Abstract
The airway epithelial cell line, 16HBE14o−, is an important cell model for studying airway disease. 16HBE14o− cells were originally generated from primary human bronchial epithelial cells by SV40‐mediated immortalization, a process that is associated with genomic instability through long‐term culture. Here, we explore the heterogeneity of these cells, with respect to expression of the cystic fibrosis transmembrane conductance regulator (CFTR) transcript and protein. We isolate clones of 16HBE14o− with stably higher and lower levels of CFTR in comparison to bulk 16HBE14o−, designated CFTRhigh and CFTRlow. Detailed characterization of the CFTR locus in these clones by ATAC‐seq and 4C‐seq showed open chromatin profiles and higher order chromatin structure that correlate with CFTR expression levels. Transcriptomic profiling of CFTRhigh and CFTRlow cells showed that the CFTRhigh cells had an elevated inflammatory/innate immune response phenotype. These results encourage caution in interpreting functional data from clonal lines of 16HBE14o− cells, generated after genomic or other manipulations.
1. INTRODUCTION
Advances in understanding human lung disease are largely dependent on the availability of robust in vitro cell culture models. Although primary airway epithelial cell cultures are preferred, their availability is always limited, so many alternative culture models were established. These include diverse lung cancer cell lines of different cellular origins, immortalized cell lines, and more recently induced pluripotent stem cell (iPSC)‐derived lung cell types. The choice of model system is largely tailored to the inherent properties of the cell line and their relevance to the topic of investigation. In the context of developing gene‐editing therapeutics, the ability to perform genomic manipulations with CRISPR/Cas9 and subsequently isolate single cell‐derived populations with extended lifespan is critical. Among cell lines frequently used for these studies are airway cells immortalized with simian virus 40 (SV40) or derivative plasmids such as origin‐defective SV40 (Fromm & Berg, 1982). Immortalization results from the stable random integration of one to a few copies of the SV40 genome and is dependent on continued expression the SV40 large T antigen (reviewed in [Pipas, 2009]). Among such immortalized airway epithelial cell lines that are routinely used in airway disease research are BEAS‐2B (adeno‐SV40 hybrid) (Reddel et al., 1988) and 16HBE14o− (SV40 ori‐ plasmid) (Cozens et al., 1994). Disease‐specific airway epithelial cell lines include the cystic fibrosis (CF) bronchial cells IB3‐1 (adeno‐SV40 hybrid) (Zeitlin et al., 1991) and ΣCFNPE14o− (SV40 ori‐ plasmid) nasal epithelial cells (Kunzelmann et al., 1995).
CF is a life‐limiting autosomal recessive disease, caused by mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) gene. Loss or aberrant function of the CFTR protein has a profound impact on multiple epithelial cell types in the airway, pancreas, intestines and male genital ducts, among other sites. In the airway, defective anion transport by mutant CFTR is associated with thickened mucus and a lung environment susceptible to persistent infection and inflammation. The 16HBE14o− cell line which has maintained a cobblestone‐like epithelial morphology and expresses abundant CFTR transcripts and lower, but readily detectable levels of CFTR protein, is a valuable resource for CF research. When polarized on permeable supports these cells can form tight junctions and thus serve as a useful model for the study of barrier function and ion transport in respiratory disease (Callaghan et al., 2020; Heijink et al., 2012; Leir et al., 2003; Wan et al., 2000). They have also been used to investigate many aspects of airway epithelial biology including drug transport (Forbes, 2000), toxicology (Feng et al., 2015) and air pollution (Zhou et al., 2015).
In previous work, we used the 16HBE14o− line, among others, to study the regulatory mechanisms governing CFTR expression in the airway epithelium. This line has the advantage over some others that it is amenable to genome editing and the derivation of single cell clones (Kerschner et al., 2022; NandyMazumdar et al., 2020; Paranjapye et al., 2022; Santos et al., 2023; Valley et al., 2019). We identified and characterized two strong airway‐selective enhancers at −35 kb and −44 kb upstream of the CFTR promoter (Kerschner et al., 2022; NandyMazumdar et al., 2020; NandyMazumdar et al., 2021; Paranjapye et al., 2022; Zhang et al., 2012, 2013, 2015). These enhancers, and many of the transcription factors and mechanisms driving their activity, are conserved in other airway cells including the lung carcinoma cell line Calu3, primary human bronchial epithelial cells, iPSC‐derived airway epithelial cells (Kerschner et al., 2021; Mutolo et al., 2018; NandyMazumdar et al., 2020, 2021; Paranjapye et al., 2022) and adult and fetal human tissues (Bergougnoux et al., 2014). DNA sequence analysis of 16HBE14o− cells indicates that at least one copy of the pSV40 (ori‐) plasmid has integrated into intron 6 of the CFTR locus, and it has been suggested that this results in mono‐allelic expression of CFTR in these cells (Valley et al., 2019). Moreover, we observed substantial recruitment of RNA polymerase II (RNAPII) to the integration site (NandyMazumdar et al., 2020), though it is unclear whether this only occurs on the SV40 integrated allele.
In the context of isolating clonal wild‐type control lines for CRISPR/Cas9 genomic manipulations in 16HBE14o− cells, we observed a wide variation in CFTR expression levels. This variation might result from off‐target gene editing or because different cells within the 16HBE14o− population had variable endogenous CFTR expression level. We investigated the latter possibility by isolating clonal cell lines from a non‐edited population culture of 16HBE14o− cells. We identified clones with reproducibly high or low levels of CFTR in relation to bulk 16HBE14o−. Here, we characterize these two populations of cells to determine their different identities, using genome‐wide transcriptional profiling and detailed analysis of the chromatin structure and 3D interactions at the CFTR locus.
2. MATERIALS AND METHODS
2.1. CFTR nomenclature
For consistency with our previous work (Gosalia & Harris, 2015), CFTR introns and exons are numbered using legacy nomenclature (Tsui & Dorfman, 2013). RefSeq conversion is published elsewhere (Yin et al., 2022).
2.2. Cell culture
Human bronchial epithelial 16HBE14o− (Cozens et al., 1994) cell lines were cultured in Dulbecco's modified Eagle's medium, low glucose supplemented with 10% fetal bovine serum. Wild‐type 16HBE14o− (RRID:CVCL_0112), of male origin, were originally a gift from Dr. D.C. Gruenert. For RNA and protein isolation, approximately 200,000 cells were seeded into individual wells of a 12‐well plate, and media were replaced at 48 h. Following two PBS washes, cells were collected at 95%–100% confluence (~96 h post plating) for protein or RNA (see below).
2.3. Subcloning of 16HBE14o −
16HBE4o− were subcloned by manual single‐cell dilution and plated into 96‐well plates. Forty‐eight clones grown from a single cell were subcultured into 48‐well plates, with 12 clones further subcultured for profiling from 12‐well plates. RNA was collected at passages 7 and 9 after subcloning (p0), and lysates were collected at passages 8 and 10 post subcloning.
