Abstract
Computational models of integrin-based adhesion complexes have revealed important insights into the mechanisms by which cells establish connections with their external environment. However, how changes in conformation and function of individual adhesion proteins regulate the dynamics of the whole adhesion complex remains largely elusive. This is because of the large separation in time and length scales between the dynamics of individual adhesion proteins (nanoseconds and nanometers) and the emergent dynamics of the whole adhesion complex (seconds and micrometers), and the limitations of molecular simulation approaches in extracting accurate free energies, conformational transitions, reaction mechanisms, and kinetic rates, that can inform mechanisms at the larger scales. In this review, we discuss models of integrin-based adhesion complexes and highlight their main findings regarding: (i) the conformational transitions of integrins at the molecular and macromolecular scales and (ii) the molecular clutch mechanism at the mesoscale. Lastly, we present unanswered questions in the field of modeling adhesions and propose new ideas for future exciting modeling opportunities.
Keywords: integrin-based adhesions, molecular dynamics simulations, coarse-graining methods, molecular clutch model
Introduction
Adhesions between cells and the extracellular matrix (ECM) are important for physiological processes, including cell survival, growth, and proliferation, and for the maintenance of tissue-level structural integrity [1]. The integrin family of transmembrane receptors creates physical connections between cells and the ECM to transmit mechanical and allosteric signals that trigger downstream biochemical events and regulate cell function. The discovery of integrins occurred in the late 1970s and was followed by the identification of cytoplasmic integrin-associated proteins, including talin, vinculin, kindlin, α-actinin, and a multitude of signaling molecules. Together, integrins and associated proteins create highly dynamic adhesion complexes that vary composition and size over time, control ECM remodeling, and adapt their ECM binding strength to regulate cell function. Today, growing interest is directed toward understanding how exactly the individual proteins of the complex, including the integrins themselves, contribute to the constant remodeling of adhesion complexes.
