Abstract
Dental calculus is a valuable resource for the reconstruction of dietary habits and oral microbiome of past populations. In 2020 the remains of Duke Alessandro Farnese and his wife Maria D’Aviz were exhumed to get novel insights into the causes of death. This study aimed to investigate the dental calculus metabolome of the noble couple by untargeted metabolomics. The pulverized samples were decalcified in a water-formic acid mixture, extracted using methanol/acetonitrile and analyzed by ultra-high performance liquid chromatography coupled to high-resolution mass spectrometry (UHPLC-HRMS) using a reversed-phase separation followed by electrospray ionization and full scan in positive and negative ion mode. Waters Synapt-G2-Si High-Definition hybrid quadrupole time-of-flight mass spectrometer was used. Significant features were then identified using MSE acquisition mode, recording information on exact mass precursor and fragment ions within the same run. This approach, together with data pre-treatment and multivariate statistical analysis allowed for the identification of compounds able to differentiate between the investigated samples. More than 200 metabolites were identified, being fatty acids, alcohols, aldehydes, phosphatidylcholines, phosphatidylglycerols, ceramides and phosphatidylserines the most abundant classes. Metabolites deriving from food, bacteria and fungi were also determined, providing information on the habits and oral health status of the couple.
Subject terms: Metabolomics, Mass spectrometry, Mass spectrometry
Introduction
Dental calculus is a mineralized microbial plaque, which accumulates at the surface of the tooth1. The mineral is deposited from crevicular fluid, but ultimately derives by precipitation of salivary calcium salts and for this reason the concentration of calculus is greater in the sites closest to the ducts of the salivary glands2. Being mainly composed of inorganic constituents, among which hydroxyapatite, fluorapatite, octacalcium phosphate and whitlockite3, dental calculus is well-preserved in archaeological samples and it can entomb biomolecules (e.g., DNA, proteins and lipids) associated to the oral microbiota, the host, and microdebris of exogenous origin4–7. Therefore, in the past two decades dental calculus has been an important resource to investigate health status, lifestyle and diet in the past populations. Optical microscopy8, scanning electron microscopy, also coupled to energy dispersive X-ray spectroscopy9,10, pyrolysis–gas chromatography-mass spectrometry11 and multi-omics techniques including proteomics, genomics and metabolomics12,13 proved to be valuable techniques to characterize exogenous debris entrapped in dental calculus, providing information on habits and health of individuals living in the past. Being able to provide a comprehensive fingerprinting of the investigated samples, mass spectrometry-based omics strategies are particularly promising thanks to their sensitivity, high-throughput and discriminating power. One of the most important advantages of these techniques relies on their untargeted nature, thus providing information on a plethora of biomolecules and allowing biomarker identification.
Combined to the use of bioinformatics and computational approaches, metabolomics plays a key role in understanding metabolome and in the identification of metabolites present in biological samples14–16. Different analytical platforms, among which high resolution mass spectrometry (HRMS) coupled to both gas- and liquid chromatography (GC-HRMS, GCxGC-HRMS and LC-HRMS) and nuclear magnetic resonance (NMR) can be applied to achieve a comprehensive covering of metabolome. Due to the enhanced specificity, sensitivity and availability of large spectral databases compared to NMR, HRMS has been widely applied for the metabolomics profiling of different biofluids, tissues and other biological samples17–19. In particular, analytical strategies involving on-line coupling of orthogonal UHPLC separations with HRMS represent the best tools to extend metabolite coverage20, because UHPLC ranks among the most efficient separation techniques21 and its combination with HRMS allows for the identification of a broad range of metabolites22.
Recently, Velsko and coauthors investigated the dental calculus metabolome in both modern and ancient specimens by using targeted and untargeted MS-based approaches23. In particular, the use of both UHPLC coupled with high-resolution tandem mass spectrometry (UHPLC-HRMS/MS) and GC-HRMS allowed the identification of metabolites belonging to different chemical classes, mainly lipids and amino acids, together with several xenobiotic compounds, providing insight into microbiome activity, metabolic processes, and degradation patterns.
