Abstract
Soft-tissue sarcomas (STS) are a rare and heterogeneous group of tumors that arise from connective tissue and can occur anywhere in the body. Among the plethora of over 50 different STS types, liposarcoma (LPS) is one of the most common. The subtypes of STS are characterized by distinct differences in tumor biology that drive responses to pharmacologic therapy and disparate oncologic outcomes. Small non-coding RNAs (sncRNAs) are a heterogeneous class of regulatory RNAs involved in the regulation of gene expression by targeting mRNAs. Among the several types of sncRNAs, microRNAs and tRNA-derived ncRNAs are the most studied in the context of tumor biology, and we are learning more about the role of these molecules as important regulators of STS tumorigenesis and differentiation. However, challenges remain in translating these findings, and no biomarkers or therapeutic approaches targeting sncRNAs have been developed for clinical use. In this review, we summarize the current landscape of sncRNAs in the context of STS with an emphasis on LPS, including the role of sncRNAs in the tumorigenesis and differentiation of these rare malignancies and their potential as novel biomarkers and therapeutic targets. Lastly, we provide an appraisal of published studies and outline future directions to study sncRNAs in STS, including tRNA-derived ncRNAs.
Introduction
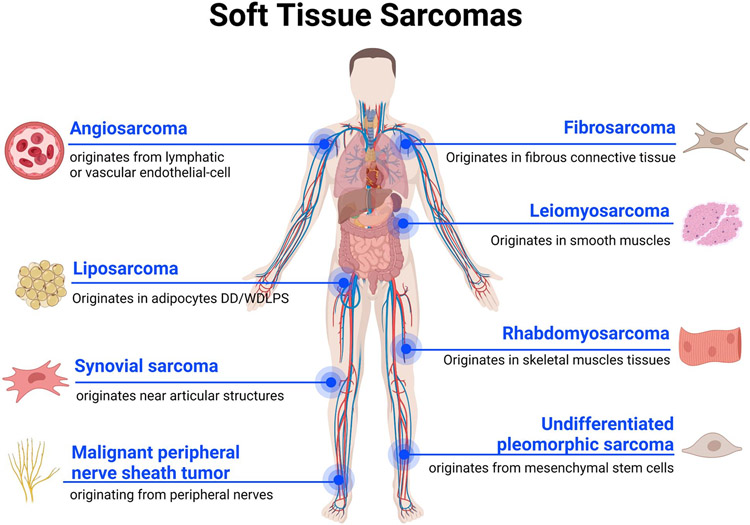
Soft-tissue sarcoma (STS) is a very heterogeneous group of tumors that accounts for 1% of all human malignancies. With approximately 12,000 new cases and 5,000 deaths in the United States annually, STS remains a lethal disease with very limited treatment options (1). More than 50 different subtypes of STS have been identified so far, with the most common types being liposarcoma (LPS), leiomyosarcoma (LMS), fibrosarcoma (FS), rhabdomyosarcoma (RMS), angiosarcoma (AS), malignant peripheral nerve sheath tumor (MPNST), undifferentiated pleomorphic sarcoma (UPS), and synovial sarcoma (SS) (2) (Figure 1). Surgical resection is the only curative approach to STS and response rates to chemotherapy and radiotherapy are less than 25% (3). Therefore, early detection of STS and the ability to predict response to systemic therapy may help to improve prognosis in this challenging patient population (3).
Figure 1.
Soft Tissue Sarcomas. Schematic representation of the main Soft Tissue Sarcoma histological subtypes and their originating tissues.
In this context, small non-coding RNAs (sncRNAs), a heterogeneous class of regulatory RNAs involved in gene expression, may play a pivotal role in orchestrating STS tumorigenesis and differentiation. Among them, microRNAs (miRNAs), a well-known class of small ncRNAs, have been studied in the setting of STS and may hold promise as novel biomarkers and therapeutic targets. However, studies are limited, and no clinical applications of sncRNAs have been reported for patients with STS. Herein we summarize the current landscape of miRNAs in STS, focusing on their roles in the tumorigenesis and differentiation of LPS, their application as novel biomarkers or therapeutic targets, and outline future directions in small ncRNA studies in STS involving tRNA-derived ncRNAs (tsRNAs).
Small non-coding RNAs
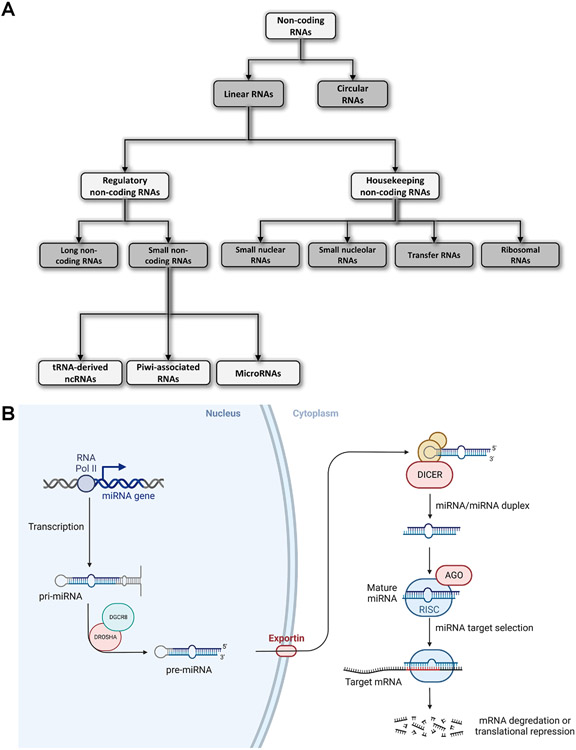
With the advent of Next Generation Sequencing (NGS) technologies, it became clear that only 2% of the human genome is transcribed into RNA messengers (mRNAs), while the rest is primarily transcribed into non-coding RNAs (ncRNAs). ncRNAs are a very heterogeneous group of different RNAs involved in multiple aspects of cell physiology (4). They are usually divided according to their size in small (< 200 nts) and long ncRNAs (≥ 200 nts) (4,5) and they can also be classified according to their function in housekeeping and regulatory ncRNAs (4,5) (Figure 2A). Housekeeping ncRNAs include small nuclear RNA (snRNA), small nucleolar RNA (snoRNA), ribosomal RNA (rRNA), and transfer RNA (tRNA) (Figure 2A). These ncRNAs are constitutively expressed in every cell type and perform several functions which are crucial for cellular physiology (4,5). On the other hand, regulatory ncRNAs include several classes of long and small RNAs, such as long non-coding RNAs (lncRNAs), miRNAs, Piwi-associated RNAs (piRNAs), and tsRNAs (4,5) (Figure 2A). The crucial roles these molecules play in regulating cell proliferation, differentiation, migration, and cell death have garnered significant interest from the scientific community (4,6-8). The expression of sncRNAs is altered in many human diseases, including cancer (4,9-12). Extracellular small ncRNAs are differentially expressed in several human cancers and have been introduced as novel diagnostic and prognostic biomarkers (4,13). To this end, sncRNAs are detectable in many different body fluids and complexed with proteins or enclosed within extracellular vesicles and are readily available (4,14).
Figure 2.
Non-coding RNA’s classification and microRNA biogenesis. A, Diagram showing the most common classification of the different classes of non-coding RNAs. B, Schematic representation of the microRNA biogenesis and their mechanism of action in regulating gene expression at the post-transcriptional level by targeting the 3’ Untranslated Region of specific mRNAs.
While the role that regulatory sncRNAs have in normal cellular physiology and the implications that dysregulation has in the development and progression of cancer is well recognized, additional studies are needed to fully understand the complex regulatory networks in which they are involved that ultimately govern these processes (15,16). Moreover, additional studies are required to translate these findings into novel applications that can be used in clinical practice. Concerning rare human tumors like STS, few studies have addressed the roles of sncRNAs in tumorigenesis and differentiation. Most studies have focused only on miRNAs, leaving other important classes, such as tsRNAs, unexplored (17).
miRNAs: biogenesis and dysregulation in cancer
miRNAs are a distinct family of sncRNAs which are around 16-26 nucleotides long and are involved in modulating gene expression at the post-transcriptional level (11). The biogenesis of miRNAs is quite articulate and starts in the nucleus, where miRNA genes are transcribed by the RNA polymerase II into a longer primary transcript named pri-miRNA, which has a 5′ cap and a 3′ poly-A tail (11). Noteworthy, some pre-miRNAs may also be transcribed by RNA polymerase III (18,19). After transcription, the pri-miRNAs are then processed into the nucleus by a complex that includes an RNA-binding protein and an RNase III named DGCR8 and Drosha, respectively, into an about 85 nucleotides stem-loop structured RNA named precursor miRNA (pre-miRNA) (20). Following this initial step of their biogenesis in the nucleus, pre-miRNAs are then transferred to the cytoplasm by the complex Ran/GTP/Exportin and processed by another RNase III called Dicer to create a miRNA duplex of 20-22 nucleotides long (20). Such duplex is then unwound, and the mature miRNA is finally incorporated into a protein complex named RNA-induced silencing complex (RISC), guiding it to target mRNAs (Figure 2B) (20). Most of the time, miRNA–mRNA interactions are mediated by the seed region, which is a 6-8 nucleotide sequence present at the 5′-end of the miRNA that binds for complementarity to the 3’ Untranslated Regions (UTR) of the mRNA target (21). However, other studies based on the CLASH-Seq technique have also identified additional non-canonical binding clusters which seem to be independent from the seed region (22). Independently from the interaction, once miRNAs bind to their mRNA targets, they can either inhibit translation or induce the degradation of the mRNA target in case of imperfect or perfect complementarity, respectively (23).
By degrading the mRNA targets or repressing their translation, miRNAs can deeply affect cellular processes, including proliferation, differentiation, and apoptosis (11). For these reasons, it is easy to think that dysregulation in the expression of such small RNA molecules may significantly affect the development of several human diseases, including cancer.
The first evidence of the involvement of miRNAs in cancer development was provided by Dr. Croce’s team from studies trying to identify novel tumor suppressors located in chromosome 13 in B-cells of patients affected by chronic lymphocytic leukemia (CLL) (24). During these studies, they found a region, frequently deleted in CLL, that, instead of containing a coding-protein gene, actually had two miRNA genes named miR-15a and miR-16-1. Further studies revealed that both miRNAs were either deleted or downregulated in the majority of CLL cases, and they were acting as tumor suppressors by targeting BCL-2, an anti-apoptotic protein overexpressed in many human cancers (25,26).
After these pioneering discoveries, the scientific community started an intense research activity to study miRNAs' role in cancers. Such studies shown that miRNA expression is affected in all human tumors, and its signatures might be used for tumor classification, diagnosis, and prognosis (11). Moreover, further studies also found that cells actively secreted miRNAs in all human biological fluids through EVs, acting as important mediators of cellular communication (27-29). Notably, their levels in biological fluids were altered in several human cancers suggesting a potential clinical application of these short RNA molecules as non-invasive diagnostic and prognostic biomarkers in oncology clinical practice (29-31). Despite the intense research, very few cases of miRNAs have been introduced in clinical practice for cancer diagnosis and treatment so far. Examples of miRNA-based cancer diagnostics already available for clinical practice include (i) miRview, a microarray panel of 64 miRNAs allowing the identification of cancers of unknown or uncertain primary origin (CUP) spanning across 42 different tumor types (32,33); (ii) ThyraMIR\ThyGeNEXT, a miRNA panel for thyroid and pancreatic cancer stratification (32,33); and (iii) RosettaGX Reveal panel which distinguishes between indeterminate or benign thyroid nodules (32,33). Concerning miRNA-based cancer treatments, many clinical trials are still ongoing. However, none of them have been approved by the FDA yet (32,33).
