Abstract
The Database of Interacting Proteins (DIP; http://dip.doe-mbi.ucla.edu ) is a database that documents experimentally determined protein–protein interactions. This database is intended to provide the scientific community with a comprehensive and integrated tool for browsing and efficiently extracting information about protein interactions and interaction networks in biological processes. Beyond cataloging details of protein–protein interactions, the DIP is useful for understanding protein function and protein–protein relationships, studying the properties of networks of interacting proteins, benchmarking predictions of protein–protein interactions, and studying the evolution of protein–protein interactions.
INTRODUCTION
The Database of Interacting Proteins (DIP) aims to integrate the diverse body of experimental knowledge about interacting proteins into a single, easily accessed database. Biological knowledge about protein–protein interactions is contained in many different scientific journals and in archives such as MEDLINE (National Library of Medicine, MD, USA). Although the literature and archives are used daily by the scientific community, retrieving specialized data from such sources requires more effort than from the DIP, which combines information from multiple observations and experimental techniques as well as providing information about networks of interacting proteins.
The primary goal of DIP is to extract and integrate the wealth of information about protein–protein interactions into a user-friendly environment. Although organism-specific databases such as YPD (1) for yeast, EcoCyc (2) for Escherichia coli, and FlyNet for Drosophila (3) often contain information regarding protein pathways and protein complexes as do pathway databases such as KEGG (4) and CNSB (5), the DIP was created to complement the existing databases and to include interacting proteins from many organisms allowing scientists to expand and complement the observations of protein–protein interactions in one organism with observations from other organisms.
DESCRIPTION AND STRUCTURE OF THE DATABASE
In its original conception (6), information on protein interaction was stored in the DIP as a single text file. To handle effectively the growing body of data, the DIP has now been implemented as a relational database written in the programming language SQL, specifically mySQL (TcX Sweden). SQL efficiently handles diverse types of data and enables rapid sorting and analysis. The database can be conveniently extended as required, without altering the existing database content, by adding new fields and tables to the data structure.
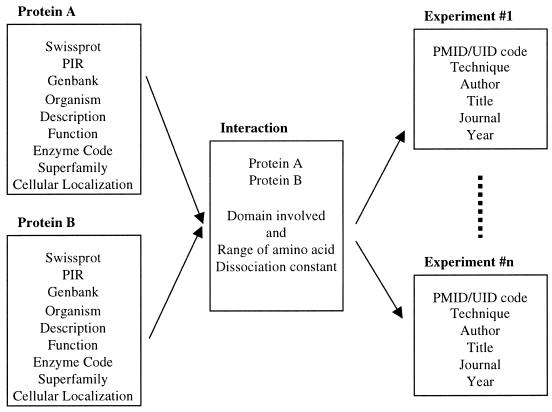
The DIP database is composed of three linked tables: a table of protein information, a table of protein–protein interactions, and a table describing details of experiments detecting the protein–protein interactions. These tables are shown schematically in Figure 1, and contain the following information.
Figure 1.
The relational structure of the DIP. The protein information (left) is linked to the interaction table (center), which in turn is linked to the experiment table (right). An interaction is a unique entry but can be linked to many different experiments.
(i) The protein information table contains protein identification codes from the SWISS-PROT (7), PIR (8) and GenBank (9) sequence databases, as well as each protein’s gene name, description, enzyme code and cellular localization, when known.
(ii) The interaction table describes proteins that interact from the protein information table, as well as the ranges of amino acids and the protein domains involved in the protein–protein interaction, when known.
(iii) The experimental article table details the experiments used to detect the interactions from the interaction table and their associated literature citations. This table includes the MEDLINE standard article code (PMID/UID), as well as the authors, title, journal and year of publication of the article. Over 20 different experimental techniques are represented in DIP, including co-immunoprecipitation, yeast two-hybrid and in vitro binding assays; for a complete list see http://dip.doe-mbi.ucla.edu/help.html . Where determined, a dissociation constant is also included.
Each interacting protein is linked to an interaction in the interaction table. Linked to the same interaction are one or more experiments from the experiment table, because some interactions are determined with many different experiments. For example, the interaction between the human proto-oncogene h-ras-1 and the ras interactor RIN1 is documented in DIP by four experimental methods (10). The scientist can therefore evaluate the quality of an interaction by the particular experiments performed.
SEARCHING THE DATABASE OF INTERACTING PROTEINS
Currently protein–protein interactions are entered into the DIP only following publication in peer-reviewed journals. Entry is done manually by the curator, followed by automated tests that show the proteins and citations exist. Interactions are double-checked by a second curator and flagged accordingly in the database.
DIP can be searched in a variety of ways. One can look for interactions involving a specific protein by entering its gene name or its accession code from GenBank, PIR or SWISS-PROT. More general searches can be performed for information such as organisms, protein superfamilies, keywords, experimental techniques or literature citations. Multiple fields can be searched simultaneously to narrow the query, and the use of wildcards and regular expressions is supported to further aid in searching. A search returns a list of protein–protein interactions, each hyperlinked to a DIP entry. Each resulting DIP entry reports information about the two interacting proteins, the protein domains and range of amino acids involved, the curator, date of entry and updating and the articles describing the interaction, and the corresponding experiments. For example, a search on a single protein returns all of the interactions recorded in DIP in which that protein participates.