In the present study, UHPLC-HRMS using a Waters hybrid quadrupole time-of-flight Synapt-G2-Si High-Definition (HD)MS QTOF mass spectrometer was applied to investigate the metabolome of dental calculus from Duke Alessandro Farnese and his wife Maria D’Aviz, who ruled the Duchy of Parma and Piacenza in the sixteenth century. Alessandro Farnese (1545–1592), a key member of the court of Philip II of Spain, was among the most influential military and political leaders of the sixteenth century and pursued numerous victories, particularly in Flanders, where he was governor for 15 years. The envy aroused by his success led to suspicion that his death could be caused by poisoning. To dispel this doubt, in 2020 the remains of both Alessandro Farnese and his wife were exhumed from the crypt of the Basilica of Santa Maria della Steccata in Parma (Italy). After opening the lead case containing the remains, the teeth were submitted to morphological analysis. The main feature in the dentition of Alessandro Farnese was severe wear, whereas the teeth of Maria D’Aviz were characterized by the presence of caries and periodontal diseases24. These findings could be associated with the different lifestyle of the noble couple, Duke Farnese spending most of his adult lifetime on battlefields all over Europe, and his wife maintaining the habits of the Portuguese court in the city of Parma. In this context, we tested the capability of UHPLC-HRMS operating in scanning MSE mode to investigate the differences in the metabolomics profile of dental calculus of the duke and his wife, with the final aim of achieving novel insights on Alessandro Farnese’s causes of death, as well on the habits, health state and diet of the noble couple.
Results and discussion
HRMS analysis was applied to achieve a highly confident identification of the metabolites present in the dental calculus of the noble couple. The capabilities of the Waters Synapt G2-Si HD mass spectrometer for complex matrix profiling were exploited by operating in MSE mode: the use of an alternate scanning acquisition of both low and high-energy profiles provides information on both precursor and fragment ions in a single run with superior duty cycle to other data independent analysis techniques.
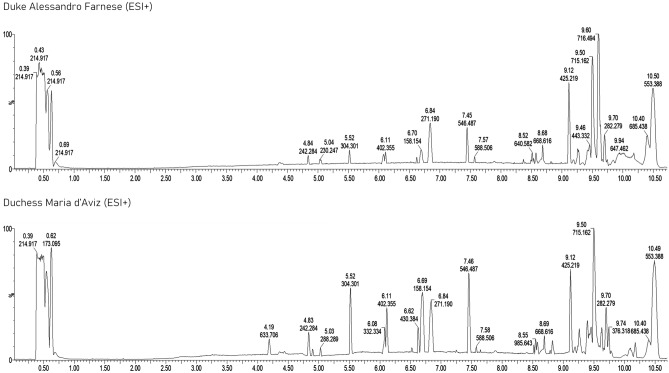
Due to the low amount of sample available, the most suitable experimental conditions for both extraction and analysis were preliminary assessed using modern dental calculus samples by operating both in the PI and NI mode. The effects of both the ratio between the aqueous and organic phase for the extraction procedure and the injection volume were investigated: a 1:3 ratio was used to induce protein precipitation, avoiding a high dilution of the extracted metabolites, while 8 µL were injected to obtain adequate signal intensities to perform measurements (Fig. 1). A good repeatability was also obtained in terms of area of selected chromatographic peaks with relative standard deviations always lower than 5 and 7% for PI and NI modes, respectively.
Figure 1.
UHPLC-HRMS chromatograms in PI mode of dental calculus samples from: top) Maria D’Aviz and bottom) Duke Alessandro Farnese. Column: Atlantis Premier BEH C18 AX 1.7 μm (2.1 × 100 mm); mobile phase: (A) water + 0.1% (v/v) formic acid and (B) acetonitrile + 0.1% (v/v) formic acid; injection volume: 8 µL.
Metabolomic analysis
The high number of features generated by HRMS is one of the most challenging aspects of untargeted metabolomics: in this context, both filtering and data reduction strategies need to be applied to select the significant m/z values for their subsequent identification based on the measured accurate mass of both precursor and fragment ions.
A total of 5918 and 5689 features were extracted in the dental calculus sample of Duke Alessandro Farnese by operating in PI and NI mode, respectively, whereas 5525 and 4728 features were recorded in the specimen of his wife Maria D’Aviz. To highlight the features responsible for the differentiation between the samples of the noble couple, only those having an intra-group variability of maximum 10% were used for subsequent processing. A minimum fold change of 3 compared to the extraction blank samples was also applied to correct the level of noise in the chromatograms of dental calculus samples. Finally, a value of 0.8 was set as reference value for statistical power analysis to detect real effects in the analyzed dataset. By applying the above-mentioned filters, a total of 4859 features was obtained.