Despite these promising examples, additional studies and clinical trials will be required to introduce miRNAs in daily clinical practice. Surely, the cheaper cost of NGS-based transcriptome experiments combined with a better understanding of the roles of these molecules in cancer development and progression will help speed up this process. However, rare malignancies, such as LPS and other STS, remain under the shadow of other common tumor types, attracting less interest from biotechnological and pharmaceutical companies. Indeed, studies about miRNAs in STS are very limited, and other classes of sncRNAs, such as tsRNAs, have not been studied so far in these malignancies.
Liposarcoma
LPS account for about 20% of all STS and can occur in the extremities or in the retroperitoneum of the abdomen and is further classified into four subtypes: (i) myxoid, (ii) well differentiated, (iii) dedifferentiated, and (iv) pleomorphic (34).
Myxoid LPS is one form of LPS that typically arises from the lower extremities. Myxoid LPS is characterized by a reciprocal translocation that generates a fusion transcription factor oncogene (35,36). Specifically, a translocation t(12;16) is present in >95% of myxoid LPS cases, fusing FUS with DDIT3, while the remaining cases present with the t(12;22) translocation. The t(12;22) translocation results in the formation of EWSR1-DDIT3 (35,36). Both FUS and EWSR1 are members of the FET heterogeneous nuclear ribonucleoprotein particle (hnRNP) protein family, which are known to bind RNA and contribute to the regulation of transcription and translation, and RNA processing (37). Noteworthy, FET proteins have been reported to enhance transcription suggesting a role in tumor progression (37). While the exact mechanism is unknown, it is hypothesized that these fusion transcription factors may exert their oncogenic effect by stimulating cellular proliferation and, at the same time, inhibiting adipogenic differentiation (36,38,39). In addition to these molecular differences, myxoid LPS presents differences from the other types of LPS in some clinical aspects as well. In fact, myxoid LPS seems to have a tendency to metastasize to additional sites where fat tissue is present such as the trunk, extremities, bone, retroperitoneum, pleura, and pericardium (40,41). Surgical excision with or without radiation therapy is the indicated treatment for patients with localized myxoid LPS. However, chemotherapy is usually recommended for patients with high grades, tumor size > 5 cm, and positive surgical margins (35).
Well-differentiated liposarcoma (WDLPS) and dedifferentiated liposarcoma (DDLPS) are the most common subtypes of LPS (34). Within the retroperitoneum, WDLPS accounts for approximately 40–45% of all LPS and is characterized as a low-grade tumor with a local recurrence rate of 60% at five years (42,43). In contrast, DDLPS is a high-grade tumor and may arise as a primary lesion (90 % of cases) or from within a WDLPS (10 % of cases) (44-46). Like WDLPS, DDLPS commonly arise in the retroperitoneum and has a characteristic high rate of local recurrence (44,47). As many as 20% of patients with DDLPS will develop metastases, and patients with DDLPS have a worse prognosis compared to WDLPS (30% vs. 90% overall survival at five years) (1). Morphologically, WDLPS and DDLPS are remarkably different. WDLPS is composed of malignant adipocytes and spindle cells showing fibroblastic/myofibroblastic differentiation with aberrantly enlarged and hyperchromatic nuclei, whereas DDLPS cells are no longer recognizably adipocytic under the microscope (1). Although morphologic differences between WDLPS and DDLPS are evident, the molecular differences that drive their disparate behavior and prognosis are largely unknown. Surgery remains the only curative treatment for patients with LPS, but the majority of patients will manifest local recurrence, and the response rate to systemic therapy is only 30% (48). Taken together, this emphasizes the need to identify novel therapeutic targets for patients with LPS. Moreover, the discovery of reliable diagnostic and prognostic biomarkers, including genetic markers to better characterize LPS, may help improve the management and outcomes for patients with this rare malignancy.
Dysregulated miRNAs in liposarcoma
Among the different types of STS, the role of miRNAs in tumorigenesis and differentiation has been most studied in LPS. In fact, multiple miRNAs were found differentially expressed in WDLPS and DDLPS with distinct roles in the biology of these cancer cells (49). Significant strides have been made in discovering biologically relevant miRNAs in LPS, and a summary of dysregulated miRNAs in WDLPS and DDLPS is summarized in Table 1. One of the first studies assessing miRNA expression in LPS was published in 2011. In this study, the authors identified more than 40 differentially expressed miRNAs in DDLPS compared to normal fat. Among them, miR-143, which was found downregulated in DDLPS, was revealed to be particularly interesting since the re-expression of such miRNA inhibited proliferation and induced apoptosis in vitro (50). However, the dedifferentiated phenotype was not completely reversed by the increased expression of miR-143 (50). In any case, this study was the first to report the pivotal part played by miRNAs in modulating LPS development and progression.
Table 1. Dysregulated microRNAs in liposarcoma.
Table showing the most relevant microRNAs found differentially expressed in WD\DDLPS.
| microRNA | Expression | LPS subtype | Reference |
|---|---|---|---|
| miR-155 | up | DDLPS | Zhang P, et al. Cancer Res. 2012. |
| miR-26a-2 | up | WD/DDLPS | Lee DH, et al. Oncogenesis. 2013. |
| miR-143 | down | DDLPS | Ugras S, et al. Cancer Res. 2011. |
| miR-193b | down | DDLPS | Mazzu YZ, et al. Cancer Res. 2017. |
| miR-133a | down | DDLPS | Yu PY, et al. Cancer Cell Int. 2018. |
| miR-145 | down | WD/DDLPS | Gits CM, et al. Int J Cancer. 2014. |
| miR-451 | down | WD/DDLPS | Gits CM, et al. Int J Cancer. 2014. |
In another study of 59 patients with LPS, Zhang et al. identified four miRNAs that were overexpressed and 31 that were downregulated in WD/DDLPS that could be used to discriminate LPS from normal fat. One of the four miRNAs that were overexpressed in the DDLPS subset was miR-155. Moreso, the knockdown of miR-155 in DDLPS cell lines (lipo246 and LPS141) inhibited cellular proliferation, reduced colony formation, and induced cell-cycle arrest in vitro (51). Such results were confirmed with in vivo experiments using mouse xenograft models that showed a stalling of tumor growth induced by the knock-down of miR-155 (51). The overexpression of miR-155 in WD/DDLPS was also observed in two additional independent studies confirming the role of miR-155 in DDLPS progression (52,53).
miR-26a-2 is another important miRNA that is overexpressed in WD/DDLPS. This miRNA is located in a chromosomal region (12q13-q22), amplified in approximately 90% of WD/DDLPS tumors, and containing important oncogenes, such as MDM2 and CDK4 (54). In vitro, miR-26a-2 enhances the growth and survival of LPS cell lines and, when expressed, results in increased cell proliferation and migration, repressed adipocyte differentiation, and resistance to apoptosis by targeting the BTB domain containing protein 1 (RCBTB1) (54). A subsequent study from the same lab also revealed that miR-26a-2 regulates cell death by targeting another gene, homeobox protein A5 (HOXA5), which is important in adipocyte differentiation and fat metabolism (55). A recent small RNA-Seq profiling of DDLPS tumors found that miRNAs of the miR-193 family were downregulated compared with normal fat. Notably, the reintroduction of miR-193b induced apoptosis in liposarcoma cell lines and promoted adipogenesis in adipose-derived stem cells (56). Another study reported that miR-133a was found downregulated in DDLPS, and the re-expression of this miRNA induced a reduction in cell proliferation in vitro (57). However, no significant effects on cell death and migration were observed. Moreover, no differences in vivo were observed in DDLPS mouse xenografts overexpressing miR-133a (57).
In clinical practice, distinguishing WDLPS and DDLPS has important implications with respect to patient management. In this context, miRNAs could be used as molecular markers to help stratify LPS subtypes. In 2014, Gits, C.M.M. et al. identified a signature of miRNAs that discriminate between different LPS subtypes (58). More precisely, the expression of miR-145 and miR-451 were reduced in LPS compared to normal fat, and their re-expression in LPS cell lines decreased cellular proliferation, impaired cell cycle progression, and induced apoptosis, indicating these two miRNAs as novel tumor suppressors in liposarcoma (58). A more recent study reported a signature of 6 miRNAs able to discriminate lipomas from WDLPS with high confidence, while a different signature of 6 miRNAs was able to stratify DDLPS from their more aggressive histologic mimics (59). While promising, further external validation of these findings is necessary, and the utility of their clinical application is unknown.
microRNAs as novel therapeutic targets for liposarcoma
Studies have shown that miRNAs are dysregulated in WDLPS and DDLPS, and some of these are responsible for the development and progression by regulating the expression of key oncogenes and tumor suppressors at the post-transcriptional level. Indeed, miRNAs can be classified as oncomiRs or tumor suppressors based on their targets. OncomiRs are typically miRNAs that are overexpressed in cancer cells and downregulate the expression of their tumor suppressor target genes. In contrast, tumor suppressor miRNAs are downregulated in cancer cells and induce the upregulation of their target oncogenes. However, because of the complexity of the regulatory networks in which miRNAs are involved, defining a miRNA as an oncogene or tumor suppressor is challenging (60). In fact, a single miRNA can target multiple mRNA targets (oncogenes and tumor suppressors), while a single mRNA can be targeted by several miRNAs (60). Since miRNAs have been proven to be important regulators of key signaling pathways, the research community has spent time and effort trying to develop new strategies for miRNA-based cancer treatment. Moreover, working with sncRNAs as therapeutic targets has some biotechnological advantages. Indeed, miRNAs can be efficiently overexpressed, transfecting cells with miRNA mimics, or they can be downregulated using antagomirs which silence the function of their complementary miRNA targets (61). miRNA-based therapeutic approaches are not without issues. Problems with toxicity and delivery systems have hampered the use of miRNAs as novel cancer therapeutic targets (61,62). Although most of the studies and clinical trials about potential miRNA-based therapeutic applications are focused on more common tumor types, some studies have reported relevant miRNAs that might be used as therapeutic targets in WD/DDLPS.
miR-155 is an oncomiR in DDLPS that plays a vital role in cell cycle progression and cell proliferation as well as colony formation by regulating Casein Kinase-1α (CK-1α) and thereby increasing β-Catenin Signaling and cyclin D1 expression (51-53). In conclusion, the authors proposed miR-155 as a novel potential target for DDLPS treatment (51).
miR-26a-2 is another example of oncomiR, which has been found upregulated in WD/DDLPS (54). As anticipated above, miR-26a-2 is located in a chromosomal region (12q13-q22), usually amplified in 90% of WD/DDLPS, which leads to the overexpression of genes present in this region, such as MDM2 and CDK4 (54). Overexpression of miR-26a-2 in three different LPS cell lines (SW872, LPS141, and LP6) boosted the growth and improved the survival, and the migration of these cells, while inhibiting adipocyte differentiation and apoptosis by post-transcriptionally regulating RCBTB1 (54). Moreover, the inhibition of miR-26a-2 in LPS cells using antagomiRs resulted in the opposite phenotype confirming the role of this miRNA in regulating such processes (54). Another study also revealed that miR-26a indirectly affects p53-independent and caspase-dependent apoptosis by negatively regulating the expression of HOXA5 (55). Indeed, inhibition of miR-26a-2 using its antagomiR or overexpressing HOXA5 induced a strong apoptotic response in LPS cells (55). After these findings, the authors concluded that the oncomiR miR-26a might be a novel therapeutic target for human LPS (54).
