Abstract
The SYSTERS (short for SYSTEmatic Re-Searching) protein sequence cluster set consists of the classification of all sequences from SWISS-PROT and PIR into disjoint protein family clusters and hierarchically into superfamily and subfamily clusters. The cluster set can be searched with a sequence using the SSMAL search tool or a traditional database search tool like BLAST or FASTA. Additionally a multiple alignment is generated for each cluster and annotated with domain information from the Pfam database of protein domain families. A taxonomic overview of the organisms covered by a cluster is given based on the NCBI taxonomy. The cluster set is available for querying and browsing at http://www.dkfz-heidelberg.de/tbi/services/cluster/systersform
INTRODUCTION
The increasing size of protein sequence databases makes a grouping of the sequences into families useful for studying and understanding their functionality. For example, searching a sequence database with a query sequence looking for homologues has become a routine operation in molecular biology. The usual strategy is to perform a database search with BLAST (1,2) or FASTA (3), the result of which is a list of sequences from the database, ordered according to the similarity to the query sequence. While hits of high significance usually correspond to correctly determined, related proteins, it is well known that many related sequences are statistically indistinguishable from unrelated sequences.
An alternative approach is to preprocess the protein database into clusters of homologous sequences, and to use the information derived from all the sequences for further analysis.
Such a clustering can help in compressing the output produced by database search programs, can aid in the automatic derivation of multiple alignments and profiles, and can provide data for evolutionary analysis.
CLASSIFICATION
The classification of the sequences of a protein sequence database into the SYSTERS (4,5) cluster set is mainly based on a traditional database search tool and done in two steps, a similarity searching step and a clustering step. First, each sequence in the database is searched against the whole sequence database using gapped BLAST down to a weak E-value of 0.05. Since database searches behave asymmetrically (the score and E-value of sequence A finding sequence B in the database and those of sequence B finding sequence A can differ significantly), the results of the database searches are not directly used for the clustering step, but taken as a hint for potential similarity of the sequences. For each entry in the resulting list of potential database hits a pairwise local alignment of the current query sequence and the database hit is re-computed using LALIGN (6). On the basis of the resulting symmetric score an E-value for each pair of sequences is re-calculated, employing the corresponding routines implemented in BLAST (7).
Based on these E-values a single-linkage clustering (8) of the whole data set is done at a conservative cutoff E-value of, e.g., 10–40. Depending on the connectivity within the resulting clusters, the clusters themselves are classified as perfect (each sequence identifies every other sequence of the cluster in a database search at this cutoff), nested (at least one sequence detects all others) or overlapping (no sequence identifies all other sequences in the cluster when used as a query).
Since a static cutoff E-value allows only a restricted view of one layer of the protein space, we constructed for the new SYSTERS release a hierarchical view to generate a further classification of the clusters into superfamilies and of the sequences within a cluster into subclusters.
In the resulting SYSTERS tree, theoretically the leaves correspond to single-sequence clusters, while the cluster located at the root of the tree includes the whole sequence space. In reality the whole data set splits into several trees, while the leaves of these trees sometimes contain a small number of nearly identical sequences.
Stepping down in the hierarchy from a static cluster usually splits off one sequence after another, but does not lead to a meaningful partitioning into subfamilies. For this purpose we used a variation of a method presented by Hartuv et al. (9). The graph built by the sequences in a cluster and the pairwise E-values connecting these sequences is iteratively split into subclusters at a minimal cut (10,11) site until a disjoint set of highly connected (non-single-sequence) subclusters is reached.
The determination of superfamilies is based solely on the branching structure of the SYSTERS tree. Close neighboring clusters in the tree are collected together to build a superfamily. We have observed that in most cases our automatic superfamily annotation corresponds surprisingly well to what one would define as a superfamily by hand.
Since the clustering step and the construction of the SYSTERS tree are based on symmetric database search results, the insertion of a new or updated sequence can be done in a consistent way using the same methods described above.
UNDERLYING DATA
The data set underlying the current SYSTERS release contains all sequences from the SWISS-PROT (12) and PIR (13) databases satisfying a minimal sequence length of 10 amino acids for a complete sequence, resp. 50 amino acids for a sequence annotated as fragmental.
Typically, single domain proteins form perfect clusters, while multi-domain proteins containing, e.g., an ATP binding site, can be found in overlapping clusters.
Sequences annotated as fragmental mostly end up in single-sequence and overlapping clusters and can be successfully included to the cluster set when completed. As a feature of the new SYSTERS release every single-sequence cluster is, if possible, linked to at least one other cluster based on percent sequence identity and overlap length, but independently of the E-value, to give a hint on potential relationships to other clusters.
