CoA, implicating that these species have the potential to produce even-numbered iso-fatty acids such as iso-C14:0 and iso-C16:0 (S1 Fig).
The genome of strain PTE2T was subjected to a comparative analysis to investigate the presence of genes involved in the fatty acid synthesis type II (FAS II) pathway, showing stain PTE2T shared a core gene set that was highly similar to that of other 14 strains (S1 Fig and S4 Table). Genomic structures of FAS II core genes are likely to be retained among described Thalassotalea species (S2 and S3 Figs), which could lead to the conclusion that the novel strain is capable of producing similar FA profiles as Thalassotalea reference strains, consisting of mainly even-numbered linear mono-unsaturated or saturated chains (i.e. C16:0, C16:1 and C18:1), odd-numbered linear mono-unsaturated or saturated chains (i.e. C15:1 and C17:0), 3-hydroxy FAs (i.e. C11:0 3-OH, C12:0 3-OH) and branched-chain FAs (i.e. iso-C14:0, iso-C16:0). However, a pathway to produce some of the major fatty acids such as C15:1 ω8c and iso-C17:1 ω5C could not be identified.
Pangenomic analysis among 15 strains also revealed a comprehensive gene set for the biosynthesis of phosphatidylglycerol (PG) and phosphatidylethanolamine (PE); plsX, plsY, plsC, cdsA, pssA, psd, pgsA and pgpA, for all the strains (S6 Table). This result indicates that PTE2T produces PG and PE as major polar lipids, similar to other 14 reference species. Interestingly, clsA/B and clsC genes, which are responsible for the production of diphosphatidylglycerol (DPG) [61], were detected in the genomes of four strains, T. piscium, T. agarivorans, T. crassostreae and T. litorea. This result suggests that these four strains are potential DPG producers.
The only respiratory quinone reported from previously described Thalassotalea species is ubiquinone-8 (Q-8) (S3 Table). Core genes of 15 strains include ubi genes responsible for the biosynthetic pathway (ubiC, ubiA, ubiD, ubiX, ubiI, ubiG, ubiH, ubiE), and ispAB. In addition to those, core genes also include three genes (ubiB, ubiJ, ubiK) coding accessory proteins also required for Q-8 biosynthesis, but with rather hypothetical functions. These results suggest that the predominant ubiquinone of PTE2T is Q-8.
Conclusions
Using the results of modern genome taxonomic studies combined with classical phenotyping, which fulfills phylogenetic, genomic, and phenotypic cohesions, we propose the strain PTE2T as Thalassotalea hakodatensis sp. nov. (PTE2T = JCM 34608T = KCTC 82592T), a novel species in the genus Thalassotalea.
Description of Thalassotalea hakodatensis sp. nov.
Thalassotalea hakodatensis sp. nov. (ha.ko.da.ten’sis. N.L. fem. adj. hakodatensis, from Hakodate, referring to the isolation site of the strain).
Gram-negative, rod-shaped and motile. Colonies on MA are yellowish-white, 0.5–0.75 mm in diameter after culture for 3 days. No pigmentation and bioluminescence are observed. The DNA G+C content is 38.4%, and genome size is 4.28 Mb. Growth occurs at 4°C, 15°C, 25°C and 30°C, with NaCl concentrations of 1%, 3%, 6%, 8% and 10%. OF-test oxidative. susceptible for ampicillin (10 μg), cefotaxime (30 μg), gatifloxacin (5 μg), carbenicillin (100 μg), clarithromycin (15 μg) and sulfamethoxazole/trimethoprim. Positive for oxidase- and catalase- test, nitrate reduction, hydrolysis of starch, alginate, tween 80, gelatin and DNA, utilization of D-mannose, D-galactose, D-fructose, succinate, xylose, D-glucose, pyruvate, L-proline, L-glutamate, L-arginine, L-asparagine, glycine, L-ornithine and L-serine. Negative for production of indole, hydrolysis of agar, utilization of sucrose, maltose, melibiose, lactose, D-gluconate, N-acetylglucosamine, fumarate, citrate, aconitate, meso-erythritol, D-mannitol, glycerol, γ-aminobutyrate, L-tyrosine, D-sorbitol, DL-malate, α-ketoglutarate, trehalose, glucuronate, acetate, D-glucosamine, δ-aminovalerate, cellobiose, putrescine, propionate and amygdalin.