Contrary to oncomiRs, other miRNAs might be downregulated in WD/DDLPS, acting as tumor suppressors. The miR-143/miR-145 cluster comprises two distinct and co-transcribed miRNAs well-known for their tumor suppressor activities (63). This cluster is present abundantly in adipose tissues, and among them, miR-143 also takes part in the tumorigenesis of LPS and the differentiation of adipocytes (64). In LPS, this cluster is barely expressed, and re-expression of miR-143 inhibited proliferation and induced apoptosis by regulating the expression levels of BCL2, type II topoisomerase, PLK1, and PRC1 (50). These findings suggest that the re-expression of miR-143 or the use of inhibitors directed at miR-143 targets may have therapeutic value in DDLPS (50).
Even miR-145 and miR-451 are reported to be poorly expressed in liposarcoma (58). The authors showed that overexpressing miR-145 and miR-451 in LPS cell lines decreased cellular proliferation, inhibited cell cycle progression, and induced apoptosis (58). These data suggest that miR-145 and miR-451 may act as tumor suppressors, and their re-expression might be used as a novel therapeutic approach for LPS (58).
Another final example of tumor suppressor miRNA in LPS is miR-193b. This miRNA is under expressed in DDLPS compared with normal fat, and its expression seems to regulate important phenotypes such as adipogenesis in human adipose-derived stem cells by targeting genes involved in the FAK–SRC–CRKL signaling. It is also reported that miR-193b mediates liposarcoma cell proliferation, differentiation, and tumorigenesis by targeting multiple genes such as PDGFR, TGFβ, SMAD4, and YAP1 (65). Moreover, in vivo experiments showed that miR-193b mimetics and FAK inhibitor (PF-562271) could inhibit LPS xenograft growth, suggesting their possible application for DDLPS treatment (56). A summary of some relevant oncomiRs and tumor suppressor miRNAs described here can be found in Table 2. Although all the aforementioned miRNAs hold the promise of new effective treatments for patients affected by LPS, additional studies will be required to confirm these findings and introduce them in clinical practice. Indeed, further in vivo experiments to confirm the therapeutic properties of these miRNAs need to be performed. Nevertheless, the implementation of more safe and effective delivery systems of miRNA-mimics and antagomiR to cancer cells needs to be developed to make miRNAs more attractive druggable targets.
Table 2. OncomiR and tumor-suppressor microRNAs in liposarcoma.
Table reporting the most relevant microRNAs acting either as oncogene or tumor-suppressor in the context of liposarcoma with their target genes and regulated biological functions.
| microRNA | microRNA type | Targets | Regulated function | LPS subtype | Reference |
|---|---|---|---|---|---|
| miR-155 | oncomiR | CK-1α | Cell cycle | DDLPS | Zhang P, et al. Cancer Res. 2012. |
| miR-26a-2 | oncomiR | RCBTB1, HOXA5 | Cell proliferation, Apoptosis, Migration, Cellular differentiation | WD/DDLPS | Lee DH, et al. Oncogenesis. 2013 Lee DH, et al. Sci Rep. 2015. |
| miR-143 | Tumor-suppressor | BCL2, PLK1, PRC1 | Cell proliferation, Apoptosis, | DDLPS | Ugras S, et al. Cancer Res. 2011. |
| miR-145 | Tumor-suppressor | - | Cell cycle, Cell proliferation, Apoptosis | WD/DDLPS | Gits CM, et al. Int J Cancer. 2014. |
| miR-451 | Tumor-suppressor | - | Cell cycle, Cell proliferation, Apoptosis | WD/DDLPS | Gits CM, et al. Int J Cancer. 2014. |
| miR-193b | Tumor-suppressor | FAK, SRC, CRKL, PDGFR, TGFβ, SMAD4, YAP1 | Cellular differentiation, Cell proliferation, Apoptosis | DDLPS | Gits CM, et al. Int J Cancer. 2014. Mazzu YZ, et al. Sci Rep. 2019. |
Circulating microRNAs as novel diagnostic biomarkers of WD/DDLPS
In the last decade, in addition to developing effective miRNA-based therapeutic approaches for several different types of human cancers, the scientific community has also turned its attention to the potential application of miRNAs as novel diagnostic and prognostic biomarkers to be used in clinical practice. Indeed, miRNAs have become attractive biomarkers given their easily detectable presence in plasma and other biological fluids such as urine and saliva. Moreover, several studies have shown unique signatures of miRNAs in the plasma or serum of patients affected by different types of malignancies, indicating their potential clinical value as novel diagnostic biomarkers (1). Currently, it is hypothesized that miRNAs enter into body fluids with three different mechanisms: (i) passive release from damaged cells, (ii) active secretion through extracellular vesicles, and (iii) active secretion in macromolecular complexes associated with AGO proteins and high-density lipoprotein (HDL) (66). Even though the biological functions of circulating miRNAs are mainly unknown, it is postulated that they may act as “hormones” implicated in cellular communication affecting, therefore, several aspects of cellular physiology (67). However, the possibility of circulating miRNAs as novel biomarkers for monitoring the stage and diagnosis of STS has yet to be extensively considered.
In this context, a recent study using LPS cell lines and plasma from patients with LPS, found that miR-25-3p and miR-92a-3p were secreted in extracellular vesicles. Interestingly, the authors were able to distinguish between patients with LPS and healthy controls (68). These results suggested that these two circulating miRNAs can effectively be used as a novel, noninvasive biomarkers for the diagnosis of LPS (68). From a mechanistic standpoint, both miR-25-3p and miR-92a-3p were shown to stimulate the secretion of proinflammatory cytokine IL-6 from tumor-associated macrophages in a TLR7/8-dependent manner within the tumor microenvironment. The end result was the promotion of LPS cell proliferation, invasion, and metastasis. These findings provided a novel and previously unreported mechanism by which LPS progression is regulated in part by miRNAs through manipulation of the tumor microenvironment (68).
Finally, a recent study showed that some miRNAs, such as miR-1246, miR-4532, miR-4454, miR-619-5p, and miR-6126, were highly expressed in serum and tissues from patients with DDLPS (69). Moreover, in vitro experiments also suggested their secretion from DDLPS cells through exosomes (69). The authors concluded that these miRNAs might serve as potential biomarkers for DDLPS diagnosis. However, further validations with a larger cohort of patients and additional in vitro and in vivo experiments about the functional activities of these miRNAs will be required. A summary of the circulating miRNAs with potential diagnostic applicability in WD/DDLPS can be found in Table 3.
Table 3. Dysregulated circulating microRNAs in liposarcoma.
Table showing the most relevant circulating microRNAs found differentially expressed in the biological fluids of patients affected by liposarcoma.
| microRNA | Expression | Bio fluid | LPS subtype | Reference |
|---|---|---|---|---|
| miR-25-3p | Up | Plasma | WD/DDLPS | Casadei L, et al. Cancer Res. 2017 |
| miR-92a-3p | Up | Plasma | WD/DDLPS | Casadei L, et al. Cancer Res. 2017 |
| miR-1246 | Up | Serum | DDLPS | Kohama I, et al. Oncol Lett. 2021. |
| miR-4532 | Up | Serum | DDLPS | Kohama I, et al. Oncol Lett. 2021. |
| miR-4454 | Up | Serum | DDLPS | Kohama I, et al. Oncol Lett. 2021. |
| miR-619-5p | Up | Serum | DDLPS | Kohama I, et al. Oncol Lett. 2021. |
| miR-6126 | Up | Serum | DDLPS | Kohama I, et al. Oncol Lett. 2021. |
Dysregulated microRNAs in other types of soft tissue sarcoma
In addition to LPS, several relevant papers about miRNAs in other STS subtypes have been published. Herein, we briefly described the miRNAs that are dysregulated in these additional STS subtypes, including LMS, FS, SS, RMS, AS, and MPNST, also commenting on their biological properties and potential clinical applicability. Moreover, a summary can be found in Table 4.
Table 4. Dysregulated microRNAs in other Soft Tissue Sarcomas.
Tables reporting the most relevant microRNAs found differentially expressed in several different histological types of Soft Tissue Sarcoma (excluding liposarcoma) with their target genes.
| microRNA | Expression | Target | STS type | Reference |
|---|---|---|---|---|
| miR-1 | DOWN | CCND2, PAX3 | RMS | Li L, et al. Lab. Investig. 2012. |
| miR-143 | DOWN | PRC1, PLK1, BCL2 | SS | Subramanian S, et al. Oncogene. 2008. |
| miR-152 | DOWN | MET, KIT | LMS | Pazzaglia L, et al. Cell. Oncol. 2017. |
| miR-199b-5p | DOWN UP |
- | LMS UPS |
Guled M, et al. Genes Chromosom. Cancer. 2014. |
| miR-206 | DOWN | CCND2, PAX3, PAX7, MET, NOTCH3 | RMS | Li L, et al. Lab. Investig. 2012. Taulli R, et al. JCI. 2009. Missiaglia E, et al. Br J Cancer. 2010. Hanna J.A, et al Cell Death & Disease. 2016. |
| miR-221 | DOWN | CCND2, CDK6, ERBB3 | RMS | Hanna J.A, et al. Oncogene. 2018. |
| miR-29 | DOWN | CCND2, PAX3, E2F7, MMP2 | FS RMS |
Kim J.H, et al. Chonnam Med. J. 2017. |
| miR-29c | DOWN | MMP2 | MPNST | Presneau N, et al. Br J Cancer. 2013. |
| miR-30d | DOWN | KPNB1 | MPNST | Zhang P, et al. J. Pathol. 2014. |
| miR-340 | DOWN | SIRT7 | AS | Wang X, et al. Biomed Pharmacother. 2018. |
| miR-34a | DOWN | - | MPNST | Subramanian S, et al. J. Pathol. 2010. |
| miR-378a-3p | DOWN | IGF1R | RMS | Megiorni F, et al. BMC Cancer. 2014. |
| miR-497-5p | DOWN | KCa3.1 | AS | Chen Y, et al. Oncotarget. 2016. |
| miR-17 | UP | CDKN1A | SS | Minami Y, et al. Cancer Sci. 2014. |
| miR-183 | UP | EGR1, PTEN | RMS SS |
Sarver A.L. et al. Cancer Res. 2010. |
| miR-21 | UP | PDCD4 | MPNST | Itani S, et al. J Cancer Res Clin Oncol. 2012. |
| miR-27a | UP | RARA, RXRA | RMS | Tombolan L, et al. PLoS ONE. 2015. |
| miR-320a | UP DOWN |
- | LMS UPS |
Guled M, et al. Genes Chromosom. Cancer. 2014. |
| miR-373 | UP | SIRT1, mTOR | FS | Liu P, et al. J. Cell Physiol. 2012. |
| miR-486-5p | UP | - | RMS | Hanna J.A, et al. Oncogene. 2018. |
| miR-520c | UP | SIRT1, mTOR | FS | Liu P, et al. J. Cell Physiol. 2012. |
| miR-9-5p | UP | - | RMS | Missiaglia E, et al. Cancer Letters. 2017. |
Leiomyosarcoma
LMS is a rare, malignant sarcoma (from 10% to 20% of all STS cases) originating from smooth muscle cells. These tumors can originate in the retroperitoneum (40–45 %), extremity (30–35 %), skin (15–20 %), or from the muscular lining of blood vessels (5%). LMS usually arises in middle-aged or older patients, even though it can also occur in young adults and children (70). Like other forms of STS, treatment options are very limited, and surgical resection remains the only curative approach. As such, the identification of novel therapeutic agents is needed in order to improve patient outcomes. A novel agent for the treatment of LMS is the tumor suppressor miR-152. This miRNA is downregulated in LMS compared to normal tissue and regulates the expression of the tyrosine kinases MET and KIT (71). Experiments in vitro showed that the re-expression of miR-152 diminished proliferation, enhanced apoptosis, inhibited the PI3K/AKT pathway, and led to halted cell cycle. Taken together, these findings support targeting the expression of miR-152 in order to induce regression in patients with LMS (79). While promising, there are challenges in drug delivery and selectively increasing the expression of a given miRNA in LMS cells. In addition, the effectiveness of this approach in vivo is unknown, and to date, this pathway has not been evaluated clinically.
