Abstract
RNA viruses always have been a serious concern for human health by causing several outbreaks, often pandemics. The excessive mortality and deaths associated with the outbreaks caused by these viruses were due to the excessive induction of pro-inflammatory cytokines leading to cytokine storm. Cytokines are important for cell-to-cell communication to maintain cell homeostasis. Disturbances of this homeostasis can lead to intricate chain reactions resulting in a massive release of cytokines. This could lead to a severe self-reinforcement of several feedback processes, which could eventually cause systemic harm, multiple organ failure, or death. Multiple inflammation-associated pathways were involved in the cytokine production and its regulation. Different RNA viruses induce these pathways through the interplay with their viral factors and host proteins and miRNAs regulating these pathways. This review will discuss the interplay of host proteins and miRNAs that can play an important role in the regulation of cytokine storm and the possible therapeutic potential of these molecules for the treatment and the challenges associated with the clinical translation.
Keywords: Cytokine storm, RNA viruses, Innate immunity, miRNAs, Antiviral therapeutics, Inflammation
Graphical abstract
Highlights
-
•
Cytokine storm (CS) is associated with multiple RNA viruses outbreaks.
-
•
Viruses dysregulate the homeostasis of Inflammatory pathways, which leads to CS.
-
•
Host proteins and miRNAs regulate the inflammatory pathways, thereby regulating CS.
-
•
Existing cytokine treatment methods have limitations, and better strategies needed.
-
•
miRNAs show high therapeutic potential against CS that needs clinical translation.
1. Introduction
RNA viruses have already caused multiple outbreaks, epidemics, and two major pandemics within the first two decades of the 21st century and thus remain a global health threat as well as a major concern for future outbreaks. Due to the lack of proof-reading ability of RNA polymerases, they continuously mutate and show excellent adaptability to challenging environments and circumvent the selective pressure acting against them, including the host's innate immune system and the existing antiviral drugs. The remarkable adaptive evolution, complemented by their potential of increased zoonotic transmissibility between the different species and the human hosts, allows the emergence of new strains of viruses of highly pathogenic potential with increased transmissibility, that can even lead to the pandemic, as in the case of current COVID-19 pandemic. These unique properties and the unpredictability of viral evolution pose a major challenge to develop proper therapeutics or vaccines against the diseases caused by RNA viruses. The last two decades saw pandemics in the form of Swine flu (H1N1 Influenza virus (IAV)), Coronavirus Disease −2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus −2 (SARS-CoV-2), epidemics, and endemics in the form of SARS-CoV, MERS-CoV, multiple outbreaks of Dengue, Ebola, Zika. Cytokine storm (CS) was associated with many of these RNA viruses and the reason for the excessive death rate.
Cytokine storm (CS) or cytokine storm syndrome has been associated with the rapid, excessive induction of inflammatory cytokines and chemokines and the over-activation of immune cells, which can ultimately lead to organ damage and death. CS is triggered by pathogens such as bacteria and viruses, autoimmune conditions, and organ transplants (Fajgenbaum and June 2020). CS patterns and the effects caused due to different pathogens differ, and thus, it is important to identify the precise diagnosis and treatment strategy associated with each condition. It is still unclear why most of the pandemics and outbreaks causing RNA viruses are associated with CS that, in turn, lead to excessive death rates. The strategies these viruses involve to escape the host immune response and divert it to cause CS is still unclear and can vary among different viruses. RNA viruses such as Human Influenza viruses (IAV), Human coronaviruses (H–CoV), and Dengue virus (DENV) are very much associated with the induction of CS. These viruses must employ some strategies to circumvent the antiviral immune responses and modulate the inflammation-related pathways to escape from cell death to achieve maximum replication. Meanwhile, there are negative regulatory mechanisms by the host immune system to avoid the excessive induction of cytokines. Previously unexplored roles of the host non-coding RNAs, such as microRNAs (miRNAs), in the regulation of the antiviral and inflammatory responses, have been demonstrated recently due to the advancements in high-throughput sequencing technologies.
Although the precise mechanism of CS induction is unclear, the review will focus on the interplay between the RNA viruses and the host's innate immune inflammation-associated pathways. This review will explain how host proteins and miRNAs interplay with virus factors and regulate the induction of CS. The possible therapeutic potential of the host factors in controlling CS during RNA virus outbreaks will be discussed.
2. Inflammation-associated pathways during RNA virus infection
The immune system's inflammatory response is a defence mechanism against various pathogens like viruses, bacteria, harmful stimuli, damaged cells, or toxic compounds. During viral infection, our immune system activates the inflammatory response through a cascade of signaling pathways to limit the virus. After encountering the pathogens in the cells, various pattern recognition receptors (PRRs) on the host cell membrane recognize the pathogen-associated molecular patterns (PAMPs) derived from the pathogens. These PRRs include Toll-like receptor (TLR), Retinoic acid-induced gene - I like receptor (RLR), Nucleotide-binding oligomerization domain (NOD)-like receptor (NLR), C-type lectin-like receptors, and cytosolic DNA sensors like cyclic GMP-AMP synthase (cGAS) (Fig. 1) (Koyama et al., 2008; Kumar et al., 2011; Yamada et al., 2021). In the case of RNA viruses, the viral genome is recognized by RLRs in the cytoplasm, TLR3, and TLR7 in the endosome of epithelial cells and immune cells. Upon recognition, these PRRs activate interferon regulatory factors (IRFs) through specific adaptor molecules such as IPS-1/MAVS, TIR-domain-containing adapter inducing IFN-β (TRIF) and MyD88, respectively (Kawai et al., 2005; Mogensen, 2018). These adaptor molecules activate kinases, like TANK binding kinase 1 (TBK1), which phosphorylate IRF3 and IRF7. The cGAS-STING pathway is associated with the recognition of cytosolic dsDNA from DNA viruses, bacteria, nucleus, and mitochondria of injured or dying cells. During RNA virus infection, the mitochondrial DNA and nuclear DNA released from damaged cells are recognized by cGAS. The cGAS molecule homodimerized after binding to dsDNA and activated cGAMP, which interacts with STING. The activated STING interacts with TBK1, which subsequently phosphorylates IRF3 (Moossavi et al., 2022). These phosphorylated IRFs get dimerized (homo or hetero dimerization), translocate into the nucleus, and induce gene expression of type I IFNs (Carty et al., 2021; Kumar et al., 2011). RNA viruses such as IAV, SARS-CoV, MERS-CoV, SARS-CoV-2, and DENV can trigger the antiviral pathways as well as inflammatory response pathways in host body. These inflammatory pathways include Nuclear factor kappa-light-chain-enhancer of activated B-cells (NF-κB) signaling, Mitogen-activated protein kinase (MAPK) signaling, Janus kinase (JAK) - signal transducer and activator of transcription (STAT) pathway, Inflammasome pathway. As a result, interferons, inflammatory cytokines, and chemokines will be produced (Liu et al., 2017).
Fig. 1.
Inflammatory pathways activated during viral infection.
During virus infection, PAMPs (Pathogen-associated molecular patterns) and DAMPs (Damage-associated molecular patterns) released from lysed cells, recognize by various PRRs (pattern recognition receptors) such as TLR (Toll-like Receptor), RLR (Retinoic Acid-Induced Gene – I like receptor), NLR (Nucleotide-binding oligomerization domain (NOD)-like receptor), cGAS (cyclic GMP–AMP synthase)-STING (stimulator of interferon genes). These PRRs recruit adaptor proteins and activate the downstream signaling which leads to the activation of different intracellular signaling pathways such as NF-κB (Nuclear Factor kappa-light-chain-enhancer of activated B-cells), IRFs (interferon regulatory factors), MAPK (Mitogen-Activated Protein Kinase), and inflammasome activation which stimulate the production of interferons, cytokines, and chemokines. Activation of inflammasome induce pyroptosis during virus infection. Disbalance of these pathways causes the dysregulated production of cytokines and chemokines which may be responsible for cytokine storm (Created with BioRender.com).
2.1. NF-κB signaling
NF-κB transcription factor plays an essential role in producing pro-inflammatory cytokines such as interleukin-6 (IL-6) and tumor necrosis factor-α (TNF-α). During viral infection, the NF-κB activity is induced by a signaling cascade of PRRs. There are five structural components of NF-κB, such as NF-κB1 (p50), NF-κB2 (p52), RelA (p65), RelB, and c-Rel, which mediate the transcription of the target genes. Under normal conditions, NF-κB is inhibited by inhibitor of NF-κB (IκB). The PRR signaling activates IκB kinase (IKK), consisting of two catalytic subunits and one regulatory subunit such as IKKα, IKKβ, and NF-κB essential modulator (NEMO) or IKKγ, respectively. IKK can be triggered by cytokines, growth factors, mitogens, microbial components, and stress stimuli (Liu et al., 2017). Upon activation, IKK phosphorylates IκB, leading to its proteasomal degradation and subsequent release of NF-κB for nuclear translocation and transcriptional activation of pro-inflammatory cytokine genes (Chen et al., 2018).
2.2. MAPK signaling
MAPKs belong to a serine/threonine protein kinase family, which induces in response to various external stimuli such as DNA damage, heat shock, and pro-inflammatory cytokines (IL-1, IL-6, TNF-α) (Chen et al., 2018). This pathway can also be activated by a wide variety of viruses, such as DENV, IAV, WNV, and Hepatitis C virus (HCV). It can either enhance or suppress viral replication (Kumar et al., 2018b). Activated MAPKs are involved in cell proliferation, differentiation, apoptosis, and immune response (Fung and Liu, 2019). The members of mammalian MAPKs, such as extracellular signal-regulated kinase 1 (ERK1), ERK2, p38α, Jun N-terminal kinase 1 (JNK1), and JNK2, are well known for their role in innate immunity milieu (Soares-Silva et al., 2016). The MAPK pathway gets activated in response to virus infection via PRR mediated pathways. To activate MAPK and inflammatory cytokine production, consecutive signal transduction through several protein kinases is required. The MAPK kinase kinase (MAP3K) gets activated by the TLR signaling process, which further activates MAPK kinase (MAP2K). Subsequently, MAP2K activates MAPKs (JNK1/2, ERK1/2, p38) and induces inflammatory cytokines production in myeloid cells (Arthur and Ley, 2013). Moreover, RANTES/CCL5 is a chemokine involved in inflammatory responses induced by MAPK pathway activation in viral infection. Reports suggest that upon IAV infection in bronchial epithelial cells, the p38 and JNK activation regulates RANTES production (Kujime et al., 2000). JNK activation induces the transcription factor, activator protein-1 (AP-1), during the early infection stage, which is crucial in producing IFN-β (Ludwig et al., 2001).
2.3. JAK/STAT signaling
JAK/STAT pathway plays a vital role in signal transduction in response to external stimuli and involved in hematopoiesis, inflammation, and immune system regulation (Calabrese et al., 2021). JAK/STAT signaling pathway gets activated in response to cytokines and growth factors by binding to their respective receptors. As a result, JAK (JAK1, JAK2, JAK3, and Tyk2) gets auto phosphorylated and interacts with STAT (STAT1, STAT2, STAT3, STAT4, STAT5a, STAT5b, and STAT6) and phosphorylate it. Conformational change of STAT leads to homodimerization (e.g., STAT1/STAT1) or heterodimerization (e.g., STAT1/STAT2), which gets transported into the nucleus and binds to specific genes for initiation of transcription (Banerjee et al., 2017). STAT1/STAT2 heterodimer bind to IRF9 and forms an Interferon Stimulated Gene Factor 3 (ISGF3) complex, which binds to IFN-stimulated response elements (ISRE), that leads to induction of hundreds of interferon stimulatory genes (ISGs), many of which have antiviral function (Ezeonwumelu et al., 2021). IL-6 activate the JAK/STAT signaling by STAT1/STAT3 dimerization for further activation of target genes. In granulocytes, this signaling initiates T-cell proliferation and IL-10 signaling (Schindler and Strehlow, 2000). The signaling cascade mediated by IL-6 induces the MAPK pathway. IL-6 also upregulates the expression of SOCS3 which is a negative regulator of the STAT1 mediated signaling pathway (Morris et al., 2018). IL-12 mediated JAK-STAT pathway involves STAT4 which induces the production of IFN-γ by NK cells and the development of Th1 cells mediated immune responses (Schindler and Strehlow, 2000). The cytokines and chemokines released from innate immune pathways recruit other immune cells such as monocytes, dendritic cells (DC), macrophages, and natural killer (NK) cells and activate the adaptive immune cells such as CD4+ and CD8+ T cells which further produce inflammatory cytokines (IL-2, IFN-γ, TNF-α, GM-CSF) (Yang et al., 2021).
2.4. Inflammasome
The NLR Family Pyrin Domain Containing 3 (NLRP3) inflammasome plays a crucial role in host antiviral immunity. Viruses like IAV and West Nile virus (WNV) induce inflammasome activation. Viral components and cytosolic Damage-associated molecular patterns (DAMPs), such as mitochondrial injury, protein aggregates, and aberrant ion concentrations, can trigger the NLRP3 inflammasome activation (Zhao and Zhao, 2020). The NLRP3 inflammasome comprises NLRP3, pro-caspase-1, and apoptosis-associated speck-like protein (ASC). These three proteins get assembled and initiate auto-cleavage of pro-caspase-1 to activated caspase-1 protease. Caspase-1 activation leads to secretion of IL-1β and IL-18 and gasdermin-D mediated pyroptosis (Ong et al., 2017). Pyroptosis is associated with inflammation-mediated programmed cell death of infected cells. This process is characterized with pore formation on plasma membrane by gasdermin-D. Gasdermin-D, a pore-forming protein, induces transmembrane pore formation, which allows K+ efflux and water influx. Thus, it disrupts the membrane potential and releases cytosolic proinflammatory cytokines, IL-1β and IL-18 from the lysed cells (Kuriakose and Kanneganti, 2019; Nguyen and Kanneganti, 2022). Study of SARS-CoV-2 infected human primary monocytes and COVID-19 patients under intensive care show that pyroptosis activated through inflammasome and increase the level of IL-1β (Ferreira et al., 2021). Reports suggest CLEC5A act as PRR during DENV infection and induce activation of NLRP3 inflammasome and pyroptosis in multiple cell types such as macrophages, platelets, DCs, and neutrophils (Nguyen and Kanneganti, 2022).
