Abstract
Extrachromosomal circular DNA (eccDNA) is a class of circular DNA molecules that originate from genomic DNA but are separate from chromosomes. They are common in various organisms, with sizes ranging from a few hundred to millions of base pairs. A special type of large extrachromosomal DNA (ecDNA) is prevalent in cancer cells. Research on ecDNA has significantly contributed to our comprehension of cancer development, progression, evolution, and drug resistance. The use of next-generation (NGS) and third-generation sequencing (TGS) techniques to identify eccDNAs throughout the genome has become a trend in current research. Here, we briefly review current advances in the biological mechanisms and applications of two distinct types of eccDNAs: microDNA and ecDNA. In addition to presenting available identification tools based on sequencing data, we summarize the most recent efforts to integrate ecDNA with single-cell analysis and put forth suggestions to promote the process.
Keywords: EcDNA, MicroDNA, Single-cell sequencing, NGS, TGS
1. Introduction
Since its discovery in wheat embryos and boar sperm in 1964, extrachromosomal circular DNA (eccDNA) has been detected in numerous cell lines and tissues across various species [1], [2]. EccDNAs are derived from genomic DNA and range in size from a few hundred bases to megabases [3]. Recent studies have classified eccDNAs into four main classes based on their sizes and sequence features: small polydispersed DNA (spcDNA), telomeric circles (t-circles), microDNA, and extrachromosomal DNA (ecDNA) [2], [4]. Recently, microDNA and ecDNA have garnered increasing attention. Due to its stability in plasma, microDNA has the potential to become a new diagnostic and prognostic biomarker for various diseases [5].
The earliest description of ecDNA dates back to 1965, when Cox et al. observed small double chromatin bodies without visible centromeres, which they named 'double minutes' (DMs) due to their presence in pairs during metaphase [6]. Currently, the term 'DMs' is being replaced by the more inclusive term 'ecDNA', encompassing both singlet ecDNA particles and double-minute pairs, with research indicating that only approximately 30% of ecDNAs occur as paired bodies [7]. EcDNA has been found in almost half of all known human cancer types and is estimated to occur in at least a quarter of all cancer patients [7], [8]. ecDNA is considered to show a size larger than 100 kb or 1 Mb, and a pan-cancer analysis with 3212 tumor samples revealed that the median size of 516 candidate ecDNAs was approximately 3.7 Mb [2], [4], [8], [9], [10], [11]. EcDNA leads to oncogene amplification and drug resistance via a different mechanism than linear chromosomes, so interventions aimed at ecDNA may improve treatment outcomes [12].
In this review, we provide a concise overview of the mechanisms and applications of microDNA and ecDNA. Subsequently, we collate various sequencing-based approaches utilized for studying these extrachromosomal elements in addition to databases. Finally, we comprehensively summarize the most recent research combining ecDNA with single-cell sequencing and providing some insights into its future development.
2. Brief overview of the progress of microDNA and ecDNA
Here, we focus on microDNA and ecDNA, which are the hotspots of current eccDNA research. On the other hand, there have been insightful reviews delving into many aspects of spcDNA and t-circles [2], [3], [13]. As research progresses, it is becoming increasingly important to redefine molecules whose sizes fall between the typical sizes of microDNA and ecDNA (i.e., between 10 kb and 100 kb), especially when they are present in noncancerous tissues [14], [15]. Since such DNA circles are currently referred to generically as eccDNA or circDNA, a more precise definition is necessary to avoid ambiguity [9], [16].
2.1. MicroDNA
In 2012, Shibata et al. discovered abundant small eccDNAs in mammalian cells referred to as microDNAs [17]. MicroDNAs are small nonrepetitive circular DNAs commonly ranging in size from 200 to 400 bp and microDNAs smaller than 10 kb may constitute over 99 % of the eccDNA population [18]. Hotspots for microDNA generation include genomic regions such as 5′UTRs, exons, and CpG islands [15], which are sites where microdeletions are more likely to occur. Differences and dynamic changes in microDNA have been noted between tumors and matched normal tissues, tissues before and after surgical resection of tumors, and tissues of fetal and maternal origins, highlighting the clinical utility of microDNA as a noninvasive biomarker (reviewed in [3], [5]). Although most microDNAs are too small to carry protein-coding genes, microDNAs can impact gene expression by producing microRNAs or small interfering RNAs (siRNAs). MicroDNAs can also serve as immunostimulants independent of their specific sequence [19], [20].
2.2. ecDNA
EcDNA has emerged as a noteworthy contributor to cancer, as it triggers massive oncogene amplification and fosters drug resistance. Many types of cancers have been reported to harbor amplified genes in the form of ecDNA, and increased ecDNA copy numbers within tumors are associated with a poorer prognosis (reviewed in [21]). Furthermore, ecDNA engenders genomic rearrangements, which may enhance oncogene expression while inhibiting tumor suppressor expression, resulting in a worse clinical outcome [22]. Despite several studies on the formation of ecDNA, the underlying mechanisms are complex and are not fully understood. Current studies suggest that homologous recombination (HR) and nonhomologous end joining (NHEJ) take part in this process, and chromothripsis is widely recognized as a cause (reviewed in [23], [24]). The random distribution of ecDNAs among daughter cells during mitosis due to their lack of centromeres leads to the rapid acquisition of numerous ecDNA copies by certain cells, explaining the fitness gain of cancer cells and the development of heterogeneity [25], [26]. EcDNAs are highly accessible and can interact with active chromatin over ultralong distances [27]. EcDNA modulates oncogenes by hijacking proximal or distal regulatory elements such as enhancers and promoters, indicating a coselection model [28], [29], [30]. It can also function as a mobile enhancer regulating gene expression on both linear chromosomes and other ecDNAs (reviewed in [9], [23]). Additionally, ecDNA hubs that promote oncogene overexpression by enabling intermolecular enhancer-gene interactions and cooperative sharing of DNA regulatory elements have been observed (reviewed in [9], [10], [12]). However, many questions remain unanswered regarding ecDNA hubs, including the conditions necessary for their formation, their frequency, and the relationship between increased transcriptional efficiency facilitated by hubs and increased transcription due to copy number gains. In conclusion, ecDNA has emerged as an attractive target for cancer therapy (reviewed in [12], [21]).