WEB SERVER
The cluster set is available for querying and browsing at http://www.dkfz-heidelberg.de/tbi/services/cluster/systersform
Clusters can be selected by cluster number, cluster size, organism, accession number [SWISS-PROT, PIR, PDB (14), ENZYME (15), PROSITE (16) or EMBL (17)], identifier (SWISS-PROT or PIR), or by searching the sequence annotations for keywords.
The sequences in every cluster have been multiply aligned using ClustalW 1.7 (18) and the alignments are collected together. A new sequence can be searched against this data using the similarity searching tool SSMAL (Shuffling Similarities with Multiple Alignments) (19) which uses features of the BLAST algorithm for scanning a database of multiple alignments.
All multiple alignments are annotated with known domains from the Pfam (20) protein family database of alignments and HMMs. Each domain annotation in a multiple alignment is linked to a list of clusters containing this domain. Vice versa, clusters can be selected directly from the list of all Pfam domains.
Additionally the clusters are linked to an unrooted tree computed with neighbor-joining (21), a set-membership matrix containing information about the density of the cluster, and the sequences in Fasta format.
New features of the SYSTERS web server include the taxonomic overview of the organisms covered by a cluster based on the NCBI taxonomy (22) and the annotation of a cluster with a function suggested by EUCLID (23), if possible.
Additionally, for each cluster and subcluster an MView (24) output is now generated and of the resulting partial multiple alignment a majority consensus sequence is calculated. All consensus sequences together build a searchable sequence database.
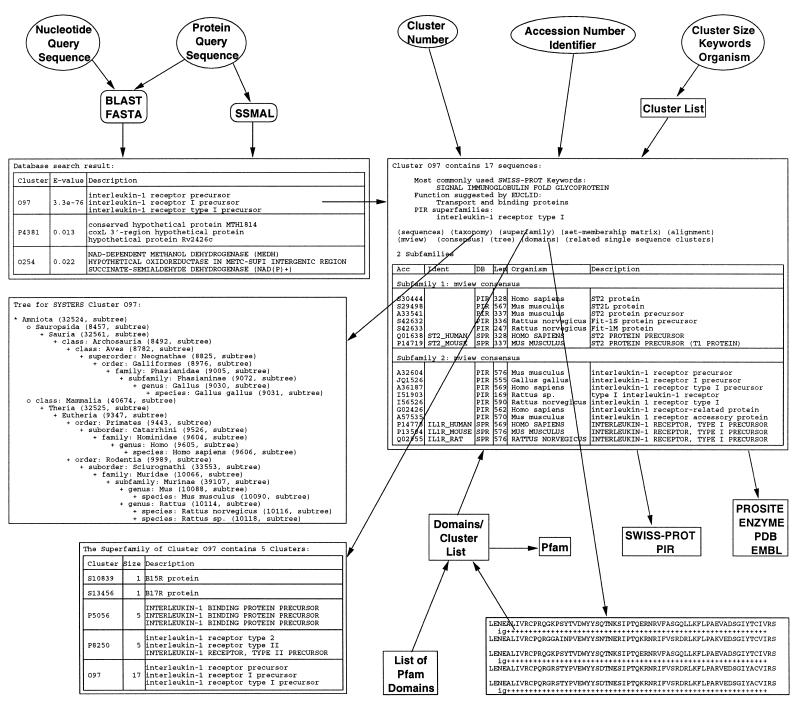
Figure 1 gives an overview of the SYSTERS Web server and its functionality shown by an example.
Figure 1.
Overview of the SYSTERS Web server. As an example, the cluster set was searched with a query sequence (top left). A more detailed insight into the overlapping cluster O97 containing 17 sequences sorted into two subfamilies is given (middle right). Additionally, the taxonomic overview of the organisms covered by the cluster (middle left), the list of clusters contained in the corresponding superfamily (bottom left), and the domain composition of the sequences in the cluster (bottom right) is shown.
CONCLUSIONS
The SYSTERS protein sequence cluster set provides an automatically generated classification of all sequences of the SWISS-PROT and PIR databases into families, superfamilies and subfamilies annotated with sequence information from various other resources. An emphasis of our method lies on the reliability of the resulting clusters. To this end we have introduced the distinction between perfect, nested and overlapping clusters. Generally the clusters in the database, in particular the perfect clusters, stay on the conservative side, i.e., we hardly ever observe that sequences that do not belong to a cluster would be included. On the other hand, this has as a side effect a fairly large number of singletons. We have refrained from assigning singletons to, e.g., the cluster they are most similar to although in a manually driven approach this would be feasible.