2.5. cGAS-STING pathway during RNA virus infection
During RNA virus infection, DAMPs like nuclear and mitochondrial DNA are released from damaged cells recognized by cGAS. The lung sample from COVID-19 patients shows the involvement of cGAS-STING pathway in tissue damage and production of IFN. Mitochondrial DNA released from SARS-CoV-2 infected endothelial cells induces the production of IFN-I and cell damage by activating the cGAS-STING pathway (Domizio et al., 2022). In the case of IAV infection, the mitochondrial DNA is released into the cytosol by the virioporin activity of the IAV M2 protein in MAVS dependent manner, which activate the cGAS-STING mediated innate immune response (Moriyama et al., 2019). However, STING also plays a critical role in RNA virus infection, independent of cGAS. The fusion of RNA virus envelops with the host cell membrane activates the STING-dependent signaling pathway and produces IFN-I (Holm et al., 2016). In COVID-19 pathogenesis, IFN-I plays a crucial role. In the early phase of infection, the rapid rise of IFN-I is associated with virus suppression, whereas in the late phase, the continuous rise of IFN-I causes hyperinflammation (Domizio et al., 2022).
2.6. Autophagy
Autophagy is an antiviral strategy of the host immune system to limit virus replication. It is the self-eating process of cells by lysosomal degradation to maintain homeostasis and clearance of pathogens. This process can synergistically interact with the PRR-induced innate immune pathways to control virus replication. It also maintains cytokine homeostasis in a prolonged immune activation state by negatively regulating various components of the innate immune pathway. During virus infection, the TLRs in the endosome are stimulated by the PAMPs in the endosomal compartments. The TLR pathway activation stimulates the interaction of MyD88/TRIF to Beclin-1 (It interacts with other proteins to form an autophagy initiation protein complex) to initiate autophagy (Choi et al., 2018). However, the TLR pathway can be inhibited by autophagy components, such as the degradation of TRIF by TAX1BP1-mediated selective autophagy (Samie et al., 2018). Downstream to the TLR-signaling pathway, NF-κB pathway components can crosstalk with autophagy. For instance, IKKβ and p65 are targets of various autophagy components for their degradation (Pradel et al., 2020). A study shows RIG-I mediated autophagy through RIG-I-MAVS-TRAF6 signaling, where Beclin-1 translocates to mitochondria and interacts with TRAF6. This interaction induces K63-polyubiquitination of Beclin-1, which induces autophagy (Lee et al., 2018). The autophagy components of the ATG5-ATG12 complex can interact with RIG-I/MAVS pathway by preventing the interaction of CARD domains and inhibiting IFN-I production (Jounai et al., 2007). Similarly, inflammasome activation can be regulated by autophagy. The poly dA:dT activation of inflammasome in macrophage trigger autophagy. This process can also act as a negative feedback regulation of inflammasome activation by polyubiquitinated lysosomal degradation of ASC (Pradel et al., 2020). Virus infection-mediated inflammation leading to severe fatal cases is well illustrated in COVID-19 cases. In such cases, autophagy could be an essential process to limit the virus and check hyper-inflammation.
These signalling pathways have a crucial role in regulating virus replication. An uncontrolled and excessive inflammatory response can harm the host, leading to a severe inflammatory condition like CS, often associated with the RNA virus infection.
3. Cytokines and Cytokine storm (CS)
3.1. Cytokines - introduction
Cytokines are small molecules produced by the immune and non-immune cells that regulate the immunological processes and facilitate the elimination of invading pathogens while maintaining immune homeostasis (Tisoncik et al., 2012). Cytokines act locally at picomolar concentrations and have a very short half-life. These molecules attach to cell surface receptors, instigating targeted cells to express distinct genes. These are classified into four structural families such as four alpha-helix bundle families (comprising IL2 subfamily, IFN subfamily, and IL-10 subfamily), IL-1 family, IL-17 family, and cysteine-knot cytokines (transforming growth factor [TGF-β] superfamily) (Ramani et al., 2015). Further, cytokines are categorized into many types, such as interferons, colony-stimulating factors (CSF), interleukins, chemokines, transforming growth factors (TGF), and TNF, which are secreted in a range of paracrine, autocrine, and endocrine pathways. These have been functionally linked with pro-inflammatory and anti-inflammatory mechanisms in various infections and immune system illnesses. Pro-inflammatory cytokines such as TNF-α, IL-1, IL-6, IL-12, IL-18, IFN-γ, granulocyte-macrophage colony-stimulating factor (GM-CSF), promote inflammation. Anti-inflammatory cytokines such as IL-1Ra, IL-10, IL-13, IL-4, TGF-β, IFN-α trigger inflammation to subside. The JAK-STAT and NF-κB pathways are two of the most widely used cytokine effector pathways. Cytokine ligands activate these pathways, which are likewise regulated and induce the release of more cytokines through adaptive immune cells. For instance, IL-10 shows the effects on other proinflammatory cytokines either by reducing or completely terminating their inflammatory response. Similarly, IL-6 involves in improving the activity of CD4+ cells by upregulating IL-21 and also helps in T-cell differentiation by suppressing IFN-γ signaling (Lordan et al., 2019). However, their dysregulation may result in an array of immuno-pathologies, such as CS, depending on the amount and duration (Karki and Kanneganti, 2021).
3.2. Cytokine storm - pathology
The distinction between a normal and a dysregulated response to a severe infection is hazy, especially since specific cytokines can be helpful and detrimental to the host. Hence the term “cytokine storm” refers to a group of immune dysregulations marked by constitutional symptoms, systemic inflammation, and multi-organ dysfunction, which can progress to multi-organ failure if left improperly treated. Almost all patients suffering from cytokine storm are febrile, and in severe instances, the temperature may rise to a high level, and headache, fatigue, anorexia, diarrhea, rash, myalgia, arthralgia, and neuropsychiatric abnormalities are also possible. These signs and symptoms might be caused by cytokine-instigated tissue damage or acute phase physiological changes, or they could be the outcome of immune-cell-mediated responses. Disseminated intravascular coagulation can develop quickly, resulting in arterial occlusion or hemorrhages, hypoxemia, dyspnea, hemostatic imbalance, vasodilatory shock, hypotension, and mortality (Fajgenbaum and June 2020). CS can be a life-threatening condition for the host due to a sudden rise in the production of various inflammatory cytokines and chemokines during viral and bacterial infection. CS is initiated by inflammatory cytokines and subsequently propagated by various inflammatory mediators, including cytokines that spread through circulation. Besides standard clinical features of inflammation, for instance, redness, swelling, edema, heat, and pain, it may cause severe failure of the vital organs. In the case of Coronavirus-associated severe pneumonia, which is commonly related to rapid replication of the virus, extensive aggregation of inflammatory cells, increased production of inflammatory mediators, and virus-induced immunopathology, all of which contribute to ARDS and acute lung injury (ALI) (Jafarzadeh et al., 2020).
3.3. Induction of cytokine storm
Depending on the stimulus, CS can be characterized as pathogen-induced (as in sepsis), auto-inflammatory or monogenic, or induced by therapeutic intervention. Pathogen-induced CS has mostly been studied with a connection to bacterial and viral respiratory infections. By attaching to PRRs, bacteria such as Streptococcus pyogenes, and Staphylococcus aureus activate nuclear NF-κB signaling and IRF3/IRF7 in innate immune cells (Karki and Kanneganti, 2021). Several factors can be responsible for CS, including triggers induced by pathogen (e.g., bacterial sepsis, influenza virus, and COVID-19), other factors like autoinflammatory disorders, monogenic disorders (e.g., primary or secondary hemophagocytic lymphohistiocytosis (HLH)). Additionally, various therapies like CAR T-cell therapy, T-cell-engaging immunotherapies, and gene therapies may also be responsible for CS (Tang et al., 2021). One of the main causes of infection-related death is sepsis, a serious clinical illness with high mortality risk. In sepsis, two phases follow one another; the first is a hyper-inflammatory phase, and the second is an immunosuppressive phase. Numerous markers, including C-reactive protein (CRP), procalcitonin, IL-1, IL-6, and TNF-ɑ, are present throughout the hyperinflammatory phase. Pro-inflammatory cytokines such as IL-1, IL-6, and TNF-ɑ are generated as an early response to injury or disease. The liver produces CRP in response to infection which is induced by IL-6. The body produces procalcitonin, which is recognized as the most useful sign of severe systemic inflammation. Several of these markers are identified as sepsis biomarkers and can be utilized to help diagnose and treat sepsis patients (Kocak Tufan, Kayaaslan, and Mer, 2021). CS triggered by viral infection is to fight or compete with immune defense systems or to escape from it (Kirsch-Volders and Fenech, 2021). IAV, DENV, and various strains of SARS-CoV infect the human host and are known to cause damage by inducing CS. Additionally, CS spreads throughout the body affecting vital parameters and causing systemic sepsis, characterized by prolonged hypotension, hyper- or hypothermia, leukocytosis, or leukopenia, and commonly thrombocytopenia thereby severely affecting vital parameters and damaging vital organs of the host (Hussell and Goulding, 2010). The exact mechanisms underlying CS in RNA virus infections are complex and multifactorial. Several factors contribute to the development of CS, including viral factors, host factors, and the interplay between the virus and the host immune system. Further research is required in this field for better understanding of induction of CS mechanism.
4. Role of RNA viruses in the stimulation of CS
In the case of RNA viruses like the influenza virus, it activates innate immunity, which is widely based on the presence of PRRs on the cell membrane of immune cells. The activation of PRRs triggers various signaling cascades leading to the production of several specific pro-inflammatory cytokines. A considerable rise in the expression of these cytokines, including TNF-α, IL-6, and IL-1β worsens the inflammatory response leading to apoptosis and reduction in mitochondrial reactive oxygen species, resulting in vascular malfunction and multiple organ failure and finally causing the demise of the host (Gu et al., 2019). Severe cases of CS uncommonly remarked in seasonal and mild influenza infections reveal the connection between the high cytokine level and the severity of the disease (Liu et al., 2016). Different cytokine waves emerge throughout an IAV infection, starting with the early responses of RANTES, IFN-α, IFN-β, IFN-κ, IL-8, and MCP-1 to the later responses of IL-1α/β, IL-6, IL-18, IFN-I, TNF-α, MCP-1, MCP-3, MIP-1α, MIP-1b, MIP-3α, RANTES, and IP-10, which are mostly produced by the infected macrophages in the lower respiratory tract (Gu et al., 2019). DENV has both structural and non-structural proteins. Among them, functionally non-structural proteins are associated with viral polyprotein processing, RNA replication, and virion assembly, and some non-structural proteins are also reported to modulate host immune response. To understand the virus strategy in vitro, studies have been conducted in a variety of cell types that are associated with DENV infection, such as epithelial cells, endothelial cells, hepatocytes, muscle cells, dendritic cells, monocytes, and mast cells. Nevertheless, immune cells are the major target for in vivo infection. The non-structural proteins of the virus have a crucial role in modifying the host's immune responses, which may ultimately lead to CS. The proteins NS2A, NS2B, NS4B, and NS5 have been demonstrated to disrupt type I IFN signaling. Also, the production of a variety of chemokines and pro-inflammatory mediators is induced by NS5 and NS4B. The cell-mediated immune system, particularly CD8+ T lymphocytes, targets NS proteins. Several studies have found that dendritic cells and macrophages express C-type lectins receptors such as DC-SIGN (CD209) and CLEC5A, which are utilized by DENV for host cell entry. When DENV interacts with its host cell receptors, various pro-inflammatory, antiviral, and immunoregulatory cytokines are expressed and released. DC-SIGN primarily function as a site of viral attachment, as viral internalization also occurs in those cells which express a mutated internalization sequence of DC-SIGN. Moreover, DENV binding to CLEC5A has been demonstrated to induce pro-inflammatory cytokines (Srikiatkhachorn et al., 2017).
As viral load correlates with disease severity, antibody-dependent enhancement (ADE) in secondary infection enhances viral entry into host cells leading to increased viral load and immune activation associated with disease severity. In DENV-infected patients, aberrant inflammatory responses have been found. In comparison to healthy controls, a number of host inflammatory biomarkers, including GM-CSF, IFN-γ, IL-15, IL-10, IL-8, IL-6, MCP-1, MIP-1β, and TNF-α, were elevated. The disease severity was reported to correlate mainly with four cytokines, including IFN-γ, IL-10, GM-CSF, and MIP-1β. Those who have a severe DENV infection frequently experience liver damage. The severity of the disease appears to be correlated with an increase in the liver enzymes aspartate transaminase (AST) and alanine transaminase (ALT). Direct viral cytopathogenic effects and immune-mediated injury are primarily involved in the pathogenesis of liver damage in DENV infection. Infected hepatocytes express IFN-β, IFN-α, RANTES, IL-8, IL-10, IL-6, and IL-12 via NF-κB. IFN-α and IFN-β are known to boost nearby cells' antiviral defenses. RANTES, IL-6, IL-8, IL-10, and IL-12 attract NK cells, CD8+, and CD4+ lymphocytes, which causes the hepatic cells to undergo apoptosis (Tarasuk et al., 2022).
