Abstract
Ferroptosis is an iron dependent form of cell death, that is triggered by the discoordination of iron, lipids, and thiols. Its unique signature that distinguishes it from other forms of cell death is the formation and accumulation of lipid hydroperoxides, particularly oxidized forms of polyunsaturated phosphatidylethanolamines (PEs), which drives cell death. These readily undergo iron-catalyzed secondary free radical reactions leading to truncated products which retain the signature PE headgroup and which can readily react with nucleophilic moieties in proteins via their truncated electrophilic acyl chains. Using a redox lipidomics approach, we have identified oxidatively-truncated PE species (trPEox) in enzymatic and non-enzymatic model systems. Further, using a model peptide we demonstrate adduct formation with Cys as the preferred nucleophilic residue and PE(26:2) +2 oxygens, as one of the most reactive truncated PE-electrophiles produced. In cells stimulated to undergo ferroptosis we identified PE-truncated species with sn-2 truncations ranging from 5 to 9 carbons. Taking advantage of the free PE headgroup, we have developed a new technology using the lantibiotic duramycin, to enrich and identify the PE-lipoxidated proteins. Our results indicate that several dozens of proteins for each cell type, are PE-lipoxidated in HT-22, MLE, and H9c2 cells and M2 macrophages after they were induced to undergo ferroptosis. Pretreatment of cells with the strong nucleophile, 2-mercaptoethanol, prevented the formation of PE-lipoxidated proteins and blocked ferroptotic death. Finally, our docking simulations showed that the truncated PE species bound at least as good to several of the lantibiotic-identified proteins, as compared to the non-truncated parent molecule, stearoyl-arachidonoyl PE (SAPE), indicating that these oxidatively-truncated species favor/promote the formation of PEox-protein adducts. The identification of PEox-protein adducts during ferroptosis suggests that they are participants in the ferroptotic process preventable by 2-mercaptoethanol and may contribute to a point of no return in the ferroptotic death process.
Keywords: Ferroptotic death, Lantibiotics, Oxidatively-truncated lipids, Protein lipidation, Oxidized phosphatidylethanolamine, 2-Mercaptoethanol
Graphical abstract
Highlights
-
•
Oxidatively truncated PE species are formed during ferroptosis.
-
•
Truncation occurred in the sn-2 chain resulting in chain lengths from 5 to 9 carbons.
-
•
We developed a new technology using the lantibiotic duramycin, to assess PE “electrophilically lipidated” protein adducts during ferroptosis.
-
•
PE-lipoxidated proteins from ferroptotic cells were identified in nuclear, cytoskeletal, mitochondrial and plasma membrane compartments.
-
•
Pretreatment of cells with 2-mercaptoethanol prevented the formation of PE-lipoxidated proteins and blocked ferroptotic death.
1. Introduction
Excessive injury to mammalian cells can lead to their death either accidently and chaotically (in an unplanned way), or via one of the regulated and genetically pre-determined death programs. Currently, over a dozen regulated cell death programs have been identified and their molecular mechanisms and machinery are being examined. Apoptosis, one of the first types of regulated cell death programs discovered, has been described as the type of cellular demise that preserves cell membrane integrity to sequester digested intracellular contents. Apoptotic cells and their “apoptotic bodies” are recognized and eliminated by several kinds of specialized receptors on professional phagocytes [[1], [2], [3], [4], [5], [6], [7]]. These features of apoptosis define its anti-inflammatory pro-resolving function in the regulation of inflammatory responses. Apoptosis is associated with a number of unique signatures, which includes, but is not limited to, activated caspases, cytochrome C release, externalized PS, nucleosomal DNA, Fas/Fas ligand, Bcl-2/Bcl-xl, and p53 [8], many which can be easily measured in patient and animal tissues. On the other hand, the common feature of several regulated necrotic programs including classical necrosis, is the loss of plasma membrane integrity, spillage of genotoxic and immunogenic intracellular contents leading to the enhanced pro-inflammatory responses [[9], [10], [11], [12], [13], [14], [15], [16], [17]]. Unique signatures for this type of necrotic cell death include RIPK1 and RIPK3 kinases, microRNAs, full length cytokeratin 18, among others [18,19].
Ferroptosis, one of latest additions to the list of death programs, is an iron dependent form of cell death, different from apoptosis, necroptosis, and classic necrosis [[20], [21], [22], [23]]. It is triggered by the discoordination of three major metabolic redox domains namely iron, lipids, and thiols [[20], [21], [22], [23]]. Its unique signature that distinguishes it from other forms of cell death is the formation and accumulation of lipid hydroperoxides which drive the death process [[20], [21], [22], [23]]. Ferroptosis is the only known death program with massive lipid peroxidation. Indeed, several links have been made between ferroptosis and pathological conditions, such as degenerative and neo-plastic diseases, ischemic injury in the liver, kidney, brain (due to stroke or intracerebral hemorrhage) and heart, as well as traumatic brain injury [[24], [25], [26], [27], [28], [29], [30]].
While the central elements of ferroptosis include oxidized polyunsaturated phospholipids and redox-active iron, the detection of these entities, and equally as important their downstream protein targets (PE-protein adducts), remains an obstacle in a variety of settings for several reasons. 1) Polyunsaturated phospholipids (PUFA-PLs) and specifically polyunsaturated phosphatidylethanolamines (PUFA-PEs) which have been identified as the selective targets of pro-ferroptotic lipid peroxidation [[20], [21], [22], [23], [24], [25], [26], [27],[31], [32], [33]], are inherently unstable and readily undergo Fe-catalyzed secondary free radical reactions, giving way to numerous truncated reactive products via beta-scission [34] and are therefore difficult to detect in the blood and other bodily fluids and tissues. 2) Further, these reactive electrophiles may subsequently undergo reactions with nucleophilic amino acids of cellular proteins forming PE-protein adducts, making them “hidden” from detection, once covalently bound. 3) Because of their low abundance, structural assessment of oxidized phospholipids, their truncated products and their downstream protein-adducts usually requires the use of highly sensitive mass spectrometry platforms. 4) Finally, short-chain leaving fragments such as HNE and HHE may confound the issue as these products, while they are good indicators of lipid oxidation, may not always be specific to ferroptosis. Thus, specific methods and reagents to definitively detect PE-protein adducts, over conditions that induce non-ferroptotic oxidative stress, are needed.