The type strain PTE2T (= JCM 34608T = KCTC 82592T) was isolated from a pentactula larvae of Apostichopus japonicus reared in a laboratory aquarium at Hokkaido University, Hokkaido, Japan. The GenBank accession number for the 16S rRNA gene sequence of the type strain is LC757706. The complete genome sequence of the strain is deposited in the DDBJ/ENA/GenBank under the accession number AP027365.
Supporting information
(PDF)
(PDF)
Strains: 1, T. sediminis KCTC 42588 T, 2, T. insulae KCTC 42588 T, 3, T. piscium JCM 18590 T, 4, T. marina KCTC 42731T, 5, T. profundi YM155T, 6, T. agarivorans JCM 13379 T, 7, T. eurytherma JCM 18482T, 8, T. atypica JCM 31894 T, 9, T. mangrovi zs-4T, 10, T. crassostreae LPB 0090T,11, T. loyana LMG 22536T, 12, T. algicola M1531T, 13, T. litorea HMF4135T, 14, T. euphylliae Eup-16T.
(PDF)
(PDF)
(PDF)
(PDF)
ACP: acyl-carrier protein; AccABCD: acetyl-CoA carboxylase complex; FabD: malonyl-CoA: ACP transacylase; FabH/FabY: 3-ketoacyl-ACP synthase Ⅲ; FabB: 3-ketoacyl-ACP synthase Ⅰ; FabF: 3-ketoacyl-ACP synthase Ⅱ; FabG: 3-ketoacyl-ACP reductase; FabA: 3-hydroxyacyl-ACP dehydratase/trans-2-decenoyl-ACP isomerase; FabV enoyl-ACP reductase; FabZ: 3-hydroxyacyl-ACP dehydratase.
(PDF)
(PDF)
Protein/enzyme name each gene is coding: fabA: 3-hydroxyacyl-ACP dehydrase/trans-2-decenoyl-ACP isomerase; fabD: malonyl-CoA: ACP transacylase; fabF: 3-ketoacyl-ACP synthase Ⅱ; fabG: 3-ketoacyl-ACP reductase; fabH: 3-ketoacyl-ACP synthase Ⅲ; fabV enoyl-ACP reductase; fabY: 3-ketoacyl-ACP synthase; fabZ: 3-hydroxyacyl-ACP dehydratase; accABCD: carboxylase complex; plsX: phosphate acyltransferase; acpP: Acyl-carrier protein.
(PDF)
Acknowledgments
We gratefully thank for Professor (Emeritus) Aharon Oren, The Hebrew University of Jerusalem, for his advice on bacterial names.
Data Availability
The GenBank accession number for the 16S rRNA gene sequence of the type strain PTE2 is LC757706. The whole genome sequence of the PTE2 strain and seven Thalassotalea type species shown in Table 1 have been deposited to DDBJ/ENA/GenBank under the accession numbers AP027361-AP027365, BSST01000001-BSST01000003, BSSU01000001-BSSU01000035, and BSSV01000001-BSSV01000017. Raw reads for genome assembly used in this study have been deposited under DRA015858.