Moreover, SARS-CoV-2, which is the devil behind the serious pandemic, caused a severe outbreak. The mortality related to COVID-19 disease is ARDS which appears to cause similar immunopathogenic features in SARS-CoV and MERS-CoV infections. It has been found that CS is one of the prominent hallmarks of ARDS. Patients with COVID-19 infection exhibit high blood levels of cytokines and chemokines such as IL-1, IL-1Ra, IL-7, IL-8, IL-9, IL-10, basic FGF2, GCSF, GMCSF, IFN, IP-10, MCP-1, MIP-1, PDGFB, TNF, and VEGFA. In extreme cases of SARS-CoV-2 infection, similar to SARS-CoV and MERS-CoV infections, the resulting CS induces a strong inflammatory immune response that contributes to ARDS, multiple organ failure, and eventually death. COVID-19-infected patients have greater leukocyte counts, aberrant respiratory findings, and higher plasma pro-inflammatory cytokine levels. The immediate cause of mortality from acute COVID-19 is CS, damaging lungs and many organs, including the heart, kidneys, and liver, which results in multiple organ depletion (Li et al., 2020; Huang et al., 2020).
The exact mechanisms by which RNA viruses stimulate CS are not fully understood, several factors have need to be identified as potential contributors. Exploration of novel findings will contribute to the understanding of the mechanisms involved in CS associated with RNA virus infections.
5. Comparative aspects of CS induction by different RNA viruses
When the host is infected with respiratory viruses like coronavirus, RSV, or IAV, a fine balance must be struck between preventing viral multiplication and preventing harm to the lung's delicate structure. Respiratory infections depend on the immune system's ability to prevent virus multiplication, yet over-activation can worsen inflammation and sickness. Although they play a role in viral pathogenesis, cytokines trigger immune responses that are antiviral. RSV, enteroviruses, adenoviruses, human metapneumovirus, IAV, and parainfluenza viruses are the main viral pathogens responsible for lower respiratory tract infections (LRTI) in humans (Forbester and Humphreys, 2021). Considering the wide prevalence of influenza subtypes, H1N1 and H5N1 are much more studied. So, these two subtypes are used as examples to focus on the function of the innate CS caused by the IAV. H1N1 has a lesser cytokine/chemokine response than H5N1, although it is greater than the seasonal H3N2 flu. IL8 is more elevated in both severe H1N1 and seasonal H3N2 cases, so it may be considered a marker of serious illness. Pro-inflammatory chemokines IP-10, MCP-1, MIG, and MIP-1 are considerably greater in H1N1 infections than in healthy controls; MCP-1 and MIG are more than twice as high in severe cases. Moreover, TNF-α and IL-6 levels in DCs following H7N9 infection were equal in H5N1 or H3N2 virus-infected cells. Other cytokines were modest and considerably lower in DCs infected with the H7N9 virus than in those infected with the H5N1 virus or even seasonal H3N2 influenza (Gu et al., 2019).
During different RNA virus infections, the disease pathogenesis is associated with the expression pattern of cytokines and chemokines. Nevertheless, similarities and differences are observed during the early and late phases of illness (see Table 1). IL-6 is a key cytokine that has a significant role in severe cases of COVID-19 and dengue hemorrhagic fever (DHF). During COVID-19 infection, rapid induction of Th1 cells activates the pro-inflammatory mediators such as IL-6. The elevated level of IL-6 leads to activation and induction of other inflammatory cytokines such as IL-8, VEGF, and MCP-1, causing an elevation in endothelial permeability and reduced expression of E-cadherin, resulting in ARDS. Conversely, IL-6 contributes less to the disease etiology of DHF than COVID-19, despite an increased vascular permeability which results in plasma leakage and shock, which are major pathological hallmarks of DHF. Additionally, the overexpression of IL-6 raises the levels of TNF-α and IL-1, and a slew of other cytokines, and leads to the infiltration of macrophages and neutrophils. This is responsible for the amplification of IL-6 following the expression of various chemokines comprising monocyte attracting protein-1 (MCP-1/CCL-2), interleukin-8 (CXCL-8), which arouse infiltration of neutrophils and macrophages into lung tissue (Dayarathna et al., 2020; Pum et al., 2021). In extreme cases of SARS-CoV, infected patients along with IL-6 show the highest proportion of serum pro-inflammatory cytokines (IFN-γ, IL-1, IL-6, IL-12, and TGF-β) and chemokines (CCL2, CXCL10, CXCL9, and IL-8) were detected compared to uncomplicated SARS patients. Likewise, the MERS study indicated disease severity was found to be linked with a higher fraction of serum pro-inflammatory cytokines (IL-6 and IFN-α) and chemokines (IL-8, CXCL- 10, and CCL5) than mild or moderate illness (Sun et al., 2020). Another crucial cytokine TNF-α which is considered to be a quintessential pro-inflammatory cytokine for the “center of the influenza cytokine storm,” is responsible for the activation of endothelial cells and upregulation of IL-1 and IL-6 production following the invasion of pathogens. In addition, the conditions like inflammation and tissue damage in cytokine release syndrome (CRS) have majorly been arbitrated by TNF-α, IL-6, IL-1, and nitric oxide (NO), which are the secretion of activated macrophages and monocytes (Morris et al., 2021).
Table-1.
Various viral infections and associated cytokine patterns.
| Virus | Cytokines and chemokines in early infection | Cytokines and chemokines in late infection | References |
|---|---|---|---|
| Influenza | RANTES, IFN-α, IFN-β, IFN-κ, IL-8, and MCP-1 | IL-1α/β, IL-6, IL-18, IFN-I, TNF-α, MCP-1, MCP-3, MIP-1α, MIP-1b, MIP-3α, RANTES, and IP-10 | (Gu et al., 2019) |
| H1N1 | IL-6, IL-12, IP-10 and TNF-a | IP-10, TNFα, IL-15, IL-12, IL-6, IL-8 and IL-9 | (Yu et al., 2011) |
| H5N1 | IL-1β, IL-6, TNF-α, IL-10 and IL-18 | IL-12 and IL-8 | (Kalaiyarasu et al., 2016) |
| H7N9 | IL-6, IL-8, MIP-1αand MIP-1β | IL-6and IL-8 | (Wang et al., 2014) |
| WNV | IFN-α, CXCL10/IP-10, CCL8/MCP-2 | CCL2/MCP-1 and CCL7/MCP-3 | (Hoffman et al., 2016) |
| RSV | IL-6 and TNF-α, CXCL10 (IP-10), CXCL8 (IL-8), CCL2 (MCP1), CCL3 (MIP-1α), and CCL5 (RANTES) | IFN-β, IFN-λ1, IFN-γ, IL-1β, TNF-α, IL-6, IL-10, CXCL8, and CXCL10 | (Lambert et al., 2014) |
| DENV | GM-CSF, IFN-γ, IL-15, IL-10, IL-8, IL-6, MCP-1, MIP-1β, and TNF-α | IFN-γ, IL-10, GM-CSF, and MIP-1β | (Tarasuk et al., 2022) |
| SARS-CoV | IFN-γ, IL-1, IL-6, IL-12, and TGF-β, CCL2, CXCL10, CXCL9, and IL-8 | IFN-γ, IL-1, IL-6, IL-12, and TGFβ, CCL2, CXCL10, CXCL9, and IL-8 | (Sun et al., 2020) |
| MERS-CoV | IL-6 and IFN-α, IL-8, CXCL- 10, and CCL5 | IL2, IL7, IL10, GSCF, IP10, MCP1, MIP1a and TNF α | (Sun et al., 2020) |
| SARS-CoV-2 | IL-10, IL-6, IL-1 and TNF-α | IL2, IL7, IL10, GSCF, IP10, MCP1, MIP1A, and TNFα. | (Costela-Ruiz et al., 2020; Hasanvand, 2022) |
Viruses evolved several methods to circumvent the host immune system, which resulted in a persistent infection that spread quickly and caused severe tissue damage and pro-inflammatory CS (Kirsch-Volders and Fenech, 2021). The studies conducted on severely ill patients with human coronaviruses (HCoVs) infection suggest that the pro-inflammatory responses have a significant role in the disease pathogenesis. In-vitro study shows that there is a delay in the release of cytokines and chemokines in respiratory epithelial cells, DCs, and macrophages at the early stage of SARS-CoV infection. In later stages, less amount of the antiviral factors (IFNs) and high levels of pro-inflammatory cytokines (IL-1β, IL-6, and TNF-α) and chemokines (CCL-2, CCL-3, and CCL-5) lead to an imbalance of cytokines and result in CS (Ye et al., 2020). In the initial stages of SARS-CoV and MERS-CoV infection, delayed production of IFNs interferes with the antiviral response (Channappanavar et al., 2019). While in later stages, delayed production of IFNs occurs and the secretion of cytokines and chemokines increases, which attracts the inflammatory immune cells. A huge inflammatory immune cluster resulting in the infiltration of the cells into the lungs causes lung inflammation. Thus, this hyperproduction of the cytokine leads to CS (Ye et al., 2020).
Some viruses enter the host cell via receptor-mediated endocytosis and other pathways, such as glycan-lectin interaction-based entry. In the case of dengue infection, the virus interacts with CLEC5A, which is a type of lectin. CLEC5A is a PRR for DENV that is highly expressed in neutrophils, monocytes, macrophages, osteoclasts, microglia, and dendritic cells. DENV activates the CLEC5A and TLR2, which induce the NET formation and release pro-inflammatory cytokines. This activation of CLEC5A along with TLR2 leads to the induction of p38 kinase and AKT kinase activation, which causes the huge NET formation in neutrophils as well as inflammasome (NALP3, NLRC4, AIM2) activation and pro-inflammatory cytokine (IL-1β, CCL2, TNF, IL-17α) production in macrophages (Sung and Hsieh, 2021). Understanding the comparative aspects of CS induction by different RNA viruses is crucial for developing targeted therapeutic strategies and interventions. Study related in this field will be helpful in the identification of potential targets for intervention and better CS treatment approaches.
6. Host proteins as regulators of inflammation and CS
To maintain the homeostasis of cytokines and protect the cells from harmful immune responses, there are some negative regulators which can target the inflammatory pathway in a negative feedback mechanism. Dysregulation of such effectors can cause severe inflammatory diseases. SOCS (Suppressor of Cytokine Signaling) proteins are the best example of negative regulators of interferons and cytokine production. SOCS1 and SOCS3 can regulate the JAK/STAT pathway by inhibiting the JAK tyrosine kinase activity, which is important to produce ISGs. SOCS1 can inhibit the NF-κB activation in response to TLR signaling. Another essential negative regulatory protein for the NF-κB pathway, A20/TNFAIP3, functions as a deubiquitinase and interacts with NEMO/IKKγ (Yoshimura et al., 2018).
Anti-inflammatory cytokines such as TGF-β, and IL-10 also take part in maintaining cytokine homeostasis. IL-10 can suppress the expression of various immune effector molecules, pro-inflammatory cytokines, and T-cell costimulatory molecules and subsequently inhibit macrophage activation. IL-10 may also increase the production of an anti-inflammatory mediator like IL-1Ra, which is an antagonist of the IL-1 receptor. TGF-β has an immunosuppressive effect on various innate and adaptive immune cells. For example, it can suppress the TLR-mediated pro-inflammatory responses in macrophages (Lee and Kim, 2007). TGF-β suppress production of IL-2 and activate the Smad family proteins, which can inhibit the proliferation of CD4+ T cells, CD8+ T cells and NK cells. However, TGF-β facilitate the proliferation and differentiation of Treg cells. (Zhang et al., 2022). Depending on the concentration of TGF-β, it may also play pro-inflammatory roles. During SARS-CoV-2 infection, the elevated expression of TGF-β can mediate ARDS, inflammation, and pulmonary fibrosis in severe conditions (Jiang et al., 2022). A study showed that the patients infected with DENV show the association of IL-10 and TGF-β levels with the disease severity (Pandey et al., 2015). Hence, the balance between pro- and anti-inflammatory effectors is crucial for immune homeostasis maintenance.
7. Host non-coding RNAs- potential role in the regulation of CS
7.1. Role of miRNAs in virus infection
The whole genome sequencing technique helps to find out the majority of non-coding genome sequences which do not take part in protein synthesis but have a significant role in gene regulation. The non-coding RNAs (ncRNAs) participate in signaling pathways by interacting with specific proteins where they can regulate the transcription or translation of genes. Reports suggest that non-coding RNAs like micro-RNA (miRNA), long non-coding RNA (lncRNA), and circular RNA (circRNA) have a potential role in the regulation of inflammation and immune system pathways (Chew et al., 2018). Among these ncRNAs, the most studied ncRNA is miRNA. miRNAs were studied in several diseases, including viral diseases. In several in vitro studies along with patient samples, it has been reported that the miRNA expression alters, which can be a potential biomarker or therapeutic target during the viral infection.
