Abstract
The effect of mutations of the catalytic dyad residues of SARS-CoV-2 main protease (MProWT) on the thermodynamics of binding of covalent inhibitors comprising nitrile [nirmatrelvir (NMV), NBH2], aldehyde (GC373), and ketone (BBH1) warheads to MPro is examined together with room temperature X-ray crystallography. When lacking the nucleophilic C145, NMV binding is ∼400-fold weaker corresponding to 3.5 kcal/mol and 13.3 °C decrease in free energy (ΔG) and thermal stability (Tm), respectively, relative to MProWT. The H41A mutation results in a 20-fold increase in the dissociation constant (Kd), and 1.7 kcal/mol and 1.4 °C decreases in ΔG and Tm, respectively. Increasing the pH from 7.2 to 8.2 enhances NMV binding to MProH41A, whereas no significant change is observed in binding to MProWT. Structures of the four inhibitor complexes with MPro1-304/C145A show that the active site geometries of the complexes are nearly identical to that of MProWT with the nucleophilic sulfur of C145 positioned to react with the nitrile or the carbonyl carbon. These results support a two-step mechanism for the formation of the covalent complex involving an initial non-covalent binding followed by a nucleophilic attack by the thiolate anion of C145 on the warhead carbon. Noncovalent inhibitor ensitrelvir (ESV) exhibits a binding affinity to MProWT that is similar to NMV but differs in its thermodynamic signature from NMV. The binding of ESV to MProC145A also results in a significant, but smaller, increase in Kd and decrease in ΔG and Tm, relative to NMV.
Keywords: SARS-CoV-2 main protease, C145A/H41A mutations, main protease inhibitors, room-temperature X-ray crystallography, inhibitor binding thermodynamics, thermal stability
The ongoing COVID-19 pandemic caused by the SARS coronavirus 2 (SARS-CoV-2) continues to create significant health problems in many countries even though vaccines and therapeutic intervention options have been available for some time (1, 2, 3, 4). Meanwhile, the virus constantly evolves through naturally occurring polymorphisms to generate new strains capable of evading vaccine-induced immunity (1, 2, 3, 4), and antiviral therapy (5, 6, 7). Therefore, new prevention and treatment regimens are constantly being sought. During SARS-CoV-2 replication, limited polyprotein processing mediated by two virally encoded proteases to produce the mature structural and functional proteins is a well-established regulatory mechanism for viral maturation (8, 9). SARS-CoV-2 belongs to enveloped, positive-sense single-stranded RNA viruses. A single copy of the main protease (MPro) is translated from an ∼30 kb genome through overlapping reading frames as part of two large polyproteins (pp) 1a and 1ab, which encompass all non-structural proteins (nsps) (10, 11, 12). MPro (nsp5) mediates its own release (autoprocessing) at its termini and the ordered processing of the polyproteins from nsp4 to nsp16 for the precise assembly of the transcription/replication complex (8, 13, 14, 15). This process together with a second papain-like protease (PLpro), encoded within nsp3 that mediates the release of nsp1-nsp3, is required for virus maturation and production of progeny virus (8, 12). Because of its indispensable role, MPro has been targeted for the development of antiviral agents, and such efforts have resulted in the discovery of several lead compounds and oral drugs for treating COVID-19 (6, 16, 17, 18).
MPro, a cysteine protease exhibiting an α-chymotrypsin-like fold (3CLpro), is fully active in its mature form as a homodimer consisting of one active site per protomer (8, 9, 19). Each protomer comprises a two-domain 3CL fold of the catalytic region (domains I and II spanning residues 1–185), connected to domain III of five alpha-helices (residues 201–306) through a long linker sequence [residues 186–200, (9, 19)]. The active site consists of a typical oxyanion loop (residues 141–145) harboring the unconventional C145-H41 catalytic dyad in each protomer (15). The nucleophilic sulfur of C145 is deprotonated by forming an ion pair with protonated H41 (20). Interestingly, falling under a picornavirus-like supercluster, which includes viruses belonging to the Picornaviridae, Caliciviridae, and Coronaviridae families, unlike 3C-proteases having only the two-domain chymotrypsin fold for catalytic function, 3CL proteases, as in MPro of SARS-CoV and SARS-CoV-2, evolved with the additional domain III (21, 22, 23, 24, 25). Domain III introduces a critical step in MPro regulation and function by enabling dimer formation. It is evident from various studies that the N-terminal autoprocessing at the nsp4/nsp5 junction to release the free N-terminus of MPro establishes the critical dimer interface contacts between the N-terminal residues and domain III and thus decreases the dimer dissociation constant (26, 27, 28). Recent studies also indicate that transitioning of the active site oxyanion loop from an inactive E conformer to an active E∗ conformer occurs in conjunction with dimer formation. Furthermore, it was shown that the dynamic E-E∗ equilibrium can be switched mainly to the E∗ form independent of dimerization upon inhibitor binding to the monomer form, although with a much weaker affinity (15). Thus, the overall binding affinity and corresponding thermal stability of a given E∗-I complex are dependent on the M-D equilibrium of the complex and relate to the dynamics and rigidity of the free enzyme and inhibitor-bound complex (15, 28, 29).
Many inhibitors of MPro are designed to mimic substrates interacting with the enzyme subsite pockets S4-S1′ for specificity (Fig. 1A, (30)), and the affinity is enhanced by positioning a reactive warhead, such as a nitrile or carbonyl group, built into the inhibitor to enable covalent bond formation with C145 (Fig. 1B). Covalent inhibitor binding to a target protein is assumed to be initially driven by non-covalent interactions, followed by the formation of a specific covalent bond between the warhead’s electrophilic carbon and the reactive thiol side chain of C145 (31). Recent studies show that nirmatrelvir (NMV)-MPro complex (18) is sufficiently stable in a mixture of MPro and NMV, allowing its detection as an imidate thioester adduct by LC/MS (15). In contrast, no hemithioacetal, and likely hemithioketal, adducts were detectable under similar conditions upon the binding of GC373 (15), or Y180 (16), to MPro. The structure of the non-covalent complex and hence, the geometrical arrangement of the active site atoms and the warhead in the complex have never been examined. Also, the role of the active site H41 in the formation of the covalent imidate thioester has not been evaluated.
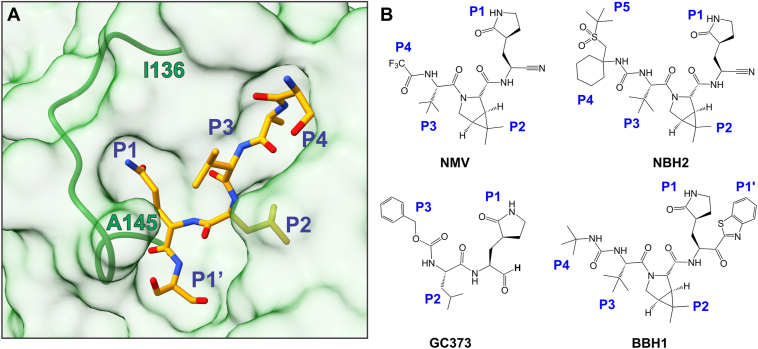
Figure 1.
MProC145A-substrate complex and chemical structures of covalent inhibitors. A, surface representation of MProC145A bound to substrate Ac-SAVLQSGF-CONH2 (7N89, (45)). Residue positions P1′ through P4 of the substrate are labeled, and the active site oxyanion loop in its E∗ conformation from I136 to A145 is shown as a green ribbon. B, chemical structures of inhibitors used in this study containing the nitrile (NMV, NBH2), aldehyde (GC373), and ketone (BBH1) groups.