Funding Statement
This study was supported by Kaken 19K22262. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Zhang Y, Tang K, Shi X, Zhang XH. Description of Thalassotalea piscium gen. nov., sp. nov., isolated from flounder (Paralichthys olivaceus), reclassification of four species of the genus Thalassomonas as members of the genus Thalassotalea gen. nov. and emended description of the genus Thalassomonas. Int J Syst Evol Microbiol. 2014; 64: 1223–1228. doi: 10.1099/ijs.0.055913-0 [DOI] [PubMed] [Google Scholar]
- 2.Yi H, Bae KS, Chun J. Thalassomonas ganghwensis sp. nov., isolated from tidal flat sediment. Int J Syst Evol Microbiol. 2004; 54:377–380. doi: 10.1099/ijs.0.02748-0 [DOI] [PubMed] [Google Scholar]
- 3.Thompson FL, Barash Y, Sawabe T, Sharon G, Swings J, Rosenberg E. Thalassomonas loyana sp. nov., a causative agent of the white plague-like disease of corals on the Eilat coral reef. Int J Syst Evol Microbiol. 2006; 56:365–368. doi: 10.1099/ijs.0.63800-0 [DOI] [PubMed] [Google Scholar]
- 4.Jean WD, Shieh WY, Liu TY. Thalassomonas agarivorans sp. nov., a marine agarolytic bacterium isolated from shallow coastal water of An-Ping Harbour, Taiwan, and emended description of the genus Thalassomonas. Int J Syst Evol Microbiol. 2006; 56:1245–1250. doi: 10.1099/ijs.0.64130-0 [DOI] [PubMed] [Google Scholar]
- 5.Park S, Choi WC, Oh TK, Yoon JH. Thalassomonas agariperforans sp. nov., an agarolytic bacterium isolated from marine sand. Int J Syst Evol Microbiol. 2011; 61: 11. doi: 10.1099/ijs.0.027821-0 [DOI] [PubMed] [Google Scholar]
- 6.Park S, Jung YT, Kang CH, Park JM, Yoon JH. Thalassotalea ponticola sp. nov., isolated from seawater, reclassification of Thalassomonas fusca as Thalassotalea fusca comb. nov. and emended description of the genus Thalassotalea. Int J Syst Evol Microbiol. 2014; 64: 3676–3682. doi: 10.1099/ijs.0.067611-0 [DOI] [PubMed] [Google Scholar]
- 7.Hou T, Liu Y, Zhong Z, Liu H, Liu Z. Thalassotalea marina sp. nov., isolated from a marine recirculating aquaculture system, reclassification of Thalassomonas eurytherma as Thalassotalea eurytherma comb. nov. and emended description of the genus Thalassotalea. Int J Syst Evol Microbiol. 2015; 65: 4710–4715. doi: 10.1099/ijsem.0.000637 [DOI] [PubMed] [Google Scholar]
- 8.Xu Z, Lu D, Liang Q, Chen G, Du ZJ. Thalassotalea sediminis sp. nov., isolated from coastal sediment. Antonie van Leeuwenhoek. 2016; 109: 371–378. doi: 10.1007/s10482-015-0639-4 [DOI] [PubMed] [Google Scholar]
- 9.Sheu S, Liu LP, Tang SL, Chen WM. Thalassotalea euphylliae sp. nov., isolated from the torch coral Euphyllia glabrescens. Int J Syst Evol Microbiol. 2016; 66: 5039–5045. doi: /10.1099/ijsem.0.001466 [DOI] [PubMed] [Google Scholar]
- 10.Chen WM, Liu LP, Chen CA, Wang JT, Sheu SY. Thalassotalea montiporae sp. nov., isolated from the encrusting pore coral Montipora aequituberculata. Int J Syst Evol Microbiol. 2016; 66: 4077–4084. doi: 10.1099/ijsem.0.001313 [DOI] [PubMed] [Google Scholar]
- 11.Choi S, Kim E, Shin S, Yi H. Thalassotalea crassostreae sp. nov., isolated from Pacific oyster. Int J Syst Evol Microbiol. 2017; 67: 988–992. doi: 10.1099/ijsem.0.001923 [DOI] [PubMed] [Google Scholar]
- 12.Kang H, Kim H, Nam IY, Joung Y, Jang TY, Joh K. Thalassotalea litorea sp. nov., isolated from seashore sand. Int J Syst Evol Microbiol. 2017; 67: 2268–2273. doi: 10.1099/ijsem.0.001938 [DOI] [PubMed] [Google Scholar]
- 13.Liu J, Sun YW, Li SN, Zhang DC. Thalassotalea profundi sp. nov. isolated from a deep-sea seamount. Int J Syst Evol Microbiol. 2017; 67: 3739–3743. doi: 10.1099/ijsem.0.002180 [DOI] [PubMed] [Google Scholar]