The miRNA are small RNA molecules of 21–25 nucleotides long. It can bind to the 3′ untranslated region (UTR) of a target mRNA and regulate its transcription. Multiple miRNAs can regulate a single gene. Various studies of miRNA expression and its effect on virus replication suggest that these small ncRNA can also regulate virus replication by targeting the virus genome. The blood sample of patients infected with H1N1 influenza virus showed altered expression of 193 miRNAs. Among these, 16 miRNAs (miR-1260, miR-1285, miR-18a, miR-185, miR-299–5p, miR-26a, miR-30a, miR-335, miR-34b, miR-519e, miR-576–3p, miR-628–3p, miR-664, miR-665, miR-765, and miR-767–5p) were highly dysregulated and able to clearly distinguish between infected and healthy individual (Tambyah et al., 2013). In a similar study of serum miRNA profile of patients’ samples infected with H7N9, four miRNAs such as miR-106a, miR-20a, miR-17, and miR-376c were highly expressed as compared to the healthy group (Zhu et al., 2014b). Another promising prognostic biomarker for HCV-mediated fibrosis is circulating miR-20a (Shrivastava et al., 2013). Difference in the expression of various miRNAs may help to distinguish the viruses which cause similar disease symptoms. A study conducted by Farr RJ et al. suggested the possibility of virus type specific expression of miRNA. Three miRNAs (miR-423–5p, miR-23a-3p, and miR-195–5p) are involved in the early stages of the SARS-CoV-2 infection but not during IAV infection (Farr et al., 2021). These findings identify the dysregulated miRNAs during viral infection that can help to distinguish between healthy and infected individuals and highlight the potential of miRNA as a prognostic biomarker during virus infection. Variations in profiles of cell-free small RNA expression, which are unique to specific virus infections, can be used as a reliable diagnostic marker for various infectious processes.
7.2. Role of miRNAs in regulating inflammation-associated pathways induced upon RNA virus infection
The miRNA can modulate the disease pathogenesis by targeting various host factors. The miRNA can target several inflammatory mediators, which can either enhance or inhibit the inflammatory response. Several miRNAs were studied in various inflammatory conditions, which can regulate cytokines, chemokines, and transcription factors. For instance, the hsa-miR-410, hsa-miR-26a, and let-7c have the potential to target IL-6 mRNA. The miRNA hsa-miR-205–3p, hsa-miR-124–3p, hsa-miR-299–3p are among the modulators of IL-6 receptor expression. Studies have shown that hsa-miR-216a, hsa-miR-138, and hsa-miR-9 can inhibit the NF-κB transcription factor (Aslani et al., 2021). The study of inflammatory lung diseases like ALI, and ARDS shows the anti-inflammatory role of miR-223 by suppression of the NLRP3 inflammasome (Yan et al., 2019). Various studies show the factors involved in inflammatory pathways are targeted by miRNA. For example, JAK2 is regulated by miR-135, STAT1 is regulated by miR-145, miR-221, and STAT3 is regulated by miR-124 and let-7 (Wu et al., 2012; Gregersen et al., 2010; Zhang et al., 2010; Koukos et al., 2013; Wang et al., 2010). These miRNAs can potentialy modulate the expression of genes which are key factors of inflammatory pathway and hence regulate inflammatory condition. Several micro RNAs are found to regulate MAPK signaling. For instance, it has been demonstrated that miR-125a-3p controls the expression of p38 MAPK in rat trigeminal ganglions experiencing orofacial inflammatory pain (Chew et al., 2018). A number of miRNAs such as hsa-miR-20b and hsa-miR-524, target ERK-2, and hsa-miR-23b binds to the 3′-UTR of TAB2/3, which can influence the activation of the MAPK/AP-1 pathway. The c-JUN is likewise targeted by the hsa-miR-216b, hsa-miR-29c, and hsa-miR-139 (Aslani et al., 2021). All these findings show the potential of miRNA to regulate inflammatory pathways, which can modulate hyperinflammatory conditions like CS.
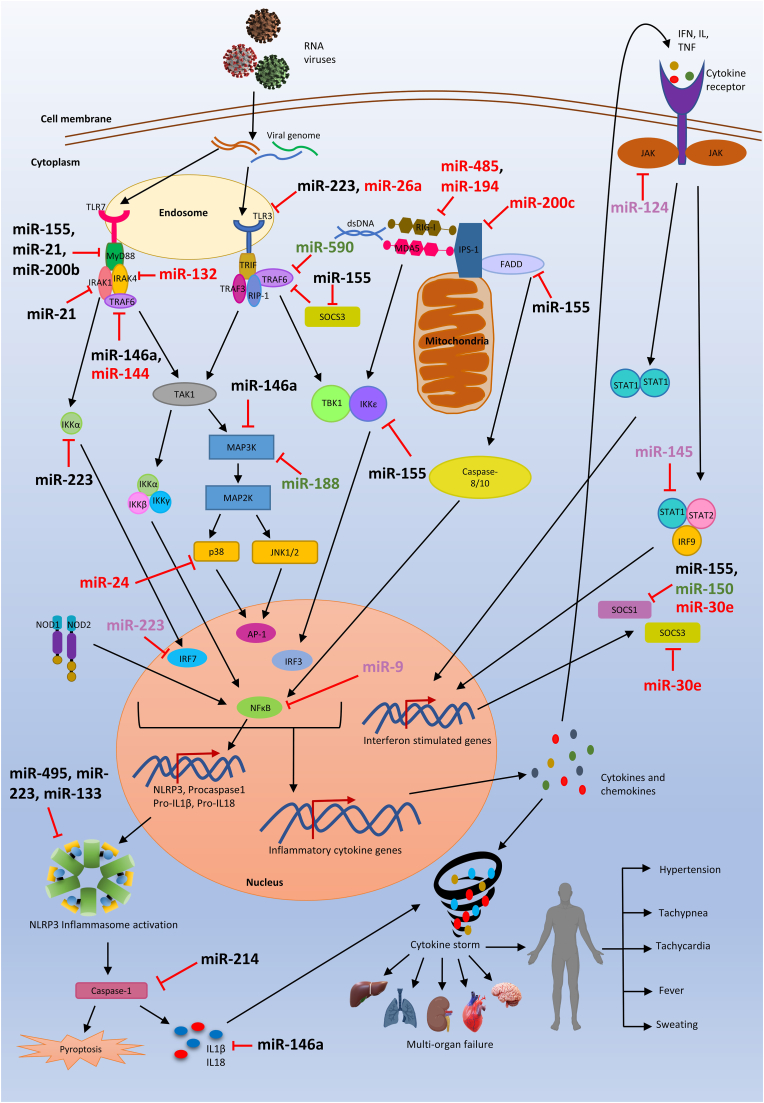
Several studies support the crucial function of miRNA in viral infection (Fig. 2; Table 2). The dysregulated miRNAs during viral infection can target the host gene as well as the viral genome either in favor of enhancement of viral genome replication or inhibition. These miRNAs can act as a positive or negative regulator of genes involved in the inflammatory pathway (Krishnamoorthy et al., 2022). For example, a study in a mouse model shows that miR-144 has a proviral role during influenza infection by targeting TRAF6. miR-144 suppresses TRAF6 level and dysregulates the downstream signaling of antiviral gene expression via IRF7 showing the complex interplay of miRNA in antiviral innate immunity (Rosenberger et al., 2017). In contrast, the antiviral activity of upregulated miR-136 was shown upon IAV H5N1 infection in vitro. This miRNA acts as an agonist of RIG-I, thereby enhancing the expression of IL-6 and IFN-β (Zhao et al., 2015). miR-324 shows a dual role during IAV infection by targeting the viral genome and the host gene CUEDC2 which is a negative regulator of the JAK-STAT pathway, hence, enhancing the host antiviral immunity (Kumar et al., 2018a). miR-155 shows antiviral effects which target SOCS1 and increase the expression of IL-6 and IFN-β during IAV infection (Izzard et al., 2014). The aberrant expression of miR-155 during viral infection can lead to the hyperactivation of proinflammatory mediators which may cause the hyperinflammatory condition. However, the miR-155 can also target essential molecules involved in immune response pathways such as MAPK/NF-κB (Krishnamoorthy et al., 2022). These findings highlight the diverse role of miRNA during viral infection. The Peripheral blood mononuclear cell (PBMC) of the influenza-infected patient sample shows the interaction of miR-302a to the 3′ UTR of IRF5. Treatment of miR-302a mimic in PBMC reduces the IRF5 expression and decreases the production of IRF5-regulated inflammatory cytokines such as IL6, IL8, TNF-α, IFN-β, CCL2, CCL5 which leads to higher virus production (Nguyen et al., 2018). The in vitro study of IAV shows the increased expression level of miR-132 and target MAPK3/ERK1 which involved in TLR, MAPK pathway (Buggele et al., 2012). The miR-30 family members are involved in the regulation of immune signaling pathways during different virus infections such as IAV, and flaviviruses (DENV, Zika virus) (Carvalho-Silva et al., 2022; Su et al., 2021). The cell lines (HeLa, U937) infected with DENV shows increased expression of miR-30e* which target IκBα which is an inhibitor of NF-κB and activates the NF-κB pathway, hence promoting expression of IFN-ꞵ and its downstream IFN stimulated genes such as MxA, OAS1, IFITM1 which inhibit the virus replication (Su et al., 2021; Zhu et al., 2014a). The miR-30e-5p is also reported to be involved in the disruption of negative feedback mechanism by targeting SOCS1 and SOCS3 and was proposed to be a potential therapeutic target to regulate inflammation (Mishra et al., 2020a; Mishra et al., 2020b). The miR-146a and miR-21 were found to be dysregulated during DENV and SARS-CoV2 infection and negatively correlated with inflammatory pathway (Maranini et al., 2022; Ouyang et al., 2016). The expression of CCL20 was found to be upregulated in inflamed airway epithelia, which can be potential targets of miR-21 (Tang et al., 2020). During DENV infection in primary human monocytes and THP-1 cells, the miR-146a act as a proviral effector by targeting TRAF6 and hence limiting host IFN-β production (Wu et al., 2013). The promoter analysis of miR-146a shows, it is dependent on NF-κB for its induction. However, the miR-146a has target sites for IRAK1 and TRAF6 which are the mediator of NF-κB signaling (Taganov et al., 2006). Hence, miR-146a can inhibit NF-κB dependent proinflammatory cytokine production by negative feedback regulation. Enterovirus study in mice model shows that the transcription factor AP-1 induces the expression of miR-146a during infection. It inhibits IFN-I production by targeting IRAK1 and TRAF6, which are involved in the TLR pathway. The expression of IRAK1 and TRAF6 was restored by the inhibition of miR-146a, which in turn increase IFN-I production. The viral replication was suppressed while there was enhancement in the survival rate of the mouse (Ho et al., 2014). The expression level of miR-150 was found elevated in the PBMC sample of DHF patients as compared to DF. This miRNA has a target site for SOCS1 and negatively correlates with the expression of SOCS1 in DHF patients (Chen et al., 2014). The aberrant expression of SOCS1 can be one of the reasons for cytokine dysregulation in DHF patients where miR-150 targeted therapeutic can be helpful. During SARS-CoV-2 infection, the upregulated expression of miR-423–5p was observed at the early stage (Farr et al., 2021). This miRNA has target site for SOCS3 which could be involved in the dysregulation of cytokines during the virus infection (Wang et al., 2019). miR-378b can bind to TBKBP1 and enhance TBK1 expression and involves the activation of the cGAS-STING pathway. It was reported that miR-29a can interact with IRF7 and activate STING/TBK1/IRF3. Additionally, miR-29a may inhibit TRAF6 expression, which could contribute to a reduction in the antiviral response (Shah et al., 2019). However, the studies related to miRNAs regulating the cGAS-STING pathway during RNA virus infection are inconclusive and this field has yet need to explore.
Fig. 2.
Regulation of inflammatory pathways by miRNA.
The management of various inflammatory pathways during RNA virus infection by miRNAs. Upon viral infection, miRNAs are known to dysregulate and can target different host factors associated with the intracellular inflammatory pathways by targeting the 3′ UTRs. This balance is maintained by miRNA that targets negative and positive regulators of these inflammatory pathways. Black colour indicates miRNAs indicating commonly found miRNA in several RNA virus infections. Red colour indicates miRNAs that are linked to IAV infection, whereas green and pink indicate miRNAs that are predicted and linked to dengue and SARS-CoV-2 infections, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Table 2.
Host factors regulated by miRNA in different RNA virus infections.
| miRNA | Virus | Target | Role in pathway | Reference |
|---|---|---|---|---|
| miR-146a | IAV | TRAF6 | Signal transducer in NF-κB pathway | (Deng et al., 2017) |
| DENV | (Ouyang et al., 2016) | |||
| SARS-CoV-2 | (Maranini et al., 2022) | |||
| miR-590 | DENV | TRAF6 | Signal transducer in NF-κB pathway | (Su et al., 2021) |
| miR-223 | SARS-CoV-2 | NLRP3 | Inflammasome complex | (Yan et al., 2019) |
| miR-155 | SARS-CoV-2 | SOCS1 | Negative regulator of cytokine signaling | (Narozna and Rubis, 2021) |
| IAV | (Izzard et al., 2014) | |||
| DENV | (Wong et al., 2020) | |||
| miR-150 | DENV | SOCS1 | Negative regulator of cytokine signaling | (Wong et al., 2020) |
| miR-124 | SARS-CoV-2 | JAK2 | Initiate JAK-STAT pathway | (Zhang et al., 2021) |
| miR-132 | IAV | MAPK3 | Inflammatory cytokine production via TLR pathway | (Buggele et al., 2012) |
| miR-423–5p | SARS-CoV-2 | SOCS3 | Negative regulator of cytokine signaling | (Narozna and Rubis, 2021) |
| miR-4776 | IAV | IκBβ | Inhibitor of NF-κB | (Othumpangat et al., 2017) |
| miR-340 | IAV | RIG-I | PRR recognize viral genome | (Zhao et al., 2019) |
| miR-451a | SARS-CoV-2 | IL6R | IL6 receptor | (Zhang et al., 2021) |
| miR-299–3p | SARS-CoV-2 | JAK1 | Initiate JAK-STAT pathway | (Zhang et al., 2021) |
| miR-24 | IAV | p38 | Activation of MAPK pathway | (Liao et al., 2021) |
| miR-21 | DENV | IL1β | Proinflammatory cytokine | (Ouyang et al., 2016) |
| SARS-CoV-2 | (Maranini et al., 2022) | |||
| miR-30e* | DENV | IκBα | Inhibitor of NF-κB | (Su et al., 2021) |
These findings indicate a significant association of miRNA in regulating the inflammatory pathways during RNA virus infection. The miRNA can interact with various components of inflammatory pathways and the transcripts of cytokines. The dysregulated expression of miRNA during RNA virus infection can affect the host's innate immune system either by enhancing or inhibiting these inflammatory responses depending on the specific miRNA targets and hence contribute to the disease severity. Thus, the understanding of the interplay between miRNA and inflammatory pathway can provide new insight into the development of the innovative therapeutic potential of miRNA to overcome the harmful effect of CS associated with RNA virus pathogenesis.