To evaluate the contribution of the reactive warheads to binding affinity, here we report the binding constants and thermodynamic parameters of four such inhibitors with either a nitrile (NMV, NBH2), aldehyde (GC373), or ketone (BBH1) warhead to MProC145A and MProH41A (Fig. 1B) along with thermal stabilities of the complexes. The room-temperature crystal structures of the four inhibitors utilizing mature MPro bearing a deletion of the two C-terminal residues 305 and 306 (MPro1-304/C145A) and an active site mutation C145A that precludes covalent bond formation between the inhibitor and enzyme are compared with MProWT. The results presented show that the enhanced binding affinity of such compounds is largely driven by covalent bond formation between the inhibitor warhead’s electrophilic carbon and C145 thiolate anion. Finally, the binding of the recently developed noncovalent inhibitor ensitrelvir, approved for emergency use for the treatment of COVID-19 in Japan, was also evaluated with MProWT and catalytic dyad mutants. Unlike NMV, which mimics the substrate and is designed to engage the S4-S1 subsites of MPro, ESV matches those corresponding to the S2-S1′ subsites across the scissile peptide bond. Thermodynamic parameters of ESV binding to MProWT and its catalytic dyad mutants allowed a direct comparison of the driving forces involved in binding both inhibitors and provided evidence of the involvement of the catalytic dyad in the binding of ESV.
Results
Rationale for the design of mature MPro1-304/C145A
In this study, we examine the contribution of the catalytic dyad residues, C145 and H41, of MProWT to the binding of inhibitors with a reactive warhead and the effect of their exclusion on the binding specificity and affinity of the inhibitor. This entails mutating the C145, thereby excluding the reactive nucleophilic thiolate anion, and its counter ion, the protonated imidazole ring of H41, to alanine. Structural studies of mature MPro from SARS-CoV and SARS-CoV-2 containing the active site C145 mutation consistently show the C-terminal residues of one subunit of the homodimer bound intermolecularly to the active site of a second dimer (32, 33, 34). Such an observation was proposed to mimic an enzyme–product complex along the reaction pathway formed by the P5-P1 residues (MPro numbering 302–306) upon cleavage of the nsp5/nsp6 site with the P1 C-terminal carboxylate positioned proximal to C145. To minimize such interaction and simplify our structural studies, the P2 and P1 residues corresponding to the C-terminal residues 305 to 306 of MPro were deleted, and the resulting construct, termed MPro1-304/C145A, was used for the structural and binding studies along with MProWT, MProC145A, and MProH41A (Fig. 1).
Dimer dissociation constant and conformational stability of Mpro1-304/C145A
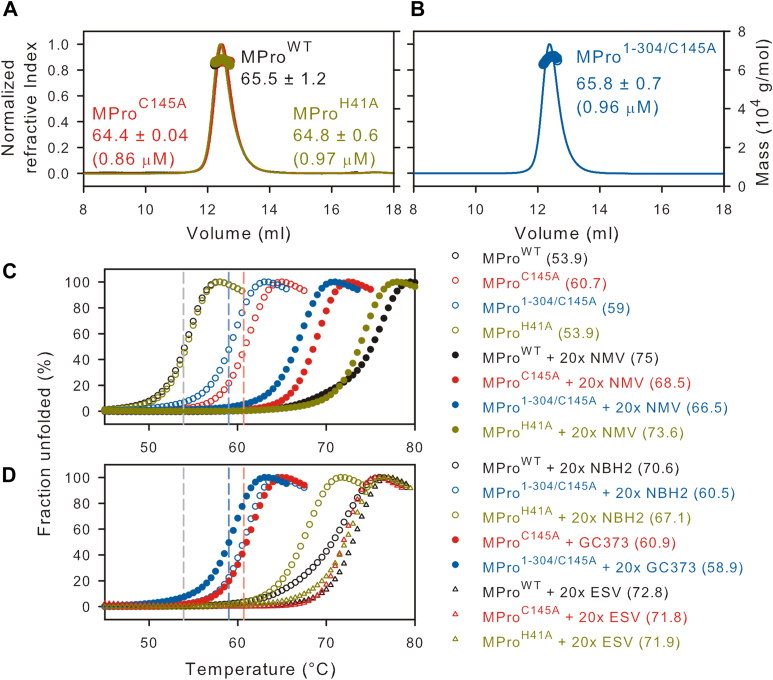
Mutations of the catalytic dyad residues, MProC145A and MProH41A, do not noticeably influence the dimer dissociation constant (Kdimer) of the mature MPro (9, 28, 29). Consistent with these results, SEC-MALS analyses indicate that MPro1-304/C145A is mainly dimeric at an estimated peak apex concentration of 0.96 μM when injecting 125 μl of the sample at 7.2 μM on Superose-12 column (1 × 30 cm) in buffer A at 25 °C (Fig. 2, A and B, see Experimental procedures) similar to MProC145A and MProH41A carried out under the same conditions. This allowed the comparison of the thermal denaturation profile of MPro1-304/C145A at 10 μM concentration where it is mainly dimeric with MProWT, MProC145A, and MProH41A.
Figure 2.
Characterization of MPro1-304/C145Aand thermal denaturation profiles. A and B, proteins were fractionated on Superose-12 column (1 × 30 cm) in buffer A at 25 °C. Eluting top of peak concentration (in parentheses) and the estimated mass (in kDa) corresponding to the monomer-dimer equilibrium boundary (circles) are indicated. C and D, thermal denaturation DSF traces at a final concentration of 10 μM in buffer B. The vertical dashed lines indicate the midpoint for the transition for MProWT (black trace), MProC145A (red), and MPro1-304/C145A (blue) serving as a reference point to compare traces acquired in the presence of inhibitor. Estimated Tm midpoints are listed in parentheses beside the construct designation, and in Table 1. 20× denotes 20 times (200 μM) the concentration of inhibitor over the protein concentration (10 μM).
As shown before (29), of the two catalytic dyad residues, surprisingly, C145A mutation significantly enhances the thermal stability of MProC145A by 6.8 °C, contrary to MProH41A, which resembles MProWT (Tm = 53.9 °C, Table 1 and Fig. 2C). Thermal denaturation profiles by circular dichroism concur with the DSF profiles (29). The high temperature (based on Tm) under which the experiments must be carried out restricts the use of conventional methods such as SV-AUC or SEC-MALS to explain the reason for the increased Tm rendered by the C145A mutation. The increased Tm of MProC145A could be due to either the stabilization of the dimer interface or just the monomer fold. In this context, it is worth noting that a predominantly monomeric form of MPro (MProM, with substitutions E290A and R298A) also exhibits a Tm nearly identical to that of MProWT (29). Like MProC145A, MPro1-304/C145A also shows an increase in Tm of 5.1 °C. The assumption that Tm signifies the conformational stability of the monomer fold with negligible contribution from the dimer interface is in accordance with the results that Kdimer of MProC145A and MPro1-304/C145A appear to be similar to that of MProWT (Table 1 and Fig. 2).
Table 1.