2.4. RNA preparation and reverse transcription quantitative PCR
Following the manufacturer's protocol, total RNA was extracted using TRIzol (Thermo Fisher, 15596018) and cDNA prepared using Taqman Reverse Transcription Reagents with random hexamers (Thermo Fisher, N8080234). Using primers listed in Table S1, qPCR performed with Taqman Fast Advanced Master Mix (Thermo Fisher, 4444557).
2.5. Western blot analysis
Cells were lysed in NET buffer (10 mM Tris–HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, 1% [v/v] Triton X‐100, 1X Protease Inhibitor Cocktail [Sigma‐Aldrich, P8430]). CFTR protein was extracted and western blots performed as described previously (Cai et al., 2015). Briefly, lysates were resolved on 4%/7% gels at 60–80 V using standard SDS‐PAGE protocols. Proteins were transferred to Immobilon‐P PVDF membranes (Millipore‐Sigma, IPVH00010) at 24 V overnight at 4°C. Membranes were probed with antibodies specific for CFTR (Cystic Fibrosis Foundation, CFF‐596, lot # TJ20200121100285, RRID:AB_2923486), β‐tubulin (Sigma‐Aldrich, T4026, lot # 128M4790V, RRID:AB_477577) and anti‐mouse‐HRP (Agilent/Dako, P0447, lot # 20051789, RRID:AB_2617137), and proteins detected with ECL Western Blotting Substrate (Pierce).
2.6. Omni assay for transposase accessible chromatin and deep sequencing
Omni assay for transposase accessible chromatin and deep sequencing (Omni‐ATAC‐seq) was performed on 50,000 cells as described previously (Corces et al., 2017) with minor modifications (NandyMazumdar et al., 2020). Library size distributions were visualized by TapeStation (Agilent) and quantified using the KAPA Library Quantification Kit (Roche). Libraries were pooled and sequenced on a NextSeq 550 at high output (Illumina) using 75 bp single reads. Data were processed by the ENCODE‐DCC/atac‐seq‐pipeline.
2.7. Chromosome conformation capture 4C‐seq
4C‐seq libraries were generated from 1 × 107 cells as described previously (Krijger et al., 2020). Cross‐linked chromatin from 1 × 107 cells was digested using NlaIII and DpnII as the primary or secondary restriction enzymes, respectively. 4C experiments were performed at least twice from independent passages. Viewpoint primer sequences and enzyme pairs are shown in Table S1. Quantification of 4C‐seq reads was generated using the pipe4C pipeline v1.1 (Krijger et al., 2020) with default parameters. Read density tracks of replicates were merged and then subtracted from bulk 16HBE14o− using the deepTools bigwigCompare tools (Ramírez et al., 2016).
2.8. RNA‐seq
RNA was isolated from triplicate samples of clonal lines or bulk populations of 16HBE14o− cells using TRIzol (Life Technologies) as described previously (Dobin et al., 2013) and RNA‐seq was performed by standard protocols (SR 50 bp) on a NovaSeq 6000 following generation of random hexamer primed cDNA libraries using TruSeq Stranded mRNA Sample Preparation Kits (Illumina). Raw reads were aligned with STAR 2.7 (https://github.com/alexdobin/STAR, RRID:SCR_004463) (Dobin et al., 2013). Aligned reads were then assigned to genomic features with featureCounts version 1.6.3 (RRID:SCR_012919) in the Subread package (http://subread.sourceforge.net/, RRID:SCR_009803) (Liao et al., 2014) and differential gene expression was analyzed using DEseq2 version 1.22.1. (https://www.bioconductor.org/packages/release/bioc/html/DESeq2.html, RRID:SCR_015687) (Love et al., 2014).
2.9. Gene ontology
Differentially expressed genes were filtered using a basemean >30, fold change ≥+1.5 and ≤−1.5 and a Benjamini–Hochberg adjusted p‐value ≤0.01, and were read into DAVID 2021 to generate GO terms (Sherman et al., 2022).
3. RESULTS
3.1. The 16HBE14o − cell line contains subpopulations of cells with differing CFTR levels
The 16HBE14o− airway epithelial line was manually diluted and expanded to generate clones of single‐cell origin, of which 12 were assayed for CFTR gene and CFTR protein expression (Figures 1 and S1). We identified clones in which CFTR levels were higher and lower than that of the bulk 16HBE14o− WT cell population (Figures 1b,c and S1). Two clonal lines each with high CFTR transcript and protein expression (clones 4 and 7) and low CFTR (clones 3 and 12) were studied further and are referred to herein as CFTRhigh and CFTRlow. CFTRhigh cells had a 1.7‐fold to 1.9‐fold increase in CFTR transcript and a substantial increase in CFTR protein compared to WT (Figures 1b,c and S1). CFTRlow cells showed a 3.1‐fold to 9.1‐fold decrease in CFTR transcript compared to WT and slightly reduced levels of CFTR protein (Figures 1b,c and S1). The CFTR transcript levels measured by reverse transcription quantitative PCR were confirmed in RNA‐seq data (see below) and were maintained over multiple passages, with passages 7 and 9 post‐subcloning assayed. These data show that within the 16HBE14o− airway epithelial cell line, there exist subpopulations of cells expressing differing amounts of CFTR.
FIGURE 1.
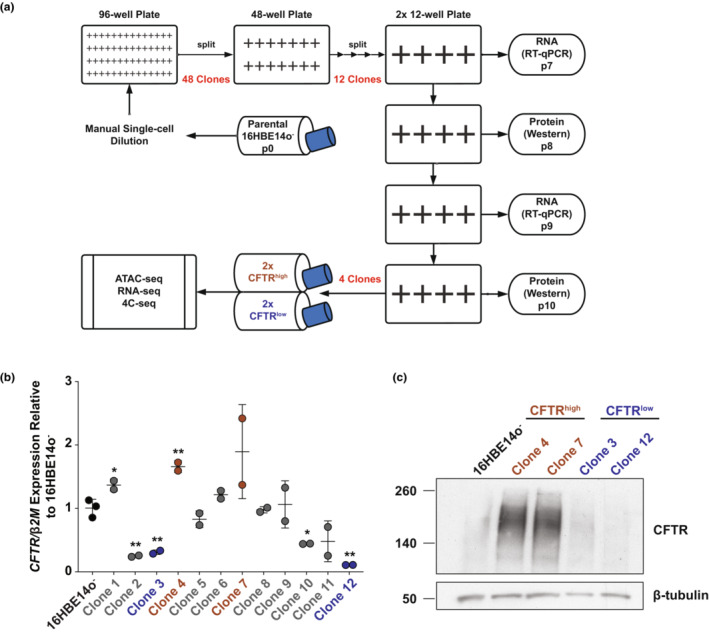
16HBE14o− cell line contains two subpopulations expressing differing amounts of CFTR transcript and protein. (a) Schema illustrates the generation of 16HBE14o− clones from single cells following manual dilution and subsequent analyses. (b) CFTR expression normalized to β2M, shown relative to 16HBE14o− WT cells (n = 2). Cells at two non‐sequential passages were evaluated for 12 clones, with the four identified for subsequent analyses shown in brown (CFTRhigh) or blue (CFTRlow). ** denotes p < 0.01 and * denotes p < 0.05, compared to 16HBE14o− using unpaired t‐tests with Welch's correction. (c) Western blot showing CFTR protein levels with an antibody specific for CFTR in 16HBE14o− cells and two clones each of CFTRhigh and CFTRlow cells. β‐tubulin provides the loading control. The data shown are representative of the two separate passages surveyed.