Undifferentiated pleomorphic sarcoma
UPS is a high-grade aggressive STS that represents about 17% of all STS and encompasses mesenchymal neoplasms that have cellular pleomorphism and lack evidence of cellular differentiation (72). The primary tumor originates from mesenchymal stem cells, usually located in soft tissues, bones, and retroperitoneum, with the ability to metastasize to other organs (73). Usually, the diagnosis is carried out through histopathology examination since imaging technologies failed to give an accurate diagnosis, while the therapy is mainly surgical excision with adjuvant radiotherapy for stage I tumors (74). Broadly, chemotherapy is reserved for large, high-grade lesions or those that are locally advanced or unresectable (74). In addition to the traditional chemotherapy, recent studies are also investigating the efficiency of immune-checkpoint inhibitors such as pembrolizumab (anti-PD1), nivolumab (anti-PD1), and ipilimumab (anti-CTLA4) to treat patients with metastatic UPS (75).
One challenge in managing patients with UPS is differentiating these tumors from patients with LMS and other forms of STS. These tumors are characterized by complex genomic profiles, and the differentiation between LMS and UPS can present a significant challenge in clinical management. In this regard, miRNAs have emerged as a novel diagnostic biomarker for STS subclassification. A relevant paper regarding this topic was published in 2014. In this paper, the authors performed a miRNA profiling on several LMS and UPS samples with the goal of identifying a signature of miRNAs that can be used for the differential diagnosis (76). In this experiment, the authors identified 38 and 46 miRNAs that were able to classify UPS and LMS samples, respectively, into separate groups compared to control samples (76). Further experiments using qPCR on a larger cohort of samples confirmed the dysregulation of two miRNAs, miR-199-5p and miR-320a. Specifically, they observed that the levels of miR-199b-5p were significantly higher in UPS as compared with LMS, while miR-320a is upregulated in LMS compared with UPS (76). Consequently, the authors concluded that these miRNAs may serve as diagnostic markers for LMS and UPS. However, additional studies of miRNAs in UPS are limited, not only due to the rarity of UPS but also due to recent changes in classification and nomenclature. Historically, these tumors were erroneously included with other soft tissue sarcomas with mixed histiocytic and fibroblastic lineage and were coined malignant fibrous histiocytoma (MFH). Over the last ten years, we have identified the unique clinical and histopathologic characteristics that define UPS, but molecular studies including sncRNAs have been delayed.
Fibrosarcoma
FS is a mesenchymal tumor, usually located on the trunk or the extremities, that derives from fibrous connective tissue and is characterized by the presence of immature proliferating fibroblasts or undifferentiated anaplastic spindle cells (77). This malignancy is extremely rare (only 500 new cases are reported each year), and it is classified as (i) infantile or congenital fibrosarcoma; and (ii) adult fibrosarcoma. The first type is characterized by the presence of the fusion gene ETV6–NTRK3 and tends to grow slowly forming benign tumors with a consequent better outcome (78). On the other hand, adult FS is much more aggressive, with a 5-year survival of 40-60%. Despite their very different clinical behaviors, histologically the two types of FS are very similar. Treatment options are limited to surgical resection and radiotherapy since chemotherapy has no definitive efficiency in patients with FS (79).
Unlike most types of STS, FS has the unique propensity to metastasize. As such, efforts are underway to identify the drivers of this phenotype. In this regard, some miRNAs have been found to regulate metastases in FS. This includes miR-29, which is downregulated in FS. However, when re-expressed, miR-29 has been shown to reduce the invasiveness of FS cell lines in vitro (80). Mechanistically, the impairment of invasion following the re-expression of miR-29 was caused in part by the downregulation of MMP2. This gene encodes for a metalloproteinase that is important for degrading the extracellular matrix and is targeted by miR-29 in FS cells (80). In addition to miR-29, miR-520c, and miR-373 have also been linked to the regulation of the extracellular matrix in FS (81). In fact, they indirectly upregulate the expression of the metalloproteinase MMP9 by targeting mTOR and SIRT1 (81). As a result, the MAPK signaling pathway is activated and phosphorylates NF-Kb, which leads to increased levels of MMP9 (81). Taken together, these events result in an increase in cellular growth and migration of FS cells (81). While promising, a significant limitation of the aforementioned studies is the lack of in vivo validation of the reported findings. These advances have likely been hampered by the limited availability of specimens and/or preclinical models of fibrosarcoma. Novel approaches to treat fibrosarcoma represent a significant unmet clinical need, and a greater understanding of the role that ncRNAs play in the regulation of fibrosarcoma tumor biology may pave the way to more efficacious therapies.
Synovial sarcoma
SS represents a unique subtype of STS that accounts for about 5% of all STS (82). Although many SS originate near articular structures, its name is misleading since these tumors do not originate from the synovial membrane presents on the inner surface of synovial joint capsules but from mesenchymal cells (82). Indeed, SS can develop in any anatomical location, even though it usually occurs in the lower extremities (82). SS also has unique genomic features, and its pathogenesis is driven by the chromosomal translocation t(X;18), which leads to the formation of the SS18-SSX fusion oncogene (82). The treatment usually consists of surgical excision combined with adjuvant radiotherapy reserved for patients with larger and deeper lesions (82). On the other hand, the use of chemotherapy in the treatment of SS remains controversial, and do not appear that its use increases the survival of patients affected with advanced disease (82). As such, the identification of novel molecular targets may help to usher in more efficacious therapeutic approaches.
A potential target recently described in SS is miR-183. This miRNA blocks the translation of two important tumor suppressors in SS cells: early growth response protein 1 (EGR1) and phosphatase and tensin homolog (PTEN) (83). The knockdown of miR-183 resulted in the upregulation of EGR1 and PTEN, which decreased tumor cell migration (83). In clinical practice, this may be translated into increased EGR1 and PTEN levels, thereby reducing tumor invasion and/or metastases. However, miR-183 has not been targeted clinically or in preclinical models of SS. In addition to miR-183, another miRNA with potential therapeutic value in SS is miR-17. This miRNA is overexpressed in SS and its transcription seems to be regulated by the fusion oncogene SS18-SSX fusion gene (84). Importantly, this miRNA acts as an oncomiR by targeting cyclin-dependent kinase inhibitor 1 (p21, CDKN1A), which inhibits the transition from the G1 into the S-phase of the cell cycle. As a result, cellular proliferation was boosted by the upregulation of miR-17, while cellular migration and invasion were not affected (84). Interestingly, antagomirs of miR-17 led to increased levels of p21 reducing cellular proliferation in SS cell lines [54]. Lastly, miR-143 is differentially expressed in SS. This miRNA, as in DDLPS, is downregulated in SS and is predicted to target SSX1. SSX1 is a gene involved in the formation of the fusion gene SYT-SSX1, a defining characteristic of SS (85). Despite the promising results described above, the small number of studies and the lack of in vivo and preclinical validation have hampered any translation into clinical practice.
Rhabdomyosarcoma
RMS is the most common type of STS in children under 15 years of age and represents 5–8 % of all pediatric malignancies (86). It typically arises from skeletal muscle but can also develop in the muscle of organs such as the bladder or uterus. RMS can be further classified into 4 major histological subtypes including (i) embryonal; (ii) alveolar; (iii) spindle cell; and (iv) pleomorphic (87). Embryonal and alveolar are the two most common subtypes and have very distinct tumor biology. Histologically, the “alveolar” subtype is composed of cells distributed around an open central space, while the “embryonal” subtype contains cells that resemble immature skeletal myoblasts (87). Molecularly, these two subtypes present remarkable differences as well. In fact, the alveolar subtype is often associated with the translocations t(2;13) and t(1;13) that lead to the formation of the fusion genes PAX3–FOXO1 and PAX7–FOXO1, respectively (87). Such fusion proteins act as oncoproteins by dysregulating multiple cellular pathways that, in turn, induce a malignant transformation by promoting the loss of contact inhibition and gain of anchorage-independent growth and proliferation (87). However, a small but significant number of patients with the “alveolar” subtype do not harbor one of these translocations. Instead, these tumors harbor mutations in other genes associated with the activation of the RAS signaling pathway (87). As such, tumors from these patients are biologically and clinically more similar to “embryonal” RMS (87).
In addition to the “alveolar” and “embryonal” subtypes, “pleomorphic” and “spindle cell” RMS have been characterized. Pleomorphic RMS is a morphological variant of RMS that typically occurs in adults and presents cytological and molecular similarities with the pediatric “embryonal” subtype, while spindle cell/sclerosing RMS usually arises in the head and/or neck region, and seems to be more likely to carry specific somatic mutations in the oncogene MYOD1 (L122R) and have a poorer prognosis (87,88). Treatment of RMS is typically multimodal and consists of a combination of chemotherapy, radiotherapy, and/or surgery depending on the size and location of the primary tumor. Adult patients with RMS who show complete response to chemotherapy have a 5-year survival rate of 57 % compared with only 7 % for non-responders (89).
As with SS, miR-183 is also overexpressed in RMS. The knockdown of this miRNA leads to increased expression of two tumor suppressors, EGR1 and PTEN in human SS cell lines (83). Moreover, cellular migration is inhibited by the knockdown of miR-183, suggesting a potential application of this miRNA as a novel target for RMS (83). Another potential oncomiR in RMS is miR-27a (90). Specifically, this miRNA is upregulated in the alveolar subtype of RMS, where it functions to promote cellular proliferation and cell cycle progression by targeting retinoic X receptor alpha (RXRA) and retinoic acid receptor alpha (RARA) (90). Indeed, such genes encode for proteins that interact forming a heterodimer that has the ability to inhibit cellular proliferation by regulating transcription via bondage to retinoic acid response elements (RAREs) (90). Overexpression of miR-27a resulted in reduced levels of RXRA and RARA in a panel of RMS cell lines and, as a consequence, in a significant increase in the proliferation rate that was paralleled by a decrease in the number of cells in the G1 phase of the cell cycle. In conclusion, these findings suggested that miR-27a may be a promising pharmacologic target to prevent the growth of RMS (90).
Two additional miRNAs with oncogenic properties in RMS include miR-9-5p and miR-486-5p (91,92). MiR-9-5p is upregulated in fusion-positive “alveolar” RMS, and its expression appears to be regulated by the PAX3-FOXO1 fusion transcription factor. Indeed, the downregulation of PAX3-FOXO1 through RNA interference (RNAi) in RMS cell lines showed a corresponding downregulation of miR-9-5p. Moreover, high levels of miR-9-5p were associated with worse survival in patients with RMS (91). Noteworthy, in vitro experiments in RMS cell lines revealed that miR-9-5p is implicated in cellular migration and metastasis (91). Like miR-9-5p, miR-486-5p is upregulated in “alveolar” RMS and is transcriptionally activated by the fusion transcription factor PAX3-FOXO1 (92). In vitro experiments also revealed that miR-486-5p promoted proliferation, invasion, and clonogenic growth in fusion-positive RMS cell lines (92). Importantly, inhibition of miR-486-5p in fusion-positive RMS xenografts decreased tumor growth, strongly indicating the importance of this miRNA in the development and progression of RMS (92). In contrast to these oncogenic miRNAs, miR-221 seems to be repressed by PAX3-FOXO1 and thereby loses its function as a tumor suppressor by targeting oncogenes such as CCND2, CDK6 and ERBB3 (92). Taken together, these studies demonstrate that the fusion transcription factor PAX3-FOXO1 regulates key miRNAs that drive the development, differentiation, and progression of fusion-positive alveolar RMS. Whether these downstream miRNA targets of PAX3-FOXO1 can be targeted for potential clinical applications is an area of active investigation.