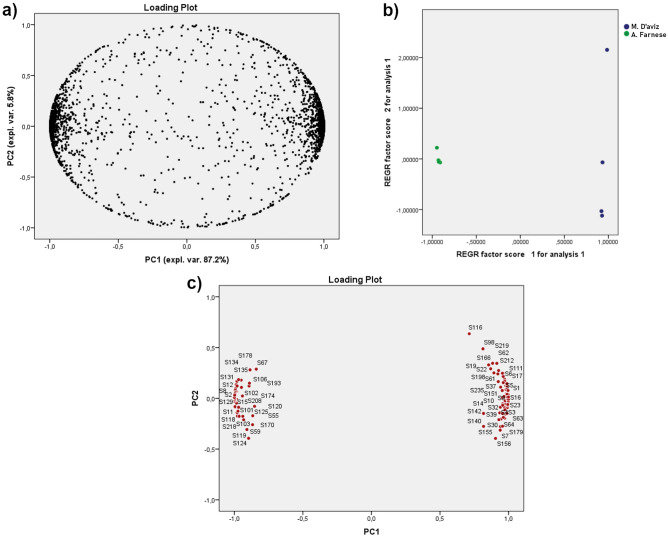
Multivariate statistical analysis was performed using principal component analysis (PCA) with 99% of the total variance explained by the first 6 PCs. As shown in Fig. 2, a good separation between the dental calculus samples of Duke Alessandro Farnese and his wife was achieved along PC1, accounting for more than 87% of the total variance. A further reduction in the number of features (4227) was achieved by submitting to the identification process only the variables having an absolute score in the loading plot higher than 0.8 on PC1 (Fig. 2c).
Figure 2.
PCA: (a) loading plot and (b) score plot of all features; (c) highlight of the features having |loading |> 0.8.
Taking into account that no a priori information was available for elucidation purposes, compounds identification was performed considering all the information deriving from accurate mass measurements, fragmentation studies, analysis of the isotopic pattern, library matching and score fit.
A total of 217 metabolites belonging to different classes were identified (Table S1). Sphingolipids, glycerophospholipids and fatty acyls were the most abundant chemical classes, accounting for 132 compounds. The presence of a high number of lipids is not surprising: in a previous study on modern and ancient metabolome of dental calculus, Velsko et al. highlighted the influence of aqueous solubility on the preservation of compounds, assuming that compounds characterized by high water solubility could be degraded more easily23. Based on these findings, the non-polar nature of lipids can favor their preservation over time. Furthermore, the presence of saturated fatty acids was almost double than of unsaturated ones: according to Rogoz et al.12, this behavior can be ascribed to the reduced oxidative stability of unsaturated compounds, thus leading to an easier degradation. Eleven amino acids were detected: these compounds could also undergo modification over time; for example, the presence of tyramine can be related to the decarboxylation of tyrosine as a result of fermentation or bacterial decomposition processes. Tyramine was mostly detected in the dental calculus of Duke Alessandro Farnese. Considering that the duke spent most of his lifetime on the battlefields in Northern Europe, the presence of tyramine could be related to the consumption of long-life foods, such as seasoned cheese, dried or smoked herring, venison, cured meats and fermented beverages25.
Among the compounds potentially ascribable to the Duke’s eating habits, cucurbitin and catechin 5-O-beta-d-glucopyranoside-4'-Me could be related to the consumption of typical foods present on the tables of Northern Europe such as Cucurbitaceae and rhubarb26,27. By contrast, 3-hydroxy-3-methyloxindole, imidazoleacetic acid riboside, 2-(methylamino)-1-phenylethanol and arginyl hydroxyproline can be related to the eating habits of Maria D’Aviz: in fact, according to Human Metabolome DataBase and Food DataBase (FooDB), these compounds could be considered potential biomarkers for the consumption of anatidaes, chickens, and domestic pigs and it is known that the Maria D’Aviz used poultry as main ingredient of several recipes described in her famous cookbook28.
Among metabolites commonly found in spices and tea, salicylaldehyde, DAG(32:4), capsiamide, estragole, phenylpyridines and eugenol quinone methide (EQM) were some of the compounds detected mostly in the dental calculus of Maria D’Aviz. The consumption of these foods is not surprising: Maria D’Aviz was a member of the Portuguese court, and the spices, which are frequently cited in the cookbook of the noblewoman, could easily be imported from both the Azores, the most important colony of the Portuguese empire and Asia29.
As for salicylates, they are contained in herbs, spices and tea30 and it is known that extracts of plants rich in salicylates, owing to their anti-inflammatory and anti-pyretic effects, have been used in ancient time to treat different human diseases31.
The presence of EQM, an oxidation derivative of eugenol32,33 can be explained not only by considering the dietary intake of spices such as cloves, nutmeg and cinnamon34, but also the anti-inflammatory and analgesic effects of eugenol-based remedies35, which have been used to treat toothache, a pathology that has affected Maria D’Aviz. However, the presence of EQM could also be ascribed to the use of essences for embalming, a practice used by the nobles in the sixteenth century36,37. Similarly, the presence of other metabolites deriving from fruits and citrus like hesperidin, neohesperidin, narirutin, naringin chalcone, dihydroxycitracridone I, amphibine H, which were observed in the dental calculus of the noble couple, could be explained taking into account both food consumption and their use as perfumes and essences used in the embalming practice.