3. Overview of eccDNA databases
The rapid advancement of high-throughput sequencing and bioinformatic analysis methods has revealed the extensive prevalence of eccDNA in humans and other species, underscoring the need to establish comprehensive databases. Here, we collated several online databases of eccDNAs, including CircleBase [31], eccDB [32], eccDNAdb [11], eccDNA Atlas [33], and TeCD [34]. We have documented the annotation and analysis modules offered by each database (as listed in Table 1). In brief, CircleBase is the first database specifically focused on human-derived eccDNA, while eccDB has the largest number of eccDNA entries from multiple species. eccDNAdb only includes amplicons identified by AmpliconArchitect (AA), while only the eccDNA Atlas categorizes eccDNA entries into microDNA, ecDNA, and spcDNA. Finally, TeCD is a database designed for studying microDNAs smaller than 1 kb from eukaryotes.
Table 1.
Summary of available eccDNA databases.
| CircleBase | eccDB | eccDNAdb | eccDNA Atlas | TeCD | |
|---|---|---|---|---|---|
| Human | 601,112 | 767,981 | 1270 | 637,241 | 200,532 |
| Average size (human) (bp) | 36,477 | 492,451 | 25,483,476 | 43,579 | 2154 |
| Median size (human) (bp) | 323 | 340 | 4,203,339 | 329 | 379 |
| eccDNAs harboring oncogenes (human) | 17,742 | 37,300 | 568 | 19,397 | 4338 |
| eccDNAs harboring oncogenes (human) (%) | 2.95 % | 4.86 % | 44.72 % | 3.04 % | 2.16 % |
| Unique oncogenes on eccDNAs (human) | 764 | 766 | 687 | 765 | 582 |
| Multiple species | √ | √ | √ | ||
| Gene expression | √ | √ | √ | ||
| Gene ontology annotation | √ | √ | √ | ||
| Pathway enrichment analysis | √ | √ | |||
| Survival analysis | √ | √ | √ | ||
| Blast | √ | √ | √ | ||
| Accessible chromatin regions | √ | √ | √ | √ | |
| Intrachromosomal interactions | √ | √ | |||
| Interchromosomal interactions | √ | ||||
| Transcription factors (TFs) | √ | √ | |||
| Enhancers | √ | √ | √ | ||
| Super enhancers | √ | √ | √ | ||
| Risk single-nucleotide polymorphisms (SNPs) | √ | √ | √ | ||
| Expression quantitative trait locus (eQTL) | √ | √ | √ | ||
| ChromHMM states | √ | √ | |||
| Histone modifications | √ | √ | |||
| DNA methylation positions | √ | √ | √ |
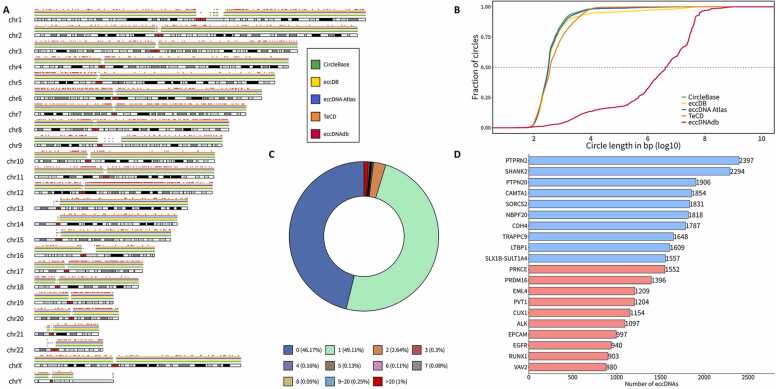
Extensive research has been conducted on the roles of eccDNAs in humans in both healthy and diseased states. We mapped the chromosomal distribution of human-derived eccDNAs, which are widely dispersed across all chromosomes (Fig. 1A). EccDNAdb is a specialized collection of circular amplicons identified by AA that contains only 1270 entries with a median entry size of 4 Mb and an average size of 25 Mb (Fig. 1B, Table 1). More than 95 % of the eccDNAs from all databases contain no or only one gene (Fig. 1C), whereas approximately 1 % of eccDNAs harbor more than 20 genes [35]. Among the oncogenes carried by eccDNAs, the most frequently occurring is PRKCE, followed by PRDM16, EML4, and PVT1. Finally, eccDNAs carrying PTPRN2 are the most common (Fig. 1D).
Fig. 1.
Overview of human-derived eccDNAs in five databases. A) Karyotype plot showing the chromosomal distribution of human-derived eccDNAs included in five databases. B) Length distribution of human-derived eccDNAs in five databases. C)Pie chart of the percentage of eccDNAs containing different numbers of genes. D)Bar plot showing the top 10 genes (blue) or oncogenes (red) most commonly carried by eccDNAs.
