Abstract
PseudoBase is a database containing structural, functional and sequence data related to RNA pseudoknots. It can be reached at http://wwwbio.Leiden Univ.nl/~Batenburg/PKB.html . This page will direct the user to a retrieval page from where a particular pseudoknot can be chosen, or to a submission page which enables the user to add pseudoknot information to the database or to an informative page that elaborates on the various aspects of the database. For each pseudoknot, 12 items are stored, e.g. the nucleotides of the region that contains the pseudoknot, the stem positions of the pseudoknot, the EMBL accession number of the sequence that contains this pseudoknot and the support that can be given regarding the reliability of the pseudoknot. Access is via a small number of steps, using 16 different categories. The development process was done by applying the evolutionary methodology for software development rather than by applying the methodology of the classical waterfall model or the more modern spiral model.
INTRODUCTION
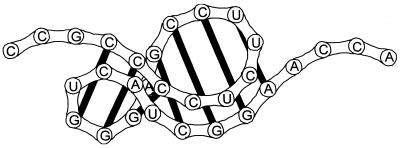
Pseudoknots are widely occurring structural motifs in RNA. First described in the early 1980s as part of tRNA-like structures in plant viral RNAs, pseudoknots were recognized as a general principle of RNA folding (1,2). The pseudoknotting in RNA involves base pairing between a loop, formed by an orthodox secondary structure and some region outside this loop. The simplest pseudoknot, the classical or so-called H-(hairpin) pseudoknot, is formed by the pairing of a region in the hairpin loop with the nucleotides downstream or upstream of the hairpin. The H-pseudoknot contains two stems and (at least) two loops (Fig. 1). If the two stems form a quasi-continuous helix, stacking of the nucleotides at the junction is possible, otherwise an additional loop might be formed. The loops may consist of hundreds of nucleotides, thereby possessing their own structure. Pseudoknots may be formed by interactions between different types of loops (e.g. bulges) and therefore may differ from the classical H-pseudoknots.
Figure 1.
Pseudoknot.
Since the first discoveries of pseudoknots, they have been found in various kinds of RNAs (reviewed in 3–5). Furthermore, their important role in a number of RNA functions has been shown, e.g. in ribosomal frameshifting, regulation of translation and splicing. Pseudoknots are also essential elements of the topology of many structural RNAs, e.g. ribosomal RNAs or ribozymes.
WHAT DATA ARE STORED?
For each pseudoknot we record 12 data items. Figure 2 shows a page for a particular pseudoknot which enumerates these items and illustrates the layout. The recorded items are:
Figure 2.

Characteristic example of a pseudoknot data page.
• PKB number: each pseudoknot submission is given a serial PKB (PseudoKnotBase) number.
• Definition: a description of the pseudoknot.
• Organism or virus.
• Abbreviation: this abbreviation appears in the list of pseudoknots on the ‘Retrieve’ page. Each abbreviation should be unique. Therefore, the abbreviation is derived from the name of the organism (e.g. virus); if necessary, additional characters or numbers are added to distinguish between different pseudoknots within one organism.
• RNA type: the category used in the Retrieve page for classification, e.g. viral tRNA-like structures, viral ribosomal frameshifting signals, mRNA, etc. (see below).
• Keywords: keywords (for example: retroviridae, gag-pro, ribosomal frameshift, aminoacylation, satellite virus, RNA 3′ end) that characterize the context or origin of the pseudoknot. Currently there is no provision to search for these keywords, but this may well be an option in the future.
• EMBL accession number of the RNA sequence. If the user clicks on this number, the search engine www.ebi.ac.uk/htbin/emblfetch? is activated. This brings the user straight to the requested sequence in the EMBL database (6).
• Submitted by: name of the submitter. If the submitter does not disagree, we add his/her email address, so clicking on it activates the ‘mailto’ option with this address. As the mailto option is not a very reliable function (discussed later), this address is also visible in order to enable the reader to copy it and paste it in his mailprogram.
• Supported by: a type of scientific evidence for this pseudoknot. We decided that a database manager cannot and should not decide about the reliability of evidence for a particular pseudoknot. Rather it should be the reader who decides. For this we added an item in the database which states on what evidence the submitted pseudoknot is based. Currently this item contains the alternatives 1. crystal structure, 2. mutagenesis, 3. NMR, 4. sequence comparison, 5. structure probing and 6. others.
• References: literature references pertinent to the pseudoknot in question.