Estimated Tm, Kd and thermodynamic parameters of binding of various inhibitors to MProWT and its catalytic dyad mutants
|
Constructs |
Tm (°C) | ΔTm (°C) | Kd (μM) | ΔH (kcal/mol) | ΔS (cal/mol/K) | ΔG (kcal/mol) |
|---|---|---|---|---|---|---|
| MProWT | 53.9 ± 0.1 | |||||
| MProC145A | 60.7 ± 0.1 | 6.8 | ||||
| MPro1-304/C145A | 59 ± 0.1 | 5.1 | ||||
| MProH41A | 53.9 ± 0.1 | 0 | ||||
| MProWT + NMV | 75 ± 0.1 | 21.1 | 0.007 ± 0.003a | −10.8 ± 0.7 | 1.57 | −11.2 |
| MProWT + NMV (8.2) | 0.012 ± 0.006 | −11.8 ± 0.1 | −2.86 | −10.9 | ||
| MProC145A + NMV | 68.5 ± 0.1 | 7.8 | 2.7 ± 0.9b | −3.9 ± 0.2 | 12.6 | −7.7 |
| MPro1-304/C145A + NMV | 66.5 ± 0.1 | 7.5 | 9.2 ± 1.7 | −4.3 ± 0.2 | 8.9 | −6.9 |
| MProH41A + NMV | 73.6 ± 0.1 | 19.7 | 0.14 ± 0.02 | −8.4 ± 0.05 | 3.5 | −9.5 |
| MProH41A + NMV (8.2) | 0.03 ± 0.02 | −8.5 ± 0.17 | 6.07 | −10.4 | ||
| MProWT + NBH2 | 70.6 ± 0.1 | 16.7 | 0.026 ± 0.016a | −8.76 ± 0.17 | 5.63 | −10.5 |
| MPro1-304/C145A + NBH2 | 60.5 ± 0.1 | 1.5 | nd | |||
| MProH41A + NBH2 | 67.1 ± 0.1 | 13.2 | 2.3 ± 0.9 | −1.47 ± 0.01 | 20.9 | 7.76 |
| MProWT + GC373c,d | 73.7 ± 0.1 | 19.8 | 0.15 ± 0.03 | −6.7 ± 0.1 | 9.1 | −9.4 |
| MProC145A + GC373 | 60.9 ± 0.1 | 0.2 | nd | |||
| MPro1-304/C145A + GC373 | 58.9 ± 0.1 | −0.1 | nd | |||
| MProH41A + GC373d | 70.8 ± 0.1 | 16.9 | 1.4 ± 0.5 | −5.6 ± 0.2 | 8.3 | −8.1 |
| MProWT + ESV | 72.9 ± 0.2 | 18.9 | 0.006 ± 0.003 | −15.4 ± 0.01 | −13.9 | −11.2 |
| MProC145A + ESV | 71.8 ± 0.2 | 11.1 | 0.226 ± 0.061 | −11.5 ± 0.2 | −7.8 | −9.2 |
| MProH41A + ESV | 71.9 ± 0.1 | 18 | 0.056 ± 0.031 | −9.53 ± 0.23 | 1.53 | −10 |
Tm values were extracted from DSF profiles shown in Figure 1, C and D. ΔTm denotes the difference in Tm in the presence and absence of inhibitor. Inhibitor binding (Kd) to MPro1-304/C145A along with corresponding MPro controls (a (38)). Not determined (nd) because of no heat response when titrating 50 μM protein in the cell and 0.5 mM inhibitor, indicative of very weak binding, relative to the binding of NMV to MProC145A and MPro1-304/C145A. DSF scans and ITC were not carried out with BBH1 because of poor solubility. b, c, d cited from references (15, 28, 29). All titrations were performed in buffer C (pH 7.2) except for two at pH 8.2, indicated in parantheses in the first column.
Binding affinities of NMV, NBH2, and GC373 to catalytic dyad mutants
It is known that increased binding affinity (i.e., decrease in inhibitor dissociation constant, Kd) to the protein correlates with an increase in the thermal stability as shown for HIV-1 protease (35, 36) and MProWT inhibitor complexes (29). Of the two catalytic dyad residues, the C145A mutation shows the largest increase on Kd, consistent with a decrease in ΔTm and ΔG (Table 1 and Fig. 2, C and D). Kd increases in the order: MProWT < MProH41A < MProC145A < MPro1-304/C145A (Table 1 and Fig. S1, A–D).
For NMV, C145A mutation results in a ∼400-fold increase in Kd (2.7 ± 0.9 μM) as compared to MProWT (0.007 ± 0.003 μM), pointing to the importance of the nitrile warhead reaction with C145 to form an imidate thioester for enhanced binding affinity. MPro1-304/C145A shows an even larger Kd increase of ∼1300-fold (Table 1). The 3-fold increase in Kd for MPro1-304/C145A relative to MProC145A points to subtle changes influencing inhibitor affinity even though the catalytic activity of SARS-CoV MPro bearing the same deletion was shown to be unaffected when assessed using an 11-mer peptide corresponding to the nsp4/nsp5 cleavage site (37). Both mutants show a ΔΔTm difference of ∼13 °C, relative MProWT (Table 1 and Fig. 2C). An increase of ∼20-fold in Kd (Fig. S1D) for NMV binding to MProH41A is consistent with a smaller decrease in ΔTm of 1.4 °C (Table 1), which likely relates to an alteration in the hydrophobic interactions of the bicyclic group at P2 placed above the imidazole ring of His41 that occupies the S2 site (29).
The Kd and ΔG values (Tables 1 and S1) point to NBH2 being the next best inhibitor of MProWT (38). The Kd values for the binding of NBH2 and GC373 to MProWT are ∼4- (0.026 ± 0.016 μM) and 21-fold (0.15 ± 0.03 μM) higher, respectively, relative to NMV binding. By ITC, no thermal response was observed for either NBH2 or GC373, with MPro1-304/C145A suggesting that the binding is weaker with a Kd that is much larger than the observed 9.2 μM for NMV binding to the same construct. Tm increases marginally by 1.5 °C for the MPro1-304/C145A-NBH2 complex and negligible change was observed for complexes of GC373 with MProC145 and MPro1-304/C145A as compared to 16.7 °C and 19.8 °C differences for the corresponding complexes with MProWT (Fig. 2D).
Noncovalent mode of binding of the covalent inhibitors to MPro1-304/C145A
To assess the mode of binding of BBH1, GC373, NBH2, and NMV inhibitors containing reactive warheads to the MPro active site lacking the catalytic cysteine, we obtained room-temperature X-ray crystal structures of the inhibitor-free MPro1-304/C145A (Fig. 3) and of its complexes with the inhibitors at 1.8 to 2.0 Å resolutions. Also, we carried out comparisons of the structures with the corresponding room temperature MProWT structures to highlight the consequences of the inhibitors’ covalent attachment to C145 on the enzyme active site. MPro1-304/C145A mainly crystallized in the monoclinic unit cell (space group I2) with one protomer in the asymmetric unit (Table S1), similar to the crystals of MProWT complexed with the nitrile and aldehyde warhead-containing inhibitors (15, 38). The exception is MPro1-304/C145A-BBH1 complex which crystallized in a triclinic unit cell (space group P1) with the entire homodimer present in the asymmetric unit. BBH1 binding is identical in both protomers of MPro1-304/C145A; therefore, the following structural analyses will be focused on protomer A.
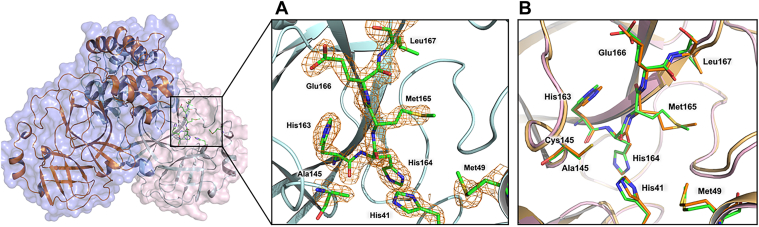
Figure 3.
MPro1-304/C145Ahomodimer in cartoon representation and structural comparison with MProWT. A, the insert shows the active site residues, indicating the point mutation C145A. The 2FO-FC electron density map is contoured at 2.0 σ. B, superposition of inhibitor-free MPro1-304/C145A on MProWT. MPro1-304/C145A is colored light pink (cartoon) and green/yellow carbon atoms. MProWT is colored light orange (cartoon) and orange carbon atoms.