4. Bulk sequencing-based methods for ecDNA/microDNA research
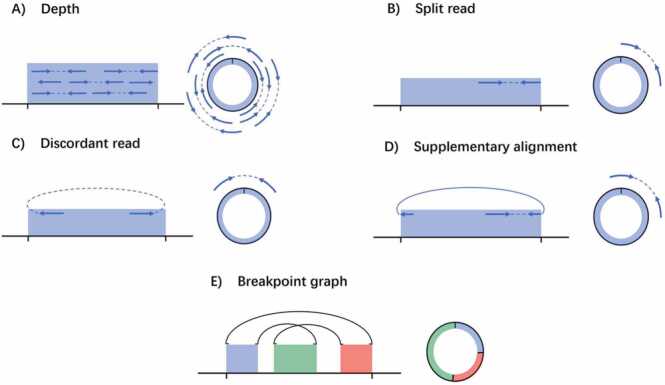
High-throughput sequencing has provided novel insights for the study of circular DNAs. In particular, the detection and structural elucidation of eccDNAs serves as the basis for further functional studies. Several tools are available for this purpose, and the main types of evidence include depths, split reads, supplementary alignments, discordant reads, and breakpoint graphs (Fig. 2). While depths provide direct evidence for the amplified region, split reads, discordant reads, and supplementary alignments provide information about the breakpoints and ligations between multiple segments. The construction of complex rearrangements is based on breakpoint graphs. We collected information on published methods, including the data type, output, experimental treatment, and main evidence (Table 2). Since each tool has distinct advantages and specific application scopes, direct comparisons between them are impractical (Table 3). AA is a powerful tool that utilizes WGS data to effectively assemble intricate ecDNAs based on breakpoint graphs [7], [8], [36]. Moreover, AmpliconReconstructor (AR) and HolistIC incorporate optical mapping (OM) and Hi-C data, respectively, to reduce ambiguity in the results obtained by AA [37], [38], [39]. The enrichment and amplification of circular DNA are commonly used techniques that can enhance the sequencing depth of circular DNA and facilitate the distinction between tandem duplication and DNA circles based on short-read data. Circle-seq is a widely used circular DNA sequencing method, but its enzymatic cleavage and rolling circle amplification (RCA) procedures may decrease the fidelity of DNA [14]. Conversely, Circulome-seq is an RCA-free technique that involves density gradient centrifugation and low levels of eccDNA initiation. Although this process may be relatively laborious, it reduces the introduction of artifacts [40], [41]. NGS-based microDNA identification tools may detect tens of thousands of candidate microDNAs within a sample, so pay special attention to false positives in the results, especially for regions that are too long (>50 kb) or too short. Circle-Map provides multiple metrics and a composite score to obtain high-confidence results. Some circular DNA detection tools have been developed based on TGS, which can reduce assembly errors and identify repeat dense regions more accurately than NGS. Nevertheless, these TGS-based tools have yet to be applied for the identification of complex ecDNA in large cancer datasets in a published study. Therefore, the selection of a specific method should be flexible and dependent on the type of eccDNA of interest, the type of data used, and the species studied.
Fig. 2.
Evidence for eccDNA construction. A) Depth indicates whether a segment shows continuity or not and can also provide approximate breakpoint information. B) Split reads offer information solely on breakpoints, typically due to a stump that is too short to form a supplementary alignment. In such cases, it may be necessary to construct a merged sequence and realign it to determine the segment linkage relationship. C) Discordant reads offer information on the link between two segments but do not provide accurate breakpoint information. D) Supplementary alignments, also known as soft-clipped reads, are perfect evidence for reads across breakpoints, suggesting the linkage between two segments and giving the exact location of the breakpoint and possible insertion and deletion at the breakpoint, etc.
E) The breakpoint graph establishes linkages between all segments, enabling the assembly of complex rearrangements. This step is vital in constructing ecDNA comprising multiple segments.
Table 2.
Sequencing-based methods for eccDNA construction.
| Method | Data type | Main result | microDNA | Complex ecDNA | Reference genome | Circle enrichment | Main research organism | NGS/TGS | Main evidence |
|---|---|---|---|---|---|---|---|---|---|
| AmpliconArchitect | WGS | ecDNA | × | √ | √ | × | Human | NGS |
|
| AmpliconReconstructor | OM + WGS | ecDNA | × | √ | √ | × | Human | NGS |
|
| HolistIC | Hi-C + WGS | ecDNA | × | √ | √ | × | Human | NGS |
|
| Circle-Map | Circle-Seq | microDNA | √ | × | √ | √ | Human, Yeast | NGS |
|
| Circle_finder | ATAC-Seq | microDNA | √ | × | √ | Optional | Human | NGS |
|
| ECCsplorer | Circle-Seq Mobilome-Seq |
microDNA | √ | × | × | √ | Plant | NGS |
|
| ecc_finder | CIDER-Seq Circle-Seq Mobilome-Seq Nanopore |
microDNA | √ | × | × | √ | Human, Plant | Optional |
|
| CIDER-Seq2 | CIDER-Seq | microDNA | √ | × | √ | √ | Virus, Plant | TGS |
|
| eccDNA_RCA_nanopore | Nanopore | microDNA | √ | × | √ | √ | Human, Mouse | TGS |
|
| CReSIL | Nanopore WGLS |
microDNA | √ | √ | √ | √ | Human, Mouse | TGS |
|
Table 3.
Descriptions and limitations of sequencing-based methods for ecDNA or microDNA construction.
| Method | Description | Advantage or limitation | Ref. |
|---|---|---|---|
| AmpliconArchitect |
|
|
[8], [36] |
| AmpliconReconstructor |
|
|
[38], [43] |
| HolistIC |
|
|
[39] |
| Circle-Map |
|
|
[60] |
| Circle_finder |
|
|
[61] |
| ECCsplorer |
|
|
[62] |
| ecc_finder |
|
|
[63], [64] |
| CIDER-Seq2 |
|
|
[65] |
| eccDNA_RCA_nanopore |
|
|
[19], [64] |
| CReSIL |
|
|
[64] |
Based on the identification of ecDNA boundaries, sequencing techniques have been employed to establish a correlation between the presence of ecDNA and changes in the transcriptome [27], [42]. Moreover, the chromatin landscape of ecDNA has been explored using ATAC-seq [27], [42], and ecDNA-mediated chromatin contacts have been characterized by using techniques such as Hi-C and ChIA-PET [27], [28], [29], [37], [43]. Although these traditional methods do not differentiate between circular and linear DNA, several innovative approaches have been developed to investigate the functions of circular DNAs in a specific manner. One of these techniques, known as CCDA-seq, involves the labeling of accessible DNA regions, followed by linear DNA removal and nanopore sequencing, to explore the chromatin status of ecDNA [44].