• Position paired: the first line specifies the four 5′- and 3′-end positions of the first stem of the pseudoknot, the second line those of the second stem. For example 6293–6295;6305–6307 (first stem) and 6300–6304;6311–6315 (second stem). The numbering corresponds to the EMBL sequence database entry as mentioned above, unless otherwise indicated. If stems contain internal loops, more than two lines are needed. For example 6300–6302;6314–6316 and 6303–6304;6311–6312 specifies the second stem with a bulge at position 6313. Pseudoknots that do not belong to the so-called H-type, for example the one in hepatitis delta ribozyme, may also require more than two lines.
• Bracket view of structure. For a simple, but effective visual representation we chose the bracket view, but amended it in a way that makes it suitable for pseudoknots. One stem is indicated by parentheses and the other one with brackets. For example:
6290 6300 6310
6789│123456789│123456789│123456789
ACUCCCGCCCCUCUUCCGAGGGUCAUCGGAACCA
– – – – – – – ( ( ( – – – – [ [ [ [ [ ) ) ) – – –] ] ] ] ] – – – –
This shows the position numbers in the first two lines, the sequence part in the third line and the structure in the last line.
HOW TO RETRIEVE PSEUDOKNOT INFORMATION
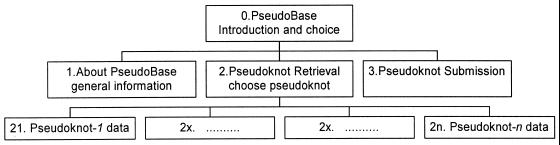
Retrieval of data, whether more general information or information about a particular pseudoknot, is a stepwise procedure through a hierarchical tree. We have tried to keep the design and the process of navigation through the site simple, so the number of hierarchical levels is as small as possible. The design hierarchy of PseudoBase is presented in Figure 3. Top of the hierarchy and also the main entry for users is the Introduction page.
Figure 3.
Design of PseudoBase. Top page is http//wwwbio.LeidenUniv.nl/~Batenburg/PKB.html
The Introduction page
The database main access is through the Introduction page. At the top of this page, as well as on all the other pages, the general structure is presented: [Intro [About...][Retrieve...][Submit...]] (Fig. 2). This is a shortcut for experienced users to jump to other pages. For new users the page itself gives a more leisurely access to the other pages.
Figure 3 shows that the Introduction page gives access to three other pages. The first one ‘About’ deals with additional information, the second one ‘Retrieve’ is the main entry to the retrieval of pseudoknot data, and the third one ‘Submit’ is designed for submitting pseudoknot data.
The About page
The About page presents the reader with auxiliary information about the PseudoBase project. Topics are: (i) purpose of PseudoBase; (ii) quality considerations; (iii) information on how to contact the authors; (iv) historic information on how this project came into existence; (v) explanations about some items in the database and a short list of literature to get acquainted with pseudoknots.
The Retrieve page
The Retrieve page presents all pseudoknots that are available in the database, one line per item. Here the user chooses the pseudoknot that is needed. Clicking on the particular line brings the page with the data about that particular pseudoknot. Currently we have organized the pseudoknots into 16 categories, based on their occurrence and function. These categories are:
1. Viral ribosomal frameshifting signals
2. Viral ribosomal readthrough signals
3. Viral tRNA-like structures
4. Other viral 5′-UTR
5. Other viral 3′-UTR
6. Viral others
7. rRNA
8. mRNA
9. tmRNA
10. snRNA
11. snoRNA
12. hnRNA
13. Ribozymes
14. Aptamers
15. Artificial molecules
16. Others
At the time of writing the database contained over 100 pseudoknots. As we intend to invest a substantial effort in filling the database in the months ahead, we expect that the database will have grown significantly by the time of publication of this article.
HOW TO CONTRIBUTE?
Our aim is to start a database that will be used and built by scientists. In particular, we hope that scientists will contribute by submitting pseudoknot data. We intend to start off with a substantial amount of pseudoknots to give the database some momentum. Afterwards we hope that this will stimulate scientists to add new data.
To enable scientists to submit new pseudoknot data we designed a form for entering pseudoknot data. The Introduction page gives access to this form on the Submission page. You can see a screen dump of this form in Figure 4.
Figure 4.