With regard to the oral microbiome, some of the compounds detected can also be ascribed to the metabolism of fungi and bacteria. An interesting result is related to the presence of the TMC-34 metabolite deriving from Actinomyces, which are involved in the development of periodontitis38–40. This metabolite was mainly present in the dental calculus of Maria D’Aviz, and it is known that the noble woman suffered from caries and periodontal diseases24.
Additional metabolites like the glycerophospholipids PIM1(37:3), LPIM4(18:2) and LPIM4(19:2) related to the bacterium Mycobacterium tuberculosis were also identified41. After his death, the autopsy of Duke Alessandro Farnese revealed the presence of lung diseases42 further confirmed by the results achieved during exhumation in 202043, thus suggesting pneumonia, a widespread disease in the fifteenth and sixteenth centuries44,45.
It has also to be highlighted the presence of different metabolites of Saccharomyces cerevisiae, namely C16 phytosphingosine, C20 phytosphingosine, MIPC 42:0;O3, MIPC 40:0;O2, PI-Cer(d46:0)46, which can be related to bread baking and fermentation processes associated with winemaking and brewing47,48. Finally, the presence of other metabolites like penipacid B (2-[[N-(2-methoxy-2-methylpropyl)-C-methylcarbonimidoyl]amino]benzoic acid), versixanthone E, asperversin G or 2,4,6,8-tetramethyl-3,4-dihydroxydec-8(9)-enolide, deriving from the fungi Penicillium paneum, Aspergillus versicolor and Botrytis cinerea, can be related to the eating habits of the noble couple. Being able to contaminate cereals, oilseeds and nuts, these molds are mainly involved in the spoilage of bread49–51.These findings suggest that also the members of upper classes living in the Renaissance consumed dried and poorly preserved bread.
Conclusions
Untargeted metabolomics using UHPLC coupled to Waters Synapt G2-Si HDMS system was successfully applied to analyze the metabolome of the dental calculus of people living in ancient time. Concerning the untargeted metabolomics of Duke Alessandro Farnese and his wife Maria D’Aviz, the ability to efficiently separate the components of such complex samples combined with data independent acquisition by high resolution MSE permits to maximize the information obtained from the samples investigated by recording both low-energy and high-collision energy profiles related to all compounds in a single run. UHPLC-HRMS followed by Principal Component Analysis was successfully used for differentiation of the samples of the noble couple allowing the identification of more than 200 metabolites able to provide novel insights on the dietary habits and health status of Duke Alessandro Farnese and his wife. From the result of metabolomics analysis, it suggested that combination of LC-HRMS with PCA offers a powerful analytical technique to differentiate dental calculus in ancient specimens and identify metabolites markers, which have essential roles in samples differentiation, According to the forensics analysis of the remains, the presence of metabolites related to Mycobacterium tuberculosis can be considered a valuable step to clarify the causes of the duke’s death, while the identification of compounds deriving from the consumption of food confirmed the different lifestyles of the noble couple.
Materials and methods
Chemicals and materials
LC–MS grade water, acetonitrile, methanol and formic acid were purchased from Honeywell Burdick & Jackson (Charlotte, NC, USA). Leucine enkephalin standard was obtained from the Waters TOF G2-S Sample Kit-1 (Waters, Milford, MA, USA).
Dental calculus collection and metabolite extraction
Historic dental calculus was collected from the remains of Duke Alessandro Farnese and Maria D’Aviz after their exhumation in 2020 from the Basilica of Santa Maria della Steccata (Parma, Italy). Notary Dr. Marco Micheli in Parma granted permission for the exhumation and for the subsequent analysis. During procedures of physical examination of teeth, the couple’s calculus was collected with the use of a sterile curette following the protocols to avoid cross-contamination, then it was stored in glass vials until analysis52,53. According to the guidelines for the identification of cases to be submitted to the Ethical Committee of the Area Vasta Emilia Nord (https://www.aou.mo.it/ComitatoEticoAVEN), no approval was required.
Modern dental calculus samples from four healthy donors to be used for assessing both the extraction and chromatographic conditions were obtained during professional oral hygiene sessions. According to the guidelines for the identification of cases to be submitted to the Ethical Committee of the Area Vasta Emilia Nord (https://www.aou.mo.it/ComitatoEticoAVEN), no approval was required. Sample collection was performed in accordance with the Declaration of Helsinki. Informed consent was obtained from the donors.