Integrins are dimers composed of an α and β chain forming a large extracellular ectodomain, followed by two single-pass transmembrane helices and two short cytoplasmic tails connecting to cytoplasmic proteins and adapters (Fig. 1). The ectodomain of the α chain contains a large β-propeller domain, followed by the thigh domain, and two calf domains (Fig. 1A). The ectodomain of the β chain consists of an N-terminal β-I domain, followed by the hybrid domain, the plexin-semaphorin-integrin domain (PSI), four cysteine-rich epidermal growth factor (EGF) modules (I-EGF 1–4) and β-T domain (Fig. 1B). In the inactive, bent conformation, the headpiece is bent against the lower legs, and the site for binding an ECM ligand, which is at the interface between β-propeller and β-I domains, is oriented toward the cell membrane (Fig. 1A). When integrin is active, its ectodomain is extended, with the ligand binding site oriented away from, rather than toward, the cell membrane (Fig. 1B). The transition of integrin from bent to extended is associated with an increase in ligand binding affinity and the recruitment of intracellular adaptors, which leads to sequestration of more integrins and formation of adhesion complexes [2–4].
Figure 1. Schematic of integrin conformation and domain organization.
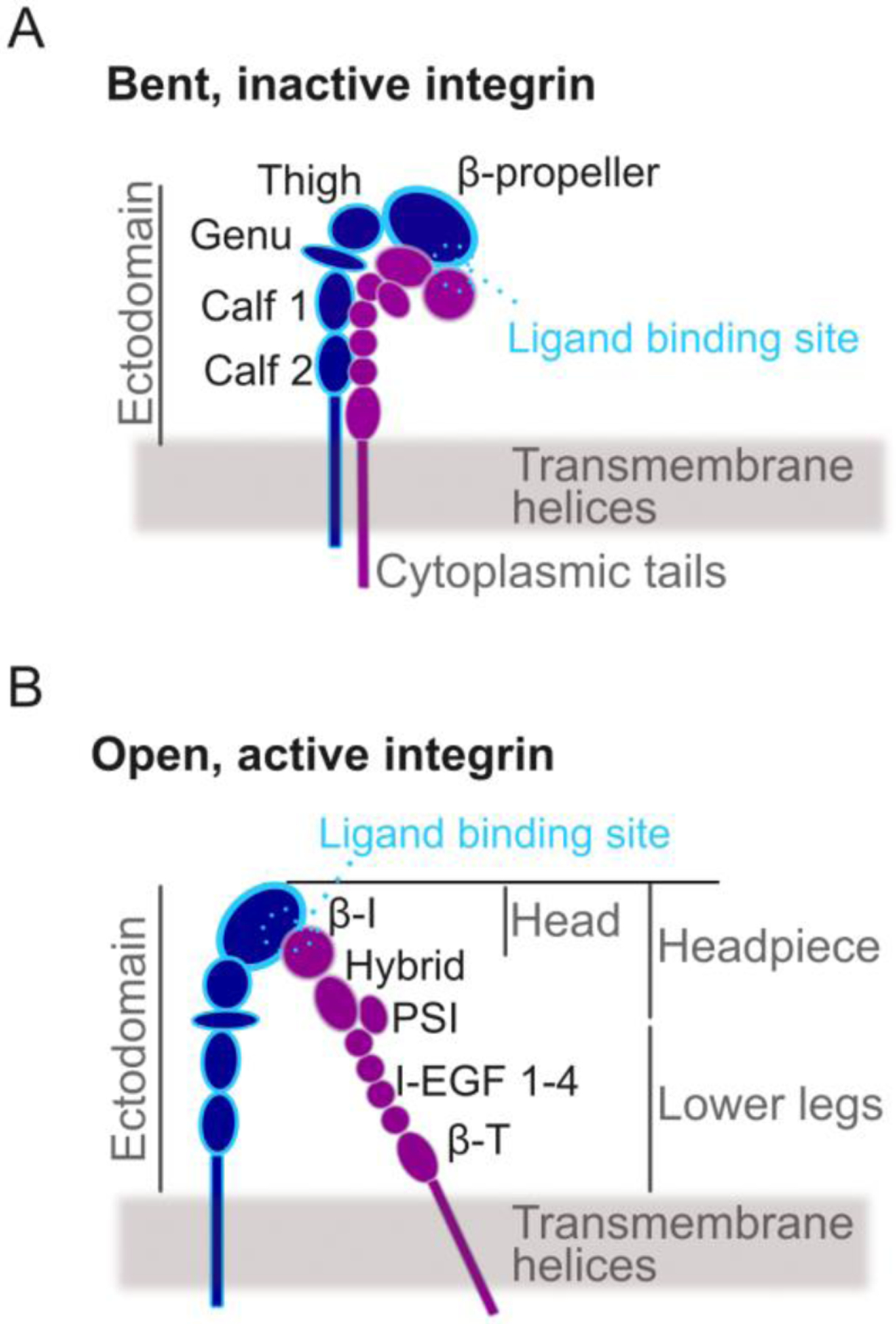
Integrins transition between: A. a bent conformation that is inactive, with low affinity for binding external ligands; B. an open conformation that is active, with high affinity for binding external ligands. The different domains of the α chain (blue) and the β chain (purple) are indicated.
Molecular modeling work has focused on how atomistic interactions of integrin drive long-range motions during its transition from bent to extended conformations [5, 6]. According to the switchblade model [7], the key pivots for integrin extension are the α chain genu, between the thigh and calf-1 domains, and the β chain knee, between I-EGF domains 1 and 2 (Fig. 2). According to the deadbolt model [8], a hairpin loop in the β-T domain acts as a deadbolt that releases the β-I domain to extend integrin (Fig. 2). These two models suggested two distinct pathways for integrin activation, but none of the current modeling approaches have fully captured these pathways because of the limited sampling of molecular simulations and the difficulty in choosing accurate collective variables for enhanced sampling methods. Mesoscale models of adhesion assembly have included experimental kinetic rates for integrin activation and deactivation and represented integrins as discrete particles switching between inactive and active conformations, without incorporating molecular details [9].
Figure 2. Models of integrin conformational activation.
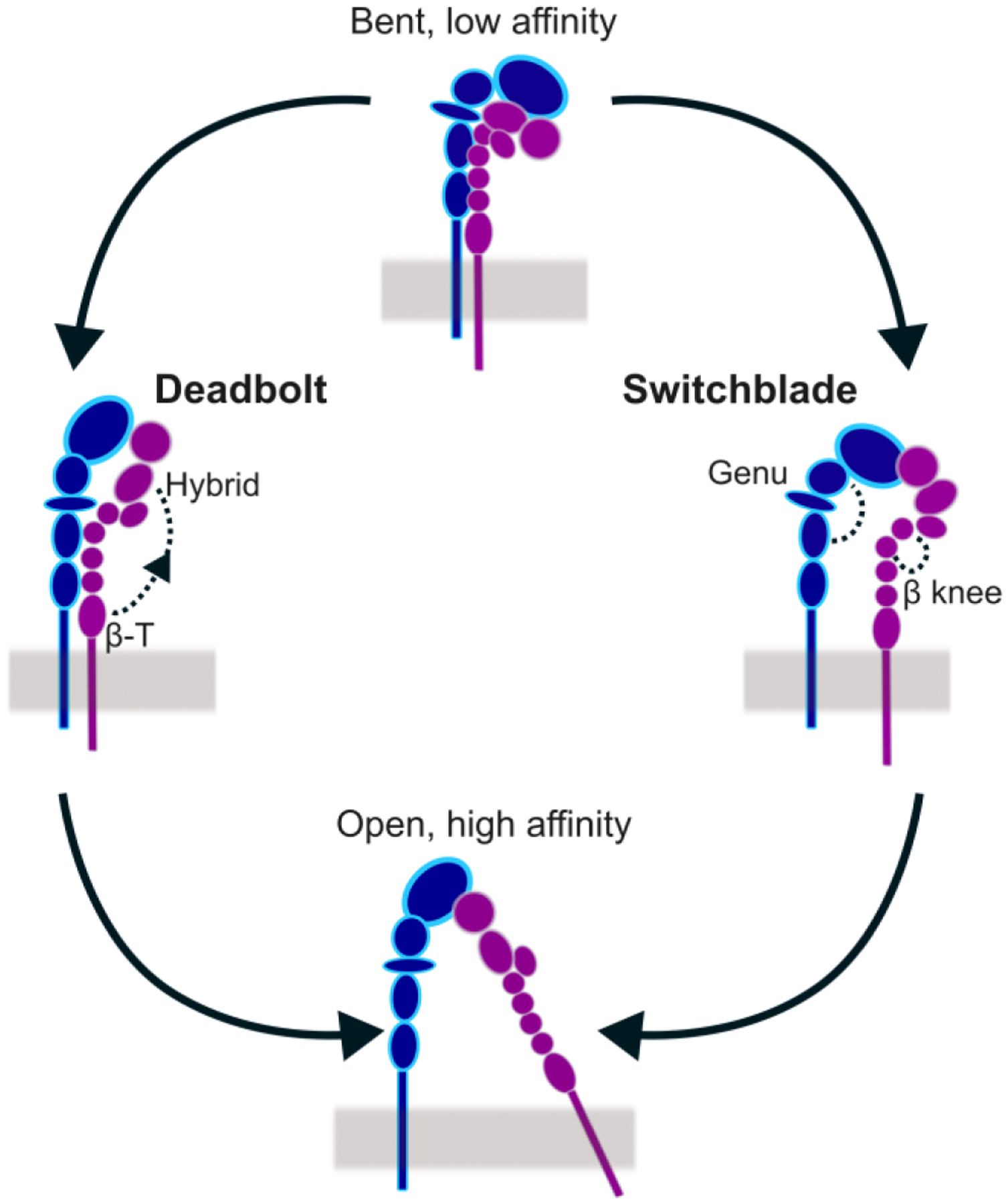
The transition from closed conformation to open conformation is characterized by intermediate integrin conformations, which differ in the degree of legs separation. According to the “deadbolt” (left) model, the headpiece extends before the legs separate laterally. According to the “switchblade” (right) model, the legs separate laterally before headpiece extension.