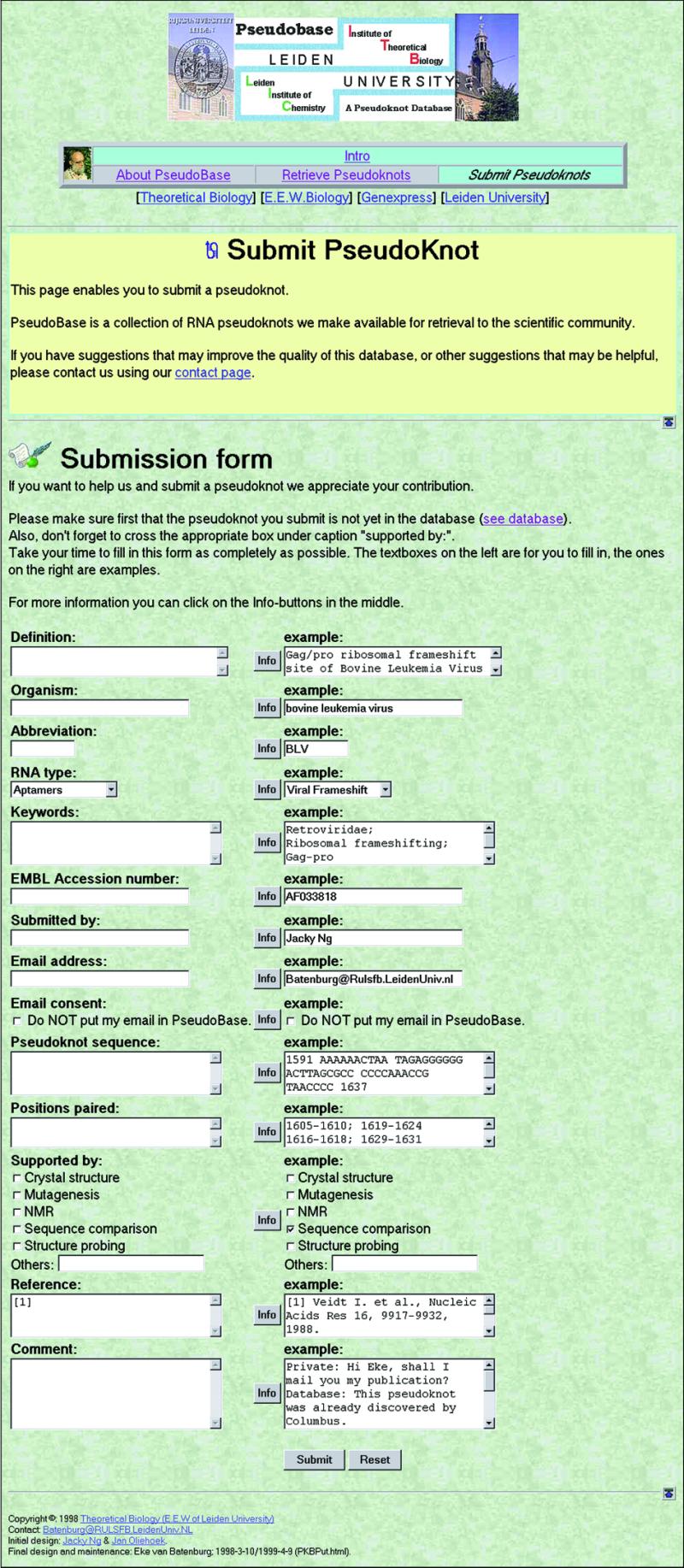
Screendump of submission form.
The figure shows that we designed the form in three vertical columns. The left column contains the fields where the user should enter his/her data. In the right column we present the same fields already filled with data so as to provide an example, which might ease the submission process. The middle column contains an [Info]-button for each field. Clicking on such a button yields information about the data to be entered in this particular field. Currently these [Info] buttons have simple explanations only; however as our experience with potential misunderstandings increases we intend to extend the information that those buttons provide.
Different pseudoknots in one RNA should be submitted separately; they are also stored on separate pages. Up to now we give such pseudoknots the same abbreviation, followed by the code PK#, where # is a serial number. Although currently not implemented, we are considering adding pointers from such pseudoknots to each other.
For extremely long loops, we have added the feature to omit part of the sequence by specifying relevant parts only. For example, instead of:
1806 AGGCGGGGCGAGCUGCAGCCCCAGUGAAUCAAAUGCA-GCAGGCGGGGCGAGCUGCAGCCCCAGUGAAUC 1874
one can enter:
1806 AGGCGGGGCGAGCUGCAG 1823 1852 GCGAGC-UGCAGCCCCAGUGAAUC 1874.
We have decided that the database should be critical about the usefulness of its information. Therefore in some cases that have well-known, strongly conserved pseudoknots, we will restrict ourselves to only a few representative sequences with references to the appropriate sequence-structure databases. For example, we have one representative for the pseudoknots in Group-I introns and one representative for ribosomal RNAs.