The preponderance of PUFA-PE oxidation in ferroptosis is most likely due to PE being the second most abundant phospholipid in eukaryotic cells, since it is critical for many cellular functions such as protein biogenesis and activity, oxidative phosphorylation, membrane fusion and autophagy [35]. PE also serves as a precursor for other lipids [35]. In addition, the PE class of phospholipids contains one of the highest degrees of polyunsaturation in their fatty acyl chains, making this class of phospholipid a prime target in ferroptosis [35]. Indeed, generation of diversified oxidized phosphatidylethanolamines (PEox) has been found typical of ferroptosis but not for other types of regulated cell death [36]. Given these facts, oxidation of PUFA-containing PEs (forming HOO-PUFA-PEs) during ferroptosis will likely generate electrophilic cleavage products capable of forming adducts with proteins. These cleavage products may be generated as electrophilic short leaving fragments without the signatures of their parental phospholipid identity or sn-2 truncated phospholipid remaining groups, retaining the parent headgroup and sn-1 fatty acyl chain [Scheme 1]. The former types of small electrophilic cleavage fragments have been intensively studied and may also be generated during general oxidative stress, making them nonunique to ferroptosis [34,[37], [38], [39]]. With this in mind, we decided to focus on the sn-2 truncated remaining groups of phosphatidylethanolamine which retained the parental headgroup, and their adducts formed with proteins as mechanistic targets of ferroptosis, as these would be exclusively associated with the ferroptotic death process. To this end, our efforts were focused on three major objectives: 1) identifying sn-2-truncated PE oxidation products using a non-enzymatic (SAPE/iron/H2O2 or SAPE/iron/ascorbate) and an enzymatic (SAPE/15LOX) model system; 2) identifying sn-2-truncated PE-peptide adducts by using the above systems with a model peptide containing nucleophilic amino acids; 3) designing a suitable probe for detecting and identifying proteins that have bound PE adducts, in a variety of cell types undergoing ferroptosis. Currently, there is a lack of information regarding the electrophilic cleavage products of PE that retain the PE-phosphatidyl-glycerol moiety with the oxidatively-truncated sn-2 PUFA chain. These truncated species can contain several electrophilic moieties capable of modifying proteins at nucleophilic sites, thereby altering protein function. We reasoned that proteins containing this modification, covalently bound truncated PE-adducts with an exposed PE headgroup, could serve as a unique signature of cells undergoing the ferroptotic death process.
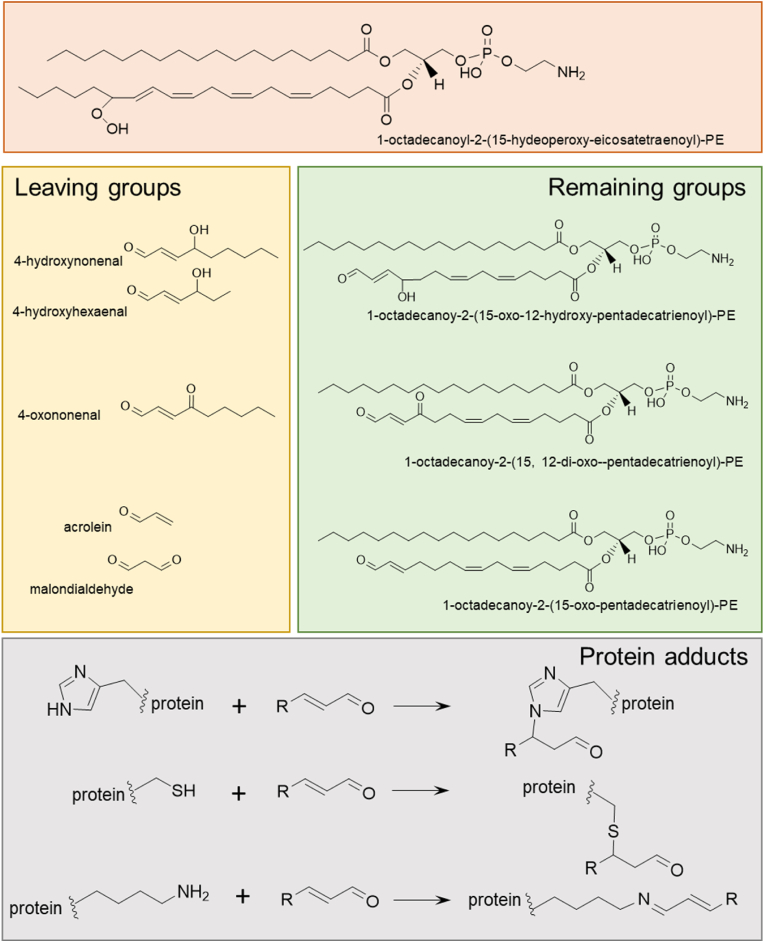
Scheme 1.
Generation of reactive PE-truncated lipids and lipid fragments and their reaction with nucleophilic amino acids. Structure of 1-octadecanoyl-2-(15-hydeoperoxy-5E,8E,11E, 13 E-eicosatetraenoyl)-sn-glycero-3- phosphoethanolamine and possible leaving groups 4-hydroxynonenal, 4-hydroxyhexaenal, 4-oxononenal, acrolein and malondialdehyde. Remaining groups include 1-octadecanoy-2-(15-oxo-5Z,8Z, 13 E-pentadecatrienoyl)-PE, 1-octadecanoy-2-(15, 12-di-oxo-5Z,8Z, 13 E-pentadecatrienoyl)-PE and 1-octadecanoy-2-(15-oxo-12-hydroxy-5Z,8Z, 13 E-pentadecatrienoyl)-PE electrophiles. Protein adducts schema showing the reaction of oxidized-PE derived electrophiles with nucleophilic amino acid side chains of histidine, cysteine and lysine.
Here using redox lipidomics, we have identified a total of 37 oxidatively-truncated PE species (trPEox) combined, which occur during non-enzymatic and enzymatic oxidation of SAPE as determined from our three model systems and our HT-22 cell biological system. Some of the truncated products in both the non-enzymatic and enzymatic systems were capable of forming adducts with a model peptide containing nucleophilic residues and we have identified Cys as the most reactive. In addition, we have identified trPEox species from HT-22 cells which occur during ferroptosis, generating sn-2 truncations from 5 to 9 carbons. Taking advantage of the free PE headgroup, we have developed a new technology, using the lantibiotic duramycin, to detect and identify protein-lipid adducts that retained the remaining truncated PE parent molecule (PE-lipoxidated proteins) in experimental models of ferroptosis [26,27,32]. It should be noted that this radical-mediated lipidation process is entirely different from other types of protein lipidation such as lipidation of LC3 by PE which involves the interaction of the amino function of the PE headgroup with a specific site in the target protein (C-terminal glycine in LC3 [40]). Whereas, under ferroptotic conditions, the ethanolamine-headgroup of the lipoxidated PE-protein adduct remains unmodified, and hence, available for specific detection. Our new protocol, described/applied here, was able to recognize and enrich the PE-lipoxidated proteins, and assess their diversity in several types of cells undergoing ferroptosis. Finally, given that our model systems demonstrated that truncated-PE species containing a C8-hydroxy-oxo-ene truncated sn-2 chain as one of the more common truncations with high reactivity toward Cys residues, we demonstrate that pretreatment of cells with the strong nucleophile, 2-mercaptoethanol, prevented the formation of PE-lipoxidated proteins and completely blocked ferroptotic death. This study serves as a first step towards elucidating PE-protein adducts as downstream targets of the ferroptotic process and indicates that PE-lipoxidated proteins are participants in ferroptotic death, preventable by 2-mercaptoethanol and likely other strong nucleophiles.