- 14.Sheu D, Sheu S, Xie P, Tang S, Chen W. Thalassotalea coralli sp. nov., isolated from the torch coral Euphyllia glabrescens. Int J Syst Evol Microbiol. 2018; 268: 185–191. doi: 10.1099/ijsem.0.002478 [DOI] [PubMed] [Google Scholar]
- 15.Wang Y, Liu T, Ming H, Sun P, Cao C, Guo M, et al. Thalassotalea atypica sp. nov., isolated from seawater, and emended description of Thalassotalea eurytherma. Int J Syst Evol Microbiol. 2018; 68: 271–276. doi: 10.1099/ijsem.0.002495 [DOI] [PubMed] [Google Scholar]
- 16.Park S, Choi J, Won M, Yoon H. Thalassotalea insulae sp. nov., isolated from tidal flat sediment. Int J Syst Evol Microbiol. 2018; 68: 1321–1326. doi: 10.1099/ijsem.0.002671 [DOI] [PubMed] [Google Scholar]
- 17.Zheng S, Zhang D, Gui J, Wang J, Zhu X, Lai Q, et al. Thalassotalea mangrovi sp. nov., a bacterium isolated from marine mangrove sediment. Int J Syst Evol Microbiol. 2019; 69: 3644–3649. doi: 10.1099/ijsem.0.003677 [DOI] [PubMed] [Google Scholar]
- 18.Lian FB, Jiang S, Ren TY, Zhou BJ, Du ZJ. Thalassotalea algicola sp. nov., an alginate-utilizing bacterium isolated from a red alga. Antonie van Leeuwenhoek. 2021; doi: 10.1007/s10482-021-01562-2 [DOI] [PubMed] [Google Scholar]
- 19.Sun C, Huo YY, Liu JJ, Pan J, Qi YZ, Zhang XQ, et al. Thalassomonas eurytherma sp. nov., a marine proteobacterium. Int J Syst Evol Microbiol. 2014; 64: 2079–2083. doi: 10.1099/ijs.0.058255-0 [DOI] [PubMed] [Google Scholar]
- 20.Jung YT, Park S, Yoon JH. Thalassomonas fusca sp. nov., a novel gammaproteobacterium isolated from tidal flat sediment. Antonie van Leeuwenhoek. 2014; 105: 81–87. doi: 10.1007/s10482-013-0055-6 [DOI] [PubMed] [Google Scholar]
- 21.Torres M, Romero M, Prado S, Dubert J, Tahrioui A, Otero A, et al. N-acylhomoserine lactone-degrading bacteria isolated from hatchery bivalve larval cultures. Microbiol Res. 2013; 168: 547–554. doi: 10.1016/j.micres.2013.04.011 [DOI] [PubMed] [Google Scholar]
- 22.Deering R, Chen J, Sun J, Ma H, Dubert J, Barja JL, et al. N-Acyl dehydrotyrosines, tyrosinase inhibitors from the marine bacterium Thalassotalea sp. PP2-459. J Nat Prod. 2016; 79: 447–450. doi: 10.1021/acs.jnatprod.5b00972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim M, Cha IT, Lee KE, Lee EY, Park SJ. Genomics reveals the metabolic potential and functions in the redistribution of dissolved organic matter in marine environments of the genus Thalassotalea. Microorganisms. 2020; 8: 1412. doi: 10.3390/microorganisms8091412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu J, Sakai Y, Mino S, Sawabe T. Characterization of microbiomes associated with the early life stages of sea cucumber Apostichopus japonicus Selenka. Front Mar Sci. 2022; 31: 37. doi: 10.3389/fmars.2022.801344 [DOI] [Google Scholar]
- 25.Al-Saari N, Gao F, Amin AKMR, Sato K, Sato K, Mino S, et al. Advanced microbial taxonomy combined with genome-based-approaches reveals that Vibrio astriarenae sp. nov., an agarolytic marine bacterium, forms a new clade in Vibrionaceae. PLoS One. 2015; 10: e0136279. doi: 10.1371/journal.pone.0136279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amin AKMR, Tanaka M, Al-Saari N, Feng G, Mino S, Ogura Y, et al. Thaumasiovibrio occultus gen. nov. sp. nov. and Thaumasiovibrio subtropicus sp. nov. within the family Vibrionaceae, isolated from coral reef seawater off Ishigaki Island, Japan. Syst Appl Microbiol. 2017; 40: 290–296. doi: 10.1016/j.syapm.2017.04.003 [DOI] [PubMed] [Google Scholar]
- 27.Sawabe T, Ogura Y, Matsumura Y, Feng G, Amin AKMR, Mino S, et al. Updating the Vibrio clades defined by multilocus sequence phylogeny: proposal of eight new clades, and the description of Vibrio tritonius sp. nov. Front Microbiol. 2013; 4: 414. doi: 10.3389/fmicb.2013.00414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanaka M, Hongyu B, Jiang C, Mino S, Milet Meirelles P, Thompson F, et al. Vibrio taketomensis sp. nov. by genome taxonomy. Syst Appl Microbiol. 2020; 43: 126048. doi: /10.1016/j.syapm.2019.126048 [DOI] [PubMed] [Google Scholar]