In cells, myosin-powered contractility and actin polymerization pushing against the cell edge drive a flow of actin filaments that is transmitted as force to the ECM through adhesion adaptors and integrins (Fig. 3). Similarly, ECM stiffness is transmitted through adhesions to the cytoskeleton. Internal and external forces change the composition of adhesions and their composition, size, and movement. A mesoscale computational model based on a coarse-grained description of adhesion complexes has described how cytoskeletal force controls the size of adhesions and physically remodel the ECM [10]. Analytical models incorporating multiple adhesion proteins into a minimal set of parameters have elucidated the relation between ECM stiffness and adhesion size, and how cytoskeletal force and adhesion velocity are connected, demonstrating a regime in which adhesion velocity depends linearly on the activation rate of integrins and ECM stiffness [11–13]. Continuum models integrating cytoskeleton architecture, adhesion adaptors, integrins and ECM stiffness have evaluated their relative contributions to the growth of adhesions [14]. In the early 2000s, the numerical implementation of a stochastic adhesion clutch model was carried out and its subsequent applications revealed the mechanisms by which adhesions sense ECM stiffness to mediate spreading and migration of cells [15–20]. The molecular clutch model is the current framework of reference for understanding how integrin-based adhesions govern cell function in response to internal and external stimuli.
Figure 3. An overview of the adhesion clutch model.
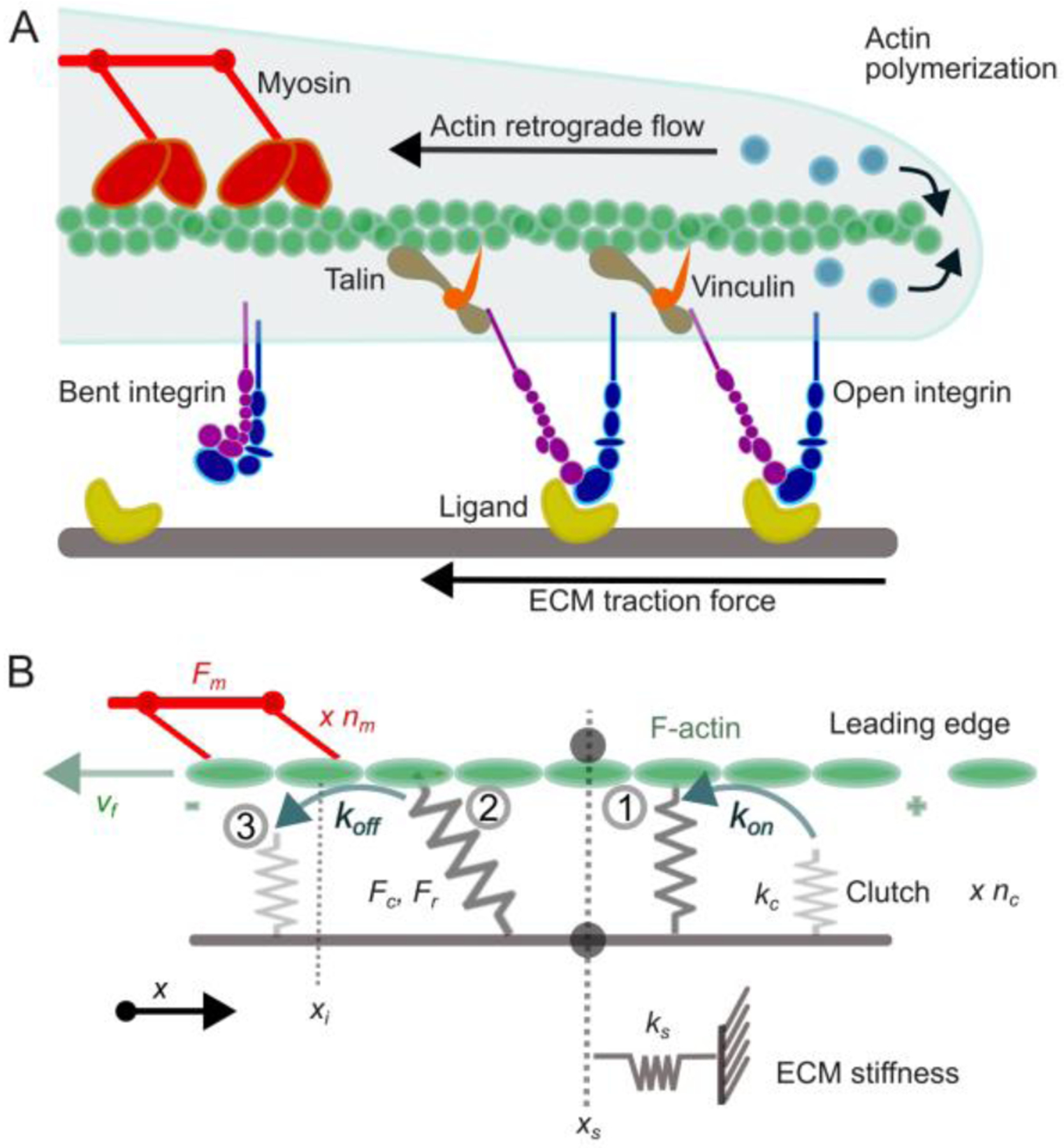
A. New actin monomers (blue) are incorporated on to the barbed end of a pre-existing actin filament (green) facing the cell leading edge membrane. Myosin motors (red) exert contractile forces on the actin filament, pulling it away from the leading edge. Transmembrane integrin dimers (blue and purple) are either bent and freely diffusive or extended and bound to ECM ligands (yellow). If integrins are not engaged to connect actin to the ECM, then actin polymerization results in rapid retrograde actin flow, no net leading edge protrusion, and no traction force on the ECM. If integrins are engaged, then talin (dark yellow) and vinculin (orange) are also recruited and the forces generated by polymerization of the actin filament and myosin motors are physically transmitted to the ECM, resulting in slowing of actin retrograde flow, traction force on the ECM, and a net edge protrusion. The complex