In addition to the aforementioned oncomiRs, other miRNAs are downregulated in RMS and function as tumor suppressors. A first example is miR-378a-3p (93). This miRNA targets insulin-like growth factor receptor 1 (IGF1R), which is a crucial receptor in the AKT signaling pathway involved in cellular proliferation and myogenic differentiation. Indeed, the overexpression of miR-378a-3p reduced the expression of IGF1R, which in turn promoted apoptosis via blockage of the AKT pathway and induction of caspase 3 (93). Taken together, these results indicate a potential clinical application of miR-378a-3p mimics to reduce cellular proliferation and enhance apoptosis.
Additional examples of miRNAs that function as tumor suppressors in RMS include miR-1 and mir-206 (94-96). These miRNAs reduce the protein expression of PAX3 in the embryonal subtype of RMS, but not in the alveolar subtype since, in the latter, the presence of the PAX3-FOXO1 fusion gene hampers the regulation of PAX3 by these miRNAs (94). Additional experiments in xenograft mouse models also revealed that the re-expression of miR-206 in RMS cells promotes myogenic differentiation and blocks tumor growth by targeting the well-known oncogene MET (95). Furthermore, low miR-206 expression has been associated with poor overall survival in patients with metastatic embryonal and alveolar RMS lacking the PAX3/7-FOXO1 fusion genes (96). A more recent study found that miR-206 targets PAX7, PAX3, NOTCH3, and CCND2, which are responsible for differentiation blockade in fusion-negative RMS but not in fusion-positive RMS (97). Specifically, they determined that PAX7 downregulation is required for cell cycle exit and myogenic differentiation induced by miR-206 (97). Importantly, the genetic deletion of miR-206 in a mouse model of fusion-negative RMS boosted tumor development, indicating that miR-206 acts as a tumor suppressor in fusion-negative RMS by downregulating PAX7 (97).
Taken together, these findings indicate that miR-206 mitigates the differentiation arrest in fusion-negative RMS and that replacement of miR-206 could be a therapeutic differentiation strategy (97). In addition to PAX3/PAX7 and MET, miR-1, miR-206, and miR-29 act as tumor suppressors by repressing the translation of cyclin-D2 (CCND2). CCND2 is a cell-cycle protein that controls how cells proceed through the cell cycle and cell division and is upregulated in several malignancies (94). In addition, miR-29 has also been shown to target the transcription factor E2F7, which is involved in DNA repair and cell cycle regulation (94). Taken together, miR-29, miR-1, and miR-206 act as tumor suppressors, and perhaps their pharmacologic mimics could be an attractive approach for the treatment of RMS.
Angiosarcoma
AS is a rare and highly aggressive malignant tumor originating from the lymphatic or vascular endothelial cell (98). It accounts for less than 2% of all STS and principally affects adult and elderly patients (99). As a clinically and genetically heterogeneous subgroup of sarcomas, AS may occur in any anatomic location, with the most common sites being cutaneous lesions usually located in the head and neck regions (99). AS is an infiltrative tumor with a characteristic high rate of local recurrence and distant metastasis. The reported rates of advanced/metastatic disease at presentation vary from 16 to 44%, and the overall survival ranges from 6 to 16 months (100). Histological examination is essential for the diagnosis of AS, and immunohistochemical confirmation is required (99). Due to the rarity of these tumors, a delay in diagnosis can make treatment more challenging. AS is characterized by aggressive tumor biology and, despite multimodal therapy (surgery/radiation/systemic therapy), is often lethal. As such, the identification of novel therapeutic agents to treat AS remains a significant unmet clinical need.
One miRNA that may hold promise in this regard is miR-497-5p. This miR-497-5p is downregulated in AS and acts as a post-transcriptional regulator of KCa3.1, a potassium intermediate conductance calcium-activated channel (101). This channel is involved in cancer cell proliferation and invasion and is upregulated in AS. Overexpression of miR-497-5p in AS cell lines resulted in decreased levels of KCa3.1 which led to impaired proliferation, reduced cellular migration and halted cell cycle (101). Mechanistically, these effects are a result of the downregulation of CCND1 and p53, which are all important to the progression through the cell cycle (101). Clinically, this may be translated by introducing miR-497-5p as a novel therapeutic agent to inhibit KCa3.1 and ultimately prevent tumor growth. However, to the best of our knowledge, this is the only study reporting the biological properties of miR-497-5p in regulating cell cycle and invasion in AS. Therefore, future studies will be necessary to attract the attention to this miRNA as a potential therapeutic agent for patients affected by AS. Another example of a novel potential therapeutic miRNA for AS comes from a more recent study published in 2018. In this paper, the authors reported a miRNA, miR-340, that was downregulated in AS when compared with normal controls (102). This miRNA seems to regulate the growth and invasion of AS cells by targeting the mRNA encoding for Sirtuin 7 (SIRT7), a gene that plays a critical role in diverse biological processes, including senescence, stress resistance, and tumorigenesis, by promoting cell proliferation and cell cycle progression while inhibiting apoptosis (102). Indeed, the induced overexpression of miR-340 in AS cell lines inhibited their growth and invasion (102). The authors concluded by saying that the miR-340/SIRT7 pathway may play an important role in the development of AS and pointed out the potential applicability of miR-340 and SIRT7 as novel therapeutic targets for the AS treatment (102). However, significant limitations exist, and like other forms of STS, in vivo models have not been used to validate the importance of this axis in AS tumorigenesis.
Noteworthy, two significant research articles pointed out the importance of DICER1, a crucial RNase involved in miRNA biogenesis, in driving the development of AS and metastasis formation (103,104). In a genetically engineered mouse model with a biallelic DICER1 deletion, the invasiveness and metastatic ability of AS was increased and, importantly, was independent from other oncogenes or tumor suppressor loss (103). Moreover, the authors found out that DICER1 functions as a strong tumor suppressor and that its deletion, in combination with either KRAS G12D expression or CDKN2A loss, was associated with the development of AS (104). These studies highlight the role of DICER1 as a crucial tumor-suppressor in AS, whereby inactivating mutations in DICER1 result in the development and metastases of AS. Importantly, these studies confirm the importance of miRNAs in regulating gene expression. In fact, when their biogenesis is altered, this results in a cancer predisposition syndrome which increases the risk of many additional tumor types (103).
Malignant peripheral nerve sheath tumor
MPNST is a rare malignancy (5% of STS) originating from the Schwann cells of peripheral nerves. It is locally aggressive and has an increased propensity to metastasize (105). While rare, they are common in individuals with the hereditary condition of neurofibromatosis type 1 (NF1). In these patients, the lifetime risk of developing MPNST approaches 10% (105). Establishing the diagnosis of MPNST can be challenging. In contrast to other types of STS, there is no genetic or immunohistochemical marker that is unique to MPNST. When a specimen clearly comes from a nerve, the diagnosis is clear. However, in other instances, a variety of immunohistochemical stains may be required to distinguish between MPNST and other sarcomas (106). Treatment of these patients is challenging as well. Indeed, the location of the tumor may limit surgical options and chemotherapy is largely ineffective (106).
A gene that drives the development of MPNST is the histone methyltransferase, enhancer of zeste homolog 2 (EZH2). Elevated expression of EZH2 has been observed in MPNST compared to healthy nerves and neurofibromas (107). EZH2 binds with two other molecules to form polycomb-repressor complex 2 (PRC2), and it functions in normal tissue to repress gene transcription (108). However, mutations in the EED and SUZ12 gene regions that encode parts of the PRC2 complex lead to the inactivation of this complex in MPNST (109,110). Given the role of the PRC2 complex in promoting MPNST tumorigenesis, additional regulators of this axis have been sought.
One such regulator is miR-30d. This miRNA is transcriptionally regulated by EZH2, which represses its transcription by binding its promoter region (107). In normal tissue, miR-30d targets the mRNA encoding for the importin subunit beta-1 (KPNB1), suppressing its translation. Interestingly, the blockage of KPNB1 resulted in apoptosis induction in MPNST cells, a similar phenomenon that is observed with miR-30d overexpression and EZH2-knockdown (107). Taken together, the authors concluded that the EZH2/miR-30d/KPNB1 signaling might be a prognostic target for patients with MPNST.
Another miRNA that is important to the tumor biology of MPNST is miR-34a. This miRNA mediates several genes that take part in the regulation of proliferation and cell cycle progression (111). However, its expression is downregulated in MPNST because of the inactive mutations in p53. In normal tissue, p53 positively regulates the expression of miR-34a. Importantly, in-vitro experiments in MPNST cell lines overexpressing miR-34a and p53 showed an enhanced apoptosis (112). Another miRNA that is less expressed in MPNST is miR-29c (113). This miRNA negatively regulates the matrix metalloproteinase 2 (MMP-2). MMP-2 is a protein that functions in the extracellular matrix by degrading glycoproteins and collagens and is important for cell migration and tumor invasion. As expected, the upregulation of miR-29c in MPNST cell lines resulted in reduced cellular invasion due to the reduced activity of MMP-2 (113). However, cellular proliferation remained unaltered (113). These results suggest a potential application of this miRNA as a way to decrease the invasiveness and metastatic ability of MPNST.
Lastly, miR-21 is a miRNA that is upregulated in MPNST and prevents apoptosis by targeting the mRNA encoding for the tumor suppressor programmed cell death protein 4 (PDCD4). In benign neurofibromas and normal peripheral nerves, the expression of PDCD4 is typically higher than in MPNST, and it regulates apoptosis via caspase cascade. However, its expression goes down in MPNST, causing resistance to apoptosis. One of the reasons for the downregulation of PDCD4 in MPNST seems to be the upregulation of miR-21. Indeed, in-vitro experiments showed the inhibition of miR-21 in MPNST cell lines resulted in elevated expression of PDCD4, which in turn induced apoptosis (114). These results seem to suggest a possible application in the clinical practice of anti-miR-21 that induces apoptosis by restoring the levels of PDCD4. In conclusion, as also in other STS subtypes, several miRNAs with relevant biological functions such as cellular proliferation, invasion, differentiation, and apoptosis have been found dysregulated in MPNST.
Future Perspectives: the role of tRNA-derived ncRNAs in soft tissue sarcoma tumor biology
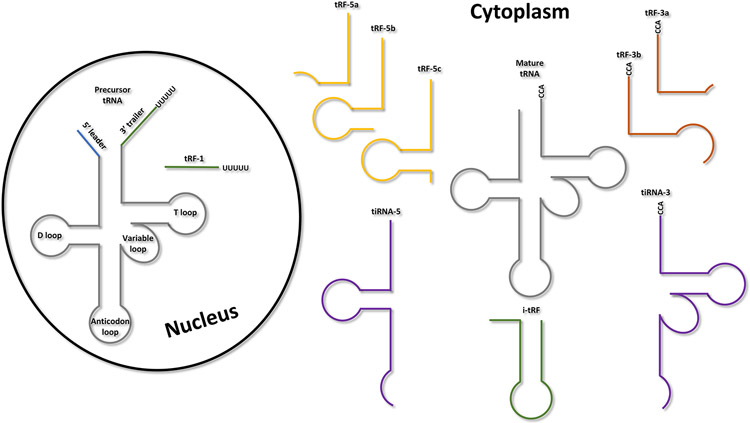
It is important to note that miRNAs are not the only regulatory sncRNAs that have been identified. To date, the studies in LPS and other STS subtypes are uniquely focused on miRNAs, while other classes of regulatory sncRNAs that might have a pivotal role in the development and progression of STS have been omitted. An emergent class of biologically important sncRNAs is the tsRNAs. These regulatory sncRNAs originate from tRNA processing and are often classified according to the location of the cleavage sites within the tRNA. More broadly, tsRNAs are divided into two main classes: (i) tRNA-derived fragments type 1 (tRF-1s), which derive from the precursor of the tRNA (pre-tRNA); and (ii) stress-induced tRNA fragments (tiRNAs), together with the tRNA-derived fragments (tRFs), which both derive from the mature tRNA (115,116). More details about their biogenesis and classification can be found in (115,116) while a schematic representation of the different classes of tsRNAs is reported in Figure 3.