After collection, both historical and modern dental calculus samples were frozen at -80 °C until metabolite extraction. Sample decalcification was performed according to Velsko et al.23 with slight modification: the samples (20 mg) were gently powdered in an agate mortar, placed in sterile glass vials and decalcified with 100 μL of 4% (v/v) formic acid in water at 4 °C for 18 days performing sonication at regular intervals. Samples to be submitted to UHPLC-HRMS analysis in ESI− mode were neutralized by the addition of 15 µL of a 5 M ammonium hydroxide solution. Finally, 300 µL of an acetonitrile/methanol solution 1:1 (v:v) were added to the samples, which were centrifuged at 12,000×g at 4 °C for 30 min. The supernatant was then collected and submitted to the UHPLC-HRMS analysis. A sample obtained by mixing the extracts of both Duke Alessandro Farnese and his wife was used as quality control sample.
Ultra-high performance liquid chromatography quadrupole high-resolution mass spectrometry
Chromatographic separation was performed on a binary Acquity UHPLC I-Class system (Waters) using an Atlantis™ Premier BEH™ C18 AX 1.7 μm (2.1 × 100 mm) column (Waters), thermostated at 40 °C. Mobile phase consisted of water (solvent A) and acetonitrile (solvent B), both containing 0.1% (v/v) formic acid. The flow rate was 0.4 mL/min and injection volume was 8 μL. A multistep linear gradient elution was carried out under the following conditions: solvent B was set at 2% for 1 min, followed by a linear gradient to 60% within 6 min, then to 95% in 1.5 min, maintained for 1.5 min before column re-equilibration (5 min). Elution was performed within 10 min.
HRMS analyses were performed on a Synapt G2-Si HDMS QTOF mass spectrometer (Waters SpA, Milan, Italy) with an electrospray ionization (ESI) Zspray™ (Waters) both in positive (PI) and negative (NI) ion modes. Mass correction during chromatographic runs was performed using leucine-enkephalin solution (50 ng/mL in acetonitrile/water, 50:50 (v/v) with 0.1% formic acid) as lock mass, by infusing it through the LockSpray™ system (Waters) with a 15 s interval. The experiments were conducted in resolution mode (20000 FWHM). Operating parameters were as follows: capillary voltage, 0.80 and 0.50 kV in ESI+ and ESI− respectively; cone voltage, 50 V; source temperature, 150 °C; source offset 80 V; desolvation temperature, 600 °C; cone gas, 50 L/h; desolvation gas, 1000 L/h; nebulizer pressure 6.5 bar. Spectra were acquired operating in data independent MSE acquisition mode by applying 5 V as collision energy for the low energy profile and using a collision energy ramp from 25 to 45 V for the high energy profile.
The UHPLC-HRMS data were recorded in *.raw files by using the MassLynx (v4.2) software (Waters).
Data analysis
Data analysis was performed by processing the .raw data in Progenesis QI software (Waters, Milford, MA, USA). The software allows for data visualization analysis, creation of 2D maps and the analysis of chromatograms and spectra. The program provides auto-alignment of signals, peak peaking, deconvolution and normalization54,55. The following adducts were considered: [M+H]+, [M+Na]+, [M+K]+, [M+NH4]+, [M+H2O+H]+, [M−H2O+H]+, [M+2H]2+, [M+2Na]2+, [M+2Na−H]+, [2M+H]+, [2M+Na]+, [M+H+Na]2+ in PI, and [M−H]−, [M+H2O−H]−, [M−H2O−H]−, [M+HCOO]−, [2M−H]−, [M+Na−2H]−, [M+K−2H]− in NI.
The data were filtered by setting a maximum intra-group variability of 10%, a power analysis value > 0.8 and a minimum fold change of 3 compared to method blank.
PCA was performed to explore the data set and to obtain the features able to differentiate samples belonging to Duke Alessandro Farnese and Maria D’Aviz. Compounds identification was performed by comparing the spectra with those stored in different ChemSpider online libraries, namely the HMDB, the FooDB, the E. coli Metabolome Database, the Yeast Metabolome Database, LipidMAPS, NPAtlas e KEGG Database, using a mass error tolerance of 5 ppm for precursor ions and 10 ppm for fragment ions.
Lipid nomenclature
The LIPID MAPS® glycerophospholipid abbreviations (PC, PE, etc.) are used here to refer to the identified analytes.
Supplementary Information
Acknowledgements
The cost of the Progenesis QI software used for this experimental investigation was supported by the Ministry of University and Research (Ministerial Decree no. 737 of 25/06/2021) and co-funded by the University of Parma through the “Scientific Instrumentation Upgrade Programme 2022”. This work has benefited from the equipment and framework of the COMP-HUB Initiative, funded by the ‘Departments of Excellence’ program of the Italian Ministry for Education, University and Research (MIUR, 2018–2022).
Author contributions
Conceptualization: F.B., N.R., M.Ma.; Investigation: N.R., F.B., M.Ma., M.P., M.Me.; Methodology: N.R., F.B., M.Ma., M.C.; Writing—original draft: N.R., F.B.; Writing—review & editing: N.R., F.B., M.Ma., M.P., M.Me., M.C.