2. Materials and Methods
2.1. Reagents
10× Tris buffered saline (TBS) and LCMS solvents (acetonitrile, 2-propanol, hexanes, methanol, water) were purchased from Fisher Scientific (Pittsburgh, PA, USA) and were all Optima™-LCMS grade except for hexanes which were Optima™ HPLC and GC grade. Ammonium formate (MS grade) was purchased from Fisher Scientific (Pittsburgh, PA, USA). Ethanol (Acros Omnisolv HPLC-Mass Spectrometry grade) and chloroform (HPLC grade with 0.75% Ethanol) were also purchased from Fisher. RSL3 and Ferrostatin-1 were purchased from R&D Systems, Inc., and Cayman Chemical (Ann Arbor, MI, USA), respectively. Lipid standards were purchased from Cayman Chemical and BSA digest standards were purchased from Thermo-Fisher Scientific (Pittsburgh, PA USA). SAPE was from Avanti Polar Lipids (Alabaster, AL, USA). 15LOX was provided by Dr. T. Holman (Chemistry and Biochemistry Dept., University of California, Santa Cruz, USA). Urea (proteomics grade) was purchased from VWR Scientific (Philadelphia, PA USA). Iodoacetamide and Duramycin were purchased from Sigma Chemicals (St. Louis, MO USA). TCEP-HCL, HEPES and protease inhibitor cocktail were purchased from Thermo-Fisher Scientific (Pittsburgh, PA USA). Tween-20 (proteomics grade) was purchased from G Biosciences (St. Louis, MO USA). Ethanolamine was purchased from Acros Organics (Fisher Scientific). Streptavidin-HRP and NHS-agarose was purchased from Pierce (Thermo-Fisher Scientific). Streptavidin coated magnetic beads were purchased from GE Healthcare (Burlington, MA USA). Sulfo–NHS–LC-LC-biotin was purchased from Pierce (ThermoFisher Scientific). Membrane lipid arrays were ordered from Echelon Biosciences (Salt Lake City, UT, USA). Fatty acid free BSA was purchased from Sigma. Microcon-10, centrifugal centrifuge units were purchased from Millipore-Sigma. A model peptide (IHYKYMCNS) was a generous gift from Dr. Walter Storkus (Department of Immunology, University of Pittsburgh, USA). Tricine gels were from Invitrogen/ThermoFisher (Waltham, MA USA) and a Pierce Silver Stain kit (Thermo Scientific, Waltham, MA USA) was used for staining. Super Block T20 blocking buffer in TBS for Western blot analysis was purchased from Thermo Fisher Scientific.
2.2. Cell culture
H9c2 cells (American Type Culture Collection (ATCC), part# ATCC CRL-1446) were pelleted and resuspended (2000 cells per ml) in media containing High glucose DMEM with l-Glutamine (ATCC part# 30–2002, lot# 80,306,190) with 10% FBS (Fisher Scientific, Part# Gibco-A31606-01, Lot# 1,992,275) and 1% penicillin streptomycin solution (Penn-Strep) also from Fisher Scientific (Part# Gibco-15140-122, Lot# 1,333,919). The HT-22 cell line was obtained from Dr. David Schubert, The Salk Institute (La Jolla, CA). They were maintained in DMEM media supplemented with 10% FBS and 1% penicillin/streptomycin. The murine lung epithelial cell line MLE-12 purchased from ATCC, was cultured at 37 °C and 5% CO2 in DMEM/F-12 (1:1) medium supplemented with 0.005 mg/ml insulin, 0.01 mg/ml transferrin, 30 nM sodium selenite (Life Technologies, Grand Island, NY), 10 nM hydrocortisone, 10 nM beta-estradiol (Sigma-Aldrich, Saint Louis, MO), 2 mM l-glutamine, 10 mM HEPES, and 10% FBS (Life Technologies, Grand Island, NY). RAW 264.7 macrophages were obtained from the American Type Culture Collection (ATCC). RAW 264.7 macrophages were cultured at 37 °C and 5% CO2 in DMEM or RPMI (ATCC) supplemented with 10% heat-inactivated fetal calf serum (FCS; Sigma-Aldrich) and 50 U ml−1 penicillin–streptomycin (ThermoFisher Scientific). RAW 264.7 macrophages were polarized to the M2 state by incubation in DMEM with 10% FBS, 50 U ml−1 penicillin–streptomycin containing IL-4 (20 ng ml−1) for 48 h, as the polarized state has been shown to be sensitive to ferroptosis [41] For ferroptotic studies, cells were incubated with 250 nM RSL3 for 3 h (M2 macrophages, HT-22 and H9c2 cells) or 18 h (MLE cells) or RSL3 (250 nM)+Ferrostatin-1 (400 nM). For ferroptotic studies of HT-22 cells in the presence of 2-mercaptoethanol, cells were incubated with 0.5 μM RSL3 for 4 h in the presence of 0, 0.5, or 5 mM of 2-mercaptoethanol.
2.3. Generation of an SAPE-truncated product model system in the presence of iron and H2O2
SAPE/iron/H2O2: SAPE (10 μM) was incubated with iron ammonium sulfate (2.5 mM) and H2O2 (5 mM) and allowed to react for 4 h at room temperature in a 5 mM ammonium bicarbonate solution. Products were assessed by C30 LC-MS/MS mass spectrometry. C30 analysis was performed on a Thermo Accucore C30 column (2.1 mm × 25 cm). Solvents consisted of the following: solvent A, acetonitrile/water (50/50, v/v); solvent B, 2-propanol/acetonitrile/water (85/10/5, v/v/v). Both solvents A and B contained 5 mM ammonium formate and 0.1% formic acid. Flow rate was maintained at 100 μl/min. The following gradient was run: 0–20 min, 30–70% B (curve 5); 20–55 min, 70–100% B (curve 5); 55–82 min, hold at 100% B (curve 5); 82–85 min, 100-30% B (curve 5); 85–95 min, hold at 30% B. Mass spectrometry was performed on a Q-Exactive mass spectrometer (Thermo Scientific) in negative ion mode. Capillary voltage, 3500; sheath, auxiliary and sweep gases were set at 6, 0 and 0, respectively; ion transfer tube was set at 320 °C. MS and MS/MS was performed in the orbitrap at 140,000/17,500 resolution, respectively. Scan range was 400–1800 m/z with 128 ms injection time. Data was collected in profile mode.
2.4. Generation of a peptide-truncated-PE adduct model system
SAPE/iron/ascorbate/peptide: SAPE vesicles were prepared by vortexing SAPE for 5 min in 20 mM ammonium bicarbonate buffer followed by a 15 min sonication in an ice bath. A model peptide (IHYKYMCNS, 200 μΜ) was incubated with SAPE vesicles (500 μM) in the presence of iron/ascorbate (100 μM FeSO4/5.0 mM) for 2 h in 20 mM ammonium bicarbonate buffer. LCMS analysis was performed on a Q-Exactive mass spectrometer using positive/negative polarity switching. Resolution was set at 140,000, AGC target 1e6, injection time 128 ms in a scan range of 300–3000 m/z. Data dependent MS/MS was performed at a resolution of 17,500 with a 256 m injection time and an isolation window of 1.0 m/z. Collision energy (HCD) was fixed at 24. Sheath and aux gases were set at 20 and 5, respectively, with a spray voltage of 4.0 kV, a capillary temperature of 320 °C and an S-lens setting of 70. Chromatography was performed on a Phenomenex C8 column (1.0 × 150 mm) using the above C30 solvent system. Flow rate was maintained at 25 μl/min using the following gradient: 0–30 min, 50–100% B; 30–45 min, hold at 100% B; 45–46 min, 50% B; 46–55 min, equilibrate at 50% B. 10–20% SDS-Tricine gels were run in parallel. Gels were fixed in 5% glutaraldehyde and stained with silver according to the manufacturer's instructions.
SAPE/iron/ascorbate/peptide in the presence of 2-mercaptoethanol: A model peptide (IHYKYMCNS, 100 μΜ) was incubated with SAPE vesicles (250 μM) in the presence of iron/ascorbate (50 μM FeSO4/2.5 mM ascorbate) for 30 min in 20 mM ammonium bicarbonate buffer in the absence or presence of 2-mercaptoethanol (5 mM). LCMS analysis was performed as indicated above.