formed by ligand-bound integrins, talins and vinculins form the adhesion clutch. B. Schematic representation of the motor-clutch model. nm myosin motors exert a maximum (a.k.a. stall) force, equal to Fmnm, and pull an F-actin bundle, resulting in F-actin sliding at velocity vf. This sliding is transmitted to the substrate through adhesions, which consist of nc clutches. Each ith clutch can be engaged with F-actin (binding rate kon) or disengaged (unbinding rate koff, that increases with force scaled to a rupture force Fr). Once engaged (1), the ith clutch is subjected to load from F-actin sliding (2) and eventually fails (3). Fc is the total force exerted by the engaged clutches, equal to Fc = ks xs, where ks is the clutch stiffness and xs is the final position of the substrate.
In this review, we discuss some of the most important findings of computational approaches used for the study of integrins and integrin-based adhesion complexes. We focus on methods that create the foundation to bridge the molecular dynamics of integrins to the scale of adhesion complexes. Lastly, we discuss unanswered questions in the field and present future modeling opportunities for bridging the gap between individual proteins and whole adhesion complexes.
All-atom and coarse-grained models of integrins
Many factors that regulate integrin conformational activation, such as ligand binding [21], divalent cations [22], and mechanical force [23], are known from experiments. However, the short lifetimes of integrin conformational transitions (< 1 s) relative to the temporal resolutions of live cell experiments (minutes) have made it challenging to capture the conformational pathway of integrin experimentally. To overcome these challenges, equilibrium and accelerated all-atom molecular dynamics (MD) simulations, combined with coarse-graining methods, are currently used.
Equilibrium and accelerated molecular dynamics simulations of integrin
Computational models based on cryo-EM or x-ray crystallographic structures of integrin have applied MD simulations to understand integrin dynamics with atomic details. These approaches rely on the application of empirical atomic force fields and use the classical Newtonian equations to calculate atomistic motions. Equilibrium MD simulations sample conformational ensembles around a local free energy minimum, and accelerated MD simulations sample a larger conformational space by decreasing the energy barriers between multiple minima. Types of accelerated MD simulations are steered MD, which applies force to perturb the atomic structure of a protein, and targeted MD, which biases an initial protein structure towards a known target, by imposing geometrical constrains.