Data availability
The datasets used and analyzed during the current study are available from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Nicolo’ Riboni, Email: nicolo.riboni@unipr.it.
Federica Bianchi, Email: federica.bianchi@unipr.it.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-36177-2.
References
- 1.Hillson S. Dental Anthropology. Cambridge University Press; 1996. [Google Scholar]
- 2.Driessens FMC, Verbeeck RMH. Recent Advances in the Study of Dental Calculus. In: Cate JM, editor. Proceedings of a Workshop 6-9 November, 1988 Noordwijkerhout, The Netherlands. Oxford University Press; 1988. pp. 7–17. [Google Scholar]
- 3.Larsen MJ, Pearce EIF. Saturation of human saliva with respect to calcium salts. Arch. Oral. Biol. 2003;48:317–322. doi: 10.1016/S0003-9969(03)00007-4. [DOI] [PubMed] [Google Scholar]
- 4.Warinner C, et al. Pathogens and host immunity in the ancient human oral cavity. Nat. Genet. 2014;46:336–344. doi: 10.1038/ng.2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Radini A, Nikita E, Buckley S, Copeland L, Hardy K. Beyond food: The multiple pathways for inclusion of materials into ancient dental calculus. Am. J. Phys. Anthropol. 2017;162:71–83. doi: 10.1002/ajpa.23147. [DOI] [PubMed] [Google Scholar]
- 6.Jersie-Christensen RR, et al. Quantitative metaproteomics of medieval dental calculus reveals individual oral health status. Nat. Commun. 2018;9:4744. doi: 10.1038/s41467-018-07148-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eerkens JW, et al. Dental calculus as a source of ancient alkaloids: Detection of nicotine by LC-MS in calculus samples from the Americas. J. Archaeol. Sci. Rep. 2018;18:509–515. [Google Scholar]
- 8.MacKenzie L, Speller CF, Holst M, Keefe K, Radini A. Dental calculus in the industrial age: Human dental calculus in the Post-Medieval period, a case study from industrial Manchester. Quatern. Int. 2021 doi: 10.1016/j.quaint.2021.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lustmann J, Lewin-Epstein J, Shteyer A. Scanning electron microscopy of dental calculus. Calcif. Tissue Res. 1976;21:47–55. doi: 10.1007/BF02547382. [DOI] [PubMed] [Google Scholar]
- 10.Fialová D, Drozdová E, Skoupý R, Mikulík P, Klíma B. Scanning electron microscopy of dental calculus from the great moravian necropolis Znojmo-Hradiště. Anthropologie. 2017;55:343–351. [Google Scholar]
- 11.Hardy K, et al. Dental calculus reveals potential respiratory irritants and ingestion of essential plant-based nutrients at Lower Palaeolithic Qesem Cave Israel. Quatern. Int. 2016;398:129–135. doi: 10.1016/j.quaint.2015.04.033. [DOI] [Google Scholar]
- 12.Rogóż J, Podbielska M, Szpyrka E, Wnuk M. Characteristics of dietary fatty acids isolated from historic dental calculus of the 17th- and 18th-century inhabitants of the Subcarpathian region (Poland) Molecules. 2021;26:2951. doi: 10.3390/molecules26102951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fotakis AK, et al. Multi-omic detection of Mycobacterium leprae in archaeological human dental calculus. Philos. Trans. R. Soc. B. 2020;375:20190584. doi: 10.1098/rstb.2019.0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Munekata PES, et al. Application of metabolomics to decipher the role of bioactive compounds in plant and animal foods. Curr. Opin. Food Sci. 2022;46:100851. doi: 10.1016/j.cofs.2022.100851. [DOI] [Google Scholar]
- 15.Pezzatti J, et al. Implementation of liquid chromatography–high resolution mass spectrometry methods for untargeted metabolomic analyses of biological samples: A tutorial. Anal. Chim. Acta. 2020;1105:28–44. doi: 10.1016/j.aca.2019.12.062. [DOI] [PubMed] [Google Scholar]
- 16.Nakabayashi R, Saito K. ScienceDirect Higher dimensional metabolomics using stable isotope labeling for identifying the missing specialized metabolism in plants. Curr. Opin. Plant Biol. 2020;55:84–92. doi: 10.1016/j.pbi.2020.02.009. [DOI] [PubMed] [Google Scholar]