8. RNA-virus derived non-coding RNAs that can regulate inflammation pathways
Several DNA viruses are known to encode miRNA which function to evade the host immune system and regulate the host or viral genes. Recent advancements in novel sequencing techniques aid in identifying the non-coding RNA fraction derived from RNA viruses, however, their function is not well studied. Several RNA viruses, such as WNV, DENV-2, EBOV, and SARS-CoV have been demonstrated to produce miRNA. Deep sequencing of DENV-infected mosquito and mammalian cell lines have identified dengue virus-encoded miRNA like small viral RNA, DENV-vsRNA-5 which autoregulates viral replication by targeting the viral non-structural protein 1 (NS1) gene. The 3′-terminal stem-loop of the WNV genome was identified to encode a small miRNA-like RNA, KUN-miR-1, which facilitates virus replication by targeting and increasing the accumulation of a transcription factor GATA4 mRNA that has been involved in lipid trafficking and immune recognition in mosquito cells (Hussain et al., 2012). H5N1 encodes microRNA-like small RNA, miR-HA-3p, which is involved in the enhanced production of cytokines in human macrophages and mice models following infection with the H5N1 virus. The miR-HA-3p has a target site in 3’ UTR of PCBP2, which is a negative regulator of RIG-I/MAVS-mediated antiviral innate immunity, and hence, contributes to CS. This study reports that the inhibition of miR-HA-3p leads to a lower concentration of IFN-β, TNF-α, IL-6, and IL-1β (Li et al., 2018). SARS-CoV-derived miRNAs have been discovered in the lungs of infected mice which are involved in pathogenesis during infection. These miRNAs are derived from nucleoprotein and non-structural protein 3 regions of the virus. The bioinformatics study of SARS-CoV-2 also revealed the putative miRNA encoded by various genomic regions of the virus which can target various signaling molecules involved in immune function, apoptosis, cell cycle, and transcription (Nanbo et al., 2021). These virus-encoded miRNAs can be used as a therapeutic target. However, experimental evaluation of their expression and biological functions in the context of viral infection requires additional research.
9. Potential therapeutics for CS treatment and associated challenges
The higher mortality of RNA virus outbreaks such as IAV and SARS-CoV-2 are associated with the higher viral load complemented with the excessive secretion of inflammatory cytokines. The most common strategy to treat the CS associated with these virus infections was to reduce the level of viruses at the early stages of infection by the drugs/molecules that can inhibit the replication and thereby prevent the CS. As evident in the ongoing pandemic caused by SARS-CoV-2, the symptoms vary between the individuals, and in many cases, the individuals already approach severity when they are diagnosed the first time. The available therapeutic methods offer hopes to treat the patients at an early stage of CS induction and have limitations in treating severe patients.
Corticosteroids are anti-inflammatory steroid hormones that were predominantly used to treat CS associated with virus infections. Timely administration of Corticosteroids was reported to control the excess CS induction and mortality and improve the clinical outcomes of severe SARS patients (Chen et al., 2006). However, improper use of glucocorticoids was associated with the aggravation of disease in SARS patients. Too early use of glucocorticoids will dampen the body's defense system, which will lead to an increase in the virus load, further worsening the disease (Zhao et al., 2020). IL-1 family antagonist drug, such as Anakinra, was shown to improve the clinical outcome of COVID-19 patients with CS (Della-Torre et al., 2021). Anakinra shows controversial results and is not recommended for patients already experiencing respiratory failure (Khani et al., 2022). IL-6 antagonists such as Tocluzimab and IL-6 inhibitors such as Chloroquine were also shown to be efficient in the treatment of CS associated with virus diseases (multicenter collaboration group of Department of, Technology of Guangdong, and Health Commission of Guangdong Province for chloroquine in the treatment of novel coronavirus 2020; Tanaka et al., 2016). TLR agonists and antagonists were also shown to have therapeutic potential against CS associated with RNA virus infection (Liu et al., 2022).
To impede the injurious effects of CS, a novel pharmacological approach is required. Recent experimental studies show the potential of miRNAs to regulate various pathological conditions by targeting the host genes or pathogen genome. The dysregulated miRNA in different pathological conditions can be used as a prospective biomarker for the early detection of the pathophysiological conditions of patients as well as a novel therapeutic target. Various miRNAs can directly or indirectly regulate the expression of inflammatory cytokines. For instance, during ARDS, miR-199 is known to have a pro-inflammatory effect in alveolar macrophages contributing to the induction of key inflammatory cytokines such as TNF-α, IL-1β, and IL6 (Desjarlais et al., 2020). It was reported that type-I IFN could be regulated by miRNA. IFN-α can be controlled by miR-466l, miR-22, and miR-122, whereas IFN-β can be regulated by miR-146a, miR-26a, miR-34, and Let-7b (Rarani et al., 2022). The miR-22 is a key regulator of MAVS gene expression during the Japanese Encephalitis virus (JEV) infection, thereby suppressing the IFN-I production and increasing viral replication (Zhao et al., 2020). Studies have shown that in SARS infection, the deregulated expression of interleukins is associated with lung damage. These can be pro-inflammatory or anti-inflammatory interleukins which can be targeted by various miRNAs such as the IL-1β expression being negatively regulated by miR-146a in human lung alveolar epithelial (Perry et al., 2008). The expression of IRAK1 and TRAF6 are inhibited by miR-146a and suppresses the NF-κB activity, thereby suppressing the target gene expression such as IL-1β, IL-6, IL-8, TNFα (Li et al., 2010). It has been shown that the anti-inflammatory interleukin IL-4 expression can be controlled through STAT6 which is a target for miR-210. The expression level of IL-8, a pro-inflammatory mediator is regulated by miR-106a, miR-520b (Rarani et al., 2022). During DENV-2 infection, the downregulation of a few miRNAs such as miR-20a, miR-30b, and miR-106b alleviates the inhibition of target genes, thereby increasing the level of the pro-inflammatory cytokine. It's possible that the inactivation of repression of CCL5 and consequently increased cytokine secretion are the results of the downregulation in miR-106b expression (Wong et al., 2020). A report suggests the inhibition of IL-10 which is an anti-inflammatory mediator by miR-27a and hence increase the production of pro-inflammatory cytokines in murine macrophages infected by Mycobacterium avium. However, the inhibition of miR-27a counteracts the effect (Hussain et al., 2017) and hence this can be a therapeutic target for controlling CS. DHF is associated with severe pathological conditions in dengue-infected patients. It has been found that miR-150 is highly expressed in DHF patient samples that have a target site for SOCS1 and there is a negative correlation between SOCS1 expression and dengue severity. SOCS1 is a suppressor of cytokine signaling and atypical expression of this gene can cause cytokine dysregulation. The report shows the suppression of SOCS1 increases the expression of IFN-I, thereby can suppress viral replication (Wong et al., 2020). Inhibition of miR-150 in such conditions could be measured to tackle the deregulated cytokine expression. These pieces of evidence support the regulation of inflammation and virus infection by specific miRNA in patients which can be a potential regulator. The repeated report of dysregulated expression of certain miRNA such as miR-21, miR-146a, miR-155, and miR-122 in various virus infection conditions as well as cancer related inflammatory conditions shows their strong potential for being a therapeutic target to regulate the inflammation-mediated severity.
Several miRNA therapeutics entered clinical trials. An artificial oligonucleotide, Miravirsen, which can bind and inhibit miR-122, entered clinical trials. miRNAs can specifically target viral transcripts and it has been considered for the generation of attenuated vaccines by incorporating the miRNA response elements (MREs) into the viral genome vaccine. Among the treatment of viral diseases, Anti-miRs and antagomiRs are undergone clinical trials for HCV and HIV. Nevertheless, none of the miRNA-based drugs have been approved yet.
Despite the fact that there have been many studies and significant advances in the understanding of the mechanisms and effectiveness of miRNA therapies, still, specific challenges to achieving maximum effectiveness remain unresolved. Some of the major concerns include targeted delivery, specificity, stability, immunological activation, and in vivo and in vitro toxicity (Segal and Slack, 2020). Moreover, the stability and uniformity of miRNAs in circulation is a hassle in miRNA therapies. Nucleases found in the serum such as RNase A-type nucleases in the blood quickly break down naked miRNAs with an unmodified 2′ OH in the ribose moiety (Segal et al., 2020). Another main problem with miRNA therapy is the off-target effect of miRNAs during release from the endosome and transported into the cytoplasm. Because incomplete hybridization with 3′ UTRs produces miRNAs that target different pathways, they may unintentionally silence other genes. Off-target gene silencing may have harmful effects and diminished therapeutic effects. The likelihood of unanticipated side effects is suggested by the research showing that a single miRNA may target many mRNAs. Even if a particular miRNA is successfully targeted, there may still be unintended target effects (Suter et al., 2017).
The host's immune system may recognize double-stranded RNAs as pathogens, and may become activated. For instance, systemic miRNA administration can stimulate the innate immune system, just like other forms of nucleic acids, which can have toxicities and serious adverse effects. Through TLR, systemic injection of miRNA duplexes can cause the release of inflammatory cytokines and type I interferons (IFNs). Single or double-stranded RNAs (dsRNAs) can activate TLRs 3, 7, and 8 to trigger innate and adaptive immune responses. IFN pathway and cytokine production are both activated when these TLRs detect dsRNA molecules in cellular endosomal and lysosomal compartments (Yu et al., 2018).
There is a gap between the understanding of the mechanism of CS-induced viral infection and their regulation. Further studies should be carried out to fill that gap and to provide robust therapeutics major to overcome the adverse effects of CS associated with RNA virus outbreaks.
10. Conclusion
RNA viruses cause multiple outbreaks and are the biggest challenge to global health due to their ever-mutating nature that makes it hard to control through any specific therapeutics or vaccines. CS is associated with severe infections of RNA viruses such as IAV and SARS-CoV-2, and hence it is important to find strategies to prevent it. The current knowledge about the mechanisms by which each virus induces CS is expanding, although there are a lot of questions left to explore further. The existing methods to treat CS have their own limitations and are not enough to efficiently control the production of inflammatory cytokines without limiting innate antiviral defense. Although causative agents were different across several virus outbreaks, CS was often associated with excessive mortality. Apart from the host protein regulators of inflammation-associated pathways leading to CS, non-coding RNAs such as miRNAs have therapeutic potential to treat or prevent CS.
CRediT authorship contribution statement
Riya Chaudhary: Concept, Conceptualization, Figures and Illustration, Manuscript writing, Manuscript Editing, Manuscript Revision, Literature search, Writing – review & editing, All authors agreed for the manuscript publication. Aparna Meher: Concept, Conceptualization, Figures and Illustration, Manuscript writing, Manuscript Editing, Manuscript Revision, Literature search, Writing – review & editing, All authors agreed for the manuscript publication. Pandikannan Krishnamoorthy: Concept, Conceptualization, Manuscript writing, Manuscript Editing, Manuscript Revision, Writing – review & editing, All authors agreed for the manuscript publication. Himanshu Kumar: Supervision, Concept, Conceptualization, Manuscript writing and editing, and critical revision, Writing – review & editing, All authors agreed for the manuscript publication.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
The authors gratefully acknowledge Science and Engineering Research Board (SERB) India for the funding. We are highly thankful to the Department of Biotechnology, India (DBT)-(BT/PR44561/MED/15/217/2021), Science and Engineering Research Board, India (SERB)-(CRG/2020/001417), and IISER Bhopal for the funding.