We reserve the right to make corrections in the submitted data to avoid inconsistencies or refuse entries that are superfluous, although we intend to enforce this right sparingly and as a last resort only.
DESIGN PROBLEM
For adding pseudoknot information to the database we designed a form that would facilitate entering and submitting data by other users. Based on our aim to keep it simple, we started to use the html feature METHOD=post and ACTION=mailto. Unfortunately, in try-out sessions we discovered that on many computers the browser unjustly reported a successful data transmission to the receiver; these data never reached the intended addressee. After analysis we found that success depended upon the correct mail-settings of the sending browser. As many of the try-out senders used a separate mailing program for mail and did not regard mail-settings in the browser relevant for them, they did not set the mail-options in the browser correctly, so the mailto option did not work.
We expected that this would be a general problem and decided not to use the mailto feature of html. Instead we implemented the public domain cgi-script CGImail v.1.12 programmed by E. Voisard, downloaded from http://www.bayside.net/sendmail/ . Once given an html-mold, this cgi-script would insert the data supplied in the form at the proper positions in that mold (in the way one inserts names in a mail-merge program). The end result was that a submission was transformed by this script into a nearly finished item.
The final sculpturing of the submitted pseudoknot data, including the development of the bracket view from the entered sequence data, is done by a specially developed program in the APL language.
FUTURE PLANS
The first priority we have is to add a substantial amount of pseudoknots. We expect to increase the total number of items considerably in the coming months.
Another plan we have is to use the database to improve our program STAR that is used for RNA structure prediction (7–9). This program already predicts simple H-pseudoknots. Recently we used STAR to investigate possible improvements for pseudoknot energy rules (10); this required extensive testing of predicted pseudoknots against reliable known pseudoknots for which we used our pseudoknot data. We intend to improve our program even more using the data in PseudoBase.
Several ideas are still under consideration. For example, adding links to sites that show the 3D representation of a pseudoknot (e.g. http://www.ebi.ac.uk/NDB/ ). Another is to add links from pseudoknot pages to other pseudoknots in the same sequence. Interesting suggestions from one referee were to add links from author to their publication in MEDLINE and to add an option to download all data. As mentioned above, we also consider the usefulness of adding a search facility to look for pseudoknot items based on keywords.
Above all, however, we want to see how the current database works and look for improvement opportunities. For this we hope to get feedback from users. If possible, we hope to improve in an evolutionary way—that is by simple small steps—in the near future; although if this is not possible we are open to revolutionary changes.
ACCESS
PseudoBase can be found at http://wwwbio.LeidenUniv.nl/~Batenburg/PKB.html . This presents the Introduction page which leads to either the About, the Retrieve or the Submit pages.
Authors would appreciate reference to this publication when readers publish work that has used data from PseudoBase.
We also invite scientists to submit pseudoknot data and to make comments on the design of the database.
REFERENCES
- 1.Rietveld K., van Poelgeest,R., Pleij,C.W.A., van Boom,J.H. and Bosch,L. (1982) Nucleic Acids Res., 10, 1929–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pleij C.W.A., Rietveld,K. and Bosch,L. (1985) Nucleic Acids Res., 13, 1717–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deiman B.A.L.M. and Pleij,C.W.A. (1997) Semin. Virol .,8, 166–175. [Google Scholar]
- 4.Pleij C.W.A. (1994) Curr. Opin. Struct. Biol., 4, 337–344. [Google Scholar]
- 5.Hilbers C.W., Michiels,P.J.A. and Heus,H.A. (1999) Biopolymers, 48, 137–153. [Google Scholar]
- 6.Stoesser G., Mosely,M.A., Sleep,J., McGowran,M., Garcia-Pastor,M. and Sterk,P. (1998) Nucleic Acids Res., 26, 8–15. Updated article in this issue, Nucleic Acids Res. (2000), 28, 19–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abrahams J.P., van den Berg,M., van Batenburg,F.H.D. and Pleij,C.W.A. (1990) Nucleic Acids Res., 18, 3035–3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Batenburg F.H.D., Gultyaev,A.P. and Pleij,C.W.A. (1995) J. Theor. Biol., 174, 269–280. [DOI] [PubMed] [Google Scholar]
- 9.Gultyaev A.P., van Batenburg,F.H.D. and Pleij,C.W.A. (1995) J. Mol. Biol., 250, 37–51. [DOI] [PubMed] [Google Scholar]
- 10.Gultyaev A.P., van Batenburg,F.H.D. and Pleij,C.W.A. (1999) RNA, 5, 609–617. [DOI] [PMC free article] [PubMed] [Google Scholar]