Figure 3.
tRNA-derived ncRNA classification. Schematic representation of the different types of tRNA-derived ncRNAs based on the location of their cleavage sites on either the precursor or mature tRNA.
Functionally, tsRNAs appear to play several important roles in the physiology of eukaryotic cells and are involved in important biological processes like ribosome biogenesis, retrotransposition, viral infection, and apoptosis (10,117-125). Furthermore, some classes of tsRNAs (e.g., tRF-1s) have been shown to bind AGO and PIWI proteins, thereby acting as post- or pre-transcriptional regulators of gene expression (122,126). In addition, other reports have also indicated the presence of functional tsRNAs in human bio-fluids of cancer patients (127-131). For these reasons, the scientific community has begun investigating the role of these regulatory sncRNAs as biomarkers and screening tools for cancer development. In this context, one of the first reports showing the involvement of tsRNAs in cancer comes from Croce et al., who noticed that two miRNAs (miR-4521 and miR-3676) were located exactly at the end of the mature sequence of two different tRNAs (132). Indeed, such miR-4521 and miR-3676 were not exactly miRNAs by tRF-1s (ts-53 and ts-101, respectively) who were able to target in a miRNA-like manner the mRNA encoding for TCL1, a critical oncogene that contributes to the pathogenesis of the aggressive form of CLL (122). Noteworthy, such tRF-1s were found to be both co-deleted with TP53, down-regulated, or mutated in CLL (122).
Following these reports, subsequent studies have been performed with the goal of defining the role of tsRNAs in human cancer. These studies have focused on (i) identifying genes targeted by tsRNAs, (ii) and reporting tsRNAs as novel diagnostic and prognostic biomarkers for multiple human cancers (17). However, no studies have been reported about tsRNAs in LPS or other STS subtypes. As a research team studying the role of sncRNAs in cancer biology, we believe that tsRNAs may play a pivotal role in the development and differentiation of LPS and other STS. Recent research has shown that tRFs can affect stemness and embryo development in mammals (133). As such, it is reasonable to speculate that such important regulatory sncRNAs may also regulate the differentiation of STS cells, thereby providing a more complete picture of the molecular differences that drive the disparate tumor biology of distinct STS subtypes.
Conclusion
In summary, given the role that miRNAs play in orchestrating gene expression at the post-transcriptional level by regulating the translation of specific mRNA targets, they have been studied in several human cancers, including LPS and other STS subtypes. Although limited in number, these studies have shown that widespread dysregulation of miRNAs in STS affects many aspects of cancer cell biology, including cell cycle progression, migration, invasion, and apoptosis. Moreover, several studies demonstrate how the expression of miRNAs can be used to distinguish between different STS subtypes. In addition, miRNAs could be used to determine prognosis and guide management. Finally, their presence in biological fluids is well-established and supports the development of circulating miRNAs as non-invasive biomarkers for the early detection of select STS subtypes. Despite promising results, the translation of these findings into novel clinical tools has been limited. In fact, no clinical trial using miRNA-based approaches for the diagnosis or treatment of STS has been conducted. In addition, additional investigation into the role of tsRNAs in STS may provide important insights into the molecular underpinnings of STS tumor biology. Indeed, a more comprehensive view of the landscape of multiple classes of regulatory sncRNAs will provide a better understanding of the complex regulatory networks in STS cells and help to identify novel biomarkers and molecular targets for the treatment of STS.
Footnotes
The authors declare no potential conflicts of interest.
References
- 1.Bill KLJ, Casadei L, Prudner BC, Iwenofu H, Strohecker AM, Pollock RE. Liposarcoma: molecular targets and therapeutic implications. Cell Mol Life Sci. 2016;73:3711–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smolle MA, Leithner A, Posch F, Szkandera J, Liegl-Atzwanger B, Pichler M. MicroRNAs in Different Histologies of Soft Tissue Sarcoma: A Comprehensive Review. Int J Mol Sci [Internet]. 2017;18. Available from: 10.3390/ijms18091960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fujiwara T, Kunisada T, Takeda K, Ozaki T. microRNAs and Soft Tissue Sarcomas. Adv Exp Med Biol. 2015;889:179–99. [DOI] [PubMed] [Google Scholar]
- 4.La Ferlita A, Battaglia R, Andronico F, Caruso S, Cianci A, Purrello M, et al. Non-Coding RNAs in Endometrial Physiopathology. Int J Mol Sci. Multidisciplinary Digital Publishing Institute; 2018;19:2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Di Bella S, La Ferlita A, Carapezza G, Alaimo S, Isacchi A, Ferro A, et al. A benchmarking of pipelines for detecting ncRNAs from RNA-Seq data. Brief Bioinform [Internet]. 2019; Available from: 10.1093/bib/bbz110 [DOI] [PubMed] [Google Scholar]
- 6.Ghafouri-Fard S, Shoorei H, Anamag FT, Taheri M. The Role of Non-Coding RNAs in Controlling Cell Cycle Related Proteins in Cancer Cells. Front Oncol. 2020;10:608975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yao S MicroRNA biogenesis and their functions in regulating stem cell potency and differentiation. Biol Proced Online. 2016;18:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Magee R, Rigoutsos I. On the expanding roles of tRNA fragments in modulating cell behavior. Nucleic Acids Res. 2020;48:9433–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anderson P, Ivanov P. tRNA fragments in human health and disease. FEBS Lett. 2014;588:4297–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balatti V, Pekarsky Y, Croce CM. Role of the tRNA-Derived Small RNAs in Cancer: New Potential Biomarkers and Target for Therapy. Adv Cancer Res. 2017;135:173–87. [DOI] [PubMed] [Google Scholar]
- 11.Peng Y, Croce CM. The role of MicroRNAs in human cancer [Internet]. Signal Transduction and Targeted Therapy. 2016. Available from: 10.1038/sigtrans.2015.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y, Kowdley KV. MicroRNAs in common human diseases. Genomics Proteomics Bioinformatics. 2012;10:246–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pardini B, Sabo AA, Birolo G, Calin GA. Noncoding RNAs in Extracellular Fluids as Cancer Biomarkers: The New Frontier of Liquid Biopsies. Cancers [Internet]. 2019;11. Available from: 10.3390/cancers11081170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abramowicz A, Story MD. The Long and Short of It: The Emerging Roles of Non-Coding RNA in Small Extracellular Vesicles. Cancers [Internet]. 2020;12. Available from: 10.3390/cancers12061445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alaimo S, Micale G, La Ferlita A, Ferro A, Pulvirenti A. Computational Methods to Investigate the Impact of miRNAs on Pathways. Methods Mol Biol. 2019;1970:183–209. [DOI] [PubMed] [Google Scholar]
- 16.La Ferlita A, Alaimo S, Ferro A, Pulvirenti A. Pathway Analysis for Cancer Research and Precision Oncology Applications. Adv Exp Med Biol. 2022;1361:143–61. [DOI] [PubMed] [Google Scholar]
- 17.Pekarsky Y, Balatti V, Croce CM. tRNA-derived fragments (tRFs) in cancer. J Cell Commun Signal [Internet]. 2022; Available from: 10.1007/s12079-022-00690-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee Y, Kim M, Han J, Yeom K-H, Lee S, Baek SH, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;13:1097–101. [DOI] [PubMed] [Google Scholar]
- 20.Macfarlane L-A, Murphy PR. MicroRNA: Biogenesis, Function and Role in Cancer. Curr Genomics. 2010;11:537–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Helwak A, Kudla G, Dudnakova T, Tollervey D. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell. 2013;153:654–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–4. [DOI] [PubMed] [Google Scholar]
- 24.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102:13944–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Calin GA, Cimmino A, Fabbri M, Ferracin M, Wojcik SE, Shimizu M, et al. MiR-15a and miR-16-1 cluster functions in human leukemia. Proc Natl Acad Sci U S A. 2008;105:5166–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–9. [DOI] [PubMed] [Google Scholar]
- 28.Mansoori B, Baradaran B, Nazari A, Gaballu FA, Cho WC-S, Mansoori B. MicroRNAs in the cancer cell-to-cell communication: An insight into biological vehicles. Biomed Pharmacother. 2022;153:113449. [DOI] [PubMed] [Google Scholar]
- 29.Mousavi SM, Amin Mahdian SM, Ebrahimi MS, Taghizadieh M, Vosough M, Sadri Nahand J, et al. Microfluidics for detection of exosomes and microRNAs in cancer: State of the art. Mol Ther Nucleic Acids. 2022;28:758–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bertoli G, Cava C, Castiglioni I. MicroRNAs: New Biomarkers for Diagnosis, Prognosis, Therapy Prediction and Therapeutic Tools for Breast Cancer. Theranostics. 2015;5:1122–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kok VC, Yu C-C. Cancer-Derived Exosomes: Their Role in Cancer Biology and Biomarker Development. Int J Nanomedicine. 2020;15:8019–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bonneau E, Neveu B, Kostantin E, Tsongalis GJ, De Guire V. How close are miRNAs from clinical practice? A perspective on the diagnostic and therapeutic market. EJIFCC. 2019;30:114–27. [PMC free article] [PubMed] [Google Scholar]
- 33.Ho PTB, Clark IM, Le LTT. MicroRNA-Based Diagnosis and Therapy. Int J Mol Sci [Internet]. 2022;23. Available from: 10.3390/ijms23137167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Casadei L, de Faria FCC, Lopez-Aguiar A, Pollock RE, Grignol V. Targetable Pathways in the Treatment of Retroperitoneal Liposarcoma. Cancers [Internet]. 2022;14. Available from: 10.3390/cancers14061362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muratori F, Bettini L, Frenos F, Mondanelli N, Greto D, Livi L, et al. Myxoid Liposarcoma: Prognostic Factors and Metastatic Pattern in a Series of 148 Patients Treated at a Single Institution. Int J Surg Oncol. 2018;2018:8928706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Graaff MA, Yu JSE, Beird HC, Ingram DR, Nguyen T, Juehui Liu J, et al. Establishment and characterization of a new human myxoid liposarcoma cell line (DL-221) with the FUS-DDIT3 translocation. Lab Invest. Nature Publishing Group; 2016;96:885–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwartz JC, Cech TR, Parker RR. Biochemical Properties and Biological Functions of FET Proteins. Annu Rev Biochem. 2015;84:355–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pérez-Mancera PA, Bermejo-Rodríguez C, Sánchez-Martín M, Abollo-Jiménez F, Pintado B, Sánchez-García I. FUS-DDIT3 prevents the development of adipocytic precursors in liposarcoma by repressing PPARgamma and C/EBPalpha and activating eIF4E. PLoS One. Public Library of Science (PLoS); 2008;3:e2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Göransson M, Andersson MK, Forni C, Ståhlberg A, Andersson C, Olofsson A, et al. The myxoid liposarcoma FUS-DDIT3 fusion oncoprotein deregulates NF-kappaB target genes by interaction with NFKBIZ. Oncogene. 2009;28:270–8. [DOI] [PubMed] [Google Scholar]