3.2. CFTR locus higher order chromatin structure and 3D organization differ in 16HBE14o − clonal lines and correlate with CFTR expression levels
CFTR expression is controlled by distal and intronic cis‐regulatory elements (CREs) and adopts cell‐type specific 3D conformations, which are required for normal CRE‐promoter interactions. We showed previously that deletion of CREs or depletion of activating transcription factors (TFs) disrupts these higher order chromatin interactions and alters CFTR expression levels (Kerschner et al., 2021; NandyMazumdar et al., 2020, 2021; Paranjapye et al., 2021, 2022; Yang et al., 2016; Yin et al., 2022). Hence, we next investigated whether the CFTR locus adopts different chromatin conformations and 3D interactions in the CFTRhigh and CFTRlow clonal cells.
First, we examined open chromatin profiles of cells from the bulk 16HBE14o− population, CFTRhigh clones, and CFTRlow clones using assay for transposase accessible chromatin with high‐throughput sequencing (ATAC‐seq) (Corces et al., 2017). In our earlier work, we identified peaks of open chromatin corresponding to CREs in multiple cell types including 16HBE14o− (Ott et al., 2009; Yang et al., 2016; Zhang et al., 2012). Similar to open chromatin in other airway cells, in addition to the topologically associated domain (TAD) boundaries at −80.1 kb and +48.9 kb, we see key CREs at −44 kb, −35 kb, and +6.8 kb, and a common peak of open chromatin of undefined function in intron 10 (legacy nomenclature intron 10c, RefSeq intron 11 iii [Yin et al., 2022]). Among airway cells, unique to 16HBE14o− cells are open chromatin peaks at −33 kb, −3.4 kb, and in introns 4 and 23 (legacy), though these sites are seen in some other cell types (NandyMazumdar et al., 2020; Ott et al., 2009; Yang et al., 2016). Compared to bulk 16HBE14o− cells, the profiles of CFTRhigh and CFTRlow clonal cells show the same open chromatin peaks across the CFTR locus; however, there are notable differences in the normalized relative height of two peaks when comparing CFTRhigh and CFTRlow profiles (Figures 2 and S2). Firstly, the −33 kb ATAC‐seq peak upstream of the CFTR promoter has increased accessibility to the transposase in the CFTRhigh cells compared to CFTRlow or 16HBE14o− bulk cells (Figures 2 and S2, brown arrow). The −33 kb element is adjacent to the strong airway‐selective enhancer at −35 kb, and like this CRE and another enhancer at −44 kb, has weak RNAPII enrichment in 16HBE14o− cells (Figure S2). In contrast, this site is not accessible in ATAC‐seq nor is it enriched for RNAPII in Calu3, a lung carcinoma cell line that also expresses CFTR (Figure S2), nor primary bronchial epithelial cells (Kerschner et al., 2021). Conversely, another CRE of unknown function, located in intron 10 (i10c) is of higher intensity in CFTRlow cells compare to 16HBE14o− or CFTRhigh cells (Figures 2 and S2, dark blue arrow).
FIGURE 2.
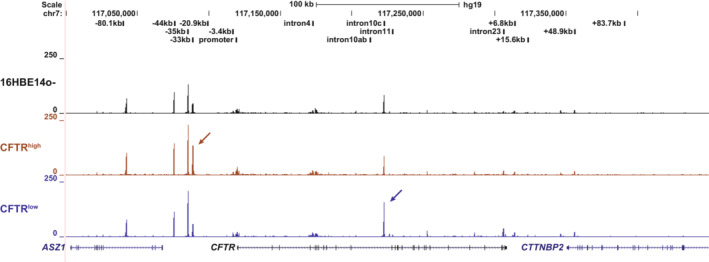
CFTRhigh and CFTRlow clones show differences in open chromatin at the CFTR locus. Open chromatin mapping of 16HBE14o− WT (black) and the mean of two clones each of CFTRhigh (clones 4 and 7, brown) and CFTRlow (clones 3 and 12, blue) cells, mapped to the hg19 genome build. Clonal cells were collected at two non‐sequential passages. Specific sites where chromatin is more open in CFTRhigh or CFTRlow clones are marked by arrows.
Next, we investigated whether the 3D organization of the CFTR locus was different in CFTRhigh or CFTRlow cells compared to 16HBE14o− cells using circular chromatin conformation capture with deep sequencing (4C‐seq) to measure long‐range chromatin contacts. We used 4C‐seq viewpoints at two key structural elements at the CFTR locus that were described previously (NandyMazumdar et al., 2020; Yang et al., 2016): the −20.9 kb CRE, a CCCTC binding factor (CTCF)‐occupied insulator element and the 3′ TAD boundary at +48.9 kb, which also binds CTCF (Blackledge et al., 2007). We first compared the −20.9 kb viewpoint interaction profiles across the CFTR locus between 16HBE14o‐ bulk cells and the CFTRhigh and CFTRlow clones (Figure 3a). In the read quantification tracks in Figure 3a, reads above the line in black denote loss of interactions compared to the 16HBE14o‐ bulk cell control interaction profile and reads below the line denote gain in interactions (in brown for CFTRhigh clones and blue for CFTRlow clones). Notably, the CFTRhigh cells have lower interaction frequencies between the −20.9 kb CRE, downstream enhancer blocking insulator elements at +6.8 kb (which binds CTCF) and +15.6 kb, as well as the 3′ TAD boundary at +48.9 kb compared to 16HBE14o− bulk cells (Figure 3a, black line). This decrease in interactions in CFTRhigh cells was confirmed reciprocally with a viewpoint at the +48.9 kb 3′ TAD boundary. Compared to bulk 16HBE14o− cells, reduced interactions were seen between this viewpoint, the 5′ TAD boundary at −80.1 kb, upstream enhancers at −44 and −35 kb, as well as the −20.9 kb CRE (Figure 3b, black line). In contrast, there was a slight increase in interactions between the −20.9 kb viewpoint, the CFTR promoter and the 5′ end of the coding region in addition to the upstream TAD boundary at −80.1 kb in CFTRhigh cells compared to 16HBE14o− (Figure 3a, brown lines). There were few changes in interactions with either the −20.9 kb CRE or the +48.9 kb site in CFTRlow cells compared to bulk 16HBE14o− cells, (Figure 3a,b). These data suggest that the higher order structure of CFTR and intra‐locus interactions correlate with high CFTR expression levels in the CFTRhigh cells.