Equilibrium, steered, and targeted MD simulations have typically used integrin αvβ3 or αIIBβ3. Steered MD simulations of integrins bound to talin or kindlin, and free energy perturbations of integrins bound to an external ligand, identified the molecular mechanisms underlining conformational activation, including affinity modifications and variations in the lifetimes of the ligand bound states [24–27].
MD-based studies identified the major energy barriers along the unbending pathway of integrin at the hybrid domain interfaces [28–30]. In the presence of an RGD sequence, which mimics ECM ligand binding, the hybrid domain opens because of molecular distortions involving reorientation of the α7 and α1 helices [28–30]. These distortions change the interaction surfaces of the hybrid domain with β propeller, β-I, PSI, β-T and EGF4 domains [28–31]. These changes, in turn, promote the swing out of the hybrid domain, followed by the opening of the hinge between the β-I and hybrid domains [32]. MD simulations also revealed that the opening of the βI/hybrid hinge is inhibited when Ca2+ is bound instead of Mn2+ [29, 33]. When force is applied to the ligand binding site, the opening of the β-I/hybrid hinge is accelerated [29, 30], suggesting that the pathway for conformational activation is the same without and with force. Once in the extended conformation, an electrostatic interaction in the α chain genu stabilizes the conformation [31]. MD simulations also demonstrated that, although these mechanisms are shared by αvβ3 and αIIBβ3 integrins, they can vary across integrin families [34].
Taken together, MD simulations provided important insights into the mechanisms underlying integrin conformational dynamics at the atomistic level. However, all-atom approaches are limited by high computational cost, low accuracy of empirical force fields, and limited sampling performance. MD simulations of integrin sample only up to tens of microseconds, a scale ~ 5 orders of magnitude shorter than the lifetime of conformational activation, and multiple independent replicates are often required to account for stochastic variability in initial conditions and for increasing the accuracy of the estimates of properties of interest, such as free energies, distribution of states, or kinetic parameters [35]. Additionally, empirical force fields inherently present low accuracy because they require predefined connectivity between atoms and are best suited to model a single protein conformation.
Coarse-grained simulations of integrin conformational activation
Capturing changes in integrin conformation with all-atom MD simulations remains a challenge because of the high computational cost, the need of multiple independent trajectories for improving sampling performance, and the low accuracy of empirical force fields. These challenges limit the access of all-atom MD simulations to collective global motions at the macromolecular scale. To address these challenges, bottom-up coarse-graining (CG) methods have provided realistic approximations of integrin macromolecular dynamics by grouping atoms into representative beads and incorporating bond breakage and formation to decrease the barriers across multiple conformations. Results identified the main directions of integrin global motions at the onset of conformational activation and the effects of residue mutations on these motions.
Bottom-up elastic and mixed elastic/plastic network models (ENMs) represent integrins as networks of beads interconnected by harmonic or mixed harmonic and anharmonic potentials, respectively [36–39]. Analysis of the normal modes of an ENM model of the bent αVβ3 integrin ectodomain has demonstrated that weakening the interactions between hybrid and β-T domains promotes the displacement of the hybrid domain away from the β leg, such that further long-range conformational changes become easier [36]. Similarly, weakening the interactions between ENM beads based on MD analysis has promoted integrin motions along a frustrated energy landscape, characterized by leg separation, headpiece extension and flattening of the α and β chain knees [38, 39]. MD-based CG models of several activating mutants have revealed the existence of mechanosensitive mutants, able to lower the force required for conformational activation [37]. The result that molecular changes in integrin confer different responses to force has been further confirmed by MD simulations on αIIBβ3 [34].
Taken together, bottom-up CG models of integrin have provided a deeper understanding of the long-range motions occurring during integrin conformational activation. However, the full pathway of integrin conformational activation remains outside the reach of current methods.
Adhesion clutch models
Mesoscale models of adhesion complexes have simplified adhesions as mechanical clutches between cells and the ECM. Adhesion clutch models have focused on the mechanisms by which adhesions convert actomyosin contractility and actin retrograde flow into ECM traction [40]. In a typical adhesion-clutch simulation, clutches have elastic properties and initially bind the ECM with a defined first-order rate constant to transmit the motion of the cytoskeleton to the ECM. As cytoskeletal force builds on the ECM-bound clutches (Fig. 3), actin flow slows and ECM traction force increases. The clutches eventually fail and release the tension, which corresponds to an increase in actin flow speed and a decrease in ECM traction force. Then, the cycle starts again, leading to a “load-and-fail” behavior [15, 17].
The adhesion clutch mechanism has been implemented in stochastic particle-based simulations, finite elements approaches, and analytical theory [17, 41–43]. The adhesion clutch model has found broad application to simulate cell behaviors due to clutch heterogeneity [19], mechanical heterogeneity and anisotropy in 1D and 2D nanofibrous environments [44], varying adhesion levels in brain tissue [45], perturbation by anticancer drugs [46], stress-relaxing environments [42], stress-relaxing cytoskeletal dynamics [47], RhoGTP signaling [48], 2D and 3D cell migration [49, 50], and negative durotaxis, i.e., migration toward softer environments [16].