Abbreviations
- ADE
Antibody-dependent enhancement
- ALI
Acute lung injury
- ALT
Alanine transaminase
- AP-1
Activator protein-1
- ARDS
Acute respiratory distress syndrome
- ASC
Apoptosis-associated speck-like protein
- AST
Aspartate transaminase
- cGAS-STING
cyclic GMP–AMP synthase (cGAS)–stimulator of interferon genes (STING)
- circRNA
circular RNA
- CRP
C-reactive protein
- CRS
Cytokine release syndrome
- CS
Cytokine storm
- DAMPs
Damage-associated molecular patterns
- DC
Dendritic cells
- DENV
Dengue virus
- DHF
Dengue hemorrhagic fever
- dsRNAs
double-stranded RNAs
- ECM
Extracellular matrix
- ERK1
Extracellular signal-regulated kinase 1
- HCV
Hepatitis C
- HIV
Human immunodeficiency virus
- HLH
Hemophagocytic lymphohistiocytosis
- HMGB1
High-mobility group box 1
- IAV
Human Influenza viruses
- IFNs
Interferons
- IKK
IκB kinase
- IL-6
Interleukin-6
- IRF3
Interferon regulatory factor - 3
- IRFs
Interferon regulatory factors
- ISGF3
Interferon Stimulated Gene Factor 3
- ISRE
IFN-stimulated response elements
- IκB
NF-κB inhibitor
- JAK
Janus kinase
- JEV
Japanese Enchephalitis virus
- JNK1
Jun N-terminal kinase 1
- lncRNA
Long noncoding RNA
- LNPs
Lipid-based nanoparticles
- LRTI
Lower respiratory tract infections
- MAP2K
MAPK kinase
- MAP3K
MAPK kinase kinase
- MAPK
Mitogen-activated protein kinase
- MAVS
Mitochondrial antiviral signaling protein
- miRNA
micro-RNA
- MREs
miRNA response elements
- ncRNAs
non-coding RNAs
- NEMO
NF-κB essential modulator
- NF-κB
Nuclear factor kappa-light-chain-enhancer of activated B-cells
- NK
Natural killer
- NLR
Nucleotide-binding oligomerization domain (NOD)-like receptor
- NLRP3
NLR Family Pyrin Domain Containing 3
- NO
Nitric oxide
- NS1
Non-structural protein 1
- PAMPs
Pathogen-associated molecular patterns
- PBMC
Peripheral blood mononuclear cell
- PCBP2
Poly(rC) binding protein 2
- PNA
Peptide nucleic acid
- PRRs
Pattern recognition receptors
- RLR
Retinoic acid-induced gene - I like receptor
- SOCS1
Suppressor of cytokine signaling 1
- STAT
Signal transducer and activator of transcription
- TBK1
TANK binding kinase 1
- TGF
Transforming growth factor
- TLR
Toll-like receptor
- TNF-α
Tumor necrosis factor-α
- TRIF
TIR-domain-containing adapter inducing IFN-β
- UTR
Untranslated region
- WNV
West Nile virus
Data availability
No data was used for the research described in the article.
References
- Arthur J.S., Ley S.C. 'Mitogen-activated protein kinases in innate immunity'. Nat. Rev. Immunol. 2013;13:679–692. doi: 10.1038/nri3495. [DOI] [PubMed] [Google Scholar]
- Aslani M., Mortazavi-Jahromi S.S., Mirshafiey A. 'Cytokine storm in the pathophysiology of COVID-19: possible functional disturbances of miRNAs'. Int. Immunopharm. 2021;101 doi: 10.1016/j.intimp.2021.108172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S., Biehl A., Gadina M., Hasni S., Schwartz D.M. 'JAK-STAT signaling as a target for inflammatory and autoimmune diseases: current and future prospects'. Drugs. 2017;77:521–546. doi: 10.1007/s40265-017-0701-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buggele W.A., Johnson K.E., Horvath C.M. 'Influenza A virus infection of human respiratory cells induces primary microRNA expression'. J. Biol. Chem. 2012;287:31027–31040. doi: 10.1074/jbc.M112.387670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese L.H., Lenfant T., Calabrese C. 'Cytokine storm release syndrome and the prospects for immunotherapy with COVID-19, part 4: the role of JAK inhibition'. Cleve. Clin. J. Med. 2021 doi: 10.3949/ccjm.87a.ccc060. [DOI] [PubMed] [Google Scholar]
- Carty M., Guy C., Bowie A.G. 'Detection of viral infections by innate immunity'. Biochem. Pharmacol. 2021;183 doi: 10.1016/j.bcp.2020.114316. [DOI] [PubMed] [Google Scholar]
- Carvalho-Silva A.C., Da Silva Junior A.R., Rigaud V.O., Martins W.K., Coelho V., Pfrimer I.A.H., Kalil J., Fonseca S.G., Cunha-Neto E., Ferreira L.R.P. 'A major downregulation of circulating microRNAs in Zika acutely infected patients: potential implications in innate and adaptive immune response signaling pathways'. Front. Genet. 2022;13 doi: 10.3389/fgene.2022.857728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R., Fehr AR, Zheng J, Wohlford-Lenane C, Abrahante JE, Mack M, Sompallae R, McCray PB, Jr., Meyerholz DK, Perlman S. IFN-I response timing relative to virus replication determines MERS coronavirus infection outcomes. J. Clin. Invest. 2019 Jul 29;129(9):3625–3639. doi: 10.1172/JCI126363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Deng H., Cui H., Fang J., Zuo Z., Deng J., Li Y., Wang X., Zhao L. 'Inflammatory responses and inflammation-associated diseases in organs'. Oncotarget. 2018;9:7204–7218. doi: 10.18632/oncotarget.23208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.C., Tang X.P., Tan S.Y., Liang B.L., Wan Z.Y., Fang J.Q., Zhong N. 'Treatment of severe acute respiratory syndrome with glucosteroids: the Guangzhou experience'. Chest. 2006;129:1441–1452. doi: 10.1378/chest.129.6.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.F., Yang K.D., Lee I.K., Liu J.W., Huang C.H., Lin C.Y., Chen Y.H., Chen C.L., Wang L. 'Augmented miR-150 expression associated with depressed SOCS1 expression involved in dengue haemorrhagic fever'. J. Infect. 2014;69:366–374. doi: 10.1016/j.jinf.2014.05.013. [DOI] [PubMed] [Google Scholar]
- Chew C.L., Conos S.A., Unal B., Tergaonkar V. 'Noncoding RNAs: master regulators of inflammatory signaling'. Trends Mol. Med. 2018;24:66–84. doi: 10.1016/j.molmed.2017.11.003. [DOI] [PubMed] [Google Scholar]
- Choi Y., Bowman J.W., Jung J.U. 'Autophagy during viral infection - a double-edged sword'. Nat. Rev. Microbiol. 2018;16:341–354. doi: 10.1038/s41579-018-0003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costela-Ruiz V.J., Illescas-Montes R., Puerta-Puerta J.M., Ruiz C., Melguizo-Rodriguez L. 'SARS-CoV-2 infection: the role of cytokines in COVID-19 disease'. Cytokine Growth Factor Rev. 2020;54:62–75. doi: 10.1016/j.cytogfr.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayarathna S., Jeewandara C., Gomes L., Somathilaka G., Jayathilaka D., Vimalachandran V., Wijewickrama A., Narangoda E., Idampitiya D., Ogg G.S., Malavige G.N. 'Similarities and differences between the 'cytokine storms' in acute dengue and COVID-19'. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-76836-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Della-Torre E., Lanzillotta M., Campochiaro C., Cavalli G., De Luca G., Tomelleri A., Boffini N., De Lorenzo R., Ruggeri A., Rovere-Querini P., Castagna A., Landoni G., Tresoldi M., Ciceri F., Zangrillo A., Dagna L. 'Respiratory impairment predicts response to IL-1 and IL-6 blockade in COVID-19 patients with severe pneumonia and hyper-inflammation'. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.675678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y., Yan Y., Tan K.S., Liu J., Chow V.T., Tao Z.Z., Wang D.Y. 'MicroRNA-146a induction during influenza H3N2 virus infection targets and regulates TRAF6 levels in human nasal epithelial cells (hNECs)'. Exp. Cell Res. 2017;352:184–192. doi: 10.1016/j.yexcr.2017.01.011. [DOI] [PubMed] [Google Scholar]
- Desjarlais M., Wirth M., Lahaie I., Ruknudin P., Hardy P., Rivard A., Chemtob S. 'Nutraceutical targeting of inflammation-modulating microRNAs in severe forms of COVID-19: a novel approach to prevent the cytokine storm'. Front. Pharmacol. 2020;11 doi: 10.3389/fphar.2020.602999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domizio J.D., Gulen M.F., Saidoune F., Thacker V.V., Yatim A., Sharma K., Nass T., Guenova E., Schaller M., Conrad C., Goepfert C., de Leval L., Garnier C.V., Berezowska S., Dubois A., Gilliet M., Ablasser A. 'The cGAS-STING pathway drives type I IFN immunopathology in COVID-19'. Nature. 2022;603:145–151. doi: 10.1038/s41586-022-04421-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezeonwumelu I.J., Garcia-Vidal E., Ballana E. 'JAK-STAT pathway: a novel target to tackle viral infections'. Viruses. 2021:13. doi: 10.3390/v13122379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajgenbaum D.C., June C.H. 'Cytokine storm'. N. Engl. J. Med. 2020;383:2255–2273. doi: 10.1056/NEJMra2026131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr R.J., Rootes C.L., Rowntree L.C., Nguyen T.H.O., Hensen L., Kedzierski L., Cheng A.C., Kedzierska K., Au G.G., Marsh G.A., Vasan S.S., Foo C.H., Cowled C., Stewart C.R. 'Altered microRNA expression in COVID-19 patients enables identification of SARS-CoV-2 infection'. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira A.C., Soares V.C., de Azevedo-Quintanilha I.G., Dias Sdsg, Fintelman-Rodrigues N., Sacramento C.Q., Mattos M., de Freitas C.S., Temerozo J.R., Teixeira L., Damaceno Hottz E., Barreto E.A., Pao C.R.R., Palhinha L., Miranda M., Bou-Habib D.C., Bozza F.A., Bozza P.T., Souza T.M.L. 'SARS-CoV-2 engages inflammasome and pyroptosis in human primary monocytes'. Cell Death Dis. 2021;7:43. doi: 10.1038/s41420-021-00428-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbester J.L., Humphreys I.R. 'Genetic influences on viral-induced cytokine responses in the lung'. Mucosal Immunol. 2021;14:14–25. doi: 10.1038/s41385-020-00355-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung T.S., Liu D.X. 'Human coronavirus: host-pathogen interaction'. Annu. Rev. Microbiol. 2019;73:529–557. doi: 10.1146/annurev-micro-020518-115759. [DOI] [PubMed] [Google Scholar]
- Gregersen L.H., Jacobsen A.B., Frankel L.B., Wen J., Krogh A., Lund A.H. 'MicroRNA-145 targets YES and STAT1 in colon cancer cells'. PLoS One. 2010;5 doi: 10.1371/journal.pone.0008836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y., Hsu A.C., Pang Z., Pan H., Zuo X., Wang G., Zheng J., Wang F. 'Role of the innate cytokine storm induced by the influenza A virus'. Viral Immunol. 2019;32:244–251. doi: 10.1089/vim.2019.0032. [DOI] [PubMed] [Google Scholar]
- Hasanvand A. 'COVID-19 and the role of cytokines in this disease'. Inflammopharmacology. 2022;30:789–798. doi: 10.1007/s10787-022-00992-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho B.C., Yu I.S., Lu L.F., Rudensky A., Chen H.Y., Tsai C.W., Chang Y.L., Wu C.T., Chang L.Y., Shih S.R., Lin S.W., Lee C.N., Yang P.C., Yu S.L. 'Inhibition of miR-146a prevents enterovirus-induced death by restoring the production of type I interferon'. Nat. Commun. 2014;5:3344. doi: 10.1038/ncomms4344. [DOI] [PubMed] [Google Scholar]
- Hoffman K.W., Sachs D., Bardina S.V., Michlmayr D., Rodriguez C.A., Sum J., Foster G.A., Krysztof D., Stramer S.L., Lim J.K. 'Differences in early cytokine production are associated with development of a greater number of symptoms following West Nile virus infection'. J. Infect. Dis. 2016;214:634–643. doi: 10.1093/infdis/jiw179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm C.K., Rahbek S.H., Gad H.H., Bak R.O., Jakobsen M.R., Jiang Z., Hansen A.L., Jensen S.K., Sun C., Thomsen M.K., Laustsen A., Nielsen C.G., Severinsen K., Xiong Y., Burdette D.L., Hornung V., Lebbink R.J., Duch M., Fitzgerald K.A., Bahrami S., Mikkelsen J.G., Hartmann R., Paludan S.R. 'Influenza A virus targets a cGAS-independent STING pathway that controls enveloped RNA viruses'. Nat. Commun. 2016;7 doi: 10.1038/ncomms10680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. 'Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China'. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain M., Torres S., Schnettler E., Funk A., Grundhoff A., Pijlman G.P., Khromykh A.A., Asgari S. 'West Nile virus encodes a microRNA-like small RNA in the 3' untranslated region which up-regulates GATA4 mRNA and facilitates virus replication in mosquito cells'. Nucleic Acids Res. 2012;40:2210–2223. doi: 10.1093/nar/gkr848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain T., Zhao D., Shah S.Z.A., Wang J., Yue R., Liao Y., Sabir N., Yang L., Zhou X. 'MicroRNA 27a-3p regulates antimicrobial responses of murine macrophages infected by Mycobacterium avium subspecies paratuberculosis by targeting interleukin-10 and TGF-beta-activated protein kinase 1 binding protein 2'. Front. Immunol. 2017;8:1915. doi: 10.3389/fimmu.2017.01915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussell T., Goulding J. 'Structured regulation of inflammation during respiratory viral infection'. Lancet Infect. Dis. 2010;10:360–366. doi: 10.1016/S1473-3099(10)70067-0. [DOI] [PubMed] [Google Scholar]