- 29.Yamano R, Yu J, Jiang C, Haditomo AHC, Mino S, Sakai Y, et al. Taxonomic revision of the genus Amphritea supported by genomic and in silico chemotaxonomic analyses, and the proposal of Aliamphritea gen. nov. PLOS ONE. 2022; 17: e0271174. doi: 10.1371/journal.pone.0271174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sawabe T, Sugimura I, Ohtsuka M, Nakano K, Tajima K, Ezura Y, et al. Vibrio halioticoli sp. nov., a non-motile alginolytic marine bacterium isolated from the gut of the abalone Haliotis discus hannai. Int J Syst Evol Microbiol. 1998; 48: 573–580, doi: 10.1099/00207713-48-2-573 [DOI] [PubMed] [Google Scholar]
- 31.Pruesse E, Peplies J, Glöckner F. SINA: Accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics. 2012; 28: 1823–1829. doi: 10.1093/bioinformatics/bts252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol. 2018; 35: 1547–1549. doi: 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stecher G, Tamura K, Kumar S. Molecular Evolutionary Genetics Analysis (MEGA) for macOS. Mol Biol Evol. 2020; 33: 1870–1874. doi: 10.1093/molbev/msz312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLOS Comput Biol. 2017; 13: e1005595. doi: 10.1371/journal.pcbi.1005595 pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanizawa Y, Fujisawa T, Nakamura Y. DFAST: a flexible prokaryotic genome annotation pipeline for faster genome publication. Bioinformatics. 2018; 34: 1037–1039. doi: 10.1093/bioinformatics/btx713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee I, Kim YO, Park SC, Chun J. OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol. 2015; 66: 1100–1103. doi: 10.1099/ijsem.0.000760 [DOI] [PubMed] [Google Scholar]
- 37.Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics. 2013; 14: 60. doi: 10.1186/1471-2105-14-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rodriguez-R LM, Konstantinidis KT. The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ. 2016; 4: e1900v1. doi: 10.7287/peerj.preprints.1900v27123377 [DOI] [Google Scholar]
- 39.Sawabe T, Kita-Tsukamoto K, Thompson FL. Inferring the evolutionary history of vibrios by means of multilocus sequence analysis. J Bacteriol. 2007; 189: 7932–7936. doi: 10.1128/JB.00693-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang C, Tanaka M, Nishikawa S, Mino S, Romalde JL, Thompson FL, et al. Vibrio Clade 3.0: new Vibrionaceae evolutionary units using genome-based approach. Curr Microbiol. 2022. a; 79: 10. doi: 10.1007/s00284-021-02725-0 [DOI] [PubMed] [Google Scholar]
- 41.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007; 23: 2947–2948, doi: 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- 42.Huson D, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006; 23: 254–267. doi: 10.1093/molbev/msj030 [DOI] [PubMed] [Google Scholar]
- 43.Eren AM, Kiefl E, Shaiber A, Veseli I, Miller SE, Schechter MS. Community-led, integrated, reproducible multi-omics with anvi’o. Nat Microbiol. 2021; 6: 3–6. doi: 10.1038/s41564-020-00834-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Delmont TO, Eren AM. Linking pangenomes and metagenomes: the Prochlorococcus metapangenome. PeerJ. 2018; 6; e4320. doi: 10.7717/peerj.4320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang C, Mino S, Sawabe T. Genomic analyses of Halioticoli clade species in Vibrionaceae reveal genome expansion with more carbohydrate metabolism genes during symbiotic to planktonic lifestyle transition. Front Mar Sci. 2022. b; 9: 844983. doi: 10.3389/fmars.2022.844983 [DOI] [Google Scholar]