- 40.Estourgie SH, Nielsen GP, Ott MJ. Metastatic patterns of extremity myxoid liposarcoma and their outcome. J Surg Oncol. 2002;80:89–93. [DOI] [PubMed] [Google Scholar]
- 41.Schwab JH, Boland P, Guo T, Brennan MF, Singer S, Healey JH, et al. Skeletal metastases in myxoid liposarcoma: an unusual pattern of distant spread. Ann Surg Oncol. 2007;14:1507–14. [DOI] [PubMed] [Google Scholar]
- 42.Dei Tos AP. Liposarcoma: new entities and evolving concepts. Ann Diagn Pathol. 2000;4:252–66. [DOI] [PubMed] [Google Scholar]
- 43.Lahat G, Anaya DA, Wang X, Tuvin D, Lev D, Pollock RE. Resectable well-differentiated versus dedifferentiated liposarcomas: two different diseases possibly requiring different treatment approaches. Ann Surg Oncol. 2008;15:1585–93. [DOI] [PubMed] [Google Scholar]
- 44.Ghadimi MP, Al-Zaid T, Madewell J, Peng T, Colombo C, Hoffman A, et al. Diagnosis, management, and outcome of patients with dedifferentiated liposarcoma systemic metastasis. Ann Surg Oncol. 2011;18:3762–70. [DOI] [PubMed] [Google Scholar]
- 45.Evans HL, Khurana KK, Kemp BL, Ayala AG. Heterologous elements in the dedifferentiated component of dedifferentiated liposarcoma. Am J Surg Pathol. 1994;18:1150–7. [DOI] [PubMed] [Google Scholar]
- 46.Henricks WH, Chu YC, Goldblum JR, Weiss SW. Dedifferentiated liposarcoma: a clinicopathological analysis of 155 cases with a proposal for an expanded definition of dedifferentiation. Am J Surg Pathol. 1997;21:271–81. [DOI] [PubMed] [Google Scholar]
- 47.Tseng WW, Madewell JE, Wei W, Somaiah N, Lazar AJ, Ghadimi MP, et al. Locoregional disease patterns in well-differentiated and dedifferentiated retroperitoneal liposarcoma: implications for the extent of resection? Ann Surg Oncol. 2014;21:2136–43. [DOI] [PubMed] [Google Scholar]
- 48.Kollár A, Benson C. Current management options for liposarcoma and challenges for the future. Expert Rev Anticancer Ther. 2014;14:297–306. [DOI] [PubMed] [Google Scholar]
- 49.Lu J, Wood D, Ingley E, Koks S, Wong D. Update on genomic and molecular landscapes of well-differentiated liposarcoma and dedifferentiated liposarcoma. Mol Biol Rep. 2021;48:3637–47. [DOI] [PubMed] [Google Scholar]
- 50.Ugras S, Brill E, Jacobsen A, Hafner M, Socci ND, Decarolis PL, et al. Small RNA sequencing and functional characterization reveals MicroRNA-143 tumor suppressor activity in liposarcoma. Cancer Res. 2011;71:5659–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Bill, Liu Young, Peng Bolshakov. MiR-155 Is a Liposarcoma Oncogene That Targets Casein Kinase-1α and Enhances β-Catenin SignalingmiRNA Deregulation in Human Liposarcoma. Cancer Res [Internet]. Available from: https://aacrjournals.org/cancerres/article-abstract/72/7/1751/578997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vincenzi B, Iuliani M, Zoccoli A, Pantano F, Fioramonti M, De Lisi D, et al. Deregulation of dicer and mir-155 expression in liposarcoma. Oncotarget. 2015;6:10586–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kapodistrias N, Mavridis K, Batistatou A, Gogou P, Karavasilis V, Sainis I, et al. Assessing the clinical value of microRNAs in formalin-fixed paraffin-embedded liposarcoma tissues: Overexpressed miR-155 is an indicator of poor prognosis. Oncotarget. 2017;8:6896–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee DH, Amanat S, Goff C, Weiss LM, Said JW, Doan NB, et al. Overexpression of miR-26a-2 in human liposarcoma is correlated with poor patient survival. Oncogenesis. 2013;2:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee DH, Forscher C, Di Vizio D, Koeffler HP. Induction of p53-independent apoptosis by ectopic expression of HOXA5 in human liposarcomas. Sci Rep. 2015;5:12580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mazzu YZ, Hu Y, Soni RK, Mojica KM, Qin L-X, Agius P, et al. miR-193b-Regulated Signaling Networks Serve as Tumor Suppressors in Liposarcoma and Promote Adipogenesis in Adipose-Derived Stem Cells. Cancer Res. 2017;77:5728–40. [DOI] [PubMed] [Google Scholar]
- 57.Yu PY, Lopez G, Braggio D, Koller D, Bill KLJ, Prudner BC, et al. miR-133a function in the pathogenesis of dedifferentiated liposarcoma. Cancer Cell Int. 2018;18:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gits CMM, van Kuijk PF, Jonkers MBE, Boersma AWM, Smid M, van Ijcken WF, et al. MicroRNA expression profiles distinguish liposarcoma subtypes and implicate miR-145 and miR-451 as tumor suppressors [Internet]. International Journal of Cancer. 2014. page 348–61. Available from: 10.1002/ijc.28694 [DOI] [PubMed] [Google Scholar]
- 59.Tan HM, Cheng H, Tang YC, Leong SM, Teo PY, Lee CK, et al. MicroRNAs as Potential Biomarkers in the Differential Diagnosis of Lipomatous Tumors and Their Mimics. Int J Mol Sci [Internet]. 2022;23. Available from: 10.3390/ijms23147804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Svoronos AA, Engelman DM, Slack FJ. OncomiR or Tumor Suppressor? The Duplicity of MicroRNAs in Cancer. Cancer Res. 2016;76:3666–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Simonson B, Das S. MicroRNA Therapeutics: the Next Magic Bullet? Mini Rev Med Chem. 2015;15:467–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fu Y, Chen J, Huang Z. Recent progress in microRNA-based delivery systems for the treatment of human disease. ExRNA. BioMed Central; 2019;1:1–14. [Google Scholar]
- 63.Almeida MI, Calin GA. The miR-143/miR-145 cluster and the tumor microenvironment: unexpected roles. Genome Med. 2016;8:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Esau C, Kang X, Peralta E, Hanson E, Marcusson EG, Ravichandran LV, et al. MicroRNA-143 regulates adipocyte differentiation. J Biol Chem. 2004;279:52361–5. [DOI] [PubMed] [Google Scholar]
- 65.Mazzu YZ, Hu Y, Shen Y, Tuschl T, Singer S. miR-193b regulates tumorigenesis in liposarcoma cells via PDGFR, TGFβ, and Wnt signaling. Sci Rep. 2019;9:3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Redis RS, Calin S, Yang Y, You MJ, Calin GA. Cell-to-cell miRNA transfer: from body homeostasis to therapy. Pharmacol Ther. 2012;136:169–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Taylor DD, Gercel-Taylor C. The origin, function, and diagnostic potential of RNA within extracellular vesicles present in human biological fluids. Front Genet. 2013;4:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Casadei L, Calore F, Creighton CJ, Guescini M, Batte K, Iwenofu OH, et al. Exosome-Derived miR-25-3p and miR-92a-3p Stimulate Liposarcoma Progression. Cancer Res. 2017;77:3846–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kohama I, Asano N, Matsuzaki J, Yamamoto Y, Yamamoto T, Takahashi R-U, et al. Comprehensive serum and tissue microRNA profiling in dedifferentiated liposarcoma. Oncol Lett. 2021;22:623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.World Health Organization. WHO Classification of Tumours of Soft Tissue and Bone. IARC Press; 2013. [Google Scholar]
- 71.Pazzaglia L, Novello C, Conti A, Pollino S, Picci P, Benassi MS. miR-152 down-regulation is associated with MET up-regulation in leiomyosarcoma and undifferentiated pleomorphic sarcoma. Cell Oncol . 2017;40:77–88. [DOI] [PubMed] [Google Scholar]
- 72.Toro JR, Travis LB, Wu HJ, Zhu K, Fletcher CDM, Devesa SS. Incidence patterns of soft tissue sarcomas, regardless of primary site, in the surveillance, epidemiology and end results program, 1978-2001: An analysis of 26,758 cases. Int J Cancer. Wiley; 2006;119:2922–30. [DOI] [PubMed] [Google Scholar]
- 73.Goldblum JR. An approach to pleomorphic sarcomas: can we subclassify, and does it matter? Mod Pathol. Nature Publishing Group; 2014;27:S39–46. [DOI] [PubMed] [Google Scholar]
- 74.von Mehren M, Kane JM, Bui MM, Choy E, Connelly M, Dry S, et al. NCCN Guidelines Insights: Soft Tissue Sarcoma, Version 1.2021: Featured Updates to the NCCN Guidelines. J Natl Compr Canc Netw. National Comprehensive Cancer Network; 2020;18:1604–12. [DOI] [PubMed] [Google Scholar]
- 75.Keung EZ, Lazar AJ, Torres KE, Wang W-L, Cormier JN, Ashleigh Guadagnolo B, et al. Phase II study of neoadjuvant checkpoint blockade in patients with surgically resectable undifferentiated pleomorphic sarcoma and dedifferentiated liposarcoma [Internet]. BMC Cancer. 2018. Available from: 10.1186/s12885-018-4829-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Guled M, Pazzaglia L, Borze I, Mosakhani N, Novello C, Benassi MS, et al. Differentiating soft tissue leiomyosarcoma and undifferentiated pleomorphic sarcoma: A miRNA analysis. Genes Chromosomes Cancer. 2014;53:693–702. [DOI] [PubMed] [Google Scholar]
- 77.Mazanet R, Antman KH. Sarcomas of soft tissue and bone. Cancer. 1991;68:463–73. [DOI] [PubMed] [Google Scholar]
- 78.Walther C, Nilsson J, von Steyern FV, Wiebe T, Bauer HCF, Nord KH, et al. Cytogenetic and single nucleotide polymorphism array findings in soft tissue tumors in infants. Cancer Genet. 2013;206:299–303. [DOI] [PubMed] [Google Scholar]
- 79.Augsburger D, Nelson PJ, Kalinski T, Udelnow A, Knösel T, Hofstetter M, et al. Current diagnostics and treatment of fibrosarcoma -perspectives for future therapeutic targets and strategies. Oncotarget. 2017;8:104638–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kim JH, Jeon S, Shin BA. MicroRNA-29 Family Suppresses the Invasion of HT1080 Human Fibrosarcoma Cells by Regulating Matrix Metalloproteinase 2 Expression. Chonnam Med J. 2017;53:161–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu P, Wilson MJ. miR-520c and miR-373 upregulate MMP9 expression by targeting mTOR and SIRT1, and activate the Ras/Raf/MEK/Erk signaling pathway and NF-κB factor in human fibrosarcoma cells. J Cell Physiol. Wiley; 2012;227:867–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gazendam AM, Popovic S, Munir S, Parasu N, Wilson D, Ghert M. Synovial Sarcoma: A Clinical Review. Curr Oncol. 2021;28:1909–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sarver AL, Li L, Subramanian S. MicroRNA miR-183 Functions as an Oncogene by Targeting the Transcription Factor EGR1 and Promoting Tumor Cell MigrationmiR--183 Functions as an Oncogene by Regulating EGR1. Cancer Res. AACR; 2010;70:9570–80. [DOI] [PubMed] [Google Scholar]
- 84.Minami Y, Kohsaka S, Tsuda M, Yachi K, Hatori N, Tanino M, et al. SS18-SSX-regulated miR-17 promotes tumor growth of synovial sarcoma by inhibiting p21WAF1/CIP1. Cancer Sci. Wiley; 2014;105:1152–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Subramanian S, Lui WO, Lee CH, Espinosa I, Nielsen TO, Heinrich MC, et al. MicroRNA expression signature of human sarcomas. Oncogene. 2008;27:2015–26. [DOI] [PubMed] [Google Scholar]
- 86.De Giovanni C, Landuzzi L, Nicoletti G, Lollini P-L, Nanni P. Molecular and cellular biology of rhabdomyosarcoma. Future Oncol. 2009;5:1449–75. [DOI] [PubMed] [Google Scholar]