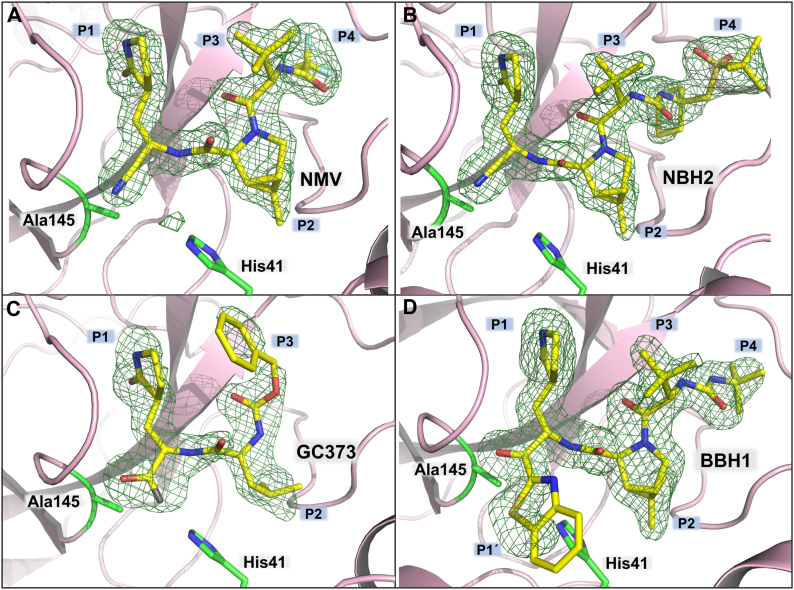
C145A mutation has minimal effect on the quaternary structure. MPro1-304/C145A superimposes on to MProWT (PDB ID 7JUN; (20)) with the r.m.s.d of 0.4 Å on the main-chain atoms, and the geometries of the two active sites are virtually identical (Fig. 3). The four inhibitors bind to the MPro1-304/C145A active site making only noncovalent interactions with the substrate-binding subsites S4-S1′ and the catalytic dyad. The warheads of the inhibitors remain intact (ketone for BBH1, aldehyde for GC373, and nitrile for NBH2 and NMV) and are clearly visible in the electron density maps (Fig. 4, A–D). The aldehyde of GC373 is observed in two alternate conformations related by a 180° flip because it can freely rotate around the C-C bond with the Cα of P1 γ-lactam before the inhibitor binds to the enzyme. In one conformation, the aldehyde oxygen interacts with the enzyme’s oxyanion hole, whereas in the other conformation, it makes a hydrogen bond with the catalytic His41. The aldehyde can react with C145 in either conformation, as has been observed previously (39, 40) in cryo-structures, but not in the room-temperature structure (15), of MProWT-GC373 complexes. Conversely, GC373 was also found in two alternate conformations in the room-temperature structure of MProH41A-GC373 complex (29). We observed weak electron density on a portion of the P1′ benzothiazole group in the MPro1-304/C145A complex with BBH1 (Fig. 4D) that did not allow us to accurately model this moiety, leading to a negative peak in the FO-FC difference electron density map. This can be due to a dynamic rotational disorder of benzothiazole. Importantly, it is unlikely that benzothiazole has been chemically altered because P1′ of BBH1 was readily visible in both electron and nuclear density maps in our previous joint X-ray/neutron structure of MProWT-BBH1 (38). Because the MPro1-304/C145A-BBH1 complex is noncovalent, it precludes its detection by mass spectrometry unlike as shown previously for the sufficiently stable covalent adduct of MProWT with NMV (15).
Figure 4.
Room-temperature X-ray structures of MPro1-304/C145Ain complex with inhibitors. NMV (A), NBH2 (B), GC373 (C) and BBH1 (D). Electron density is shown as Polder omit maps (green mesh contoured at 3.5 σ).
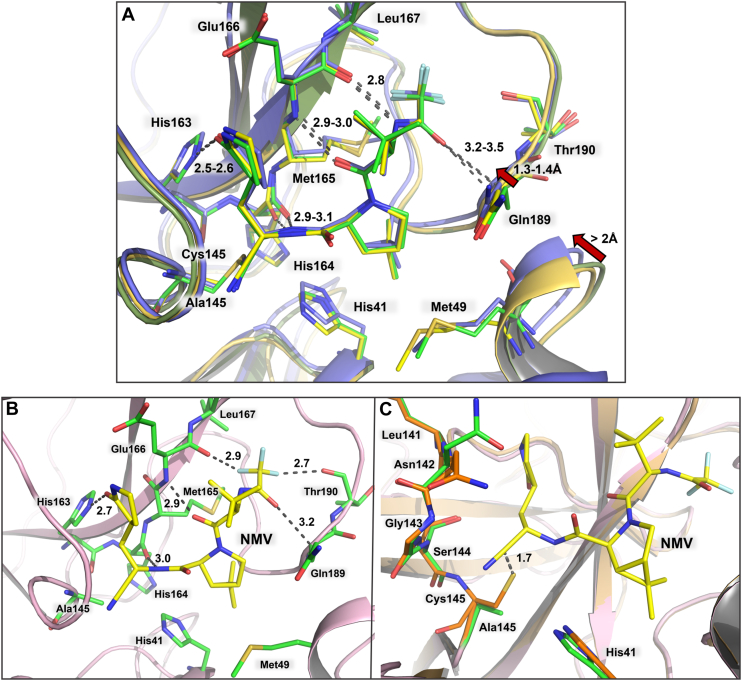
The four studied inhibitors bind to MPro1-304/C145A in the same manner as they do to MProWT. For direct structural comparison, we chose X-ray structures of MPro complexes with the studied inhibitors obtained at room temperature. Our previous structural comparisons of MProWT-NMV complexes obtained at room- [7SI9, (38)] and cryo-temperatures [8DZ2, (18)] revealed conformational differences of the MPro active sites. However, H-bonding by NMV is similar at both temperatures [Fig. 5A, (38)]. MPro1-304/C145A-NMV superimposes on 7SI9 and 8DZ2 with the root mean square deviation (RMSD) of 0.16 and 0.77 Å, respectively, on the main-chain atoms. Significant shifts of the S2 helix of ∼ 2 Å, containing residue M49, and S5 loop of 1.3 to 1.4 Å, including residue Q189, are detectable between MPro1-304/C145A-NMV and 8DZ2 which could be attributable to the differences in data collection temperatures. The inhibitors make several hydrogen bonding and hydrophobic interactions, as illustrated for NMV, with the active site residues in MPro1-304/C145A (Fig. 5, A and B). These contacts are identical to those made by the inhibitors with MProWT (15, 38). The structures thus agree with the inhibitors first binding noncovalently to the enzyme active site, followed by the formation of a covalent bond with the C145 thiolate. In fact, the distances between the reactive carbon atoms of the warheads and the C145 sulfur atoms are 1.7 to 1.8 Å for NMV, NBH2, and GC373 when the MPro1-304/C145A-inhibitor complexes are aligned with inhibitor-free MProWT, with the r.m.s.d’s of 0.5 Å for all structures (Figs. 5C and S2, A and B). This observation demonstrates that the warheads are poised for the nucleophilic attack by C145 immediately after the inhibitors are properly situated in the enzyme active site by noncovalent interactions. BBH1 is an exception, showing a significantly longer distance of 2.2 Å from the warhead carbonyl carbon to the C145 sulfur (Fig. S2C). This is probably due to the presence of a large hydrophobic benzothiazole P1′ group in BBH1 that other inhibitors lack, requiring significant structural rearrangements to accommodate the P1′ substituent in the active site after the covalent bond formation with C145. Indeed, substantial distortions of the active site geometry were previously observed in the neutron structure of MProWT-BBH1 (38) and in the cryo-X-ray structure of MProWT complexed with the inhibitor 5h, also having the benzothiazole P1′ group (41), compared to the inhibitor-free enzyme. When we superimposed MPro1-304/C145A-BBH1 complex with the structure of MProWT-BBH1, we could appreciate the significance of the structural rearrangements necessary for the hemithioketal formation between C145 and the BBH1 ketone warhead (Fig. S2D). The large steric size of the benzothiazole moiety results in the apparent pushing of H41 away from C145 into the active site interior that cascades further to remove the conserved water molecule (Wcat) normally coordinated by His41, His164, and Glu187 completely out of the active site.
Figure 5.
Binding of NMV to MPro1-304/C145Aand structural comparison with MProWT. A, superposition MPro1-304/C145A-NMV complex (colored as yellow-orange cartoon and yellow carbons) on MProWT-NMV complex structures obtained at room- and cryo-temperatures with PDB IDs 7SI9 (colored as smudge-green cartoon and green carbon atoms) and 8DZ2 (colored as purple cartoon and carbon atoms), respectively. B, hydrogen bonding and unconventional F…O interactions made by NMV. C, superposition of MPro1-304/C145A-NMV complex on MProWT (PDB ID 7JUN). MPro1-304/C145A is colored light pink (cartoon) and green/yellow carbon atoms. MProWT is colored light orange (cartoon) and orange carbon atoms. Distances are shown in Å.