The other consequence of the conservative nature of this clustering is that a manually curated database like Pfam tends to link several of the SYSTERS clusters. To account for this additional information we have introduced the links between our more family oriented clusters and the Pfam domains. This allows for very interesting analysis about the distribution of domains over families that we are currently working out. The other direction of current activity aims at a visualisation of the hierarchical structure of the clustering. This will in the future allow to navigate through a large tree of protein sequences where the current version of SYSTERS is just one level.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Heiko Schmidt for providing the interface to the taxonomic classification. We acknowledge financial support from BMBF (Bundesministerium für Bildung, Wissenschaft, Forschung und Technologie).
REFERENCES
- 1.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S.F., Madden,T.L., Schäffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pearson W.R. and Lipman,D.J. (1988) Proc. Natl Acad. Sci. USA, 85, 2444–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krause A. and Vingron,M. (1998) Bioinformatics, 14, 430–438. [DOI] [PubMed] [Google Scholar]
- 5.Krause A., Nicodème,P., Bornberg-Bauer,E., Rehmsmeier,M. and Vingron,M. (1999) Bioinformatics, 15, 262–263. [DOI] [PubMed] [Google Scholar]
- 6.Huang X. and Miller,W. (1991) Adv. Appl. Math., 12, 337–357. [Google Scholar]
- 7.Altschul S.F. and Gish,W. (1996) Methods Enzymol., 266, 460–480. [DOI] [PubMed] [Google Scholar]
- 8.Sokal R.R. and Sneath,P.H.A. (1963) Principles of Numerical Taxonomy. Freeman, London.
- 9.Hartuv E., Schmitt,A., Lange,J., Meier-Ewert,S., Lehrach,H. and Shamir,R. (1999) Proceedings of the Third Annual International Conference on Computational Molecular Biology (RECOMB), 188–194.
- 10.Stoer M. and Wagner,F. (1994) Algorithms–ESA ‘94, LNCS 855, 141–147.
- 11.Mehlhorn K. and Näher,S. (1995) Comm. ACM, 38, 96–102. [Google Scholar]
- 12.Bairoch A. and Apweiler,R. (1999) Nucleic Acids Res., 27, 49–54. Updated article in this issue: Nucleic Acids Res. (2000) 28, 45–48.9847139 [Google Scholar]
- 13.Barker W.C., Garavelli,J.S., McGarvey,P.B., Marzec,C.R., Orcutt,B.C., Srinivasarao,G.Y., Yeh,L.L., Ledley,R.S., Mewes,H.-W., Pfeiffer,F., Tsugita,A. and Wu,C. (1999) Nucleic Acids Res., 27, 39–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernstein F.C., Koetzle,T.F., Williams,G.J., Meyer,E.E.,Jr, Brice,M.D., Rodgers,J.R., Kennard,O., Shimanouchi,T. and Tasumi,M. (1977) J. Mol. Biol., 112, 535–542. [DOI] [PubMed] [Google Scholar]
- 15.Bairoch A. (1999) Nucleic Acids Res., 27, 310–311. Updated article in this issue: Nucleic Acids Res. (2000) 28, 304–305.9847212 [Google Scholar]
- 16.Hofmann K., Bucher,Ph., Falquet,L. and Bairoch,A. (1999) Nucleic Acids Res., 27, 215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stoesser G., Tuli,M.A., Lopez,R. and Sterk,P. (1999) Nucleic Acids Res., 27, 18–24. Updated article in this issue: Nucleic Acids Res. (2000) 28, 19–23.9847133 [Google Scholar]
- 18.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nicodème P. (1998) Bioinformatics, 14, 508–515. [DOI] [PubMed] [Google Scholar]
- 20.Bateman A., Birney,E., Durbin,R., Eddy,S.R., Finn,R.D. and Sonnhammer,E.L.L. (1999) Nucleic Acids Res., 27, 260–262. Updated article in this issue: Nucleic Acids Res. (2000) 28, 263–266.9847196 [Google Scholar]
- 21.Saitou N. and Nei,M. (1987) Mol. Biol. Evol., 4, 406–425. [DOI] [PubMed] [Google Scholar]
- 22.Benson D.A., Boguski,M.S., Lipman,D.J., Ostell,J., Ouellette,B.F.F., Rapp,B.A. and Wheeler,D.L. (1999) Nucleic Acids Res., 27, 12–17. Updated article in this issue: Nucleic Acids Res. (2000) 28, 15–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tamames J., Ouzounis,Ch., Casari,G., Sander,C. and Valencia,A. (1998) Bioinformatics, 14, 542–543. [DOI] [PubMed] [Google Scholar]
- 24.Brown N.P., Leroy,C. and Sander,C. (1998) Bioinformatics, 14, 380–381. [DOI] [PubMed] [Google Scholar]