SAPE/LOX/peptide: A model peptide (IHYKYMCNS, 200 μΜ) was incubated with SAPE vesicles (1.0 mM) in the presence of 15LOX (1 μM) for 1.5 h in 20 mM ammonium bicarbonate buffer. LCMS analysis and SDS gel electrophoresis was performed as described above.
2.5. Synthesis and purification of the duramycin probe
Ten mg of Duramycin was dissolved in 1.0 ml of 10 mM HEPES buffer, pH 8.5. To this mixture, 40 mg of sulfo–NHS–LC-LC-biotin was added. The reaction was allowed to proceed with stirring for 2 h at room temperature, followed by an additional 2 h at 4 °C. The mixture was first purified using a C8 reverse-phase column (2.0 mm i. d. X 15 cm, Phenomenex C8, 0.2 ml/min) and scaled up to a larger C8 column (4.6 mm i. d. X 25 cm, Phenomenex C8, 1.0 ml/min) on a Waters HPLC system consisting of two 515 pumps, a 717 plus autosampler and a 2996 PDA detector (Waters, Milford, MA USA). The solvent system consisted of the following: solvent A: 2% acetonitrile containing 0.1% formic acid; solvent B: 100% acetonitrile containing 0.1% formic acid. The gradient was as follows: 10–100% B, 0–30 min (linear); 100-10% B, 30–35 min; hold at 10% B, 35–45 min for equilibration. The effluent was monitored at 215 nm. Three major species were detected which consisted of unreacted duramycin, and duramycin containing one and two LC-LC-biotins. The product containing one-LC-LC-biotin was collected. The product was confirmed by mass spectrometry. Mass spectrometry was performed on a Waters Q-TOF Premier operating in positive ion mode. Spectra were scanned from m/z 400–3000 with the following conditions: scan time = 1 s; cone = 25; extraction cone = 5.0; ion guide = 2.0; desolvation = 400 L/h; resolution = 15; collision cell = 0.45; source = 80 °C and further confirmed by high resolution LCMS on a Q-Exactive mass spectrometer and C8 chromatography using the method described above for the peptide/SAPE model systems.
2.6. Specificity testing of the duramycin-biotin probe
Specificity testing of the duramycin-LC-LC-biotin probe was performed on a membrane lipid array consisting of the following lipids: diacylglycerol (DAG), phosphatidic acid (PA), phosphatidylserine (PS), phosphatidylinositol (PI), phosphatidylethanolamine (PE), phosphatidylcholine (PC), phosphatidylglycerol (PG) and sphingomyelin (SM). All lipids contained saturated (16:0) fatty acyl chains from 100 pmole/spot to 1.56 pmole/spot in doubling dilutions. Duramycin-LC-LC-biotin was incubated overnight with the lipid array in 50 mM Tris-HCl/150 mM NaCl/0.1% Tween 20 containing 2 mg/ml of fatty acid free BSA at 4 °C. After incubation, the lipid array was washed in the above buffer and incubated with Streptavidin-HRP (1:10,000 dilution) at room temperature for 1 h. The arrays were washed again and processed for imaging on an Amersham Imager 600 (GE Healthcare, Chicago, IL).
2.7. Preparation of a duramycin affinity resin
Ten mg of duramycin was dissolved in 10 ml of 0.1 M phosphate buffer containing 0.15 M NaCl, pH 7.5. One hundred fifty mg of dry NHS-activated Sepharose was added to the above solution and the reaction was allowed to proceed overnight at 4 °C with gentle rocking. The resin was then centrifuged at 300×g and the supernatant was removed. The resin was washed 3× with 0.1 M sodium phosphate/0.15 M NaCl buffer. The unreacted sites on the resin were blocked with 1 M Tris, pH 7.4 (10 ml) for 20 min at room temperature. The resin was washed extensively in sodium phosphate buffer to remove the Tris and stored in phosphate buffer containing 0.05% azide.
2.8. Disruption of the Duramycin-PE interaction
Duramycin-sepharose resin: Two types of small unilamellar vesicles (SUVs) were prepared: dioleoyl phosphatidylcholine (DOPC) alone or DOPC/SAPE (stearoyl-arachidonoyl PE, SAPE at a 9:1, mole/mole DOPC:SAPE ratio), respectively. SUVs were prepared by evaporating lipids to dryness followed by sonication in aqueous solution containing 20 mM HEPES, pH 7.4, 100 mM NaCl and 10 mM KCl. Two aliquots of 0.5 ml of swollen duramycin affinity resin were placed in individual affinity columns and washed free of azide with 20 mM HEPES buffer, pH 7.2. Washed resins were then transferred to individual Eppendorf tubes. Three hundred microliter of DOPC or DOPC/SAPE SUVs were added to their respective Eppendorf tubes and placed on a rotating mixer for 1 h at room temperature. Individual resins were then transferred back to their respective columns and washed with 5 ml of 20 mM HEPES buffer pH 7.2 and 1 ml fractions were collected. The DOPC vesicles eluted in the wash and the DOPC/SAPE SUVs were only able to be eluted in the presence of 200 mM ethanolamine solution in HEPES buffer.
Streptavidin-coated magnetic beads: Fifty microliters of a 1 h reaction mixture containing SAPE (1 mM) and Fe/ascorbate, 100 μM/0.5 mM) in 20 mM ammonium bicarbonate buffer was added to 60 μl of the duramycin-biotin probe (0.19 μmole) and the solution was adjusted to 1.0 ml with ammonium bicarbonate buffer. The incubation was allowed to proceed with mixing for 2 h at room temperature. One hundred microliters of washed streptavidin beads were added and the incubation was continued for an additional 2 h with mixing. The beads were loaded onto an empty column and 1.0 ml of the flow-through was collected. The beads were washed with 1.0 ml of ammonium bicarbonate buffer. Finally, 1.0 ml of buffer containing 0.2 M ethanolamine was added and collected. The 1.0 ml fractions were concentrated in a speed vac system and brought up in 100 μl of solvent B and 5 μl was injected for C8 LCMS analysis as described above.
2.9. Cell death assay
Raw 264.7 macrophages, HT-22 cells and MLE-12 cells were plated in a 24-well plate and seeded at a density of 15,000–20,000 cells per well. H9c2 cells were seeded at a density of 8000 cells per well. Adherent cells, in fresh media, were treated with RSL3 (250 nM) for 3 h (Raw 264.7 macrophages, HT-22 cells and H9C2 cells) and for 18 h (MLE cells) in the absence or presence of ferrostatin-1 (400 nM). Cell death was assessed by measuring the released lactate dehydrogenase (LDH) activity. LDH activity was quantified using the CytoTox-ONE™ Cyto-toxicity Detection Kit (Promega, Madison, WI, USA) according to the manufacturer's instructions.
2.10. Western blot analysis using the duramycin-biotin probe
HT-22 cells were induced to undergo ferroptosis using the GPX4 inhibitor, RSL3 (0.5 μΜ) for 4 h in the absence or presence of varying concentrations of 2-mercaptoethanol (0, 0.5 and 5.0 mM) or with RSL3 plus ferrostatin-1 (0.4 μΜ). Proteins were extracted in RIPA buffer and electrophoresed on 4–20% gradient gels. Proteins were transferred to nitrocellulose membranes and the membrane was blocked using SuperBlock (T20) according to the manufacturer's instructions. The membrane was then incubated with the duramycin-biotin probe (5 μg/ml) in the same buffer overnight at 4 °C. Finally, after washing to remove any excess probe, Streptavidin-HRP (1:10,000 dilution) in blocking buffer was added to the membrane and incubated for 1 h at room temperature. The blot was washed again and processed for imaging on an Amersham Imager 600 (GE Healthcare, Chicago, IL).