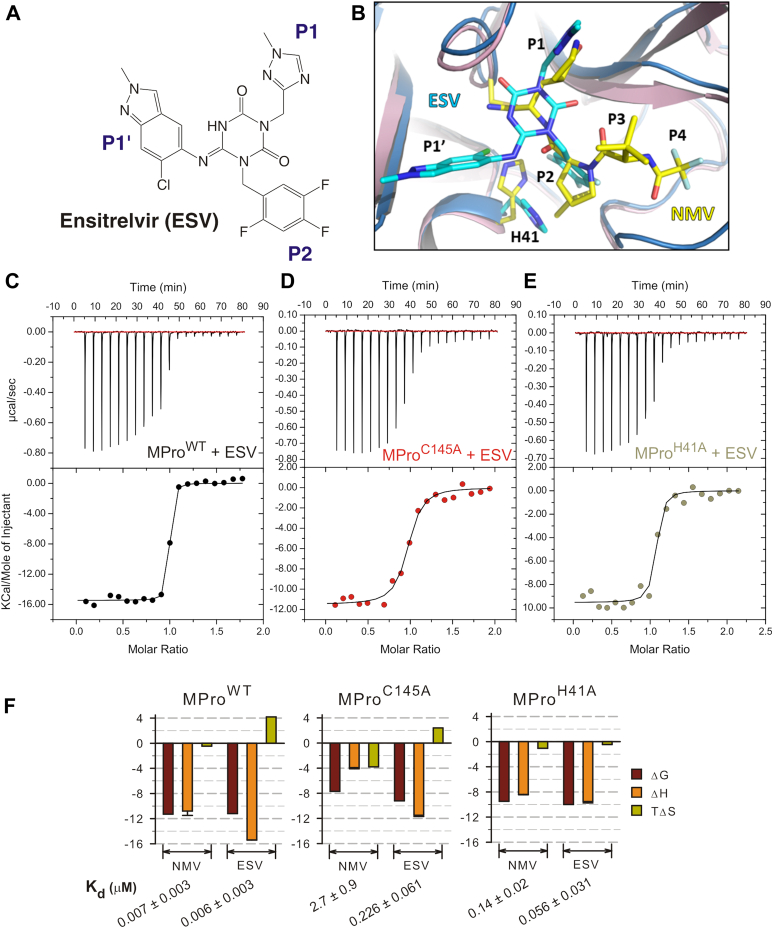
Binding thermodynamics of ensitrelvir to MProWT and catalytic dyad mutants
A comparison of the binding affinity and thermodynamics of a recently discovered noncovalent inhibitor (6, 17), to differentiate the contribution of the catalytic dyad residues for binding from NMV, is shown in Table 1 and Figure 6. Ensitrelvir (ESV) differs in its interactions by recognizing the S2-S1′ subsites of MPro, unlike the covalent compounds which span the S4-S1 subsites. ESV is similar in its binding affinity to NMV with a Kd of ∼6 nM and ΔG of −11.2 kcal/mol but differs in its thermodynamic profile from NMV. The improved binding enthalpy for ESV binding is offset by a positive entropy change, which contrasts NMV, resulting in a net ΔG being similar to NMV. H41A mutation appears to reverse the positive entropy change, thus improving the overall -ΔG to offset the decrease in enthalpy. A large Tm increase is observed upon the binding of ESV, similar to that for the binding of the covalent inhibitors to MProWT. A decrease in the binding affinity for MProC145A and MProH41A with ESV by 38- and 9-fold corresponds to a ΔΔTm of 7.8 and 0.9 °C, respectively (Table 1 and Fig. 2D).
Figure 6.
Thermodynamics of ESV binding to MProWTand its catalytic dyad mutants.A, chemical structure of ensitrelivir (ESV). B, superposition of the crystal structures of MProWT-NMV (light pink cartoon, yellow carbon atoms) and MProWT-ESV (blue cartoon, cyan carbon atoms) complexes. H41 rotates ∼135° around the Cα-Cβ bond away from C145 to accommodate the chlorine atom present in P1′ group of ESV. C–E, binding isotherms of ESV to MProWT and catalytic dyad mutants. Titrations were performed in buffer C at 28 °C with 30 μM protein in the cell and inhibitor in the syringe. F, comparison of the Kd and enthalpy/entropy compensation upon binding of NMV and ESV to MProWT and catalytic dyad mutants (Table 1).
Discussion
Both nitrile and carbonyl inhibitors, such as NMV and GC373, respectively, bind to MProWT through a two-step mechanism. The first step involves the formation of a non-covalent complex followed by the reaction of the nucleophilic sulfur of C145 with the electrophilic nitrile carbon and carbonyl carbon, respectively, to form a covalent complex. The covalent complex of NMV is a relatively stable imidate thioester which is a homolog of the thioester intermediate in peptide hydrolysis. In contrast, the covalent complex of the carbonyl inhibitor is a hemithioacetal or -ketal, a tetrahedral intermediate that mimics the transition state of the hydrolytic reaction (28). The C145A mutation of MPro has a profound effect on the binding of both the nitrile- and the carbonyl-based inhibitors. NMV binding constants to MProC145A and MPro1-304/C145A increase by ∼400-fold and 1300-fold, respectively, compared to that of MProWT. While NBH2 has a binding constant of 0.026 μM to MProWT, the binding constant of NBH2 to MPro1-304/C145A is expected to be weak because of the absence of meaningful thermal response in ITC up to 50 μM protein concentration. Thus, the relatively stable imidate thioester bond contributes 3.5 kcal/mol to the binding energy of NMV. A similar trend could be predicted for the binding of the aldehyde inhibitor GC373 to the C145A mutant. Both NMV and GC373 bind to MProH41A with a Kd of ∼20- (0.14 ± 0.02 μM) and 9-fold (1.4 ± 0.5 μM) larger, than to those of MProWT, corresponding to 1.7 and 1.3 kcal/mol of binding energy, respectively. These results are consistent with the involvement of the catalytic dyad of the enzyme in binding the inhibitors. In inhibitor-free MProWT, the catalytic dyad is ionized, having a positively charged protonated imidazole ring of H41 forming an ion pair with the negatively charged thiolate anion of the side chain of C145 (20). In the absence of the imidazole ring, the sulfur atom of C145 is expected to be partially or fully protonated with a pKa of about 8.0 (42). At pH 7.2, about 16% of the thiol of C145 of MProH41A would be in the thiolate form. Thus, the differences observed for Kd listed in Table 1 may be accounted for by the difference in the nucleophilicity of the thiolate anion of MProWT and MProH41A as well as the amount of thiolate anion at pH 7.2. Pearson et al. (43, 44) defined the nucleophilicity constant (n): n = log (ky/kmethanol), where ky is the second-order rate constant of the reaction of a nucleophile (y) with methyl iodide in methanol, and kmethanol is the second order rate constant of the solvolysis of methyl iodide in methanol. The reported n values for sulfide ion and diethyl thioether are 8 and 5.34, respectively. The difference in the n value shows that the thiolate anion reacts about 450 times faster than the uncharged thiol. Thus, the predominant reaction for adduct formation appears to be from a nucleophilic attack by the thiolate anion and not the neutral thiol. This conclusion is further strengthened by the observed 79% decrease in Kd for the binding of NMV to MProH41A at pH 8.2 where the thiol of C145 is about 61% in the thiolate anion form. In contrast, no significant change is observed in Kd for MProWT under the same conditions (see Table 1 and Fig. S3). The absence of the nucleophilic thiol/thiolate side chain of C145 obviates the possibility of forming any covalent bond. These results are consistent with the previously observed inverse correlation between Kd and catalytic activities of MPro mutants as well as the observation of the thioimidate ester of MProWT and not MProC145A (15). They also illustrate the critical role of the thiolate anion in binding covalent inhibitors to MProWT.
The recent discovery of the noncovalent inhibitor ESV enabled a timely comparison with NMV. For ease of comparison, plots of the enthalpy/entropy compensation upon binding of NMV and ESV are shown in Figure 6F. Loss in enthalpic contribution when lacking the nucleophilic C145 or H41 is offset by a gain in entropy resulting in a net decrease in ΔG for NMV. Interestingly, the larger enthalpy change observed upon binding of ESV results in an unfavorable entropy change, which contrasts NMV binding, resulting in a binding affinity similar to that of NMV. This trend is also seen for the binding of ESV to the C145A mutant but with a smaller ΔG decrease of 2 kcal/mol, roughly half the effect of the C145A mutation on NMV binding. Reversing to a favorable entropy upon H41A mutation may indicate a relieving of a steric constraint induced by the large P1′ group of ESV containing Cl that clashes with H41 in MProWT, forcing it to rotate ∼180° around Cα-Cβ bond (Fig. 6B).