FIGURE 3.
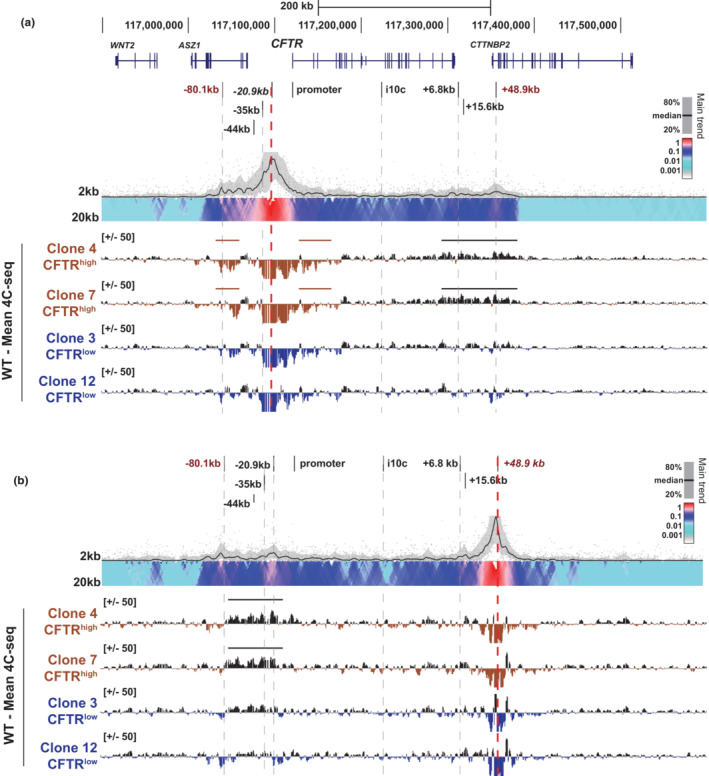
The CFTR locus 3D structure in 16HBE14o− cells correlates with CFTR expression levels. = 4C‐seq analysis of 16HBE14o− WT (black), CFTRhigh clones (brown), and CFTRlow clones (blue) with viewpoints (red dotted lines) at the −20.9 kb CRE (a) or the +48.9 kb 3’ TAD boundary (b). Key CFTR CREs are shown at the top of each panel. For each viewpoint, a 16HBE14o− WT domainogram is shown at the top and below is the subtraction of the read quantification tracks of each CFTRhigh or CFTRlow clone 4C‐seq interaction profile, from 16HBE14o− WT cells in log2 scale. Losses and gains in interactions from 16HBE14o− WT are above or below the y‐axis, respectively. Regions of specific interest are marked by horizontal bars.
3.3. The transcriptome of CFTRhigh and CFTRlow cells suggests different cellular identities
To determine whether the 16HBE14o− CFTRhigh and CFTRlow cells represented different cell lineages in the bronchial epithelium, we investigated their transcriptome. RNA‐seq was performed in triplicate on the same two clones each of CFTRhigh and CFTRlow cells that were used in the genomic analysis of CFTR described above and cells. Principal component analysis (PCA) showed strong correlation between replicates and confirmed clear transcriptomic differences between CFTRhigh and CFTRlow cells (Figure 4a). Differentially expressed genes (DEGs) between the CFTRhigh (n = 6) and CFTRlow (n = 6) phenotypes were identified by pairwise comparisons, filtering for genes with a fold change ≥1.5 or ≤−1.5, an adjusted p‐value ≤0.01, and a basemean >30 (Figure 4b, Table S2). Using these cutoffs, a total of 480 genes were differentially regulated, with 311 genes upregulated in CFTRhigh cells compared to CFTRlow cells and in the reciprocal comparison 169 genes were upregulated in CFTRlow cells (i.e., downregulated in CFTRhigh cells). GO term enrichment was performed on the DEGs upregulated in each set of cells. Among processes upregulated in CFTRhigh cells, were those associated with the inflammatory response, cellular response to lipopolysaccharide, cell migration, positive regulation of gene expression, the plasma membrane, and extracellular regions (Figure 4c). Genes associated with these terms included cytokines (CXCL6 and CXCL10), interleukins (IL6, IL1B, and IL18R1), integrin alpha 2 (ITGA2), and transcription factors such as ETS homologous factor (EHF), GATA binding protein 6 (GATA6), ETS proto‐oncogene 1, transcription factor (ETS1), and KLF transcription factor 4 (KLF4) (Table S3). Processes upregulated in CFTRlow cells included those associated with cell adhesion, the plasma membrane, cytoplasmic vesicle, and calcium ion binding (Figure 4d). Genes included in the cell adhesion biological process include protocadherins (PCDHB2, PCDHB3, PCDHGA2, and PCDHGA4) (Table S4). Protein channels with roles in epithelial cells are encoded by genes such as ATP‐binding cassette subfamily C member 2 (ABCC2), chloride voltage‐gated channel Kb (CLCNKB), transient receptor potential cation channel subfamily V member 6 (TRPV6), and the solute carriers SLC16A14, SLC40A1, and SLC9A4 are included in cellular component terms for the plasma membrane and cytoplasmic vesicle (Table S4). In combination, these transcriptomic profiles suggest that the phenotype of the CFTRhigh cells is more closely related to secretory epithelial cells in the airway, having functions in the innate immune response and inflammation, while the CFTRlow cells do not share a strong identity with any differentiated lung epithelial cell type and are likely adapted to prolonged culture in vitro.
FIGURE 4.