2.11. Assessment of truncated PE species from HT-22 cells
Lipid extraction of ferroptotic cells was performed as described previously [29,30]. C30 analysis was performed on a Thermo Accucore C30 column (2.1 mm × 25 cm). Solvents consisted of the following: solvent A, acetonitrile/water (50/50, v/v); solvent B, 2-propanol/acetonitrile/water (85/10/5, v/v/v). Both solvents A and B contained 5 mM ammonium formate and 0.1% formic acid. Flow rate was maintained at 100 μl/min. The following gradient was run: 0–20 min, 30–70% B (curve 5); 20–55 min, 70–100% B (curve 5); 55–82 min, hold at 100% B (curve 5); 82–85 min, 100-30% B (curve 5); 85–95 min, hold at 30% B. Mass spectrometry was performed on a Fusion Lumos mass spectrometer (Thermo Scientific) in negative ion mode. Capillary voltage, 3500; sheath, auxiliary and sweep gases were set at 35, 17 and 0, respectively; ion transfer tube was set at 300 °C. MS and MS/MS was performed in the orbitrap at 120,000/35,000 resolution, respectively. Scan range was 400–1200 m/z with 100 ms injection time. Data was collected in profile mode with a targeted mass list for PEox and truncated PE species.
Quantitative differences in ions detected from HT-22 lipid extracts were assessed by Compound Discoverer (Thermo Fisher, version 2.0) and graphed using Prism (GraphPad Software LLC, version 9.2.0) as previously described [30]. In brief, the LCMS data files in RAW format (Thermo Fisher, Xcalibur version 4.1.0.0) were loaded onto a custom workflow in Compound Discoverer. Truncated PE species were identified using Compound Discoverer v.2.0 software (ThermoFisher Scientific, San Jose, CA) with a custom workflow as described earlier [33]. We generated a database of oxidized and truncated SAPE species based on molecular structure and identified LCMS peaks with S/N > 3 and intensity >5000 that matched to < 7 ppm mass error. The database of masses of potential structures for PEox and truncated PE species was utilized for identification with the “Search Mass List” function. Peak areas were determined from extracted ion chromatograms of the LCMS data files by Compound Discoverer. Output data was then exported to an Excel spreadsheet. Select species that matched the exact mass of a truncated PE species to within 1 ppm were chosen, filtered by retention time, and their structures were verified from the MS/MS data.
2.12. Affinity isolation of PE-protein adducts using the duramycin affinity probe
Cells were harvested and cell pellets were washed three times with PBS. The cells were then resuspended in 150–200 μl of TBS with protease inhibitors followed by an equal volume of 2× lysis buffer (280 mM SDS, 115 mM TCEP-HCl in PBS). The lysate was sonicated on ice with a probe tip sonicator for 15 s at 25% power, followed by a 1-min rest period. This was repeated two additional times. Eight volumes of urea buffer (8.5 M urea, 20 mM TCEP in PBS) were added and the lysate was incubated at room temperature for 30 min with shaking. Five hundred μL of the lysate were added to a 10,000 MWCO filtration device (Millipore-Sigma) and centrifuged at 14,000×g for 30 min. The filtrate was discarded and this was repeated several times until the remaining lysate was finished. Five hundred microliters of urea buffer were then added to the retentate and centrifuged at 14,000×g for 30 min. This was repeated once. Five hundred microliters of iodoacetamide (50 mM in urea buffer) were then added to the retentate and the centrifuge units were incubated with shaking in the dark for 30 min at room temperature. The solution was then centrifuged at 14,000×g. The filtrate was discarded and the retentate was washed with 500 μl TBS containing 1.0% Tween 20 and centrifuged as above for 30 min. This step was repeated 3 times. The volume of the retentate was adjusted to 500 μl and the duramycin-biotin probe was added at a final concentration of 10 μg/ml. The solution was incubated overnight at 4 °C with shaking. After incubation, the solution was added to Sera-Mag beads (100 μl of the Sera-Mag bead solution was previously pre-washed with 500 μl of TBS-1.0% Tween 20 three times) and incubated for 2 h at room temperature with shaking. The supernatant was then removed, and the beads were washed four times with 500 μl of 20 mM ammonium bicarbonate buffer. The beads were then incubated in ammonium bicarbonate buffer containing 0.2 M ethanolamine at room temperature for 1 h with shaking. The supernatant was collected, and the incubation was repeated. Five hundred microliters of the combined eluates were added to microcentrifugation units (10,000 MWCO) and centrifuged at 14,000×g for 30 min. The remaining portion of the eluate was treated in the same way. The retentate was then washed four times with 20 mM ammonium bicarbonate buffer. The retentate volume was reduced to 50 μl and proteomics grade trypsin was added to the microcentrifuge units and the solution was incubated overnight with shaking at 37 °C. The solution was then centrifuged into a fresh Eppendorf tube at 14,000×g for 10 min. One hundred microliters of water were added to the centrifuge unit and centrifuged until the filter was completely dry. The resulting eluate was dried in a speed vac and the samples were resuspended in 30 μl water containing 0.1% formic acid. 2.5 μl was injected for LC-MS/MS analysis.
2.13. LC-MS/MS proteomic analysis
Peptides were separated on a Pep Map C18 column (Thermo Scientific, 300-μm x 15 cm containing 2-μm particles) at a flow rate of 12 μl/min on a Thermo Ultimate 3000 HPLC system (Thermo Scientific). The column was maintained at 35 °C. The solvent system consisted of the following: solvent A, 2% acetonitrile containing 0.1% formic acid; solvent B, 100% acetonitrile containing 0.1% formic acid. The gradient was as follows: 1% B to 40% B in 110 min, 110–115 min 40–85% B, 115–130 min 85% B, 130–135 min then returned to 1% B. All gradients were linear. Mass spectrometry was performed on a Fusion Lumos Tribrid mass spectrometer (Thermo Scientific) in positive ion mode. Conditions were as follows: spray voltage, 3.2 kV; sheath gas, aux gas and sweep gas were 25, 3 and 1 arbitrary units, respectively; ion transfer tube = 300 deg. C; resolution, 120,000; scan range 400–1500 m/z; Rf lens 30%; charge state 2–7 selected with a dynamic exclusion of 60 s; an isolation window of 1.6 m/z was selected for MS/MS analysis in the ion trap using CID at 35 with a rapid scan rate. Protein identification and sequence analysis was performed using Proteome Discoverer Software (Thermo Scientific) using the SEQUEST-HT engine which included the subcellular locations of the identified proteins. Two missed cleavages were allowed. Analyses were performed in triplicate and PE-lipoxidated proteins involved in ferroptosis were identified by duramycin affinity capture based on their ability to increase significantly after RSL3-treatment and decrease significantly in ferrostatin-1 treatment in the various cell types used.