The availability of the complexes of NMV (38) and ESV (17) permits a comparison of the role of the catalytic dyad residues for binding affinity. In terms of H-bonds, both inhibitors have four H-bonds. ESV makes a 3.1 Å H-bond with the C145 main chain amide NH, whereas NMV’s imidate’s N is not positioned to make such an H-bond, being directed away from C145 NH with a 3.4 Å N…N distance. In addition, H41 rotation in MProWT–ESV complex leads to the formation of π-π stacking interactions between ESV’s P2 group and H41 imidazole, interactions replaced with weaker C-H…π contacts in MProWT-NMV. The H41 rotation caused by the bulky P1′ group of ESV allows for an H bond formation with the T25 main chain NH that is not possible for NMV lacking a P1′. Thus, the presence of the P1′ in ESV can be essential for this inhibitor to retain most of its affinity when C145 is mutated to A, unlike NMV which drastically loses affinity when the covalent bond formation is not possible. These results highlight the importance of aligning the reacting atoms of the warhead to the thiolate anion for retaining potency.
To conclude, we engineered an MPro construct MPro1-304/C145A bearing a C145A mutation that limits inter-dimer association at concentrations necessary for crystallization due to the binding of the C-terminal residues (302–306) to the active site of the second homodimer. Covalent inhibitors NMV, NBH2, GC373, and BBH1 bind exclusively through noncovalent interactions to MPro1-304/C145A according to the room-temperature X-ray structures of the complexes. The binding affinity of these inhibitors is shown to be unequivocally and largely affected by the absence of C145 thiolate anion and thus, their inability to form covalent bonds leads to increasing Kd by more than two orders of magnitude for NMV. Our results demonstrate that covalent bond formation is a major driving force for the enhanced affinity of inhibitors that mimic enzyme substrates. The above results validate the two-step mechanism, the first step involves the specific network of noncovalent interactions of the inhibitor to the active site, followed by covalent bond formation. For NMV, a favorable enthalpy contribution of −7.9 kcal/mol upon covalent bond formation is offset by an entropy loss resulting in a net ΔG increase of 3.5 kcal/mol equivalent to ∼400-fold increase in binding affinity. Contrary to NMV, ESV decreases in its binding affinity by about 38- and 9-fold to MProC145A, and MProH41A, respectively, indicating a less significant contribution of its interaction with catalytic C145 and H41 residues for binding affinity. These analyses will facilitate comparisons of how evolving drug-resistant mutations may impair binding specificity and reactivity of covalent and noncovalent inhibitors in conjunction with the oxyanion loop and monomer-dimer conformational equilibria regulating MPro function (15, 28).
Experimental procedures
General information
Ni-NTA columns were purchased from Cytiva (Piscataway, New Jersey, USA). His-tagged Human Rhinovirus (HRV) 3C protease was purchased from Sigma. Crystallization reagents and supplies were purchased from Hampton Research. Crystallographic supplies for crystal mounting and X-ray diffraction data collection at room temperature were purchased from MiTeGen. NMV (CAS # 2628280–40–8) and ESV were purchased from MedChemExpress and Millipore Sigma, respectively. The synthetic procedures for BBH1 and NBH2 were published elsewhere (38). GC376 (a prodrug of GC373) was purchased from Selleckchem.
Plasmid construction, expression, and purification of MPro constructs
The new construct MPro1-304/H41A prepared for this work by omitting the last two residues (305–306) of MPro was synthesized and cloned into pJ414 vector (ATUM, Newark, CA). Expression and purification of MPro1-304/H41A were carried out as described for MProWT (GenBank ID: MN908947.3), MProC145A, and MProH41A (15, 29, 45). Purity was verified both by SDS-PAGE on 4% to 20% gradient mini-protean TGX precast gel (Bio-Rad) and reverse-phase liquid chromatography with in-line electrospray ionization mass spectrometry (15). Protein concentrations were measured before storage and prior to the experiment at least in duplicate based on the extinction coefficient at 280 nm.
Size exclusion chromatography with multi-angle light scattering
Molecular mass was estimated by analytical SEC with in-line MALS (DAWN Heleos-II, Wyatt Technology Inc), refractive index (Optilab T-rEX, Wyatt Technology Inc.), and UV (Waters 2487, Waters Corporation) detectors. Sample (125 μl) was applied onto a pre-equilibrated Superose-12 column (1.0 × 30 cm, Cytiva) and eluted at a flow rate of 0.5 ml/min in buffer A (25 mM Tris-HCL, pH 7.6, 150 mM NaCl, 1 mM TCEP) at 25°C. The injection concentrations were 30 μM of MProWT and 7.2 μM each of MProC145A, MProH41A, and MPro1-304/C145A. Molecular mass was calculated using the Astra software provided with the instrument. Estimated Kdimer of MProWT are reported in references (9, 28).
Differential scanning fluorimetry
Samples in duplicate were prepared with SYPRO orange dye (5000×, Millipore Sigma product number S5692) to yield a final concentration of 10 μM protein and 5× dye in 25 μl of buffer B (25 mM Tris-HCl, pH 7, 50 mM NaCl and 1 mM TCEP) and 0.1% DMSO. The fluorescence signal was monitored as a function of temperature in a Bio-Rad C1000 Touch Thermal Cycler, and data were processed with the provided software and plotted using Sigmaplot (Systat Software Inc.). Experiments were repeated at least twice.
Isothermal titration calorimetry
Purified proteins were diluted from a stock solution to slightly above the desired concentration and dialyzed extensively against buffer C (25 mM Tris-HCl, pH 7.2, 20 mM NaCl and 1 mM TCEP) or buffer D (25 mM Tris-HCl, pH 8.2, 20 mM NaCl and 1 mM TCEP). Concentrations were estimated after dialysis based on their 280 nm absorbance at least twice. Stock solutions of inhibitors in buffer C were diluted in the same buffer to the desired concentration. Titrations were performed with proteins (30–50 μM) kept in the cell and inhibitors at 10 times the concentration of the protein in the syringe at 28°C on iTC200 microcalorimeter (Malvern Instruments Inc., Westborough, MA). Titrations were repeated when needed. Nineteen injections of data were processed using the Origin software provided with the instrument.
Protein crystallization and room-temperature X-ray crystallography
For crystallization, MPro1-304/C145A protein sample was concentrated to ∼6 mg/ml. GC376 stock was prepared at 10 mM GC376 in buffer C (25 mM Tris-HCl, pH 7.2, 20 mM NaCl and 1 mM TCEP) for crystallization purposes and stored at −30 °C. GC376 is converted to the reactive aldehyde GC373 when dissolved in an aqueous solution. Stocks of BBH1, NBH2, and NMV inhibitors were prepared at 50 mM in 100% dimethyl sulfoxide and stored at −30 °C. For co-crystallization, MPro1-304/C145A was mixed with BBH1, NBH2, or GC373 at 1:5 M ratio and allowed to incubate at room temperature for at least 30 min before setting up crystallization trays. Crystals of inhibitor-free MPro1-304/C145A and of the BBH1-, NBH2-, and GC373-bound complexes were grown at 14 °C by sitting drop vapor diffusion methodology with 15 to 18% PEG3350, 0.1 M Bis-Tris pH 6.5 or pH 7.0 (1 ml) as the precipitant solution by mixing protein and well solution at 1:1 ratio to give 20 μl crystallization drops. To ensure the growth of quality crystals, crystallization drops were seed struck using the crystals of MProWT in complex with a covalent ligand NBH2 as previously described (38). Similar to the crystallization attempts using MProWT (9), crystals of MPro1-304/C145A in a complex with NMV could not be grown by co-crystallization. Instead, MPro1-304/C145A-NMV complex was obtained by soaking with NMV (38). The crystals suitable for X-ray diffraction measurements were mounted in MiTeGen room-temperature capillary setups for data collection.