5. Single-cell sequencing and ecDNA research
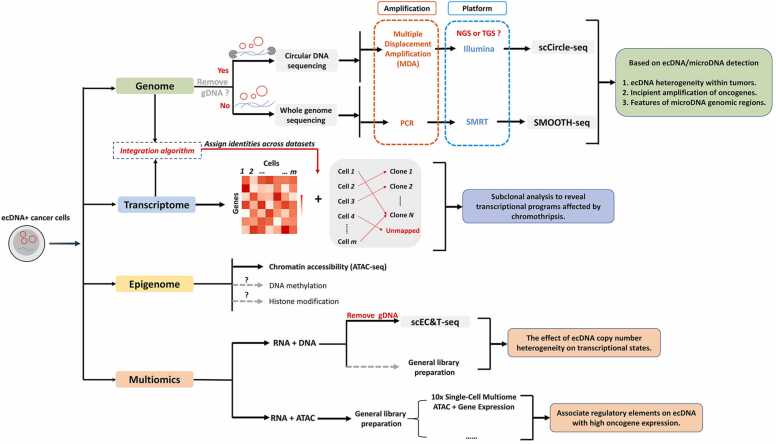
Bulk sequencing methods have effectively revealed the presence and features of ecDNA, but they may require up to millions of cells as input, and the diversity of the cells is thus unavoidably averaged. The sequencing signals obtained from cancer samples may originate from cells that harbor ecDNA as well as those that do not. Although in cancer cells with a high number of ecDNA copies, the sequencing signals are primarily attributed to ecDNA, the interpretation of bulk data is more challenging when ecDNAs are present in lower copy numbers or in only a small subset of cells in a heterogeneous population [9]. Additionally, some cells may contain multiple ecDNAs, and distinguishing these rare cells is important to study their selective advantages [30], [45]. Although the recently proposed CRISPR-CATCH technique enables the isolation and separate construction of multiple structurally distinct ecDNAs, it can only enrich ecDNAs containing known sequences, such as oncogenes, and is therefore a targeted method [45]. Single-cell sequencing provides different types of information within individual cells in an unbiased way encompassing genomic alterations (such as mutations and CNVs), DNA methylation loci, accessible chromatin regions, and mRNA or protein abundance and has been widely used in cancer research [46], [47]. While there are a few currently available studies on this topic, we expect that single-cell sequencing holds great potential for ecDNA research (Fig. 3).
Fig. 3.
Current strategies for applying single-cell sequencing to study ecDNA or microDNA. Identification of ecDNA through single-cell whole genome sequencing or circular DNA sequencing can shed light on the heterogeneity of ecDNA copies within cells as well as early oncogene amplification. Furthermore, through the use of multiomics to analyze the impact of ecDNA on transcription or how regulatory elements on ecDNA affect oncogene expression, we can gain a deeper understanding of the role of ecDNA in cancer. Additionally, current research suggests that microDNA production occurs in a cell-type-specific manner and that microDNA has potential applications in cancer diagnosis.
5.1. Single-cell genomics
Advancements in single-cell DNA sequencing methods have significantly facilitated the identification of ecDNA. For instance, Fan et al. proposed the SMOOTH-seq approach and demonstrated its efficacy in detecting ecDNA, identifying 125 candidate ecDNAs with a median size of approximately 100 kb [48]. However, the presence of linear DNA may hinder circle identification. scCircle-seq and scEC&T-seq are two other techniques that have been developed, in which linear DNA is digested before sequencing to avoid interference from genomic DNA. These two methods, which are based on NGS, are more easily applicable than TGS-based SMOOTH-seq [16], [49], [50]. Using scCircle-seq, Chen et al. detected numerous small microDNAs and some large ecDNAs from 156 cells. They reported that genomic regions that produce circles are cell type-specific and enable the clustering of cells with a shared origin [16]. Nevertheless, some challenges remain, such as the assembly of complex circles comprising multiple segments, the high cost of these methods, and the limited number of cells that can be sequenced (usually a few hundred), with amplification biased toward smaller, more abundant circles [15].
Single-cell technologies offer a higher resolution than bulk WGS for analyzing patterns of heterogeneity within tumors, revealing subclones, and facilitating the understanding of cancer evolution. Stöber et al. conducted a study on cells from neuroblastoma (NB) cell lines and patients and revealed that MYCN copy numbers among cells varied significantly, by orders of magnitude, and that such extensive heterogeneity was only observed in the ecDNA region [49]. Spain et al. also reported the discovery of heterogeneous KIT copies in melanoma patients carrying ecDNA through single-cell whole genome sequencing (scWGS-seq) [51]. In another study, Parra et al. found that in chromothriptic medulloblastoma (MB), most tumor cells carried 10–20 copies of ecDNA, while very few cells harbored more than 100 copies [42]. Moreover, they identified six major clones from primary tumors at the single-cell level based on copy number variation (CNV) clustering. They further compared clonal compositions, finding that only a few or none of the six clones were present in recurrent and patient-derived xenograft (PDX) samples. Interestingly, their study showed significant heterogeneity in ecDNA numbers within and between subclones. Pongor et al. studied MYC and MYCL ecDNAs in small-cell lung cancer (SCLC) by targeting 196 amplicons, revealing copies that differed by over an order of magnitude between cells. The correlation between MYC and MYCL copy numbers indicated that some cells may carry multiple ecDNAs. Furthermore, heterogeneity has been observed across metastasis sites, with ecDNA-positive cells displaying a wider range of MYC copies than HSR-positive cells [30]. These studies emphasize the advantages of single-cell DNA sequencing in exploring ecDNA, as it allows the occurrence of ecDNA to be described at the level of individual cells, rather than entire samples, rendering it a potent tool for elucidating ecDNA-mediated heterogeneity.