Adhesion clutch models have also incorporated the effects of talin and vinculin as adaptors between integrins and actin filaments (Fig. 3), using experimentally extracted kinetic rate constants [51]. Further modeling work based on Brownian dynamics implemented the catch bond kinetics of individual integrin-ECM bonds, again using experimental lifetime versus force relations of the bonds [52]. These studies, collectively, have demonstrated that adhesion clutches aid in membrane protrusion depending on the proteins involved and bond dynamics, regulate force transmission, mechanosensing and filopodia dynamics, and can also govern cycles of membrane oscillations.
Given its ability to explain a large variety of cell phenomena, the adhesion clutch model is the framework of reference to understand adhesion-mediated processes in cells. However, given the complexity of the dynamic changes in conformation and function of individual adhesion proteins in the clutch, the exact relation between the dynamics of individual protein components and the clutch mechanism remains largely unexplored. It remains a major challenge to convert atom-level structural information into parameter values, such as clutch stiffness, on- and off-rate constants, and characteristic bond rupture forces, that inform the motor-clutch model at the mesoscale.
Concluding remarks
Cells in almost any physiological setting, from neurons within the brain to immune cells crossing tissues, form adhesions with the ECM. The development and application of MD simulations and CG models for the study of adhesion proteins, combined with mesoscale and continuum models of whole adhesion complexes, have allowed the field to gain a fundamental understanding of how cell-ECM adhesions operate under internal and external stimuli. MD and CG methods have revealed the molecular underpinnings for integrins conformational activation, which are inaccessible by current experimental approaches. Mesoscale and continuum models of adhesion clutches have revealed how integrin-based adhesions regulate force transmission, cell edge protrusion, cell mechanosensing, and migration by mediating internal and external force transmission. However, these models are still used in isolation. Because of the lack of experimental approaches with sufficient spatial and temporal resolution to monitor how the changes in structure and function of multiple adhesion proteins govern the dynamics of whole adhesion complexes, it will be interesting in the future to develop multiscale models that rigorously relate molecular mechanisms with mesoscale properties of adhesions and the clutch mechanism.
Today, while this effort is ongoing, several outstanding questions remain to be addressed. At the molecular scale, how force is transmitted and distributed across adhesion proteins remains largely elusive. Addressing this question will require developing computational methods able to track force distribution across protein domains and residues, while simulating multiple interacting proteins. Second, how exactly the emergent global motions of adhesion proteins emerge from atomistic rearrangements remains largely elusive. Developing new methods able to directly access molecular events such as rearrangements of secondary structure elements, changes in interaction surfaces between domains, and their relationship with long-range structural motion will allow us to relate molecular with macromolecular dynamics of integrins and other adhesion proteins. For these studies, multiple atomistic MD replica will be required to decrease the error of the average of the estimates and infer functional mechanisms. Lastly, it is highly likely that atomistic changes and long-range global motions of integrins and other adhesion proteins are correlated with the assembly and stabilization of whole adhesion complexes, and with the efficiency of force transmission to and from the ECM. Conformational and affinity changes of adhesion proteins can also affect the movement, size, and lifetimes of the whole adhesion. Developing new multiscale methods exploring relationships between the molecular and macromolecular properties of adhesion proteins and the mesoscale properties of adhesion complexes will allow the field to address these open questions. Multiscale methods for the study of adhesion complexes will further enable the testing and refining of novel clutch models to elucidate how the clutch mechanisms more precisely mediates cell function. Addressing these and other open questions is thus likely to lead to exciting new model developments in the coming years.
Acknowledgements
This work was supported by: NIGMS R35 GM 147491-01, NCI U54 CA210190, NCI P01 CA254849, NCI U54 CA268069, NCI R37 CA240846, NHLBI R01 HL132906, NIGMS R35 GM 136302, NIGMS R21 GM147898, NINDS R01 NS117968
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest
The authors declare no conflict of interest.