Room temperature X-ray crystallographic data were collected on a Rigaku HighFlux HomeLab instrument equipped with a MicroMax-007 HF X-ray generator, Osmic VariMax optics, and a DECTRIS Eiger R 4M hybrid photon counting detector. X-ray diffraction data were integrated using the CrysAlis Pro software suite (Rigaku Inc., The Woodlands, TX), then reduced and scaled using Aimless (46) from the CCP4 suite (47). Structures were solved by molecular replacement using Phaser (48). Inhibitor-free MProWT [PDB code 7JUN, (20)] was used as a search model to solve the structure of inhibitor-free MPro1-304/C145A. MProWT-NMV complex structure [PDB code 7SI9, (38)] was used to solve the structures of all the inhibitor complexes. Each structural model was iteratively refined with phenix.refine from the PHENIX suite (49) and COOT (50). Geometry validation was aided by Molprobity (51). Restraints for the inhibitors were generated with eLBOW (52) using geometry optimized by quantum chemical calculations in Gaussian16 at B3LYP/6 to 31g(d,p) level of theory (53). Final data collection and refinement statistics for the five determined structures can be found inTable S1.
Data availability
All data that support the findings of this study are contained within this article and its accompanying files. PDB accession numbers are listed in Supporting Information, Table S1.
Supporting information
This article contains supporting information (15).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
This research used resources at the Spallation Neutron Source, the High Flux Isotope Reactor, and the Center for Nanophase Material Sciences for the synthesis of BBH1 and NBH2, which are DOE Office of Science User Facilities operated by the Oak Ridge National Laboratory. The Office of Biological and Environmental Research supported research at ORNL's Center for Structural Molecular Biology (CSMB), a DOE Office of Science User Facility. ORNL is managed by UT-Battelle LLC for DOE’s Office of Science, the single largest supporter of basic research in the physical sciences in the United States. We thank John Lloyd and the NIDDK mass spectrometry core facility. This work was supported by the Intramural Research Program of National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), NIH, United States.
Author contributions
A. Y. K and J. M. L. Conceptualization; A. Y. K., J. M. L., A. A., L. C., P. V. B., and N. T. N. Methodology; A. Y. K., J. M. L., A. A., L. C., P. V. B. and N. T. N. investigation; A. Y. K and J. M. L. Writing – original draft. A. A., L. C., P. V. B. and N. T. N. writing – review and editing.
Funding and additional information
This work was funded by NIDDK (Project number: DK075166-01) to J. M. L.
Reviewed by members of the JBC Editorial Board. Edited by Joseph Jez
Contributor Information
Andrey Kovalevsky, Email: kovalevskyay@ornl.gov.
John M. Louis, Email: johnl@niddk.nih.gov.
Supporting information
References
- 1.Therapeutics and COVID-19: living guideline, J. G. W. H. O . World Health Organization; Geneva: 2023. Therapeutics and COVID-19: Living Guideline, 13 January 2023. [Google Scholar]
- 2.Bebenek I., Bannister R., Dubinion J., Fortin M., Liu M., Motter A.L., et al. COVID-19 therapeutics and vaccines: a race to save lives. Toxicol. Sci. 2022;185:119–127. doi: 10.1093/toxsci/kfab130. [DOI] [PubMed] [Google Scholar]
- 3.Uraki R., Ito M., Furusawa Y., Yamayoshi S., Iwatsuki-Horimoto K., Adachi E., et al. Humoral immune evasion of the omicron subvariants BQ.1.1 and XBB. Lancet Infect. Dis. 2023;23:30–32. doi: 10.1016/S1473-3099(22)00816-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Willett B.J., Grove J., MacLean O.A., Wilkie C., De Lorenzo G., Furnon W., et al. SARS-CoV-2 Omicron is an immune escape variant with an altered cell entry pathway. Nat. Microbiol. 2022;7:1161–1179. doi: 10.1038/s41564-022-01143-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noske G.D., de Souza Silva E., de Godoy M.O., Dolci I., Fernandes R.S., Guido R.V.C., et al. Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS-CoV-2 Main Protease. J. Biol. Chem. 2023;299:103004. doi: 10.1016/j.jbc.2023.103004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eltayb W.A., Abdalla M., Rabie A.M. Novel investigational anti-SARS-CoV-2 agent ensitrelvir "S-217622": a very promising potential universal broad-spectrum antiviral at the therapeutic frontline of coronavirus species. ACS Omega. 2023;8:5234–5246. doi: 10.1021/acsomega.2c03881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou Y., Gammeltoft K.A., Ryberg L.A., Pham L.V., Tjornelund H.D., Binderup A., et al. Nirmatrelvir-resistant SARS-CoV-2 variants with high fitness in an infectious cell culture system. Sci. Adv. 2022;8 doi: 10.1126/sciadv.add7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Groneberg D.A., Hilgenfeld R., Zabel P. Molecular mechanisms of severe acute respiratory syndrome (SARS) Respir. Res. 2005;6:8. doi: 10.1186/1465-9921-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mariano G., Farthing R.J., Lale-Farjat S.L.M., Bergeron J.R.C. Structural characterization of SARS-CoV-2: where we are, and where we need to Be. Front. Mol. Biosci. 2020;7 doi: 10.3389/fmolb.2020.605236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang C., Liu Z., Chen Z., Huang X., Xu M., He T., et al. The establishment of reference sequence for SARS-CoV-2 and variation analysis. J. Med. Virol. 2020;92:667–674. doi: 10.1002/jmv.25762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoshimoto F.K. The proteins of severe acute respiratory syndrome coronavirus-2 (SARS CoV-2 or n-COV19), the cause of COVID-19. Protein J. 2020;39:198–216. doi: 10.1007/s10930-020-09901-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen S., Jonas F., Shen C., Hilgenfeld R. Liberation of SARS-CoV main protease from the viral polyprotein: N-Terminal autocleavage does not depend on the mature dimerization mode. Protein Cell. 2010;1:59–74. doi: 10.1007/s13238-010-0011-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muramatsu T., Kim Y.T., Nishii W., Terada T., Shirouzu M., Yokoyama S. Autoprocessing mechanism of severe acute respiratory syndrome coronavirus 3C-like protease (SARS-CoV 3CLpro) from its polyproteins. FEBS J. 2013;280:2002–2013. doi: 10.1111/febs.12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nashed N.T., Kneller D., Coates L., Ghirlando R., Aniana A., Kovalevsky A., et al. Autoprocessing and oxyanion loop reorganization upon GC373 and nirmatrelvir binding of monomeric SARS-CoV-2 main protease catalytic domain. Commun. Biol. 2022;5:976. doi: 10.1038/s42003-022-03910-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quan B.X., Shuai H., Xia A.J., Hou Y., Zeng R., Liu X.L., et al. An orally available M(pro) inhibitor is effective against wild-type SARS-CoV-2 and variants including Omicron. Nat. Microbiol. 2022;7:716–725. doi: 10.1038/s41564-022-01119-7. [DOI] [PubMed] [Google Scholar]
- 17.Unoh Y., Uehara S., Nakahara K., Nobori H., Yamatsu Y., Yamamoto S., et al. Discovery of S-217622, a noncovalent oral SARS-CoV-2 3CL protease inhibitor clinical candidate for treating COVID-19. J. Med. Chem. 2022;65:6499–6512. doi: 10.1021/acs.jmedchem.2c00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Owen D.R., Allerton C.M.N., Anderson A.S., Aschenbrenner L., Avery M., Berritt S., et al. An oral SARS-CoV-2 M(pro) inhibitor clinical candidate for the treatment of COVID-19. Science. 2021;374:1586–1593. doi: 10.1126/science.abl4784. [DOI] [PubMed] [Google Scholar]