2.14. Docking simulations
ETE-PE, a phospholipid with stearic acid (SA) and arachidonic acid (AA or ETE) at the respective sn-1 and sn-2 chains (also called SAPE) and its oxidized and truncated forms at carbon C5 (TrPE_C5) and at C9 (TrPE_C9) of the sn-2 chain, were docked onto ten structurally-resolved [[42], [43], [44], [45], [46], [47]] or homology-modeled proteins (see Supplementary Table 2) randomly selected from amongst those identified by duramycin affinity capture. Ligands were first prepared, and energy minimized using Maestro [48]. Five runs were performed using SMINA [49] for each combination of protein-lipid complex, with each run yielding 10 highly scoring poses for binding ETE-PE, TrPE_C5 or TrPE_C9 onto each of the selected proteins. Overall, this series of docking simulations yielded 50 phospholipid-bound protein conformations for each of the 30 combinations (of ten proteins and three phospholipids), which we analyzed using our in-house scripts in ProDy API [50,51].
2.15. Statistical analysis
All statistics that are presented as means±SD, were performed through Excel software. SEM and a two-tailed t-test comparing two groups of equal variance was used for comparison of truncated species between control and treated groups. An unpaired t-test was used for cell death analysis of as assessed by LDH release or PI uptake.
3. Results
3.1. Identification of oxidatively-truncated PE species in non-enzymatic and enzymatic model systems
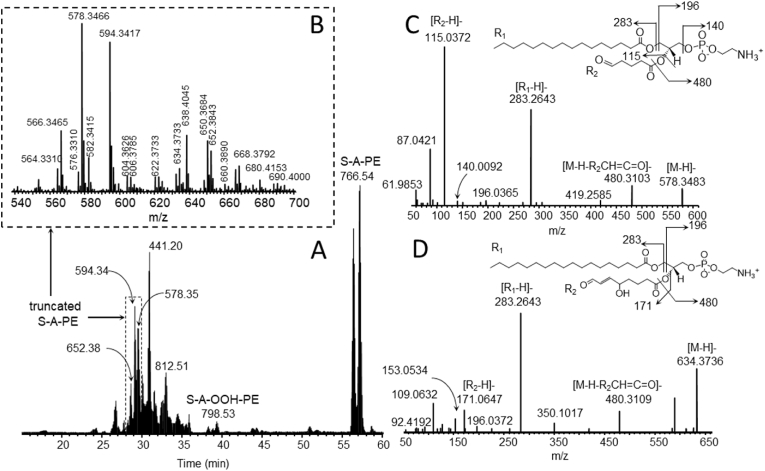
In order to identify the oxidatively truncated phospholipid species, we constructed a database of potential oxidatively-truncated PE species and developed a C30 reverse-phase LC-MS/MS protocol as described in the Methods to detect these species. To test our database, consisting of over 300 potential species, we generated truncated products of SAPE by incubating SAPE with iron ammonium sulfate and H2O2. The resulting LCMS analysis indicated that numerous reactive truncated electrophilic products were formed in the model system as identified with Compound Discoverer [Fig. 1A and B]. Out of sixty unique precursor species detected as potential truncated species, 22 species were confirmed by MS/MS analysis. The length of the truncated chains ranged from 4 to 11 carbons and contained 1–3 oxygens [See Supplementary Table 1]. MS/MS analysis of two truncated products, a C5 truncated species (PE(18:0/C5-oxo-anoyl), m/z 578.3466) and a C8 truncated species (PE(18:0/C8-hydroxy-oxo-ene) m/z 634.3736) are shown in Fig. 1C and D, respectively. The former species is one of the shortest and most abundant truncated species, while the latter is one of the longer and lower abundance truncated species detected.
Fig. 1.
Generation of SAPE-truncated species in an iron/H2O2/SAPE model system. SAPE was incubated with iron ammonium sulfate and H2O2 and allowed to react for 4 h at room temperature as described in Materials and Methods. SAPE and its truncated products were assessed by C30-ESI-MS/MS analysis. 1A, TIC C30 chromatogram displaying the truncated SAPE region. 1B, m/z 540–700 expanded range indicating the numerous truncated species. 1C, MS/MS analysis of a C5 truncated species (m/z 578.3466). 1D, MS/MS analysis of a C8 truncated species (m/z 634.3736).
3.2. Identification of peptide-PE adducts with an SAPE/iron/ascorbate system
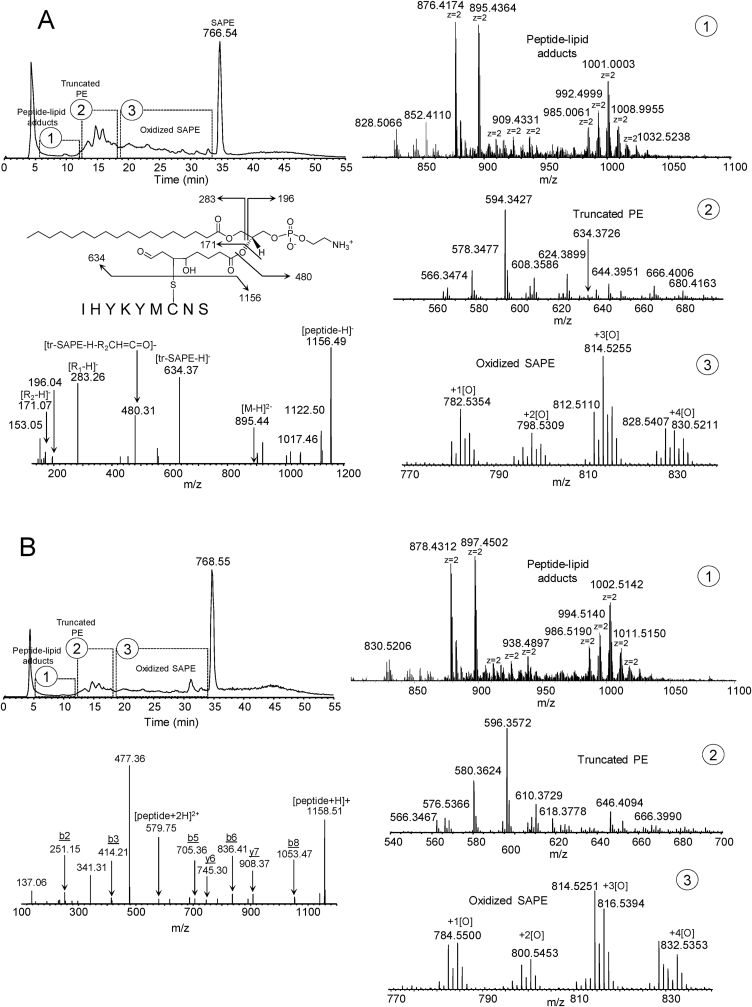
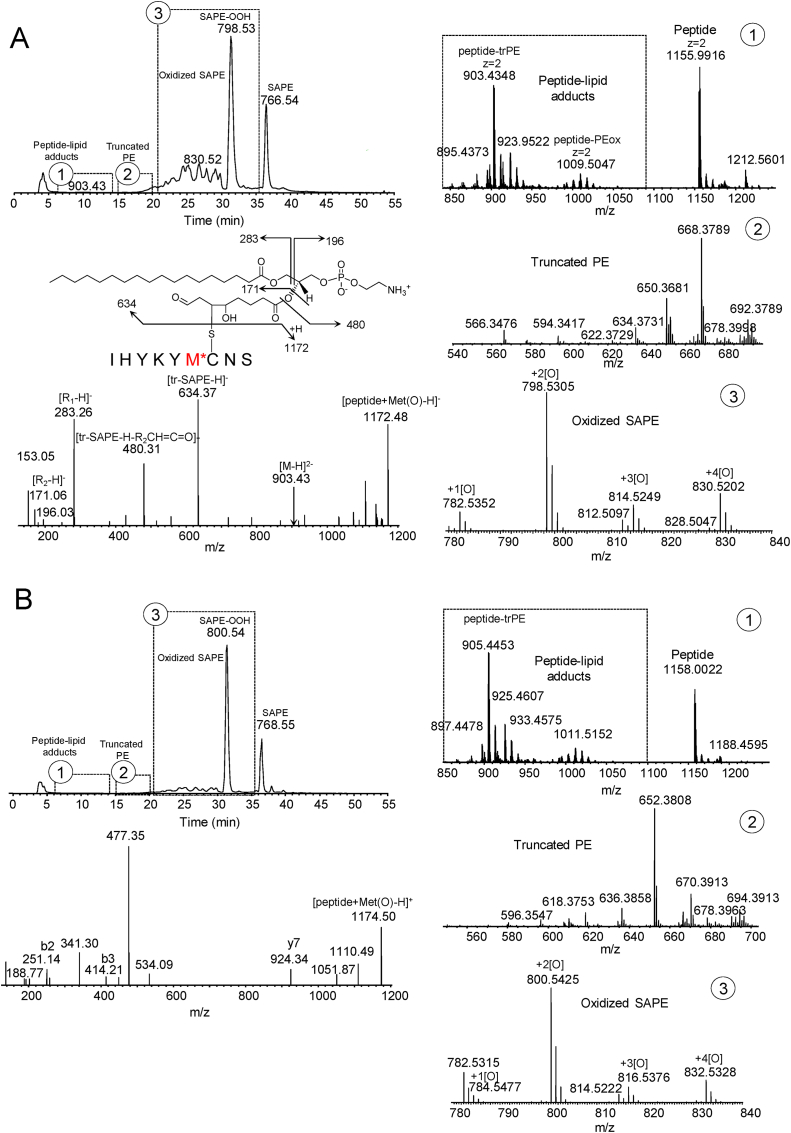
We next sought to determine the types of adducts that are formed in a model system containing the peptide with sequence IHYKYMCNS, with SAPE and iron/ascorbate [Fig. 2]. The peptide was reacted with SAPE/iron/ascorbate for 30 min at 37 °C as described in Materials and Methods and the products separated and assessed by C8-LC-ESI-MS analysis [Fig. 2A]. Three regions on the TIC chromatogram were noted: region 1 containing the peptide-lipid adducts, region 2 containing the truncated PE species, and region 3, containing the oxidized SAPE species. Multiple doubly-charged PE-peptide adduct species (z = 2) were formed which consisted of two types: adduct species with the model peptide covalently bound to truncated-PE species (m/z range ∼850–950) and adduct species with the model peptide covalently bound