- Izzard L., Ye S., Jenkins K., Xia Y., Tizard M., Stambas J. 'miRNA modulation of SOCS1 using an influenza A virus delivery system'. J. Gen. Virol. 2014;95:1880–1885. doi: 10.1099/vir.0.063834-0. [DOI] [PubMed] [Google Scholar]
- Jafarzadeh A., Chauhan P., Saha B., Jafarzadeh S., Nemati M. 'Contribution of monocytes and macrophages to the local tissue inflammation and cytokine storm in COVID-19: lessons from SARS and MERS, and potential therapeutic interventions'. Life Sci. 2020;257 doi: 10.1016/j.lfs.2020.118102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Zhao T., Zhou X., Xiang Y., Gutierrez-Castrellon P., Ma X. 'Inflammatory pathways in COVID-19: mechanism and therapeutic interventions'. MedComm (2020) 2022;3:e154. doi: 10.1002/mco2.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jounai N., Takeshita F., Kobiyama K., Sawano A., Miyawaki A., Xin K.Q., Ishii K.J., Kawai T., Akira S., Suzuki K., Okuda K. 'The Atg5 Atg12 conjugate associates with innate antiviral immune responses'. Proc. Natl. Acad. Sci. U. S. A. 2007;104:14050–14055. doi: 10.1073/pnas.0704014104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalaiyarasu S., Kumar M., Senthil Kumar D., Bhatia S., Dash S.K., Bhat S., Khetan R.K., Nagarajan S. 'Highly pathogenic avian influenza H5N1 virus induces cytokine dysregulation with suppressed maturation of chicken monocyte-derived dendritic cells'. Microbiol. Immunol. 2016;60:687–693. doi: 10.1111/1348-0421.12443. [DOI] [PubMed] [Google Scholar]
- Karki R., Kanneganti T.D. 'The 'cytokine storm': molecular mechanisms and therapeutic prospects'. Trends Immunol. 2021;42:681–705. doi: 10.1016/j.it.2021.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T., Takahashi K., Sato S., Coban C., Kumar H., Kato H., Ishii K.J., Takeuchi O., Akira S. 'IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction'. Nat. Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- Khani E., Shahrabi M., Rezaei H., Pourkarim F., Afsharirad H., Solduzian M. 'Current evidence on the use of anakinra in COVID-19'. Int. Immunopharm. 2022;111 doi: 10.1016/j.intimp.2022.109075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsch-Volders M., Fenech M. 'Inflammatory cytokine storms severity may be fueled by interactions of micronuclei and RNA viruses such as COVID-19 virus SARS-CoV-2. A hypothesis'. Mutat. Res. Rev. Mutat. Res. 2021;788 doi: 10.1016/j.mrrev.2021.108395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocak Tufan Z., Kayaaslan B., Mer M. 'COVID-19 and sepsis'. Turk. J. Med. Sci. 2021;51:3301–3311. doi: 10.3906/sag-2108-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koukos G., Polytarchou C., Kaplan J.L., Morley-Fletcher A., Gras-Miralles B., Kokkotou E., Baril-Dore M., Pothoulakis C., Winter H.S., Iliopoulos D. 'MicroRNA-124 regulates STAT3 expression and is down-regulated in colon tissues of pediatric patients with ulcerative colitis'. Gastroenterology. 2013;145:842–852 e2. doi: 10.1053/j.gastro.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama S., Ishii K.J., Coban C., Akira S. 'Innate immune response to viral infection'. Cytokine. 2008;43:336–341. doi: 10.1016/j.cyto.2008.07.009. [DOI] [PubMed] [Google Scholar]
- Krishnamoorthy P., Raj A.S., Kumar P., Das N., Kumar H. 'Host and viral non-coding RNAs in dengue pathogenesis'. Rev. Med. Virol. 2022;32 doi: 10.1002/rmv.2360. [DOI] [PubMed] [Google Scholar]
- Kujime K., Hashimoto S., Gon Y., Shimizu K., Horie T. 'p38 mitogen-activated protein kinase and c-jun-NH2-terminal kinase regulate RANTES production by influenza virus-infected human bronchial epithelial cells'. J. Immunol. 2000;164:3222–3228. doi: 10.4049/jimmunol.164.6.3222. [DOI] [PubMed] [Google Scholar]
- Kumar H., Kawai T., Akira S. 'Pathogen recognition by the innate immune system'. Int. Rev. Immunol. 2011;30:16–34. doi: 10.3109/08830185.2010.529976. [DOI] [PubMed] [Google Scholar]
- Kumar R., Khandelwal N., Thachamvally R., Tripathi B.N., Barua S., Kashyap S.K., Maherchandani S., Kumar N. ‘Role of MAPK/MNK1 signaling in virus replication’. Virus Res. 2018;253:48–61. doi: 10.1016/j.virusres.2018.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Kumar A., Ingle H., Kumar S., Mishra R., Verma M.K., Biswas D., Kumar N.S., Mishra A., Raut A.A., Takaoka A., Kumar H. ‘MicroRNA hsa-miR-324-5p suppresses H5N1 virus replication by targeting the viral PB1 and host CUEDC2’. J. Virol. 2018:92. doi: 10.1128/JVI.01057-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriakose T., Kanneganti T.D. 'Pyroptosis in antiviral immunity'. Curr. Top. Microbiol. Immunol. 2019 doi: 10.1007/82_2019_189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert L., Sagfors A.M., Openshaw P.J., Culley F.J. 'Immunity to RSV in early-life'. Front. Immunol. 2014;5:466. doi: 10.3389/fimmu.2014.00466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.S., Kim Y.J. 'Signaling pathways downstream of pattern-recognition receptors and their cross talk'. Annu. Rev. Biochem. 2007;76:447–480. doi: 10.1146/annurev.biochem.76.060605.122847. [DOI] [PubMed] [Google Scholar]
- Lee N.R., Ban J., Lee N.J., Yi C.M., Choi J.Y., Kim H., Lee J.K., Seong J., Cho N.H., Jung J.U., Inn K.S. 'Activation of RIG-I-mediated antiviral signaling triggers autophagy through the MAVS-TRAF6-Beclin-1 signaling Axis'. Front. Immunol. 2018;9:2096. doi: 10.3389/fimmu.2018.02096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Chen X.P., Li Y.J. 'MicroRNA-146a and human disease'. Scand. J. Immunol. 2010;71:227–231. doi: 10.1111/j.1365-3083.2010.02383.x. [DOI] [PubMed] [Google Scholar]
- Li X., Fu Z., Liang H., Wang Y., Qi X., Ding M., Sun X., Zhou Z., Huang Y., Gu H., Li L., Chen X., Li D., Zhao Q., Liu F., Wang H., Wang J., Zen K., Zhang C.Y. 'H5N1 influenza virus-specific miRNA-like small RNA increases cytokine production and mouse mortality via targeting poly(rC)-binding protein 2'. Cell Res. 2018;28:157–171. doi: 10.1038/cr.2018.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Geng M., Peng Y., Meng L., Lu S. 'Molecular immune pathogenesis and diagnosis of COVID-19'. J Pharm Anal. 2020;10:102–108. doi: 10.1016/j.jpha.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Guo S., Liu G., Qiu Z., Wang J., Yang D., Tian X., Qiao Z., Ma Z., Liu Z. 'Host non-coding RNA regulates influenza A virus replication'. Viruses. 2021:14. doi: 10.3390/v14010051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Zhou Y.H., Yang Z.Q. 'The cytokine storm of severe influenza and development of immunomodulatory therapy'. Cell. Mol. Immunol. 2016;13:3–10. doi: 10.1038/cmi.2015.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Zhang L., Joo D., Sun S.C. 'NF-kappaB signaling in inflammation'. Signal Transduct. Targeted Ther. 2017;2 doi: 10.1038/sigtrans.2017.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z.M., Yang M.H., Yu K., Lian Z.X., Deng S.L. 'Toll-like receptor (TLRs) agonists and antagonists for COVID-19 treatments'. Front. Pharmacol. 2022;13 doi: 10.3389/fphar.2022.989664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lordan Ronan, Tsoupras Alexandros, Zabetakis Ioannis. In: The Impact of Nutrition and Statins on Cardiovascular Diseases. Zabetakis Ioannis, Lordan Ronan, Tsoupras Alexandros., editors. Academic Press; 2019. Chapter 2 - inflammation. [Google Scholar]
- Ludwig S., Ehrhardt C., Neumeier E.R., Kracht M., Rapp U.R., Pleschka S. 'Influenza virus-induced AP-1-dependent gene expression requires activation of the JNK signaling pathway'. J. Biol. Chem. 2001;276:10990–10998. [PubMed] [Google Scholar]
- Maranini B., Ciancio G., Ferracin M., Cultrera R., Negrini M., Sabbioni S., Govoni M. vol. 12. A Narrative Review; Life (Basel): 2022. (microRNAs and Inflammatory Immune Response in SARS-CoV-2 Infection). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra R., Bhattacharya S., Rawat B.S., Kumar A., Kumar A., Niraj K., Chande A., Gandhi P., Khetan D., Aggarwal A., Sato S., Tailor P., Takaoka A., Kumar H. 'MicroRNA-30e-5p has an integrated role in the regulation of the innate immune response during virus infection and systemic lupus erythematosus'. iScience. 2020;23 doi: 10.1016/j.isci.2020.101322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra R., Krishnamoorthy P., Kumar H. 'MicroRNA-30e-5p regulates SOCS1 and SOCS3 during bacterial infection'. Front. Cell. Infect. Microbiol. 2020;10 doi: 10.3389/fcimb.2020.604016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogensen T.H. 'IRF and STAT transcription factors - from basic Biology to roles in infection, protective immunity, and primary immunodeficiencies'. Front. Immunol. 2018;9:3047. doi: 10.3389/fimmu.2018.03047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moossavi M., Rastegar M., Moossavi S.Z., Khorasani M. 'Molecular function of cGAS-STING in SARS-CoV-2: a novel approach to COVID-19 treatment'. BioMed Res. Int. 2022;2022 doi: 10.1155/2022/6189254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama M., Koshiba T., Ichinohe T. 'Influenza A virus M2 protein triggers mitochondrial DNA-mediated antiviral immune responses'. Nat. Commun. 2019;10:4624. doi: 10.1038/s41467-019-12632-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris G., Bortolasci C.C., Puri B.K., Marx W., O'Neil A., Athan E., Walder K., Berk M., Olive L., Carvalho A.F., Maes M. 'The cytokine storms of COVID-19, H1N1 influenza, CRS and MAS compared. Can one sized treatment fit all?'. Cytokine. 2021;144 doi: 10.1016/j.cyto.2021.155593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris R., Kershaw N.J., Babon J.J. 'The molecular details of cytokine signaling via the JAK/STAT pathway'. Protein Sci. 2018;27:1984–2009. doi: 10.1002/pro.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- multicenter collaboration group of Department of Science. Province Technology of Guangdong and pneumonia Health Commission of Guangdong Province for chloroquine in the treatment of novel coronavirus '[Expert consensus on chloroquine phosphate for the treatment of novel coronavirus pneumonia]'. Zhonghua Jiehe He Huxi Zazhi. 2020;43:185–188. doi: 10.3760/cma.j.issn.1001-0939.2020.03.009. [DOI] [PubMed] [Google Scholar]
- Nanbo A., Furuyama W., Lin Z. 'RNA virus-encoded miRNAs: current insights and future challenges'. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.679210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narozna M., Rubis B. 'Anti-SARS-CoV-2 strategies and the potential role of miRNA in the assessment of COVID-19 morbidity, recurrence, and therapy'. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms22168663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L.N., Kanneganti T.D. 'PANoptosis in viral infection: the missing puzzle piece in the cell death field'. J. Mol. Biol. 2022;434 doi: 10.1016/j.jmb.2021.167249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T.H., Liu X., Su Z.Z., Hsu A.C., Foster P.S., Yang M. 'Potential role of MicroRNAs in the regulation of antiviral responses to influenza infection'. Front. Immunol. 2018;9:1541. doi: 10.3389/fimmu.2018.01541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong J.D., Mansell A., Tate M.D. 'Hero turned villain: NLRP3 inflammasome-induced inflammation during influenza A virus infection'. J. Leukoc. Biol. 2017;101:863–874. doi: 10.1189/jlb.4MR0616-288R. [DOI] [PubMed] [Google Scholar]
- Othumpangat S., Bryan N.B., Beezhold D.H., Noti J.D. 'Upregulation of miRNA-4776 in influenza virus infected bronchial epithelial cells is associated with downregulation of NFKBIB and increased viral survival'. Viruses. 2017;9 doi: 10.3390/v9050094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang X., Jiang X., Gu D., Zhang Y., Kong S.K., Jiang C., Xie W. 'Dysregulated serum MiRNA profile and promising biomarkers in dengue-infected patients'. Int. J. Med. Sci. 2016;13:195–205. doi: 10.7150/ijms.13996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey N., Jain A., Garg R.K., Kumar R., Agrawal O.P., Lakshmana Rao P.V. 'Serum levels of IL-8, IFNgamma, IL-10, and TGF beta and their gene expression levels in severe and non-severe cases of dengue virus infection'. Arch. Virol. 2015;160:1463–1475. doi: 10.1007/s00705-015-2410-6. [DOI] [PubMed] [Google Scholar]