5.2. Single-cell epigenomics
Single-cell ATAC-seq (scATAC-seq) has great potential for detecting ecDNA. The use of Tn5 transposase to cleave circular DNA produces chimeric reads in the ATAC-seq library that correspond to circles. Nevertheless, scATAC-seq presents a challenge due to its lower sequencing depth compared to bulk sequencing. To address this issue, a common approach is to create pseudobulk samples by merging data from approximately 100 cells and then apply algorithms that are developed for bulk sequencing [47]. In a recent study, ecc_finder, which is based on bulk sequencing data, was used to identify circular DNA in scATAC-seq data from glioblastoma (GBM) [52]. However, current tools do not fully capitalize on ATAC-seq accessibility data to detect highly accessible ecDNA regions, and the pseudobulk approach may overlook peaks associated with low levels of open chromatin due to a lack of information, even in the integrated data. Thus, there is a pressing need for novel algorithms specifically designed for the single-cell detection of ecDNA. In this context, calculating amplicon copy numbers from scATAC-seq data has proven to be an effective method that provides an additional genomic dimension [43]. While mapping ecDNA methylation and histone modifications at the single-cell level are potential avenues for future research, isolating specific ecDNAs followed by single-molecule sequencing is currently a more practical approach for describing the ecDNA epigenomic landscape [45].
5.3. Single-cell multiomics sequencing
Single-cell multiomic analysis allows the combination of various experimental approaches by integrating data or subjecting the same cell to multiple assays, thereby enabling the study of the functional consequences or regulatory role of ecDNA, going beyond the mere detection of ecDNA [53].
5.3.1. Integrative analysis of genome and transcriptome data
The integration of genome and transcriptome data has facilitated insightful analysis of the association between ecDNA copies and RNA expression within cells. Hung et al. employed joint scATAC-seq and scRNA-seq to analyze two colorectal cancer cell lines in which MYC was amplified in the form of ecDNA or HSR separately. Based on amplicon copy numbers inferred from scATAC-seq data, their analysis revealed a stronger correlation between MYC copy numbers and expression in ecDNA-positive cells than in HSR-positive cells [43]. The results provided more specific evidence than bulk sequencing that ecDNA amplification is a more effective mechanism driving high oncogene expression than linear amplification. Similarly, Chen et al. utilized scCircle-seq to demonstrate that while there was no correlation between the copies of small microDNAs and gene expression, a clear correlation was observed for genes carried by large ecDNA, such as MYC, in COLO320DM cells [16]. Their studies underscore the critical distinction between clonal ecDNA and nonclonal microDNA within cancer cells for the first time at the single-cell level. In another study, Stöber et al. proved that ecDNA-mediated high MYCN expression in NB is causally associated with elevated MYCN target gene expression and changes in pathways including ribosome biogenesis and cell-cell interactions. In contrast to previous studies that have focused on samples or cell lines with different levels of MYCN expression, they revealed cell subpopulations with different transcriptional states due to the intercellular heterogeneity of ecDNA [49].
5.3.2. Integrative analysis of epigenome and transcriptome data
The groundbreaking study of Hung et al. provides a paradigm for the multiomics analysis of ecDNA to investigate the impact of regulatory elements of ecDNA on oncogene expression. They also integrated genomic information based on amplicon copies inferred by scATAC-seq, resulting in a comprehensive study of ecDNA covering the genome, transcriptome, and epigenome. Based on combinatorial barcoding to differentiate between mRNA and open chromatin in individual cell nuclei, followed by sequencing, Hung et al. scrutinized ecDNA regions and identified 47 ecDNA regulatory elements associated with high MYC expression, only two of which were also active in the HSR-amplified cell line. Subsequently, they delved into the five most significantly variable elements of ecDNA and observed that the high accessibility of these elements was associated with high MYC expression in ecDNA-positive cells. Additionally, the higher accessibility of these elements in ecDNA-positive cells than in HSR-positive cells suggests that ecDNA elements control the elevated expression of oncogenes [43].
6. Summary and outlook
Due to the numerous unexplored features of microDNA and ecDNA, it is inadequate to classify eccDNAs based solely on their origin and size. Thus, refining the defining characteristics of eccDNAs is essential for future research. Currently, eccDNA research is burgeoning, but some critical areas require more attention. First, there is an urgent need to develop tools capable of leveraging accumulated NGS data. It is a challenging task for most published tools to make inferences about complex ecDNA. Long-read data or OM data are necessary to confirm the actual structure of ecDNA. In addition, the newest methods for detecting structural variation, such as single-tube long fragment read (stLFR) and linked read sequencing, may provide inspiration for detecting ecDNA [54], [55]. Second, exonuclease treatment inevitably damages large ecDNAs [14], [44], while RCA favors the amplification of small, abundant circles, as obtained in Circle-seq and CCDA-seq [56]. Recently, researchers have proposed the CRISPR-mediated specific removal of mitochondrial DNA as an alternative to the application of restriction enzymes, which may damage ecDNA [57]. Therefore, there is significant potential for improving current circular DNA sequencing methods. Third, it is imperative to improve library preparation methods and develop novel algorithms to advance single-cell techniques for eccDNA research. Moreover, when analyzing large-sized ecDNA, single-nucleus rather than single-cell sequencing may be a better choice due to its advantages in capturing malignant cells and allowing the use of frozen samples [58], [59].
EcDNA has become the focus of cancer research. Despite significant progress, many questions regarding ecDNA are still not fully answered, including its formation and maintenance mechanisms, sequence and structural features, relationship with tumor evolution, and potential applications in early diagnosis, targeted therapy, and prognosis. Addressing these challenges calls for the development of powerful tools and methods. In the era of high-throughput sequencing, we anticipate that extrachromosomal circular DNA will continue to attract increasing attention, providing new opportunities for advancing clinical diagnosis and treatment.
Funding
This work was supported by the National Natural Science Foundation of China (31971117), the National Key R&D Program of China [2018YFA0801100], the Natural Science Foundation of Suzhou [SYS201517], and the Priority Academic Program Development of Jiangsu Higher Education Institutions.