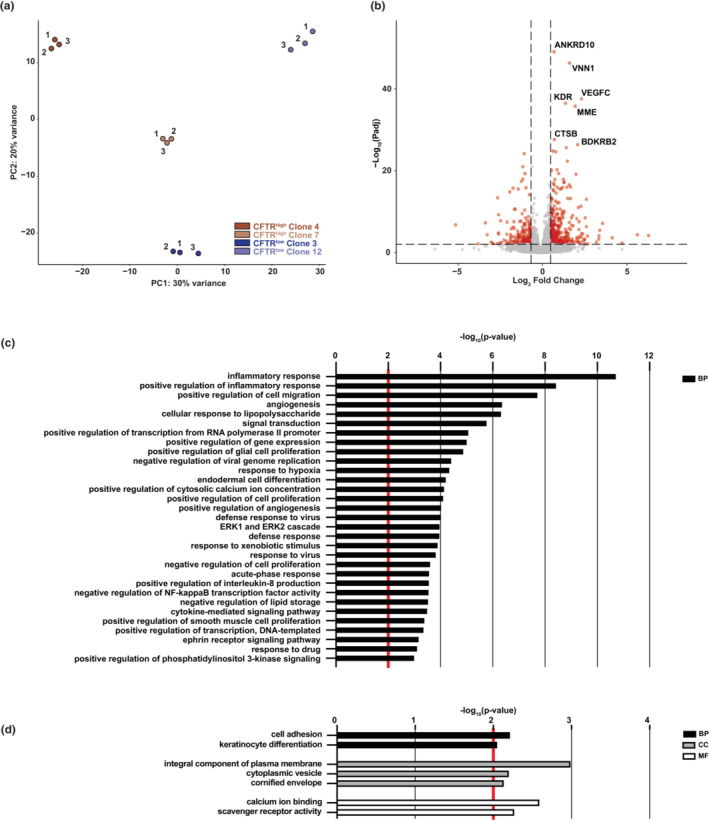
Transcriptomic differences of CFTRhigh and CFTRlow cells. (a) Principal component analysis (PCA) plot comparing RNA‐seq data from individual replicates of CFTRhigh and CFTRlow clones (n = 3 of each). (b) Volcano plot of RNA‐seq data comparing CFTRhigh and CFTRlow cells (−log10 adjusted p‐value vs. log2 fold change) with genes upregulated in CFTRlow and in CFTRhigh cells shown to the left and right, respectively. Shown in red are genes with an absolute fold change of ≥1.5 and an adjusted p‐value of ≤0.01. (c,d) Gene ontology process enrichment analysis of 311 genes upregulated in CFTRhigh cells (c) and 169 genes upregulated in CFTRlow cells (d), when comparing CFTRhigh and CFTRlow. Terms with a p‐value of ≤0.01 (denoted by red line) for biological process (BP), cellular component (CC), and molecular function (MF) are shown, with only the top 20 biological process (BP) terms listed in (c).
4. DISCUSSION
The 16HBE14o− cell line has been widely used as a surrogate for primary HBE cells in many studies of epithelial function in airway disease. However, like other immortalized human cell lines, it has limitations, mainly arising from sequelae of the immortalization procedure and subsequent dedifferentiation in prolonged culture in vitro. Here, we investigated the heterogeneity of the 16HBE14o− cell line, particularly with respect to CFTR/CFTR expression since this line and its gene‐edited derivatives are frequently used as the model system of choice in CF research (Bednarski et al., 2016; Erwood et al., 2020; Ko et al., 2022; Michaels et al., 2022; Santos et al., 2022, 2023; Valley et al., 2019). We performed single‐cell cloning to isolate clonal populations of 16HBE14o−‐derived cells and found clones that stably express high or low levels of CFTR mRNA and protein. We further characterized two clones each phenotype (Figure 1).
Focusing first on the CFTR locus and its CREs, open chromatin profiling revealed accessibility differences at −33 kb upstream of the CFTR promoter and at a site within intron 10 (legacy, Refseq intron 11iii) of the gene (Figure 2). The −33 kb CRE, which to date is functionally uncharacterized, is only observed in 16HBE14o− cells and not in the CFTR‐expressing lung adenocarcinoma cell line Calu3 (Figure S2) or primary human bronchial epithelial cells (Kerschner et al., 2021; Suzuki et al., 2020). This site is more open in CFTRhigh cells compared to parental 16HBE14o− or CFTRlow cells and it is enriched for RNAPII and H3K27ac in bulk 16HBE14o− cells (NandyMazumdar et al., 2020). Of note, its open chromatin state is not dependent on the presence or position of the strong enhancer at −35 kb in these cells (Kerschner et al., 2022; NandyMazumdar et al., 2020). Inspecting the ENCODE DNaseI hypersensitivity 125 cell types track (Thurman et al., 2012), shows open chromatin at this site only in primary human epidermal keratinocytes from a single donor (Lonza, NHEK), suggesting that this CRE has a highly specialized function in 16HBE14o− cells.
The i10c CRE (Refseq i11iii) was first identified as a site of open chromatin in Caco2 cells, a colorectal adenocarcinoma cell line which expresses CFTR (Smith et al., 2000 ), however, it lacked enhancer activity in these cells (Phylactides et al., 2002); moreover, it was evident in skin fibroblasts that do not express CFTR (Yang et al., 2016). The i10c CRE chromatin is also open in many cell types profiled in the ENCODE database which do not express CFTR. In addition to being a site of open chromatin in 16HBE14o− cells (NandyMazumdar et al., 2020; Zhang et al., 2013), i10c interacts with the transcription factor orthodenticle homeobox 2 (OTX2) where it represses CFTR in definitive endoderm cells (Kerschner et al., 2021). Consistent with the earlier data, the accessibility of this element is inversely correlated to CFTR expression, again suggesting a repressive role in these cells. Of note, interactions across the CFTR locus including the i10c region were diminished in airway cells when CFTR expression was de‐repressed (Paranjapye et al., 2022).
While the 3D looping structure of CFTR in the CFTRlow cells was not substantially different from bulk 16HBE14o− cells, we observed multiple changes to the 3D organization of the locus in CFTRhigh cells (Figure 3). Many of the same alterations were observed in Calu3 and 16HBE14o− cells in which CFTR expression increased following transient depletion or stable deletion of the CFTR repressor, Krüppel‐like factor 5 (KLF5) (Paranjapye et al., 2022). In the CFTRhigh cells, as well as the KLF5‐modulated Calu3 and 16HBE14o− cells, a decrease in the association between the 5′ and 3’ CFTR TAD boundaries was observed. Concurrently, increased interactions were detected between the −20.9 kb CTCF‐bound insulator, the CFTR promoter and the 5’ TAD boundary. This suggests that in some airway epithelial cells, the unperturbed 3D locus structure supports higher levels of CFTR expression.
Examination of the RNA‐seq data comparing CFTRhigh and CFTRlow cells did not reveal DEG profiles that enable clear assignment of a known airway epithelial cell identity (Figure 4). Furthermore, there was no obvious difference in cell morphology when comparing CFTRhigh and CFTRlow cells (data not shown). However, the most significantly upregulated processes: inflammatory response and innate immunity (response to bacteria and viruses) indicate that the CFTRhigh cells may be more closely related to surface secretory cells in the airway than to other cell types. In addition to experiments on CFTR regulation and CFTR function, 16HBE14o− cells are frequently used as a model to study infection and the innate immune response in airway disease. Our data would caution against using clonal isolates of these cells to study inflammation or the immune response, as different responses may reflect the parental cell of origin and not the genetic manipulation under study.
While loss of functional CFTR protein is associated with chronic inflammation in the lung, this is likely due to the recurrent cycles of infection (Cantin et al., 2015; Cohen‐Cymberknoh et al., 2013). None of the cells examined here were exposed to inflammatory agonists during culture; hence, the transcriptomic profiles of the clonal cell lines likely reflect the intrinsic properties of the cell of origin and not merely the levels of CFTR protein.