- Perry M.M., Moschos S.A., Williams A.E., Shepherd N.J., Larner-Svensson H.M., Lindsay M.A. 'Rapid changes in microRNA-146a expression negatively regulate the IL-1beta-induced inflammatory response in human lung alveolar epithelial cells'. J. Immunol. 2008;180:5689–5698. doi: 10.4049/jimmunol.180.8.5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradel B., Robert-Hebmann V., Espert L. 'Regulation of innate immune responses by autophagy: a goldmine for viruses'. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.578038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pum A., Ennemoser M., Adage T., Kungl A.J. 'Cytokines and chemokines in SARS-CoV-2 infections-therapeutic strategies targeting cytokine storm'. Biomolecules. 2021;11 doi: 10.3390/biom11010091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramani T., Auletta C.S., Weinstock D., Mounho-Zamora B., Ryan P.C., Salcedo T.W., Bannish G. 'Cytokines: the good, the Bad, and the deadly'. Int. J. Toxicol. 2015;34:355–365. doi: 10.1177/1091581815584918. [DOI] [PubMed] [Google Scholar]
- Rarani F.Z., Rashidi B., Jafari Najaf Abadi M.H., Hamblin M.R., Reza Hashemian S.M., Mirzaei H. 'Cytokines and microRNAs in SARS-CoV-2: what do we know?'. Mol. Ther. Nucleic Acids. 2022;29:219–242. doi: 10.1016/j.omtn.2022.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberger C.M., Podyminogin R.L., Diercks A.H., Treuting P.M., Peschon J.J., Rodriguez D., Gundapuneni M., Weiss M.J., Aderem A. 'miR-144 attenuates the host response to influenza virus by targeting the TRAF6-IRF7 signaling axis'. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samie M., Lim J., Verschueren E., Baughman J.M., Peng I., Wong A., Kwon Y., Senbabaoglu Y., Hackney J.A., Keir M., McKenzie B., Kirkpatrick D.S., van Lookeren Campagne M., Murthy A. 'Selective autophagy of the adaptor TRIF regulates innate inflammatory signaling'. Nat. Immunol. 2018;19:246–254. doi: 10.1038/s41590-017-0042-6. [DOI] [PubMed] [Google Scholar]
- Schindler C., Strehlow I. 'Cytokines and STAT signaling'. Adv. Pharmacol. 2000;47:113–174. doi: 10.1016/s1054-3589(08)60111-8. [DOI] [PubMed] [Google Scholar]
- Segal M., Biscans A., Gilles M.E., Anastasiadou E., De Luca R., Lim J., Khvorova A., Slack F.J. 'Hydrophobically modified let-7b miRNA enhances Biodistribution to NSCLC and downregulates HMGA2 in vivo'. Mol. Ther. Nucleic Acids. 2020;19:267–277. doi: 10.1016/j.omtn.2019.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal M., Slack F.J. 'Challenges identifying efficacious miRNA therapeutics for cancer'. Expet Opin. Drug Discov. 2020;15:987–992. doi: 10.1080/17460441.2020.1765770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah A.U., Cao Y., Siddique N., Lin J., Yang Q. vol. 7. Vaccines; Basel): 2019. (miR29a and miR378b Influence CpG-Stimulated Dendritic Cells and Regulate cGAS/STING Pathway). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastava S., Petrone J., Steele R., Lauer G.M., Di Bisceglie A.M., Ray R.B. 'Up-regulation of circulating miR-20a is correlated with hepatitis C virus-mediated liver disease progression'. Hepatology. 2013;58:863–871. doi: 10.1002/hep.26296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares-Silva M., Diniz F.F., Gomes G.N., Bahia D. 'The mitogen-activated protein kinase (MAPK) pathway: role in immune evasion by Trypanosomatids'. Front. Microbiol. 2016;7:183. doi: 10.3389/fmicb.2016.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikiatkhachorn A., Mathew A., Rothman A.L. 'Immune-mediated cytokine storm and its role in severe dengue'. Semin. Immunopathol. 2017;39:563–574. doi: 10.1007/s00281-017-0625-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y., Lin T., Liu C., Cheng C., Han X., Jiang X. 'microRNAs, the link between dengue virus and the host genome'. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.714409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Wang T., Cai D., Hu Z., Chen J., Liao H., Zhi L., Wei H., Zhang Z., Qiu Y., Wang J., Wang A. 'Cytokine storm intervention in the early stages of COVID-19 pneumonia'. Cytokine Growth Factor Rev. 2020;53:38–42. doi: 10.1016/j.cytogfr.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P.S., Hsieh S.L. 'C-type lectins and extracellular vesicles in virus-induced NETosis'. J. Biomed. Sci. 2021;28:46. doi: 10.1186/s12929-021-00741-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter S.R., Ball-Jones A., Mumbleau M.M., Valenzuela R., Ibarra-Soza J., Owens H., Fisher A.J., Beal P.A. 'Controlling miRNA-like off-target effects of an siRNA with nucleobase modifications'. Org. Biomol. Chem. 2017;15:10029–10036. doi: 10.1039/c7ob02654d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taganov K.D., Boldin M.P., Chang K.J., Baltimore D. 'NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses'. Proc. Natl. Acad. Sci. U. S. A. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tambyah P.A., Sepramaniam S., Mohamed Ali J., Chai S.C., Swaminathan P., Armugam A., Jeyaseelan K. 'microRNAs in circulation are altered in response to influenza A virus infection in humans'. PLoS One. 2013;8 doi: 10.1371/journal.pone.0076811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Narazaki M., Kishimoto T. 'Immunotherapeutic implications of IL-6 blockade for cytokine storm'. Immunotherapy. 2016;8:959–970. doi: 10.2217/imt-2016-0020. [DOI] [PubMed] [Google Scholar]
- Tang H., Gao Y., Li Z., Miao Y., Huang Z., Liu X., Xie L., Li H., Wen W., Zheng Y., Su W. 'The noncoding and coding transcriptional landscape of the peripheral immune response in patients with COVID-19'. Clin. Transl. Med. 2020;10 doi: 10.1002/ctm2.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X.D., Ji T.T., Dong J.R., Feng H., Chen F.Q., Chen X., Zhao H.Y., Chen D.K., Ma W.T. 'Pathogenesis and treatment of cytokine storm induced by infectious diseases'. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms222313009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarasuk M., Songprakhon P., Chieochansin T., Choomee K., Na-Bangchang K., Yenchitsomanus P.T. 'Alpha-mangostin inhibits viral replication and suppresses nuclear factor kappa B (NF-kappaB)-mediated inflammation in dengue virus infection'. Sci. Rep. 2022;12 doi: 10.1038/s41598-022-20284-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisoncik J.R., Korth M.J., Simmons C.P., Farrar J., Martin T.R., Katze M.G. 'Into the eye of the cytokine storm'. Microbiol. Mol. Biol. Rev. 2012;76:16–32. doi: 10.1128/MMBR.05015-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Teng Y., Zhao G., Li F., Hou A., Sun B., Kong W., Gao F., Cai L., Jiang C. 'Exosome-Mediated delivery of inducible miR-423-5p enhances resistance of MRC-5 cells to rabies virus infection'. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20071537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Lu Y., Toh S.T., Sung W.K., Tan P., Chow P., Chung A.Y., Jooi L.L., Lee C.G. 'Lethal-7 is down-regulated by the hepatitis B virus x protein and targets signal transducer and activator of transcription 3'. J. Hepatol. 2010;53:57–66. doi: 10.1016/j.jhep.2009.12.043. [DOI] [PubMed] [Google Scholar]
- Wang Z., Zhang A., Wan Y., Liu X., Qiu C., Xi X., Ren Y., Wang J., Dong Y., Bao M., Li L., Zhou M., Yuan S., Sun J., Zhu Z., Chen L., Li Q., Zhang Z., Zhang X., Lu S., Doherty P.C., Kedzierska K., Xu J. 'Early hypercytokinemia is associated with interferon-induced transmembrane protein-3 dysfunction and predictive of fatal H7N9 infection'. Proc. Natl. Acad. Sci. U. S. A. 2014;111:769–774. doi: 10.1073/pnas.1321748111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong R.R., Abd-Aziz N., Affendi S., Poh C.L. 'Role of microRNAs in antiviral responses to dengue infection'. J. Biomed. Sci. 2020;27:4. doi: 10.1186/s12929-019-0614-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Huang M., Cao P., Wang T., Shu Y., Liu P. 'MiR-135a targets JAK2 and inhibits gastric cancer cell proliferation'. Cancer Biol. Ther. 2012;13:281–288. doi: 10.4161/cbt.18943. [DOI] [PubMed] [Google Scholar]
- Wu S., He L., Li Y., Wang T., Feng L., Jiang L., Zhang P., Huang X. 'miR-146a facilitates replication of dengue virus by dampening interferon induction by targeting TRAF6'. J. Infect. 2013;67:329–341. doi: 10.1016/j.jinf.2013.05.003. [DOI] [PubMed] [Google Scholar]
- Yamada T., Sato S., Sotoyama Y., Orba Y., Sawa H., Yamauchi H., Sasaki M., Takaoka A. 'RIG-I triggers a signaling-abortive anti-SARS-CoV-2 defense in human lung cells'. Nat. Immunol. 2021;22:820–828. doi: 10.1038/s41590-021-00942-0. [DOI] [PubMed] [Google Scholar]
- Yan Y., Lu K., Ye T., Zhang Z. 'MicroRNA-223 attenuates LPS-induced inflammation in an acute lung injury model via the NLRP3 inflammasome and TLR4/NF-kappaB signaling pathway via RHOB'. Int. J. Mol. Med. 2019;43:1467–1477. doi: 10.3892/ijmm.2019.4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Xie X., Tu Z., Fu J., Xu D., Zhou Y. 'The signal pathways and treatment of cytokine storm in COVID-19'. Signal Transduct. Targeted Ther. 2021;6:255. doi: 10.1038/s41392-021-00679-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q., Wang B., Mao J. 'The pathogenesis and treatment of the ;Cytokine Storm' in COVID-19'. J. Infect. 2020;80:607–613. doi: 10.1016/j.jinf.2020.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura A., Ito M., Chikuma S., Akanuma T., Nakatsukasa H. 'Negative regulation of cytokine signaling in immunity'. Cold Spring Harbor Perspect. Biol. 2018;10 doi: 10.1101/cshperspect.a028571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H.R., Huang L.H., Li S.C. 'Roles of microRNA in the immature immune system of neonates'. Cancer Lett. 2018;433:99–106. doi: 10.1016/j.canlet.2018.06.014. [DOI] [PubMed] [Google Scholar]
- Yu X., Zhang X., Zhao B., Wang J., Zhu Z., Teng Z., Shao J., Shen J., Gao Y., Yuan Z., Wu F. 'Intensive cytokine induction in pandemic H1N1 influenza virus infection accompanied by robust production of IL-10 and IL-6'. PLoS One. 2011;6 doi: 10.1371/journal.pone.0028680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Han L., Zhang A., Yang W., Zhou X., Pu P., Du Y., Zeng H., Kang C. 'Global changes of mRNA expression reveals an increased activity of the interferon-induced signal transducer and activator of transcription (STAT) pathway by repression of miR-221/222 in glioblastoma U251 cells'. Int. J. Oncol. 2010;36:1503–1512. doi: 10.3892/ijo_00000637. [DOI] [PubMed] [Google Scholar]
- Zhang H.Q., Yang S.W., Fu Y.C., Chen M.C., Yang C.H., Yang M.H., Liu X.D., He Q.N., Jiang H., Zhao M.Y. 'Cytokine storm and targeted therapy in hemophagocytic lymphohistiocytosis'. Immunol. Res. 2022;70:566–577. doi: 10.1007/s12026-022-09285-w. [DOI] [PubMed] [Google Scholar]
- Zhang S., Amahong K., Sun X., Lian X., Liu J., Sun H., Lou Y., Zhu F., Qiu Y. 'The miRNA: a small but powerful RNA for COVID-19'. Briefings Bioinf. 2021;22:1137–1149. doi: 10.1093/bib/bbab062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Zhao W. 'NLRP3 inflammasome-A key player in antiviral responses'. Front. Immunol. 2020;11:211. doi: 10.3389/fimmu.2020.00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J.P., Hu Y., Du R.H., Chen Z.S., Jin Y., Zhou M., Zhang J., Qu J.M., Cao B. [Expert consensus on the use of corticosteroid in patients with 2019-nCoV pneumonia] Zhonghua Jiehe He Huxi Zazhi. 2020;43:183–184. doi: 10.3760/cma.j.issn.1001-0939.2020.03.008. [DOI] [PubMed] [Google Scholar]
- Zhao L., Zhang X., Wu Z., Huang K., Sun X., Chen H., Jin M. 'The downregulation of MicroRNA hsa-miR-340-5p in IAV-infected A549 cells suppresses viral replication by targeting RIG-I and OAS2'. Mol. Ther. Nucleic Acids. 2019;14:509–519. doi: 10.1016/j.omtn.2018.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Zhu J., Zhou H., Zhao Z., Zou Z., Liu X., Lin X., Zhang X., Deng X., Wang R., Chen H., Jin M. 'Identification of cellular microRNA-136 as a dual regulator of RIG-I-mediated innate immunity that antagonizes H5N1 IAV replication in A549 cells'. Sci. Rep. 2015;5 doi: 10.1038/srep14991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., He Z., Hu Y., Wen W., Lin C., Yu J., Pan J., Li R., Deng H., Liao S., Yuan J., Wu J., Li J., Li M. ‘MicroRNA-30e* suppresses dengue virus replication by promoting NF-kappaB-dependent IFN production’. PLoS Neglected Trop. Dis. 2014;8 doi: 10.1371/journal.pntd.0003088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Qi Y., Ge A., Zhu Y., Xu K., Ji H., Shi Z., Cui L., Zhou M. ‘Comprehensive characterization of serum microRNA profile in response to the emerging avian influenza A (H7N9) virus infection in humans’. Viruses. 2014;6:1525–1539. doi: 10.3390/v6041525. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.