CRediT authorship contribution statement
Rong Jiang, Manqiu Yang and Moli Huang: Conceptualization. Rong Jiang and Manqiu Yang: Investigation. Rong Jiang, Manqiu Yang, and Shufan Zhang: Writing- Original Draft. Moli Huang and Rong Jiang: Writing- Review & Editing. Moli Huang: Supervision, Project administration.
Declaration of Competing Interest
The authors declare no potential conflicts of interest.
References
- 1.Hotta Y., Bassel A. Molecular size and circularity of dna in cells of mammals and higher plants. Proc Natl Acad Sci. 1965;53:356–362. doi: 10.1073/pnas.53.2.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liao Z., Jiang W., Ye L., Li T., Yu X., Liu L. Classification of extrachromosomal circular DNA with a focus on the role of extrachromosomal DNA (ecDNA) in tumor heterogeneity and progression. Biochim Biophys Acta BBA - Rev Cancer. 2020;1874 doi: 10.1016/j.bbcan.2020.188392. [DOI] [PubMed] [Google Scholar]
- 3.Paulsen T., Kumar P., Koseoglu M.M., Dutta A. Discoveries of extrachromosomal circles of DNA in normal and tumor cells. Trends Genet. 2018;34:270–278. doi: 10.1016/j.tig.2017.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang T., Zhang H., Zhou Y., Shi J. Extrachromosomal circular DNA: a new potential role in cancer progression. J Transl Med. 2021;19:257. doi: 10.1186/s12967-021-02927-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao Y., Yu L., Zhang S., Su X., Zhou X. Extrachromosomal circular DNA: current status and future prospects. ELife. 2022;11 doi: 10.7554/eLife.81412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cox D., Yuncken C., Spriggs A.. Minute chromatin bodies in malignant tumours of childhood. Lancet. 1965;286:55–58. doi: 10.1016/S0140-6736(65)90131-5. [DOI] [PubMed] [Google Scholar]
- 7.Turner K.M., Deshpande V., Beyter D., Koga T., Rusert J., Lee C., et al. Extrachromosomal oncogene amplification drives tumour evolution and genetic heterogeneity. Nature. 2017;543:122–125. doi: 10.1038/nature21356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim H., Nguyen N.-P., Turner K., Wu S., Gujar A.D., Luebeck J., et al. Extrachromosomal DNA is associated with oncogene amplification and poor outcome across multiple cancers. Nat Genet. 2020;52:891–897. doi: 10.1038/s41588-020-0678-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hung K.L., Mischel P.S., Chang H.Y. Gene regulation on extrachromosomal DNA. Nat Struct Mol Biol. 2022 doi: 10.1038/s41594-022-00806-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weiser N.E., Hung K.L., Chang H.Y. Oncogene convergence in extrachromosomal DNA hubs. Cancer Disco. 2022;12:1195–1198. doi: 10.1158/2159-8290.CD-22-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peng L., Zhou N., Zhang C.-Y., Li G.-C., Yuan X.-Q. eccDNAdb: a database of extrachromosomal circular DNA profiles in human cancers. Oncogene. 2022;41:2696–2705. doi: 10.1038/s41388-022-02286-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yi E., Chamorro González R., Henssen A.G., Verhaak R.G.W. Extrachromosomal DNA amplifications in cancer. Nat Rev Genet. 2022 doi: 10.1038/s41576-022-00521-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tomaska L., Nosek J., Kramara J., Griffith J.D. Telomeric circles: universal players in telomere maintenance. Nat Struct Mol Biol. 2009;16:1010–1015. doi: 10.1038/nsmb.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Møller H.D., Parsons L., Jørgensen T.S., Botstein D., Regenberg B. Extrachromosomal circular DNA is common in yeast. Proc Natl Acad Sci. 2015:112. doi: 10.1073/pnas.1508825112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Møller H.D., Mohiyuddin M., Prada-Luengo I., Sailani M.R., Halling J.F., Plomgaard P., et al. Circular DNA elements of chromosomal origin are common in healthy human somatic tissue. Nat Commun. 2018;9:1069. doi: 10.1038/s41467-018-03369-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J.P., Bouwman B., Wu H., Chen C., Bienko M., Crosetto N. scCircle-seq unveils the diversity and complexity of circular DNAs in single cells. Review. 2023 doi: 10.21203/rs.3.rs-2617401/v1. [DOI] [Google Scholar]
- 17.Shibata Y., Kumar P., Layer R., Willcox S., Gagan J.R., Griffith J.D., et al. Extrachromosomal microDNAs and chromosomal microdeletions in normal tissues. Science. 2012;336:82–86. doi: 10.1126/science.1213307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paulsen T., Malapati P., Shibata Y., Wilson B., Eki R., Benamar M., et al. MicroDNA levels are dependent on MMEJ, repressed by c-NHEJ pathway, and stimulated by DNA damage. Nucleic Acids Res. 2021;49:11787–11799. doi: 10.1093/nar/gkab984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y., Wang M., Djekidel M.N., Chen H., Liu D., Alt F., et al. EccDNAs are apoptotic products with high innate immunostimulatory activity. Nature. 2021;599:308–314. doi: 10.1038/s41586-021-04009-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paulsen T., Shibata Y., Kumar P., Dillon L., Dutta A. Small extrachromosomal circular DNAs, microDNA, produce short regulatory RNAs that suppress gene expression independent of canonical promoters. Nucleic Acids Res. 2019;47:4586–4596. doi: 10.1093/nar/gkz155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Z., Wang B., Liang H., Han L. Pioneering insights of extrachromosomal DNA (ecDNA) generation, action and its implications for cancer therapy. Int J Biol Sci. 2022;18:4006–4025. doi: 10.7150/ijbs.73479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koche R.P., Rodriguez-Fos E., Helmsauer K., Burkert M., MacArthur I.C., Maag J., et al. Extrachromosomal circular DNA drives oncogenic genome remodeling in neuroblastoma. Nat Genet. 2020;52:29–34. doi: 10.1038/s41588-019-0547-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu S., Bafna V., Chang H.Y., Mischel P.S. Extrachromosomal DNA: an emerging hallmark in human cancer. Annu Rev Pathol Mech Dis. 2022;17:367–386. doi: 10.1146/annurev-pathmechdis-051821-114223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bafna V., Mischel P.S. Extrachromosomal DNA in Cancer. Annu Rev Genom Hum Genet. 2022;23 doi: 10.1146/annurev-genom-120821-100535. annurev-genom-120821-100535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yi E., Gujar A.D., Guthrie M., Kim H., Zhao D., Johnson K.C., et al. Live-cell imaging shows uneven segregation of extrachromosomal DNA elements and transcriptionally active extrachromosomal DNA hubs in cancer. Cancer Disco. 