It is unclear from the original reports whether the 16HBE14o− cell line available to researchers today was truly a clonal isolate (Cozens et al., 1994). Irrespective of this, the line has evolved over many decades in culture to become a heterogeneous population of cells. This is not unusual as a consequence of SV40‐mediated immortalization, which may result in loss of allelic heterozygosity and/or increased ploidy (Meisner et al., 1988). In the experiments presented here, we provide evidence for the heterogeneity of the 16HBE14o− cell line when grown as standard 2D cultures. It is possible that further transcriptional divergence may arise if cells are grown under different conditions for example at air–liquid interface on permeable supports, or in response to cellular perturbations or stimuli. It would also be of interest to investigate the proteome of the cells, though in other 16HBE14o− derived cell lines transcriptome and proteome differences were positively correlated (Santos et al., 2023). However, our data encourage caution when interpreting experiments performed in 16HBE14o− cells, and suggest that multiple different clonal populations should be assayed following any genetic manipulation of this line.
AUTHOR CONTRIBUTIONS
Alekh Paranjapye and Ann Harris conceived the project. Alekh Paranjapye and Jenny L. Kerschner performed experiments. Alekh Paranjapye and Jenny L. Kerschner analyzed data. Alekh Paranjapye, Jenny L. Kerschner, and Ann Harris interpreted results. Jenny L. Kerschner and Ann Harris wrote the manuscript.
FUNDING INFORMATION
This work was supported by the National Institutes of Health (R01 HL094585 [A.H.]; T32 GM008056 [A. P.]) and the Cystic Fibrosis Foundation (Harris 18P0, Davis 19XX0). The funders had no role in the design of the study, in the collection, analysis, or interpretation of the data; in the writing of the manuscript; or in the decision to publish the results. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
ETHICS STATEMENT
This study did not involve human or animal subjects.
DISCLOSURES
The authors declare no conflict of interest.
Supporting information
Table S1.
Table S2.
Table S3.
Table S4.
ACKNOWLEDGMENTS
We thank the University of Chicago Genomics Core and CWRU School of Medicine Genomics Core for sequencing and Dr. G.C. Coatti for assistance.
Kerschner, J. L. , Paranjapye, A. , & Harris, A. (2023). Cellular heterogeneity in the 16HBE14o − airway epithelial line impacts biological readouts. Physiological Reports, 11, e15700. 10.14814/phy2.15700
Jenny L. Kerschner and Alekh Paranjapye contributed equally to this study.
DATA AVAILABILITY STATEMENT
The deep sequencing data supporting the findings of this study are available at GEO accession GSE220855.
REFERENCES
- Bednarski, C. , Tomczak, K. , Vom, H. B. , Weber, W. M. , & Cathomen, T. (2016). Targeted integration of a super‐exon into the CFTR locus leads to functional correction of a cystic fibrosis cell line model. PLoS One, 11, e0161072. 10.1371/JOURNAL.PONE.0161072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergougnoux, A. , Rivals, I. , Liquori, A. , Raynal, C. , Varilh, J. , Magalhães, M. , Perez, M. J. , Bigi, N. , Des Georges, M. , Chiron, R. , Squalli‐Houssaini, A. S. , Claustres, M. , & De Sario, A. (2014). A balance between activating and repressive histone modifications regulates cystic fibrosis transmembrane conductance regulator (CFTR) expression in vivo. Epigenetics, 9, 1007–1017. 10.4161/EPI.28967/SUPPL_FILE/KEPI_A_10928967_SM0001.ZIP [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackledge, N. P. , Carter, E. J. , Evans, J. R. , Lawson, V. , Rowntree, R. K. , & Harris, A. (2007). CTCF mediates insulator function at the CFTR locus. The Biochemical Journal, 408, 267–275. 10.1042/BJ20070429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, Z. , Palmai‐Pallag, T. , Khuituan, P. , Mutolo, M. J. , Boinot, C. , Liu, B. , Scott‐Ward, T. S. , Callebaut, I. , Harris, A. , & Sheppard, D. N. (2015). Impact of the F508del mutation on ovine CFTR, a Cl− channel with enhanced conductance and ATP‐dependent gating. The Journal of Physiology, 593, 2427–2446. 10.1113/JP270227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaghan, P. J. , Ferrick, B. , Rybakovsky, E. , Thomas, S. , & Mullin, J. M. (2020). Epithelial barrier function properties of the 16HBE14o‐ human bronchial epithelial cell culture model. Bioscience Reports, 40, 20201532. 10.1042/BSR20201532/226530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantin, A. M. , Hartl, D. , Konstan, M. W. , & Chmiel, J. F. (2015). Inflammation in cystic fibrosis lung disease: Pathogenesis and therapy. Journal of Cystic Fibrosis, 14, 419–430. 10.1016/J.JCF.2015.03.003 [DOI] [PubMed] [Google Scholar]
- Cohen‐Cymberknoh, M. , Kerem, E. , Ferkol, T. , & Elizur, A. (2013). Airway inflammation in cystic fibrosis: Molecular mechanisms and clinical implications. Thorax, 68, 1157–1162. 10.1136/THORAXJNL-2013-203204 [DOI] [PubMed] [Google Scholar]
- Corces, M. R. , Trevino, A. E. , Hamilton, E. G. , Greenside, P. G. , Sinnott‐Armstrong, N. A. , Vesuna, S. , Satpathy, A. T. , Rubin, A. J. , Montine, K. S. , Wu, B. , Kathiria, A. , Cho, S. W. , Mumbach, M. R. , Carter, A. C. , Kasowski, M. , Orloff, L. A. , Risca, V. I. , Kundaje, A. , Khavari, P. A. , … Chang, H. Y. (2017). An improved ATAC‐seq protocol reduces background and enables interrogation of frozen tissues. Nature Methods, 14(10), 959–962. 10.1038/nmeth.4396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozens, A. L. , Yezzi, M. J. , Kunzelmann, K. , Ohrui, T. , Chin, L. , Eng, K. , Finkbeiner, W. E. , Widdicombe, J. H. , & Gruenert, D. C. (1994). CFTR expression and chloride secretion in polarized immortal human bronchial epithelial cells. American Journal of Respiratory Cell and Molecular Biology, 10, 38–47. 10.1165/AJRCMB.10.1.7507342 [DOI] [PubMed] [Google Scholar]