CURRENT STATE OF THE DIP
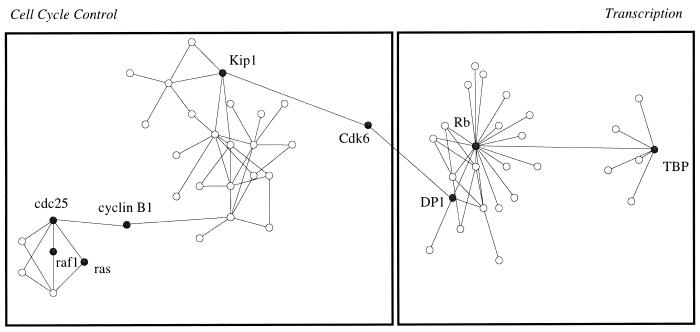
As of August 1999, the DIP contains 1089 unique proteins and 1269 pairwise interactions. Numerous large networks of protein–protein interactions are represented in DIP, as illustrated in Figure 2 for 57 proteins controlling cell cycle and transcription. The largest such network of proteins in DIP contains 514 proteins involved in cell cycle and transcription; each protein is connected to the network by at least one protein–protein interaction.
Figure 2.
Diagram of 57 interacting proteins functioning in cell cycle and transcription control contained in the DIP database. Each open circle represents a protein; each line is an experimentally determined interaction. Cell division protein kinase 6 (Cdk6) bridges cell cycle control proteins with transcription related proteins by interacting with both cyclin dependent kinase inhibitor (Kip1) and E2F related transcription factor (DP1). The highlighted proteins are the following: Rb (retinoblastoma associated protein), Kip1 (cyclin dependent kinase inhibitor), TBP (TATA binding protein associated factor II), cyclin B1 (G2/mitotic specific cyclin B1), cdc25 (M phase inducer phosphatase 1), raf1 (proto-oncogene ser/thr protein kinase), ras (transforming protein p21/ras).
FUTURE DIRECTIONS
Although the DIP has grown to its current state by manual entry of interaction data, we plan to implement automatic literature search and text extraction methods, such as those described by Blaschke et al. (11), to target interactions for inclusion in the database, followed by manual expert review. In the near future, we hope to include tools to visualize and analyze interaction network properties, as well as add a measure of interaction quality determined by the number and type of experiments defining the interaction. Finally, we hope to interactively link the DIP with the various sequence databases, such as SWISS-PROT, GenBank and PIR, as well to other databases containing interacting proteins such as KEGG, CNSB, Ecocyc, Flynet and YPD.
DATA SUBMISSION
As for SWISS-PROT, an example of a well-curated database, we would like expert curators to screen each entry to the DIP. For this reason, scientists are invited to visit and contribute to this database, which can be edited directly over the World Wide Web by obtaining a user account. To obtain an account, please contact us at dip@mbi.ucla.edu. Help for editing and submission is available online; questions can also be directed to dip@mbi.ucla.edu . Please feel free to send Email containing published protein–protein interactions, and a curator will enter this information in the DIP.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Rob Grothe for discussions at the beginning of the project and DOE for support. I.X. is a fellow of the Swiss National Fund for Research (SNFR).
REFERENCES
- 1.Hodges P.E., McKee,A.H., Davis,B.P., Payne,W.E. and Garrels,J.I. (1999) Nucleic Acids Res., 27, 69–73. Updated article in this issue: Nucleic Acids Res. (2000), 28, 73–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karp P.D., Riley,M., Paley,S.M., Pellegrini-Toole,A. and Krummenacker,M. (1999) Nucleic Acids Res., 27, 55–58. Updated article in this issue: Nucleic Acids Res. (2000), 28, 56–59.9847140 [Google Scholar]
- 3.Sanchez C., Lachaize,C., Janody,F., Bellon,B., Roder,L., Euzenat,J., Rechenmann,F. and Jacq,B. (1999) Nucleic Acids Res., 27, 89–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ogata H., Goto,S., Sato,K., Fujibuchi,W., Bono,H. and Kanehisa,M. (1999) Nucleic Acids Res., 27, 29–34. Updated article in this issue: Nucleic Acids Res. (2000), 28, 27–30.9847135 [Google Scholar]
- 5.Takai-Igarashi T., Nadaoka,Y. and Kaminuma,T. (1998) J. Comput. Biol., 5, 747–754. [DOI] [PubMed] [Google Scholar]
- 6.Marcotte E.M., Pellegrini,M., Ng,H.L., Rice,D.W., Yeates,T.O. and Eisenberg,D. (1999) Science, 285, 751–753. [DOI] [PubMed] [Google Scholar]
- 7.Bairoch A. and Apweiler,R. (1999) Nucleic Acids Res., 27, 49–54. Updated article in this issue: Nucleic Acids Res. (2000), 28, 45–48.9847139 [Google Scholar]
- 8.Barker W.C., Garavelli,J.S., McGarvey,P.B., Marzec,C.R., Orcutt,B.C., Srinivasarao,G.Y., Yeh,L.S., Ledley,R.S., Mewes,H.W., Pfeiffer,F., Tsugita,A. and Wu,C. (1999) Nucleic Acids Res., 27, 39–43. Updated article in this issue: Nucleic Acids Res. (2000), 28, 41–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benton D. (1990) Nucleic Acids Res., 18, 1517–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han L., Wong,D., Dhaka,A., Afar,D., White,M., Xie,W., Herschman,H., Witte,O. and Colicelli,J. (1997) Proc. Natl Acad. Sci. USA, 94, 4954–4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blaschke C., Andrade,M.A., Ouzounis,C. and Valencia,A. (1990) ISMB, 99, 60–67. [PubMed] [Google Scholar]