References
- 1.Schwartz MA, Schaller MD, and Ginsberg MH, Integrins: emerging paradigms of signal transduction. Annu Rev Cell Dev Biol, 1995. 11: p. 549–99. [DOI] [PubMed] [Google Scholar]
- 2.Li J, et al. , Conformational equilibria and intrinsic affinities define integrin activation. EMBO J, 2017. 36(5): p. 629–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lock JG, Wehrle-Haller B, and Strömblad S. Cell–matrix adhesion complexes: master control machinery of cell migration. in Seminars in cancer biology. 2008. Elsevier. [DOI] [PubMed] [Google Scholar]
- 4.Puklin-Faucher E and Sheetz MP, The mechanical integrin cycle. Journal of cell science, 2009. 122(2): p. 179–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kulke M and Langel W, Molecular dynamics simulations to the bidirectional adhesion signaling pathway of integrin alphaV beta3. Proteins, 2020. 88(5): p. 679–688. [DOI] [PubMed] [Google Scholar]
- 6.Kalli AC, Campbell ID, and Sansom MS, Multiscale simulations suggest a mechanism for integrin inside-out activation. Proc Natl Acad Sci U S A, 2011. 108(29): p. 11890–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luo BH, Carman CV, and Springer TA, Structural basis of integrin regulation and signaling. Annu Rev Immunol, 2007. 25: p. 619–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arnaout MA, Mahalingam B, and Xiong JP, Integrin structure, allostery, and bidirectional signaling. Annu Rev Cell Dev Biol, 2005. 21: p. 381–410. [DOI] [PubMed] [Google Scholar]
- 9.Bidone TC, et al. , Multiscale model of integrin adhesion assembly. PLoS Comput Biol, 2019. 15(6): p. e1007077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao X, et al. , Multiscale model predicts increasing focal adhesion size with decreasing stiffness in fibrous matrices. Proc Natl Acad Sci U S A, 2017. 114(23): p. E4549–E4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walcott S and Sun SX, A mechanical model of actin stress fiber formation and substrate elasticity sensing in adherent cells. Proc Natl Acad Sci U S A, 2010. 107(17): p. 7757–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harland B, Walcott S, and Sun SX, Adhesion dynamics and durotaxis in migrating cells. Phys Biol, 2011. 8(1): p. 015011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walcott S, et al. , Nucleation and decay initiation are the stiffness-sensitive phases of focal adhesion maturation. Biophys J, 2011. 101(12): p. 2919–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu Z, et al. , Two Distinct Actin Networks Mediate Traction Oscillations to Confer Focal Adhesion Mechanosensing. Biophys J, 2017. 112(4): p. 780–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan CE and Odde DJ, Traction dynamics of filopodia on compliant substrates. Science, 2008. 322(5908): p. 1687–91. [DOI] [PubMed] [Google Scholar]
- 16.*.Isomursu A, et al. , Directed cell migration towards softer environments. Nat Mater, 2022. 21(9): p. 1081–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]; The adhesion clutch model predicted, and experiments verified, that cell migration occurs towards area of ‘optimal stiffness’, including towards soft ECM. This ‘optimal stiffness’ is regulated by the number of clutches and corresponds to maximal cell traction.
- 17.Sens P, Stick-slip model for actin-driven cell protrusions, cell polarization, and crawling. Proc Natl Acad Sci U S A, 2020. 117(40): p. 24670–24678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bangasser BL, et al. , Shifting the optimal stiffness for cell migration. Nat Commun, 2017. 8: p. 15313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elosegui-Artola A, et al. , Rigidity sensing and adaptation through regulation of integrin types. Nat Mater, 2014. 13(6): p. 631–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.**.Andreu I, et al. , The force loading rate drives cell mechanosensing through both reinforcement and cytoskeletal softening. Nat Commun, 2021. 12(1): p. 4229. [DOI] [PMC free article] [PubMed] [Google Scholar]; The adhesion clutch model captured the experimental finding that the rate of force application (loading rate) govern mechanosensing of cells.
- 21.Takagi J, et al. , Structure of integrin alpha5beta1 in complex with fibronectin. EMBO J, 2003. 22(18): p. 4607–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mould AP, Akiyama SK, and Humphries MJ, Regulation of integrin alpha 5 beta 1-fibronectin interactions by divalent cations. Evidence for distinct classes of binding sites for Mn2+, Mg2+, and Ca2+. J Biol Chem, 1995. 270(44): p. 26270–7. [DOI] [PubMed] [Google Scholar]
- 23.Friedland JC, Lee MH, and Boettiger D, Mechanically activated integrin switch controls alpha5beta1 function. Science, 2009. 323(5914): p. 642–4. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, et al. , Prediction of Catch-Slip Bond Transition of Kindlin2/beta3 Integrin via Steered Molecular Dynamics Simulation. J Chem Inf Model, 2020. 60(10): p. 5132–5141. [DOI] [PubMed] [Google Scholar]
- 25.*.Su S, et al. , Force-enhanced biophysical connectivity of platelet beta3 integrin signaling through Talin is predicted by steered molecular dynamics simulations. Sci Rep, 2022. 12(1): p. 4605. [DOI] [PMC free article] [PubMed] [Google Scholar]; Steered MD simulations identified a force-enhanced linkage of talin with integrin, demonstrating the moelcular origin of catch bond kinetics between these proteins.