- Dobin, A. , Davis, C.A. , Schlesinger, F., Drenkow, J. , Zaleski, C. , Jha, S. , Batut, P. , Chaisson, M. , Gingeras, T.R. , (2013). STAR: ultrafast universal RNA‐seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwood, S. , Laselva, O. , Bily, T. M. I. , Brewer, R. A. , Rutherford, A. H. , Bear, C. E. , & Ivakine, E. A. (2020). Allele‐specific prevention of nonsense‐mediated decay in cystic fibrosis using homology‐independent genome editing. Molecular Therapy‐Methods & Clinical Development, 17, 1118–1128. 10.1016/J.OMTM.2020.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, W. , Guo, J. , Huang, H. , Xia, B. , Liu, H. , Li, J. , Lin, S. , Li, T. , Liu, J. , & Li, H. (2015). Human Normal bronchial epithelial cells: A novel In vitro cell model for toxicity evaluation. PLoS One, 10, e0123520. 10.1371/JOURNAL.PONE.0123520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes, B. (2000). Human airway epithelial cell lines for in vitro drug transport and metabolism studies. Pharmaceutical Science & Technology Today, 3, 18–27. 10.1016/S1461-5347(99)00231-X [DOI] [PubMed] [Google Scholar]
- Fromm, M. , & Berg, P. (1982). Deletion mapping of DNA regions required for SV40 early region promoter function in vivo. Journal of Molecular and Applied Genetics, 1, 457–481. [PubMed] [Google Scholar]
- Gosalia, N. , & Harris, A. (2015). Chromatin dynamics in the regulation of CFTR expression. Genes (Basel), 6, 543–558. 10.3390/GENES6030543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijink, I. H. , Brandenburg, S. M. , Postma, D. S. , & van Oosterhout, A. J. M. (2012). Cigarette smoke impairs airway epithelial barrier function and cell–cell contact recovery. European Respiratory Journal, 39, 419–428. 10.1183/09031936.00193810 [DOI] [PubMed] [Google Scholar]
- Kerschner, J. L. , Paranjapye, A. , NandyMazumdar, M. , Yin, S. , Leir, S. H. , & Harris, A. (2021). OTX2 regulates CFTR expression during endoderm differentiation and occupies 3′ cis‐regulatory elements. Developmental Dynamics, 250, 684–700. 10.1002/DVDY.293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerschner JL, Paranjapye A, Vaghela N, Wilson MD, Harris A. (2022). An ectopic enhancer restores CFTR expression through de Novo chromatin looping. Gene therapy. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko, W. , Porter, J. J. , Sipple, M. T. , Edwards, K. M. , & Lueck, J. D. (2022). Efficient suppression of endogenous CFTR nonsense mutations using anticodon‐engineered transfer RNAs. Molecular Therapy‐Nucleic Acids, 28, 685–701. 10.1016/J.OMTN.2022.04.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krijger, P. H. L. , Geeven, G. , Bianchi, V. , Hilvering, C. R. E. , & de Laat, W. (2020). 4C‐seq from beginning to end: A detailed protocol for sample preparation and data analysis. Methods, 170, 17–32. 10.1016/J.YMETH.2019.07.014 [DOI] [PubMed] [Google Scholar]
- Kunzelmann, K. , Lei, D. C. , Eng, K. , Escobar, L. C. , Koslowsky, T. , & Gruenert, D. C. (1995). Epithelial cell specific properties and genetic complementation in a ΔF508 cystic fibrosis nasal polyp cell line. In Vitro Cellular & Developmental Biology‐Animal, 31(8), 617–624. 10.1007/BF02634315 [DOI] [PubMed] [Google Scholar]
- Leir, S. H. , Holgate, S. T. , & Lackie, P. M. (2003). Inflammatory cytokines can enhance CD44‐mediated airway epithelial cell adhesion independently of CD44 expression. American Journal of Physiology. Lung Cellular and Molecular Physiology, 285, 1305–1311. 10.1152/ajplung.00255.2002 [DOI] [PubMed] [Google Scholar]
- Liao, Y. , Smyth, G. K. , & Shi, W. (2014). featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics, 30, 923–930. 10.1093/BIOINFORMATICS/BTT656 [DOI] [PubMed] [Google Scholar]
- Love, M. I. , Huber, W. , & Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biology, 15, 1–21. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisner, L. F. , Wu, S.‐Q. , Christian, B. J. , & Reznikoff, C. A. (1988). Cytogenetic instability with balanced chromosome changes in an SV40 transformed human uroepithelial cell line. Cancer Research, 48, 3215–3220. [PubMed] [Google Scholar]
- Michaels, W. E. , Pena‐Rasgado, C. , Kotaria, R. , Bridges, R. J. , & Hastings, M. L. (2022). Open reading frame correction using splice‐switching antisense oligonucleotides for the treatment of cystic fibrosis. Proceedings of the National Academy of Sciences of the United States of America, 119, e2114886119. 10.1073/PNAS.2114886119/SUPPL_FILE/PNAS.2114886119.SAPP.PDF [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutolo, M. J. , Leir, S. H. , Fossum, S. L. , Browne, J. A. , & Harris, A. (2018). A transcription factor network represses CFTR gene expression in airway epithelial cells. Biochemical Journal, 475, 1323–1334. 10.1042/BCJ20180044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- NandyMazumdar, M. , Paranjapye, A. , Browne, J. , Yin, S. , Leir, S. H. , & Harris, A. (2021). BACH1, the master regulator of oxidative stress, has a dual effect on CFTR expression. The Biochemical Journal, 478, 3741–3756. 10.1042/BCJ20210252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- NandyMazumdar, M. , Yin, S. , Paranjapye, A. , Kerschner, J. L. , Swahn, H. , Ge, A. , Leir, S. H. , & Harris, A. (2020). Looping of upstream cis‐regulatory elements is required for CFTR expression in human airway epithelial cells. Nucleic Acids Research, 48, 3513–3524. 10.1093/NAR/GKAA089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott, C. J. , Blackledge, N. P. , Kerschner, J. L. , Leir, S.‐H. , Crawford, G. E. , Cotton, C. U. , & Harris, A. (2009). Intronic enhancers coordinate epithelial‐specific looping of the active CFTR locus. Proceedings of the National Academy of Sciences of the United States of America, 106, 19934–19939. 10.1073/pnas.0900946106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paranjapye, A. , NandyMazumdar, M. , Browne, J. A. , Leir, S. H. , & Harris, A. (2021). Krüppel‐like factor 5 regulates wound repair and the innate immune response in human airway epithelial cells. Journal of Biological Chemistry, 297, 100932. 10.1016/J.JBC.2021.100932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paranjapye, A. , NandyMazumdar, M. , & Harris, A. (2022). Krüppel‐like factor 5 regulates CFTR expression through repression by maintaining chromatin architecture coupled with direct enhancer activation. Journal of Molecular Biology, 434, 167561. 10.1016/J.JMB.2022.167561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phylactides, M. , Rowntree, R. , Nuthall, H. , Ussery, D. , Wheeler, A. , & Harris, A. (2002). Evaluation of potential regulatory elements identified as DNase I hypersensitive sites in the CFTR gene. European Journal of Biochemistry, 269, 553–559. 10.1046/J.0014-2956.2001.02679.X [DOI] [PubMed] [Google Scholar]