- 17.Chen Y, Li E-M, Xu L-Y. Guide to metabolomics analysis: A bioinformatics workflow. Metabolites. 2022;12:357. doi: 10.3390/metabo12040357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Letertre MPM, Giraudeau P, de Tullio P. Nuclear magnetic resonance spectroscopy in clinical metabolomics and personalized medicine: Current challenges and perspectives. Front. Mol. Biosci. 2021;8:1–25. doi: 10.3389/fmolb.2021.698337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhong AB, et al. Multiplatform metabolomics studies of human cancers with NMR and mass spectrometry imaging. Front. Mol. Biosci. 2022;9:1–12. doi: 10.3389/fmolb.2022.785232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ortmayr K, Causon TJ, Hann S, Koellensperger G. Increasing selectivity and coverage in LC-MS based metabolome analysis. TrAC Trends Anal. Chem. 2016;82:358–366. doi: 10.1016/j.trac.2016.06.011. [DOI] [Google Scholar]
- 21.Mattarozzi M, et al. Reversed-phase and weak anion-exchange mixed-mode stationary phase for fast separation of medium-, long- and very long chain free fatty acids by ultra-high-performance liquid chromatography-high resolution mass spectrometry. J. Chromatogr. A. 2021;1648:462209. doi: 10.1016/j.chroma.2021.462209. [DOI] [PubMed] [Google Scholar]
- 22.Kaufmann A. Combining UHPLC and high-resolution MS: A viable approach for the analysis of complex samples? TrAC, Trends Anal. Chem. 2014;63:113–128. doi: 10.1016/j.trac.2014.06.025. [DOI] [Google Scholar]
- 23.Velsko IM, et al. The dental calculus metabolome in modern and historic samples. Metabolomics. 2017;13:134. doi: 10.1007/s11306-017-1270-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peracchia M, et al. Oral status of a noble European couple from the 16th century: A morphologic analysis of the teeth of Alessandro Farnese and his wife Maria D’Aviz. Anthropol. Anz. 2022;79:69–81. doi: 10.1127/anthranz/2021/1423. [DOI] [PubMed] [Google Scholar]
- 25.Andersen G, Marcinek P, Sulzinger N, Schieberle P, Krautwurst D. Food sources and biomolecular targets of tyramine. Nutr. Rev. 2019;77:107–115. doi: 10.1093/nutrit/nuy036. [DOI] [PubMed] [Google Scholar]
- 26.Blagrove RJ, Lilley GG. Characterisation of cucurbitin from various species of the cucurbitaceae. Eur. J. Biochem. 1980;103:577–584. doi: 10.1111/j.1432-1033.1980.tb05982.x. [DOI] [PubMed] [Google Scholar]
- 27.Nonaka G-I, Ezaki E, Hayashi K, Nishioka I. Flavanol glucosides from rhubarb and Rhaphiolepis umbellata. Phytochemistry. 1983;22:1659–1661. doi: 10.1016/0031-9422(83)80105-8. [DOI] [Google Scholar]
- 28.O livro de cozinha da Infanta D. Maria de Portugal: Primeira edição integral do codice português 1. E 33 da Biblioteca Nacional de Napoles. (Offic. da grafica de Coimbra, 1967).
- 29.Meilink-Roelofsz MAP. Asian Trade and European Influence. Springer; 1962. [Google Scholar]
- 30.Swain AR, Dutton SP, Truswell AS. Salicylates in foods. J. Am. Diet Assoc. 1985;85:950–960. doi: 10.1016/S0002-8223(21)03743-3. [DOI] [PubMed] [Google Scholar]
- 31.Duthie GG, Wood AD. Natural salicylates: Foods, functions and disease prevention. Food Funct. 2011;2:515. doi: 10.1039/c1fo10128e. [DOI] [PubMed] [Google Scholar]
- 32.Thompson D, et al. Peroxidase-catalyzed oxidation of eugenol: Formation of a cytotoxic metabolite(s) J. Biol. Chem. 1989;264:1016–1021. doi: 10.1016/S0021-9258(19)85046-9. [DOI] [PubMed] [Google Scholar]
- 33.Bolton JL, Dunlap T. Formation and biological targets of quinones: Cytotoxic versus cytoprotective effects. Chem. Res. Toxicol. 2017;30:13–37. doi: 10.1021/acs.chemrestox.6b00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chaieb K, et al. The chemical composition and biological activity of clove essential oil, Eugenia caryophyllata (Syzigium aromaticum L. Myrtaceae): A short review. Phytother. Res. 2007;21:501–506. doi: 10.1002/ptr.2124. [DOI] [PubMed] [Google Scholar]
- 35.Chung G, Oh SB. Eugenol as local anesthetic. In: Ramawat KG, Mérillon J-M, editors. Natural Products. Springer; 2013. pp. 4001–4015. [Google Scholar]
- 36.Azeredo CMO, Soares MJ. Combination of the essential oil constituents citral, eugenol and thymol enhance their inhibitory effect on Crithidia fasciculata and Trypanosoma cruzi growth. Rev. Bras. 2013;23:762–768. [Google Scholar]