- 26.Ji Y, Fang Y, and Wu J, Tension Enhances the Binding Affinity of beta1 Integrin by Clamping Talin Tightly: An Insight from Steered Molecular Dynamics Simulations. J Chem Inf Model, 2022. [DOI] [PubMed] [Google Scholar]
- 27.Guest EE, et al. , Molecular Simulation of alphavbeta6 Integrin Inhibitors. J Chem Inf Model, 2020. 60(11): p. 5487–5498. [DOI] [PubMed] [Google Scholar]
- 28.Gaillard T, Dejaegere A, and Stote RH, Dynamics of beta3 integrin I-like and hybrid domains: insight from simulations on the mechanism of transition between open and closed forms. Proteins, 2009. 76(4): p. 977–94. [DOI] [PubMed] [Google Scholar]
- 29.Puklin-Faucher E and Vogel V, Integrin activation dynamics between the RGD-binding site and the headpiece hinge. J Biol Chem, 2009. 284(52): p. 36557–36568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Puklin-Faucher E, et al. , How the headpiece hinge angle is opened: New insights into the dynamics of integrin activation. J Cell Biol, 2006. 175(2): p. 349–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen W, et al. , Molecular dynamics simulations of forced unbending of integrin alpha(v)beta(3). PLoS Comput Biol, 2011. 7(2): p. e1001086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Provasi D, et al. , Targeted molecular dynamics reveals overall common conformational changes upon hybrid domain swing-out in beta3 integrins. Proteins, 2009. 77(2): p. 477–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li N, et al. , Comparison of Linear vs. Cyclic RGD Pentapeptide Interactions with Integrin alphavbeta3 by Molecular Dynamics Simulations. Biology (Basel), 2021. 10(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pijning AE, et al. , An alternate covalent form of platelet alphaIIbbeta3 integrin that resides in focal adhesions and has altered function. Blood, 2021. 138(15): p. 1359–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knapp B, Ospina L, and Deane CM, Avoiding False Positive Conclusions in Molecular Simulation: The Importance of Replicas. J Chem Theory Comput, 2018. 14(12): p. 6127–6138. [DOI] [PubMed] [Google Scholar]
- 36.Matsumoto A, et al. , Key interactions in integrin ectodomain responsible for global conformational change detected by elastic network normal-mode analysis. Biophys J, 2008. 95(6): p. 2895–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.**.Driscoll TP, et al. , Integrin-based mechanosensing through conformational deformation. Biophys J, 2021. 120(20): p. 4349–4359. [DOI] [PMC free article] [PubMed] [Google Scholar]; A new MD-based CG elastic network model (ENM) allowed capturing differeneces in integrin extension between integrin activating mutants and wild type under low forces. These results were validated experimentally using FRET sensors and traction force microscopy.
- 38.Bidone TC, et al. , Coarse-Grained Simulation of Full-Length Integrin Activation. Biophys J, 2019. 116(6): p. 1000–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.*.Kanada R, et al. , Enhanced Conformational Sampling with an Adaptive Coarse-Grained Elastic Network Model Using Short-Time All-Atom Molecular Dynamics. J Chem Theory Comput, 2022. 18(4): p. 2062–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]; A new MD-based CG elastic network model (ENM) allowed sampling various conformational ensembles of integrin while significantly reducing computational cost.
- 40.Elosegui-Artola A, Trepat X, and Roca-Cusachs P, Control of Mechanotransduction by Molecular Clutch Dynamics. Trends Cell Biol, 2018. 28(5): p. 356–367. [DOI] [PubMed] [Google Scholar]
- 41.Ghosh D, Ghosh S, and Chaudhuri A, Deconstructing the role of myosin contractility in force fluctuations within focal adhesions. Biophys J, 2022. 121(9): p. 1753–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.*.Adebowale K, et al. , Enhanced substrate stress relaxation promotes filopodia-mediated cell migration. Nat Mater, 2021. 20(9): p. 1290–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]; The adhesion clutch model predicts, and experiments confirm, that substrate relaxation confer robust cell migration on soft substrates.
- 43.Petit C, et al. , Regulation of SMC traction forces in human aortic thoracic aneurysms. Biomech Model Mechanobiol, 2021. 20(2): p. 717–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Estabridis HM, et al. , Cell Migration in 1D and 2D Nanofiber Microenvironments. Ann Biomed Eng, 2018. 46(3): p. 392–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klank RL, et al. , Biphasic Dependence of Glioma Survival and Cell Migration on CD44 Expression Level. Cell Rep, 2017. 19(3): p. 668. [DOI] [PubMed] [Google Scholar]
- 46.Prahl LS, et al. , Microtubule-Based Control of Motor-Clutch System Mechanics in Glioma Cell Migration. Cell Rep, 2018. 25(9): p. 2591–2604 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tan SJ, et al. , Regulation and dynamics of force transmission at individual cell-matrix adhesion bonds. Sci Adv, 2020. 6(20): p. eaax0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee SH, et al. , A molecular clock controls periodically driven cell migration in confined spaces. Cell Syst, 2022. 13(7): p. 514–529 e10. [DOI] [PubMed] [Google Scholar]
- 49.Prahl LS, et al. , Predicting Confined 1D Cell Migration from Parameters Calibrated to a 2D Motor-Clutch Model. Biophys J, 2020. 118(7): p. 1709–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eichinger JF, et al. , A computational framework for modeling cell-matrix interactions in soft biological tissues. Biomech Model Mechanobiol, 2021. 20(5): p. 1851–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Elosegui-Artola A, et al. , Mechanical regulation of a molecular clutch defines force transmission and transduction in response to matrix rigidity. Nat Cell Biol, 2016. 18(5): p. 540–8. [DOI] [PubMed] [Google Scholar]
- 52.Oakes PW, et al. , Lamellipodium is a myosin-independent mechanosensor. Proc Natl Acad Sci U S A, 2018. 115(11): p. 2646–2651. [DOI] [PMC free article] [PubMed] [Google Scholar]