2022;12:468–483. doi: 10.1158/2159-8290.CD-21-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lange J.T., Rose J.C., Chen C.Y., Pichugin Y., Xie L., Tang J., et al. The evolutionary dynamics of extrachromosomal DNA in human cancers. Nat Genet. 2022 doi: 10.1038/s41588-022-01177-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu S., Turner K.M., Nguyen N., Raviram R., Erb M., Santini J., et al. Circular ecDNA promotes accessible chromatin and high oncogene expression. Nature. 2019;575:699–703. doi: 10.1038/s41586-019-1763-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morton A.R., Dogan-Artun N., Faber Z.J., MacLeod G., Bartels C.F., Piazza M.S., et al. Functional enhancers shape extrachromosomal oncogene amplifications. Cell. 2019;179:1330–1341. doi: 10.1016/j.cell.2019.10.039. e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chapman O.S., Luebeck J., Wani S., Tiwari A., Pagadala M., Wang S., et al. The landscape of extrachromosomal circular DNA in medulloblastoma. Cancer Biol. 2021 doi: 10.1101/2021.10.18.464907. [DOI] [Google Scholar]
- 30.Pongor L.Sandor, Schultz C.W., Rinaldi L., Wangsa D., Redon C.E., Takahashi N., et al. Extrachromosomal DNA amplification contributes to small cell lung cancer heterogeneity and is associated with worse outcomes. Cancer Disco. 2023;(CD):22–0796. doi: 10.1158/2159-8290.CD-22-0796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao X., Shi L., Ruan S., Bi W., Chen Y., Chen L., et al. CircleBase: an integrated resource and analysis platform for human eccDNAs. Nucleic Acids Res. 2021;50:D72–D82. doi: 10.1093/nar/gkab1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang M., Qiu B., He G.-Y., Zhou J.-Y., Yu H.-J., Zhang Y.-Y., et al. eccDB: a comprehensive repository for eccDNA-mediated chromatin contacts in multi-species. Bioinformatics. 2022 doi: 10.1101/2022.09.22.509011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhong T., Wang W., Liu H., Zeng M., Zhao X., Guo Z. eccDNA Atlas: a comprehensive resource of eccDNA catalog. Bioinformatics. 2022 doi: 10.1101/2022.11.06.515328. [DOI] [PubMed] [Google Scholar]
- 34.Guo J., Zhang Z., Li Q., Chang X., Liu X. TeCD: the eccDNA collection database for extrachromosomal circular DNA. BMC Genom. 2023;24:47. doi: 10.1186/s12864-023-09135-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y., Sun J., Zhao M. ONGene: a literature-based database for human oncogenes. J Genet Genom. 2017;44:119–121. doi: 10.1016/j.jgg.2016.12.004. [DOI] [PubMed] [Google Scholar]
- 36.Deshpande V., Luebeck J., Nguyen N.-P.D., Bakhtiari M., Turner K.M., Schwab R., et al. Exploring the landscape of focal amplifications in cancer using AmpliconArchitect. Nat Commun. 2019;10:392. doi: 10.1038/s41467-018-08200-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu Y., Gujar A.D., Wong C.-H., Tjong H., Ngan C.Y., Gong L., et al. Oncogenic extrachromosomal DNA functions as mobile enhancers to globally amplify chromosomal transcription. Cancer Cell. 2021;39(694–707) doi: 10.1016/j.ccell.2021.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luebeck J., Coruh C., Dehkordi S.R., Lange J.T., Turner K.M., Deshpande V., et al. AmpliconReconstructor integrates NGS and optical mapping to resolve the complex structures of focal amplifications. Nat Commun. 2020;11:4374. doi: 10.1038/s41467-020-18099-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hayes M., Nguyen A., Islam R., Butler C., Tran E., Mullins D., et al. HolistIC: leveraging Hi-C and whole genome shotgun sequencing for double minute chromosome discovery. Bioinforma Oxf Engl. 2021;(btab816) doi: 10.1093/bioinformatics/btab816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shoura M.J., Gabdank I., Hansen L., Merker J., Gotlib J., Levene S.D., et al. Intricate and cell type-specific populations of endogenous circular DNA (eccDNA) in Caenorhabditis elegans and Homo sapiens. G3 Genes. 2017;7:3295–3303. doi: 10.1534/g3.117.300141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lanciano S., Carpentier M.-C., Llauro C., Jobet E., Robakowska-Hyzorek D., Lasserre E., et al. Sequencing the extrachromosomal circular mobilome reveals retrotransposon activity in plants. PLOS Genet. 2017;13 doi: 10.1371/journal.pgen.1006630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parra R.G., Przybilla M.J., Simovic M., Susak H., Ratnaparkhe M., Wong J.K., et al. Single cell multi-omics analysis of chromothriptic medulloblastoma highlights genomic and transcriptomic consequences of genome instability. Cancer Biol. 2021 doi: 10.1101/2021.06.25.449944. [DOI] [Google Scholar]
- 43.Hung K.L., Yost K.E., Xie L., Shi Q., Helmsauer K., Luebeck J., et al. ecDNA hubs drive cooperative intermolecular oncogene expression. Nature. 2021;600:731–736. doi: 10.1038/s41586-021-04116-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen W., Weng Z., Xie Z., Xie Y., Zhang C., Chen Z., et al. Sequencing of methylase-accessible regions in integral circular extrachromosomal DNA reveals differences in chromatin structure. Epigenetics Chromatin. 2021;14:40. doi: 10.1186/s13072-021-00416-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hung K.L., Luebeck J., Dehkordi S.R., Colón C.I., Li R., Wong I.T.-L., et al. Targeted profiling of human extrachromosomal DNA by CRISPR-CATCH. Nat Genet. 2022;54:1746–1754. doi: 10.1038/s41588-022-01190-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lei Y., Tang R., Xu J., Wang W., Zhang B., Liu J., et al. Applications of single-cell sequencing in cancer research: progress and perspectives. J Hematol OncolJ Hematol Oncol. 2021;14:91. doi: 10.1186/s13045-021-01105-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee J., Hyeon D.Y., Hwang D. Single-cell multiomics: technologies and data analysis methods. Exp Mol Med. 2020;52:1428–1442. doi: 10.1038/s12276-020-0420-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fan X., Yang C., Li W., Bai X., Zhou X., Xie H., et al. SMOOTH-seq: single-cell genome sequencing of human cells on a third-generation sequencing platform. Genome Biol. 2021;22:195. doi: 10.1186/s13059-021-02406-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stöber M.C., González R.C., Brückner L., Conrad T., Szymansky A., Eggert A., et al. Intercellular extrachromosomal DNA copy number heterogeneity drives cancer cell state diversity n.d. [DOI] [PubMed]
- 50.Parallel sequencing of extrachromosomal circular DNAs and transcriptomes in single cancer cells. Nat Genet n.d.