to intact oxidized PE species in various oxidation states (m/z range ∼ 980–1050) [Fig. 2A1]. Expanded m/z regions for the truncated PE species [Fig. 2A2] and oxidized SAPE species [Fig. 2A3] are shown. A total of 6 adduct species were confirmed by MS/MS [Supplementary Table 2A and associated excel file]. The truncated species at m/z 634.3726 (PE(26:2) +2 oxygens) appeared to be the most common/reactive among the identified adducts. Fragmentation in negative mode of the doubly-charged m/z 895.43 species yielded the intact peptide at m/z 1156.49, the truncated PE species at m/z 634.37, the lyso-PE species at m/z 480.31, the 18:0 fatty acid species at m/z 283.26 as well as the diagnostic truncated fragment at m/z 171.07. The same TIC chromatogram pattern was seen in positive mode [Fig. 2B], consisting of an adduct, a truncated and an oxidized SAPE region. The same types of doubly-charged adduct species are seen in positive mode, running at 2 mass units higher [Fig. 2B1]. The expanded mass regions for the truncated PE and oxidized PE species are shown in Fig. 2B2 and 2B3, respectively. Fragmentation of the corresponding doubly-charged 897.45 species in positive mode yielded the intact peptide, several b and y peptide ions including a m/z 477.36 fragment [Fig. 2B]. The latter fragment corresponds to positive mode fragmentation of the truncated species that loses the PE headgroup as a neutral (m/z 141) forming a diacylglycerol species with an additional loss of water most likely from the OH group from the truncated fatty acyl chain [52]. The fragment at m/z 341.31 represents the sn-1 chain which loses the head group as a neutral with a loss of the truncated fatty acyl chain at the carbonyl-oxygen bond [52]. Given the reactivity of Cys residues over His and Lys residues, the labile nature of Michael adducts with Cys residues [53], and the fact that intense ions of unmodified peptide are generated in positive mode from either truncated or intact PE adducts, it is suggested that the adduct formation is preferentially with the Cys residue of the peptide as shown. Fragmentation of the peptide alone (positive mode) is shown in Supplementary Fig. 1 and indicates that mostly b-ion series fragments are formed. The exact nature of the linkage of the covalently bound intact-PEox-peptide adducts could not be determined at this time (see below concerning the SAPE/15LOX model system).
Fig. 2.
Identification of peptide-PE adducts in a SAPE/iron/ascorbate model system. Fig. 2A, negative mode C8-LC-ESI-MS chromatogram of peptide-PE adducts displaying the adduct, truncated and oxidized SAPE regions. 2A1-3, expanded m/z regions for the adduct, truncated and oxidized-SAPE regions, respectively. Oxidized SAPE species in negative mode includes m/z 782.5363 + 1[O], m/z 798.5318 + 2[O], m/z 814.5261 + 3[O], and m/z 830.5218 + 4[O]. Negative mode MS/MS analysis of the truncated peptide-PE adduct, m/z 895.44, is shown. 2 B, positive mode C8-LC-ESI-MS chromatogram of peptide-PE adducts displaying the adduct, truncated and oxidized SAPE regions. 2B1-3, expanded m/z regions for the adduct, truncated and oxidized-PE regions, respectively. Oxidized SAPE species in positive mode includes m/z 784.5500 + 1[O], m/z 800.5453 + 2[O], m/z 816.5394 + 3[O], and m/z 832.5354 + 4[O]. Positive mode MS/MS analysis of the corresponding truncated peptide-SAPE adduct, m/z 897.45, is shown.
The truncated-PE-peptide adducts as well as the intact oxidized PE-peptide adducts had an early to intermediate retention time window (approximately 6–12 min) when compared to peptide alone (4.5 min), lyso-PE species and truncated PE species (approximately 13–18 min), oxidized PE species (approximately 19–33 min) and SAPE (35 min, Fig. 2A and B). The peak shape and retention time was extremely broad for the truncated or intact oxidized PE-peptide adducts, owing to their dual hydrophobic/hydrophilic nature and may also relate to the variety of different types of adducts that are formed. This was further confirmed by low molecular weight gel analysis using a 2 h reaction for the peptide/iron/ascorbate/SAPE system (Supplementary Fig. 2) which displayed the adducted peptide forming a higher molecular weight diffuse broad band.