- 87.Skapek SX, Ferrari A, Gupta AA, Lupo PJ, Butler E, Shipley J, et al. Rhabdomyosarcoma. Nat Rev Dis Primers. 2019;5:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rekhi B, Upadhyay P, Ramteke MP, Dutt A. MYOD1 (L122R) mutations are associated with spindle cell and sclerosing rhabdomyosarcomas with aggressive clinical outcomes. Mod Pathol. 2016;29:1532–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Esnaola NF, Rubin BP, Baldini EH, Vasudevan N, Demetri GD, Fletcher CD, et al. Response to chemotherapy and predictors of survival in adult rhabdomyosarcoma. Ann Surg. 2001;234:215–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tombolan L, Zampini M, Casara S, Boldrin E, Zin A, Bisogno G, et al. MicroRNA-27a Contributes to Rhabdomyosarcoma Cell Proliferation by Suppressing RARA and RXRA. PLoS One. 2015;10:e0125171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Missiaglia E, Shepherd CJ, Aladowicz E, Olmos D, Selfe J, Pierron G, et al. MicroRNA and gene co-expression networks characterize biological and clinical behavior of rhabdomyosarcomas. Cancer Lett. 2017;385:251–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hanna JA, Garcia MR, Lardennois A, Leavey PJ, Maglic D, Fagnan A, et al. PAX3-FOXO1 drives miR-486-5p and represses miR-221 contributing to pathogenesis of alveolar rhabdomyosarcoma. Oncogene. 2018;37:1991–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Megiorni F, Cialfi S, McDowell HP, Felsani A, Camero S, Guffanti A, et al. Deep Sequencing the microRNA profile in rhabdomyosarcoma reveals down-regulation of miR-378 family members. BMC Cancer. 2014;14:880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li L, Sarver AL, Alamgir S, Subramanian S. Downregulation of microRNAs miR-1, -206 and -29 stabilizes PAX3 and CCND2 expression in rhabdomyosarcoma [Internet]. Laboratory Investigation. 2012. page 571–83. Available from: 10.1038/labinvest.2012.10 [DOI] [PubMed] [Google Scholar]
- 95.Taulli R, Bersani F, Foglizzo V, Linari A, Vigna E, Ladanyi M, et al. The muscle-specific microRNA miR-206 blocks human rhabdomyosarcoma growth in xenotransplanted mice by promoting myogenic differentiation. J Clin Invest. 2009;119:2366–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Missiaglia E, Shepherd CJ, Patel S, Thway K, Pierron G, Pritchard-Jones K, et al. MicroRNA-206 expression levels correlate with clinical behaviour of rhabdomyosarcomas. Br J Cancer. 2010;102:1769–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hanna JA, Garcia MR, Go JC, Finkelstein D, Kodali K, Pagala V, et al. PAX7 is a required target for microRNA-206-induced differentiation of fusion-negative rhabdomyosarcoma. Cell Death Dis. 2016;7:e2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Young RJ, Brown NJ, Reed MW, Hughes D, Woll PJ. Angiosarcoma. Lancet Oncol. 2010;11:983–91. [DOI] [PubMed] [Google Scholar]
- 99.Cao J, Wang J, He C, Fang M. Angiosarcoma: a review of diagnosis and current treatment. Am J Cancer Res. 2019;9:2303–13. [PMC free article] [PubMed] [Google Scholar]
- 100.Buehler D, Rice SR, Moody JS, Rush P, Hafez G-R, Attia S, et al. Angiosarcoma outcomes and prognostic factors: a 25-year single institution experience. Am J Clin Oncol. 2014;37:473–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chen Y, Kuang D, Zhao X, Chen D, Wang X, Yang Q, et al. miR-497-5p inhibits cell proliferation and invasion by targeting KCa3.1 in angiosarcoma. Oncotarget. 2016;7:58148–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang X, Song Y. MicroRNA-340 inhibits the growth and invasion of angiosarcoma cells by targeting SIRT7. Biomed Pharmacother. 2018;103:1061–8. [DOI] [PubMed] [Google Scholar]
- 103.Hanna JA, Drummond CJ, Garcia MR, Go JC, Finkelstein D, Rehg JE, et al. Biallelic Dicer1 Loss Mediated by aP2-Cre Drives Angiosarcoma. Cancer Res. 2017;77:6109–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hanna JA, Langdon CG, Garcia MR, Benton A, Lanman NA, Finkelstein D, et al. Genetic context of oncogenic drivers dictates vascular sarcoma development in aP2-Cre mice. J Pathol. 2022;257:109–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Knight SWE, Knight TE, Santiago T, Murphy AJ, Abdelhafeez AH. Malignant Peripheral Nerve Sheath Tumors—A Comprehensive Review of Pathophysiology, Diagnosis, and Multidisciplinary Management. Children. Multidisciplinary Digital Publishing Institute; 2022;9:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Prudner BC, Ball T, Rathore R, Hirbe AC. Diagnosis and management of malignant peripheral nerve sheath tumors: Current practice and future perspectives [Internet]. Neuro-Oncology Advances. 2020. page i40–9. Available from: 10.1093/noajnl/vdz047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhang P, Garnett J, Creighton CJ, Al Sannaa GA, Igram DR, Lazar A, et al. EZH2-miR-30d-KPNB1 pathway regulates malignant peripheral nerve sheath tumour cell survival and tumourigenesis. J Pathol. 2014;232:308–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Simon JA, Lange CA. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat Res. 2008;647:21–9. [DOI] [PubMed] [Google Scholar]
- 109.De Raedt T, Beert E, Pasmant E, Luscan A, Brems H, Ortonne N, et al. PRC2 loss amplifies Ras-driven transcription and confers sensitivity to BRD4-based therapies. Nature. 2014;514:247–51. [DOI] [PubMed] [Google Scholar]
- 110.Lee W, Teckie S, Wiesner T, Ran L, Prieto Granada CN, Lin M, et al. PRC2 is recurrently inactivated through EED or SUZ12 loss in malignant peripheral nerve sheath tumors. Nat Genet. 2014;46:1227–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.He X, He L, Hannon GJ. The guardian’s little helper: microRNAs in the p53 tumor suppressor network. Cancer Res. 2007;67:11099–101. [DOI] [PubMed] [Google Scholar]
- 112.Subramanian S, Thayanithy V, West RB, Lee C-H, Beck AH, Zhu S, et al. Genome-wide transcriptome analyses reveal p53 inactivation mediated loss of miR-34a expression in malignant peripheral nerve sheath tumours [Internet]. The Journal of Pathology. 2010. page 58–70. Available from: 10.1002/path.2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Presneau N, Eskandarpour M, Shemais T, Henderson S, Halai D, Tirabosco R, et al. MicroRNA profiling of peripheral nerve sheath tumours identifies miR-29c as a tumour suppressor gene involved in tumour progression. Br J Cancer. 2013;108:964–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Itani S, Kunisada T, Morimoto Y, Yoshida A, Sasaki T, Ito S, et al. MicroRNA-21 correlates with tumorigenesis in malignant peripheral nerve sheath tumor (MPNST) via programmed cell death protein 4 (PDCD4). J Cancer Res Clin Oncol. 2012;138:1501–9. [DOI] [PubMed] [Google Scholar]
- 115.La Ferlita A, Alaimo S, Veneziano D, Nigita G, Balatti V, Croce CM, et al. Identification of tRNA-derived ncRNAs in TCGA and NCI-60 panel cell lines and development of the public database tRFexplorer. Database [Internet]. 2019;2019. Available from: 10.1093/database/baz115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Filetti V, La Ferlita A, Di Maria A, Cardile V, Graziano ACE, Rapisarda V, et al. Dysregulation of microRNAs and tRNA-derived ncRNAs in mesothelial and mesothelioma cell lines after asbestiform fiber exposure. Sci Rep. 2022;12:9181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kim HK, Fuchs G, Wang S, Wei W, Zhang Y, Park H, et al. A transfer-RNA-derived small RNA regulates ribosome biogenesis. Nature. 2017;552:57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Schorn AJ, Gutbrod MJ, LeBlanc C, Martienssen R. LTR-Retrotransposon Control by tRNA-Derived Small RNAs. Cell. 2017;170:61–71.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ivanov P Emerging Roles of tRNA-derived Fragments in Viral Infections: The Case of Respiratory Syncytial Virus. Mol. Ther 2015. page 1557–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Saikia M, Jobava R, Parisien M, Putnam A, Krokowski D, Gao X-H, et al. Angiogenin-cleaved tRNA halves interact with cytochrome c, protecting cells from apoptosis during osmotic stress. Mol Cell Biol. 2014;34:2450–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Balatti V, Nigita G, Veneziano D, Drusco A, Stein GS, Messier TL, et al. tsRNA signatures in cancer. Proc Natl Acad Sci U S A. 2017;114:8071–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Pekarsky Y, Balatti V, Palamarchuk A, Rizzotto L, Veneziano D, Nigita G, et al. Dysregulation of a family of short noncoding RNAs, tsRNAs, in human cancer. Proc Natl Acad Sci U S A. 2016;113:5071–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Slack FJ. Tackling Tumors with Small RNAs Derived from Transfer RNA. N Engl J Med. 2018;378:1842–3. [DOI] [PubMed] [Google Scholar]
- 124.Huang B, Yang H, Cheng X, Wang D, Fu S, Shen W, et al. tRF/miR-1280 Suppresses Stem Cell-like Cells and Metastasis in Colorectal Cancer. Cancer Res. 2017;77:3194–206. [DOI] [PubMed] [Google Scholar]
- 125.Shao Y, Sun Q, Liu X, Wang P, Wu R, Ma Z. tRF-Leu-CAG promotes cell proliferation and cell cycle in non-small cell lung cancer. Chem Biol Drug Des. 2017;90:730–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kuscu C, Kumar P, Kiran M, Su Z, Malik A, Dutta A. tRNA fragments (tRFs) guide Ago to regulate gene expression post-transcriptionally in a Dicer-independent manner. RNA. 2018;24:1093–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Li S, Xu Z, Sheng J. tRNA-Derived Small RNA: A Novel Regulatory Small Non-Coding RNA. Genes [Internet]. 2018;9. Available from: 10.3390/genes9050246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhao C, Tolkach Y, Schmidt D, Kristiansen G, Müller SC, Ellinger J. 5′-tRNA Halves are Dysregulated in Clear Cell Renal Cell Carcinoma. J Urol. 2018;199:378–83. [DOI] [PubMed] [Google Scholar]
- 129.Yeri A, Courtright A, Reiman R, Carlson E, Beecroft T, Janss A, et al. Total Extracellular Small RNA Profiles from Plasma, Saliva, and Urine of Healthy Subjects. Sci Rep. 2017;7:44061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Dhahbi JM, Spindler SR, Atamna H, Boffelli D, Martin DIK. Deep Sequencing of Serum Small RNAs Identifies Patterns of 5′ tRNA Half and YRNA Fragment Expression Associated with Breast Cancer. Biomark Cancer. SAGE Publications Ltd STM; 2014;6:BIC.S20764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Godoy PM, Bhakta NR, Barczak AJ, Cakmak H, Fisher S, MacKenzie TC, et al. Large Differences in Small RNA Composition Between Human Biofluids. Cell Rep. 2018;25:1346–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Balatti V, Rizzotto L, Miller C, Palamarchuk A, Fadda P, Pandolfo R, et al. TCL1 targeting miR-3676 is codeleted with tumor protein p53 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2015;112:2169–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Guzzi N, Bellodi C. Novel insights into the emerging roles of tRNA-derived fragments in mammalian development. RNA Biol. 2020;17:1214–22. [DOI] [PMC free article] [PubMed] [Google Scholar]