Author response image 1. (a) RAD21mAC cells stained for S9.
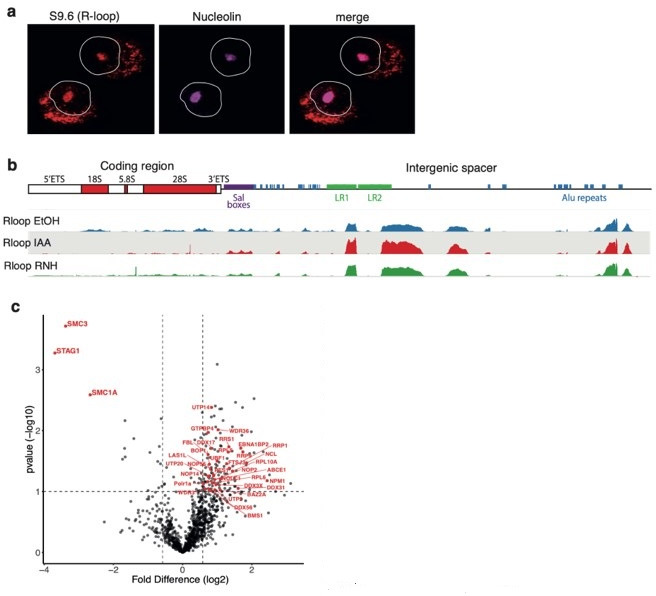
6 representing R-loops and Nucleolin. White line denotes the nucleus based on DAPI. NB. the expected nucleolar R-loop signal. (b) Top, cartoon of the consensus hg19 ribosomal DNA (rDNA), showing the ribosomal genes and the intergenic spacer (IGS) region which contains several Alu elements (blue). Bottom, S9.6 DRIP-seq from RAD21mAC cells treated with EtOH, IAA and RNaseH (RNH) and aligned to the consensus rDNA. NB. R-loops within the coding region are sensitive to RNH and IAA while those within the IGS are less affected. (C) Volcano plot displaying the statistical significance (-log10 p-value) versus magnitude of change (log2 fold change) from SA1 IPMS data produced from EtOH or IAA-treated RAD21mAC cells. Data as in Figure 2b from the manuscript except the proteins that are part of the enriched ‘Ribosome biogenesis’ functional category are highlighted in red.