- 46.Jiang C, Kasai H, Mino S, Romalde JL, Sawabe T. The pan-genome of Splendidus clade species in the family Vibrionaceae: Insights into evolution, adaptation, and pathogenicity. Environ Microbiol. 2022. c; 24: 4587–4606. doi: 10.1111/1462-2920.16209 [DOI] [PubMed] [Google Scholar]
- 47.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015; 10: 845–858. doi: 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yarza P, Yilmaz P, Pruesse E. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat Rev Microbiol. 2014; 12: 635–645. doi: 10.1038/nrmicro3330 [DOI] [PubMed] [Google Scholar]
- 49.Kim M, Oh HS, Park SC, Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol. 2014; 64: 346–351. doi: 10.1099/ijs.0.059774-0 [DOI] [PubMed] [Google Scholar]
- 50.Konstantinidis KT, Tiedje JM. Towards a genome-based taxonomy for prokaryotes. J Bacteriol Res. 2005; 187: 6258–6264. doi: 10.1128/JB.187.18.6258-6264.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Galloway WRJD, Hodgkinson JT, Bowden SD, Welch M, Spring DR. Quorum sensing in Gram-negative bacteria: small-molecule modulation of AHL and AI-2 quorum sensing pathways. Chem Rev. 2011; 111: 28–67. doi: 10.1021/cr100109t [DOI] [PubMed] [Google Scholar]
- 52.Chen F, Gao Y, Chen X, Yu Z, Li X. Quorum quenching enzymes and their application in degrading signal molecules to block quorum sensing-dependent infection. Int J Mol Sci. 2013; 14: 17477–17500. doi: 10.3390/ijms140917477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feng QM, Ru XS, Zhang LB, Zhang SY, Yang HS. Differences in feeding behavior and intestinal microbiota may relate to different growth rates of sea cucumbers (Apostichopus japonicus). Aquaculture. 2022; 559: 738368. doi: 10.1016/j.aquaculture.2022.738368 [DOI] [Google Scholar]
- 54.Janßen HJ, Steinbüchel A. Fatty acid synthesis in Escherichia coli and its applications towards the production of fatty acid based biofuels. Biotechnol Biofuels. 2014; 7: 7. doi: 10.1186/1754-6834-7-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lopez-Lara IM, Geiger O. Formation of fatty acids. In: Timmis KN. Handbook of hydrocarbon and lipid microbiology. Springer-Verlag Berlin Heidelberg; 2010. pp. 386–393. doi: 10.1007/978-3-540-77587-4_26 [DOI] [Google Scholar]
- 56.Kaya K. Odd carbon fatty acids. Oleoscience. 2020; 20: 337–340. In Japanese. [Google Scholar]
- 57.Coleman J, Raetz CR. First committed step of lipid A biosynthesis in Escherichia coli: sequence of the lpxA gene. J Bacteriol. 1998; 170: 1268–1274. doi: 10.1128/jb.170.3.1268-1274.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Buetow L, Smith TK, Dawson A, Fyffe S, and Hunter WN. Structure and reactivity of LpxD, the N-acyltransferase of lipid A biosynthesis. PNAS. 2007; 104: 4321–4326. doi: 10.1073/pnas.0606356104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mansilla MC, de Mendoza D. The Bacillus subtilis desaturase: a model to understand phospholipid modification and temperature sensing. Arch Microbiol. 2005; 183: 229–235. doi: 10.1007/s00203-005-0759-8 [DOI] [PubMed] [Google Scholar]
- 60.Clarke SD, Nakamura MT. Fatty acid structure and synthesis. In: Lennarz WJ, Lane DM. Encyclopedia of biological chemistry. Second edition. Cambridge: Academic Press; 2013. pp. 285–289. doi: 10.1016/B978-0-12-378630-2.00038-4 [DOI] [Google Scholar]
- 61.Tan BK, Bogdanov M, Zhao J, Dowhan W, Raetz CRH, Guan Z. Discovery of a cardiolipin synthase utilizing phosphatidylethanolamine and phosphatidylglycerol as substrates. Proc Natl Acad Sci. USA. 2012; 109: 16504–16509. doi: 10.1073/pnas.1212797109 [DOI] [PMC free article] [PubMed] [Google Scholar]