- 51.Spain L., Coulton A., Lobon I., Rowan A., Schnidrig D., Shepherd S.T.C., et al. Late-stage metastatic melanoma emerges through a diversity of evolutionary pathways. Cancer Disco. 2023;(CD):22–1427. doi: 10.1158/2159-8290.CD-22-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kang J., Dai Y., Li J., Fan H., Zhao Z. Investigating cellular heterogeneity at the single-cell level by the flexible and mobile extrachromosomal circular DNA. Comput Struct Biotechnol J. 2023;21:1115–1121. doi: 10.1016/j.csbj.2023.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cosenza M.R., Rodriguez-Martin B., Korbel J.O. Structural variation in cancer: role, prevalence, and mechanisms. Annu Rev Genom Hum Genet. 2022;23:123–152. doi: 10.1146/annurev-genom-120121-101149. [DOI] [PubMed] [Google Scholar]
- 54.Wang O., Chin R., Cheng X., Wu M.K.Y., Mao Q., Tang J., et al. Efficient and unique cobarcoding of second-generation sequencing reads from long DNA molecules enabling cost-effective and accurate sequencing, haplotyping, and de novo assembly. Genome Res. 2019;29:798–808. doi: 10.1101/gr.245126.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.De Coster W., Van Broeckhoven C. Newest methods for detecting structural variations. Trends Biotechnol. 2019;37:973–982. doi: 10.1016/j.tibtech.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 56.Norman A., Riber L., Luo W., Li L.L., Hansen L.H., Sørensen S.J. An improved method for including upper size range plasmids in metamobilomes. PLoS One. 2014;9 doi: 10.1371/journal.pone.0104405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Feng W., Arrey G., Zole E., lv W., Liang X., Han P., et al. Targeted removal of mitochondrial DNA from mouse and human extrachromosomal circular DNA with CRISPR-Cas9. Comput Struct Biotechnol J. 2022;20:3059–3067. doi: 10.1016/j.csbj.2022.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu H., Kirita Y., Donnelly E.L., Humphreys B.D. Advantages of single-nucleus over single-cell RNA sequencing of adult kidney: rare cell types and novel cell states revealed in fibrosis. J Am Soc Nephrol JASN. 2019;30:23–32. doi: 10.1681/ASN.2018090912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Slyper M., Porter C.B.M., Ashenberg O., Waldman J., Drokhlyansky E., Wakiro I., et al. A single-cell and single-nucleus RNA-Seq toolbox for fresh and frozen human tumors. Nat Med. 2020;26:792–802. doi: 10.1038/s41591-020-0844-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cen Y., Fang Y., Ren Y., Hong S., Lu W., Xu J. Global characterization of extrachromosomal circular DNAs in advanced high grade serous ovarian cancer. Cell Death Dis. 2022;13:342. doi: 10.1038/s41419-022-04807-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kumar P., Kiran S., Saha S., Su Z., Paulsen T., Chatrath A., et al. ATAC-seq identifies thousands of extrachromosomal circular DNA in cancer and cell lines. Sci Adv. 2020;6(eaba2489) doi: 10.1126/sciadv.aba2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mann L., Seibt K.M., Weber B., Heitkam T. ECCsplorer: a pipeline to detect extrachromosomal circular DNA (eccDNA) from next-generation sequencing data. BMC Bioinforma. 2022;23:40. doi: 10.1186/s12859-021-04545-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang P., Peng H., Llauro C., Bucher E., Mirouze M. ecc_finder: a robust and accurate tool for detecting extrachromosomal circular DNA from sequencing data. Front Plant Sci. 2021;12 doi: 10.3389/fpls.2021.743742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wanchai V., Jenjaroenpun P., Leangapichart T., Arrey G., Burnham C.M., Tümmler M.C., et al. CReSIL: accurate identification of extrachromosomal circular DNA from long-read sequences. Brief Bioinform. 2022 doi: 10.1093/bib/bbac422. bbac422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mehta D., Cornet L., Hirsch-Hoffmann M., Zaidi S.S.-A., Vanderschuren H. Full-length sequencing of circular DNA viruses and extrachromosomal circular DNA using CIDER-Seq. Nat Protoc. 2020;15:1673–1689. doi: 10.1038/s41596-020-0301-0. [DOI] [PubMed] [Google Scholar]