- Pipas, J. M. (2009). SV40: Cell transformation and tumorigenesis. Virology, 384, 294–303. 10.1016/J.VIROL.2008.11.024 [DOI] [PubMed] [Google Scholar]
- Ramírez, F. , Ryan, D. P. , Grüning, B. , Bhardwaj, V. , Kilpert, F. , Richter, A. S. , Heyne, S. , Dündar, F. , & Manke, T. (2016). deepTools2: A next generation web server for deep‐sequencing data analysis. Nucleic Acids Research, 44, W160–W165. 10.1093/NAR/GKW257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddel, R. R. , Ke, Y. , Gerwin, B. I. , McMenamin, M. G. , Lechner, J. F. , Su, R. T. , Brash, D. E. , Park, J.‐B. , Rhim, J. S. , & Harris, C. C. (1988). Transformation of human bronchial epithelial cells by infection with SV40 or adenovirus‐12 SV40 hybrid virus, or transfection via strontium phosphate coprecipitation with a plasmid containing SV40 early region genes. Cancer Research, 48, 1904–1909. [PubMed] [Google Scholar]
- Santos, L. , Mention, K. , Cavusoglu‐Doran, K. , Sanz, D. J. , Bacalhau, M. , Lopes‐Pacheco, M. , Harrison, P. T. , & Farinha, C. M. (2022). Comparison of Cas9 and Cas12a CRISPR editing methods to correct the W1282X‐CFTR mutation. Journal of Cystic Fibrosis, 21, 181–187. 10.1016/J.JCF.2021.05.014 [DOI] [PubMed] [Google Scholar]
- Santos, L. , Nascimento, R. , Duarte, A. , Railean, V. , Amaral, M. D. , Harrison, P. T. , Gama‐Carvalho, M. , & Farinha, C. M. (2023). Mutation‐class dependent signatures outweigh disease‐associated processes in cystic fibrosis cells. Cell & Bioscience, 13, 1–22. 10.1186/S13578-023-00975-Y/TABLES/2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, B. T. , Hao, M. , Qiu, J. , Jiao, X. , Baseler, M. W. , Lane, H. C. , Imamichi, T. , & Chang, W. (2022). DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Research, 50, W216–W221. 10.1093/NAR/GKAC194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, D. J. , Nuthall, H. N. , Majetti, M. E. , & Harris, A. (2000). Multiple potential intragenic regulatory elements in the CFTR gene. Genomics, 64, 90–96. 10.1006/GENO.1999.6086 [DOI] [PubMed] [Google Scholar]
- Suzuki, S. , Crane, A. M. , Anirudhan, V. , Barillà, C. , Matthias, N. , Randell, S. H. , Rab, A. , Sorscher, E. J. , Kerschner, J. L. , Yin, S. , Harris, A. , Mendel, M. , Kim, K. , Zhang, L. , Conway, A. , & Davis, B. R. (2020). Highly efficient gene editing of cystic fibrosis patient‐derived airway basal cells results in functional CFTR correction. Molecular Therapy, 28, 1684–1695. 10.1016/j.ymthe.2020.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurman, R. E. , Rynes, E. , Humbert, R. , Vierstra, J. , Maurano, M. T. , Haugen, E. , Sheffield, N. C. , Stergachis, A. B. , Wang, H. , Vernot, B. , Garg, K. , John, S. , Sandstrom, R. , Bates, D. , Boatman, L. , Canfield, T. K. , Diegel, M. , Dunn, D. , Ebersol, A. K. , … Crawford, G. E. (2012). Stamatoyannopoulos JA. The accessible chromatin landscape of the human genome. Nature, 489(7414), 75–82. 10.1038/nature11232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui, L. C. , & Dorfman, R. (2013). The cystic fibrosis gene: A molecular genetic perspective. Cold Spring Harbor Perspectives in Medicine, 3, a009472. 10.1101/CSHPERSPECT.A009472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valley, H. C. , Bukis, K. M. , Bell, A. , Cheng, Y. , Wong, E. , Jordan, N. J. , Allaire, N. E. , Sivachenko, A. , Liang, F. , Bihler, H. , Thomas, P. J. , Mahiou, J. , & Mense, M. (2019). Isogenic cell models of cystic fibrosis‐causing variants in natively expressing pulmonary epithelial cells. Journal of Cystic Fibrosis, 18, 476–483. 10.1016/J.JCF.2018.12.001 [DOI] [PubMed] [Google Scholar]
- Wan, H. , Winton, H. , Soeller, C. , Stewart, G. , Thompson, P. , Gruenert, D. , Cannell, M. , Garrod, D. , & Robinson, C. (2000). Tight junction properties of the immortalized human bronchial epithelial cell lines Calu‐3 and 16HBE14o−. European Respiratory Journal, 15, 1058–1068. [DOI] [PubMed] [Google Scholar]
- Yang, R. , Kerschner, J. L. , Gosalia, N. , Neems, D. , Gorsic, L. K. , Safi, A. , Crawford, G. E. , Kosak, S. T. , Leir, S. H. , & Harris, A. (2016). Differential contribution of cis‐regulatory elements to higher order chromatin structure and expression of the CFTR locus. Nucleic Acids Research, 44, 3082–3094. 10.1093/NAR/GKV1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, S. , NandyMazumdar, M. , Paranjapye, A. , & Harris, A. (2022). Cross‐talk between enhancers, structural elements and activating transcription factors maintains the 3D architecture and expression of the CFTR gene. Genomics, 114, 110350. 10.1016/J.YGENO.2022.110350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlin, P. L. , Lu, L. , Rhim, J. , Cutting, G. , Stetten, G. , Kieffer, K. A. , Craig, R. , & Guggino, W. B. (1991). A cystic fibrosis bronchial epithelial cell line: Immortalization by adeno‐12‐SV40 infection. American Journal of Respiratory Cell and Molecular Biology, 4, 313–319. 10.1165/AJRCMB/4.4.313 [DOI] [PubMed] [Google Scholar]
- Zhang, Z. , Leir, S. H. , & Harris, A. (2015). Oxidative stress regulates CFTR gene expression in human airway epithelial cells through a distal antioxidant response element. American Journal of Respiratory Cell and Molecular Biology, 52, 387–396. 10.1165/RCMB.2014-0263OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z. , Leir, S.‐H. , & Harris, A. (2013). Immune mediators regulate CFTR expression through a bifunctional airway‐selective enhancer. Molecular and Cellular Biology, 33, 2843–2853. 10.1128/MCB.00003-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z. , Ott, C. J. , Lewandowska, M. A. , Leir, S. H. , & Harris, A. (2012). Molecular mechanisms controlling CFTR gene expression in the airway. Journal of Cellular and Molecular Medicine, 16, 1321–1330. 10.1111/J.1582-4934.2011.01439.X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Z. , Liu, Y. , Duan, F. , Qin, M. , Wu, F. , Sheng, W. , Yang, L. , Liu, J. , & He, K. (2015). Transcriptomic analyses of the biological effects of airborne PM2.5 exposure on human bronchial epithelial cells. PLoS One, 10, e0138267. 10.1371/JOURNAL.PONE.0138267 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
Table S2.
Table S3.
Table S4.
Data Availability Statement
The deep sequencing data supporting the findings of this study are available at GEO accession GSE220855.