- 37.Corbineau R, et al. Plants and aromatics for embalming in Late Middle Ages and modern period: A synthesis of written sources and archaeobotanical data (France, Italy) Veg. Hist. Archaeobot. 2018;27:151–164. doi: 10.1007/s00334-017-0620-4. [DOI] [Google Scholar]
- 38.Pujic P, Beaman BL, Ravalison M, Boiron P, Rodríguez-Nava V. Nocardia and actinomyces. In: Ram R, editor. Molecular Medical Microbiology. Elsevier; 2015. pp. 731–752. [Google Scholar]
- 39.Moore WEC, Moore LVH. The bacteria of periodontal diseases. Periodontology. 1994;2000(5):66–77. doi: 10.1111/j.1600-0757.1994.tb00019.x. [DOI] [PubMed] [Google Scholar]
- 40.Kaldas M, Barghorn A, Schmidlin PR. Actinomycosis as a rare local manifestation of severe periodontitis. Case Rep. Dent. 2020;2020:1–7. doi: 10.1155/2020/5961452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sartain MJ, Dick DL, Rithner CD, Crick DC, Belisle JT. Lipidomic analyses of Mycobacterium tuberculosis based on accurate mass measurements and the novel “Mtb LipidDB”. J. Lipid Res. 2011;52:861–872. doi: 10.1194/jlr.M010363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Follini, N. Alessandro Farnese III Duca di Parma e Piacenza : 1545–1592. (Tip. A. Bellocchio, 1932).
- 43.Zaniboni Mattioli, A. et al. Alessandro Farnese. Il corpo del potere, un caso irrisolto del Rinascimento. (Grafiche Step, 2022).
- 44.Bates JH, Stead WW. The history of tuberculosis as a global epidemic. Med. Clin. North Am. 1993;77:1205–1217. doi: 10.1016/S0025-7125(16)30188-2. [DOI] [PubMed] [Google Scholar]
- 45.Hatcher J. Mortality in the fifteenth century: Some new evidence. Econ. Hist. Rev. 1986;39:19. doi: 10.2307/2596099. [DOI] [PubMed] [Google Scholar]
- 46.Guan XL, Wenk MR. Mass spectrometry-based profiling of phospholipids and sphingolipids in extracts fromSaccharomyces cerevisiae. Yeast. 2006;23:465–477. doi: 10.1002/yea.1362. [DOI] [PubMed] [Google Scholar]
- 47.Lahue C, Madden AA, Dunn RR, Smukowski Heil C. History and domestication of Saccharomyces cerevisiae in bread baking. Front. Genet. 2020;11:584718. doi: 10.3389/fgene.2020.584718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cavalieri D, McGovern PE, Hartl DL, Mortimer R, Polsinelli M. Evidence for S. cerevisiae fermentation in ancient wine. J. Mol. Evol. 2003;57:S226–S232. doi: 10.1007/s00239-003-0031-2. [DOI] [PubMed] [Google Scholar]
- 49.Hocking AD. Aspergillus and related teleomorphs. In: Ram R, editor. Food Spoilage Microorganisms. Elsevier; 2006. pp. 451–487. [Google Scholar]
- 50.Saranraj P, Sivasakthivelan P, et al. Microorganisms involved in spoilage of bread and its control measures. In: Rossel CM, et al., editors. Bread and its Fortification: Nutrition and Health Benefits. CRC Press; 2015. pp. 132–149. [Google Scholar]
- 51.Hamdache A, Ezziyyani M, Lamarti A. Study of growth and production of botrytis cinerea conidia of some Morrocan isolates in different nutrients media. Adv. Intell. Syst. Comput. 2019;911:62–68. [Google Scholar]
- 52.Weyrich LS, Dobney K, Cooper A. Ancient DNA analysis of dental calculus. J. Hum. Evol. 2015;79:119–124. doi: 10.1016/j.jhevol.2014.06.018. [DOI] [PubMed] [Google Scholar]
- 53.Warinner C, Speller C, Collins MJ. A new era in palaeomicrobiology: Prospects for ancient dental calculus as a long-term record of the human oral microbiome. Philos. Trans. R. Soc. B. 2015;370:20130376. doi: 10.1098/rstb.2013.0376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang J, et al. The profiling of the metabolites of hirsutine in rat by ultra-high performance liquid chromatography coupled with linear ion trap Orbitrap mass spectrometry: An improved strategy for the systematic screening and identification of metabolites in multi-samp. J. Pharm. Biomed. Anal. 2017;134:149–157. doi: 10.1016/j.jpba.2016.11.034. [DOI] [PubMed] [Google Scholar]
- 55.Progenesis QI | Waters. https://www.waters.com/waters/en_US/Progenesis-QI/nav.htm?locale=en_US&cid=134790652
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author upon reasonable request.