- 19.Kneller D.W., Phillips G., O'Neill H.M., Jedrzejczak R., Stols L., Langan P., et al. Structural plasticity of SARS-CoV-2 3CL M(pro) active site cavity revealed by room temperature X-ray crystallography. Nat. Commun. 2020;11:3202. doi: 10.1038/s41467-020-16954-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kneller D.W., Phillips G., Weiss K.L., Pant S., Zhang Q., O'Neill H.M., et al. Unusual zwitterionic catalytic site of SARS-CoV-2 main protease revealed by neutron crystallography. J. Biol. Chem. 2020;295:17365–17373. doi: 10.1074/jbc.AC120.016154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi J., Han N., Lim L., Lua S., Sivaraman J., Wang L., et al. Dynamically-driven inactivation of the catalytic machinery of the SARS 3C-like protease by the N214A mutation on the extra domain. PLoS Comput. Biol. 2011;7:e1001084. doi: 10.1371/journal.pcbi.1001084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hussey R.J., Coates L., Gill R.S., Erskine P.T., Coker S.F., Mitchell E., et al. A structural study of norovirus 3C protease specificity: binding of a designed active site-directed peptide inhibitor. Biochemistry. 2011;50:240–249. doi: 10.1021/bi1008497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim Y., Lovell S., Tiew K.C., Mandadapu S.R., Alliston K.R., Battaile K.P., et al. Broad-spectrum antivirals against 3C or 3C-like proteases of picornaviruses, noroviruses, and coronaviruses. J. Virol. 2012;86:11754–11762. doi: 10.1128/JVI.01348-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kidera A., Moritsugu K., Ekimoto T., Ikeguchi M. Functional dynamics of SARS-CoV-2 3C-like protease as a member of clan PA. Biophys. Rev. 2022;14:1473–1485. doi: 10.1007/s12551-022-01020-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anand K., Palm G.J., Mesters J.R., Siddell S.G., Ziebuhr J., Hilgenfeld R. Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra alpha-helical domain. EMBO J. 2002;21:3213–3224. doi: 10.1093/emboj/cdf327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goyal B., Goyal D. Targeting the dimerization of the main protease of coronaviruses: a potential broad-spectrum therapeutic strategy. ACS Comb. Sci. 2020;22:297–305. doi: 10.1021/acscombsci.0c00058. [DOI] [PubMed] [Google Scholar]
- 27.Xia B., Kang X. Activation and maturation of SARS-CoV main protease. Protein Cell. 2011;2:282–290. doi: 10.1007/s13238-011-1034-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nashed N.T., Aniana A., Ghirlando R., Chiliveri S.C., Louis J.M. Modulation of the monomer-dimer equilibrium and catalytic activity of SARS-CoV-2 main protease by a transition-state analog inhibitor. Commun. Biol. 2022;5:160. doi: 10.1038/s42003-022-03084-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kovalevsky A., Coates L., Kneller D.W., Ghirlando R., Aniana A., Nashed N.T., et al. Unmasking the conformational stability and inhibitor binding to SARS-CoV-2 main protease active site mutants and miniprecursor. J. Mol. Biol. 2022;434:167876. doi: 10.1016/j.jmb.2022.167876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.MacDonald E.A., Frey G., Namchuk M.N., Harrison S.C., Hinshaw S.M., Windsor I.W. Recognition of divergent viral substrates by the SARS-CoV-2 main protease. ACS Infect. Dis. 2021;7:2591–2595. doi: 10.1021/acsinfecdis.1c00237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghosh A.K., Samanta I., Mondal A., Liu W.R. Covalent inhibition in drug discovery. ChemMedChem. 2019;14:889–906. doi: 10.1002/cmdc.201900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noske G.D., Nakamura A.M., Gawriljuk V.O., Fernandes R.S., Lima G.M.A., Rosa H.V.D., et al. A crystallographic snapshot of SARS-CoV-2 main protease maturation process. J. Mol. Biol. 2021;433 doi: 10.1016/j.jmb.2021.167118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muramatsu T., Takemoto C., Kim Y.T., Wang H., Nishii W., Terada T., et al. SARS-CoV 3CL protease cleaves its C-terminal autoprocessing site by novel subsite cooperativity. Proc. Natl. Acad. Sci. U. S. A. 2016;113:12997–13002. doi: 10.1073/pnas.1601327113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee J., Worrall L.J., Vuckovic M., Rosell F.I., Gentile F., Ton A.T., et al. Crystallographic structure of wild-type SARS-CoV-2 main protease acyl-enzyme intermediate with physiological C-terminal autoprocessing site. Nat. Commun. 2020;11:5877. doi: 10.1038/s41467-020-19662-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sayer J.M., Liu F., Ishima R., Weber I.T., Louis J.M. Effect of the active site D25N mutation on the structure, stability, and ligand binding of the mature HIV-1 protease. J. Biol. Chem. 2008;283:13459–13470. doi: 10.1074/jbc.M708506200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Louis J.M., Aniana A., Weber I.T., Sayer J.M. Inhibition of autoprocessing of natural variants and multidrug resistant mutant precursors of HIV-1 protease by clinical inhibitors. Proc. Natl. Acad. Sci. U. S. A. 2011;108:9072–9077. doi: 10.1073/pnas.1102278108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin P.Y., Chou C.Y., Chang H.C., Hsu W.C., Chang G.G. Correlation between dissociation and catalysis of SARS-CoV main protease. Arch. Biochem. Biophys. 2008;472:34–42. doi: 10.1016/j.abb.2008.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kneller D.W., Li H., Phillips G., Weiss K.L., Zhang Q., Arnould M.A., et al. Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease. Nat. Commun. 2022;13:2268. doi: 10.1038/s41467-022-29915-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma C., Sacco M.D., Hurst B., Townsend J.A., Hu Y., Szeto T., et al. Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res. 2020;30:678–692. doi: 10.1038/s41422-020-0356-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fu L., Ye F., Feng Y., Yu F., Wang Q., Wu Y., et al. Both Boceprevir and GC376 efficaciously inhibit SARS-CoV-2 by targeting its main protease. Nat. Commun. 2020;11:4417. doi: 10.1038/s41467-020-18233-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hattori S.I., Higashi-Kuwata N., Hayashi H., Allu S.R., Raghavaiah J., Bulut H., et al. A small molecule compound with an indole moiety inhibits the main protease of SARS-CoV-2 and blocks virus replication. Nat. Commun. 2021;12:668. doi: 10.1038/s41467-021-20900-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walsh C. Enzymatic Reaction Mechanisms. W H Freeman & Co; New York, NY: 1979. [Google Scholar]
- 43.Pearson R.G., Sobel H.R., Songstad J. Nucleophilic reactivity constants toward methyl iodide and trans-dichlorodi(pyridine)platinum(II) J. Am. Chem. Soc. 1968;90:319–326. [Google Scholar]
- 44.Lowry T.H., Richardson K.S. Harper & Row; New York, NY: 1976. Mechanism and Theory in Organic Chemistry. [Google Scholar]
- 45.Kneller D.W., Zhang Q., Coates L., Louis J.M., Kovalevsky A. Michaelis-like complex of SARS-CoV-2 main protease visualized by room-temperature X-ray crystallography. IUCrJ. 2021;8:973–979. doi: 10.1107/S2052252521010113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Evans P.R., Murshudov G.N. How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liebschner D., Afonine P.V., Baker M.L., Bunkoczi G., Chen V.B., Croll T.I., et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in phenix. Acta Crystallogr. D Struct. Biol. 2019;75:861–877. doi: 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Casanal A., Lohkamp B., Emsley P. Current developments in coot for macromolecular model building of electron cryo-microscopy and crystallographic data. Protein Sci. 2020;29:1069–1078. doi: 10.1002/pro.3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen V.B., Arendall W.B., 3rd, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moriarty N.W., Grosse-Kunstleve R.W., Adams P.D. Electronic Ligand Builder and Optimization Workbench (eLBOW): a tool for ligand coordinate and restraint generation. Acta Crystallogr. D Biol. Crystallogr. 2009;65:1074–1080. doi: 10.1107/S0907444909029436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Frisch M.J., Trucks G.W., Schlegel H.B., Scuseria G.E., Robb M.A., Cheeseman J.R., et al. Gaussian Inc.; Wallingford, CT: 2016. Gaussian 16, Revision B.01. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data that support the findings of this study are contained within this article and its accompanying files. PDB accession numbers are listed in Supporting Information, Table S1.