3.3. Identification of peptide-PE adducts with an SAPE/15LOX model system
We next sought to determine the type of adducts formed in an SAPE/15LOX model system. Similar to the iron/ascorbate system, three regions on the TIC chromatogram were noted as determined by C8-LC-ESI/MS analysis [Fig. 3A]: region 1 containing the peptide-lipid adducts, region 2 containing the truncated PE species, and region 3, containing the oxidized SAPE species. Again, two types of adduct species were generated: those with a covalently bound truncated-PE species (m/z range ∼ 850–950) as well as an intact oxidized-PE adduct species (m/z range ∼ 980–1040), all as doubly-charged ions [Fig. 3A1]. Expanded m/z regions for truncated PE products as well as oxidized PE species are displayed in Fig. 3A2 and 3A3, respectively. A total of 6 adduct species were confirmed by MS/MS analysis [Supplementary Table 2B and associated excel file]. In addition, a peptide dimer (m/z 1155.9916, [M − 2H]2-) was also formed [Fig. 3A1]. Fragmentation of the m/z 903.43 species (negative mode) yielded the same truncated PE adduct (PE(26:2) +2 oxygens) as seen in the iron/ascorbate system with an additional oxygen associated with the peptide itself, most likely a Met oxidation (m/z 1172.48). The same TIC chromatogram pattern was seen in positive mode [Fig. 3B], consisting of an adduct, a truncated and an oxidized SAPE region. The same types of doubly-charged adduct species are seen in positive mode, running at 2 mass units higher [Fig. 3B1]. The expanded mass regions for the truncated PE and oxidized PE species are shown in Fig. 3B2 and 3B3, respectively. Positive mode fragmentation of m/z 905.45 yielded similar fragments as seen with the iron/ascorbate system except for the parent peptide running at a m/z of 1174.50 due to the Met-oxidation (Fig. 3B) as well as the characteristic m/z ions at 341.30 and 477.35 for the positive mode fragmentation of the truncated PE species as noted above. The dual hydrophobic/hydrophilic and potential variable character of the adducts were evidenced by broad peak shapes, early to intermediate retention times as well as diffuse bands on low molecular weight gels [Fig. 3A and B and Supplementary Fig. 3]. Multimers of 15LOX itself were also noted in the LOX/SAPE system [Supplementary Fig. 3]. As with the iron/ascorbate/SAPE system, the SAPE/LOX system also yielded intact PEox-peptide adducts (m/z range 980–1040), in which the exact nature of the linkage of the covalently bound intact-PEox-peptide adducts could not be determined. Fragmentation of these species in positive mode yielded the intact peptide species with some additional b ions (not shown). In negative mode, fragmentation resulted in ions of the intact PE-oxidized species as well as the intact peptide species with very little additional structural information (not shown).
Fig. 3.
Identification of peptide-PE adducts in an SAPE/LOX model system. Fig. 3A, negative mode C8-LC-ESI-MS chromatogram of peptide-PE adducts displaying the adduct, truncated and oxidized SAPE regions. 3A1-3, expanded m/z regions for the adduct, truncated and oxidized-SAPE regions, respectively. Oxidized SAPE species in negative mode includes m/z 782.5351 + 1[O], m/z 798.5305 + 2[O], m/z 814.5249 + 3[O], and m/z 830.5202 + 4[O]. Negative mode MS/MS analysis of the truncated peptide-PE adduct, m/z 903.43, is shown. 3 B, positive mode C8-LC-ESI-MS chromatogram of peptide-PE adducts displaying the adduct, truncated and oxidized SAPE regions. 3B1-3, expanded m/z regions for the adduct, truncated and oxidized-SAPE regions, respectively. Oxidized SAPE species in positive mode includes m/z 784.5315 + 1[O], m/z 800.5424 + 2[O], m/z 816.5376 + 3[O], and m/z 832.5328 + 4[O]. Positive mode MS/MS analysis of the corresponding truncated peptide-PE adduct, m/z 905.45, is shown.
3.4. Identification of oxidatively-truncated PE species in ferroptotic HT-22 cells
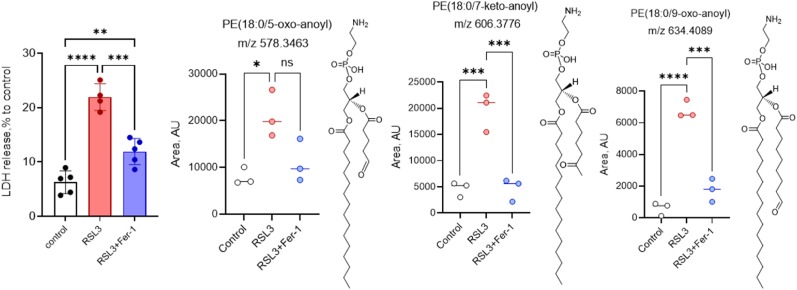
Using these tools and the above information, we identified several truncated PE lipid species in HT-22 cells that underwent ferroptosis triggered by RSL3, an inhibitor of glutathione peroxidase 4 (GPX4) and was sensitive to ferrostatin treatment [Fig. 4]. Three “free” (unbound to proteins) species, (out of a total of 63 truncated species identified by our in-house database) were shown to increase significantly in the presence of RSL3 namely ions of m/z 578.3463, p = 0.013; m/z 606.3776, p = 0.003; and m/z 634.4089, p = 0.001. Their structures were confirmed by exact mass as well as MS/MS analysis which indicated that the sn-2 truncation ranged from 5 carbons to 9 carbons for these particular species [Fig. 4, Supplementary Fig. 4]. The remaining truncated species identified had truncations ranging from 4 to 17 carbons (not shown). Given that truncated PE species can be formed during ferroptosis, we rationalized that these truncated remaining groups could form adducts with proteins. With this in mind, we sought to develop a method to identify these PE-protein adducts.
Fig. 4.
Truncated PE species identified in HT-22 ferroptotic cells. HT-22 cells were induced to undergo ferroptosis in the presence of RSL3 (250 nm, 3 h) or RSL3+Ferrostatin (0.4 μM). Cell death was determined by LDH release (first panel). Cells were harvested and processed for lipid extraction. LCMS analysis was performed using C30 reverse-phase chromatography as described in Material and Mehtods. Quantitative differences in relative areas of individual truncated species as determined by Compound Discoverer were obtained for PE(18:0/C5-oxo-anoyl, m/z 578.3463), PE(18:0/C7-keto-anoyl, m/z 606.3776), and PE(18:0/C9-oxo-anoyl, m/z 634.4089). White = control; red = RLS3; blue = RSL3+Ferrostatin-1. *p < 0.05; **p < 0.01; ***p < 0.005; ****p < 0.001. (For interpretation of the references to color in this figure legend the reader is referred to the web version of this article).
3.5. Lantibiotics as affinity probes to identify PE-lipoxidated proteins
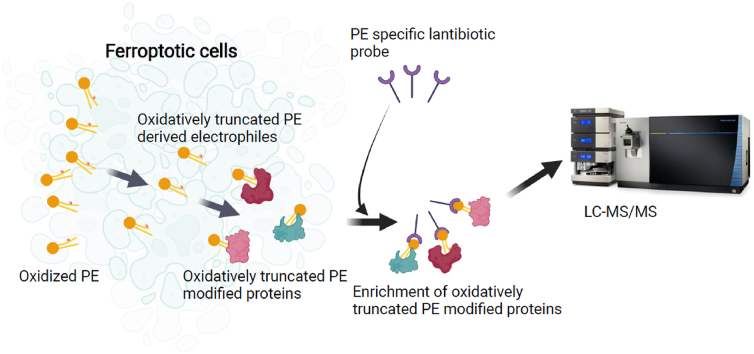
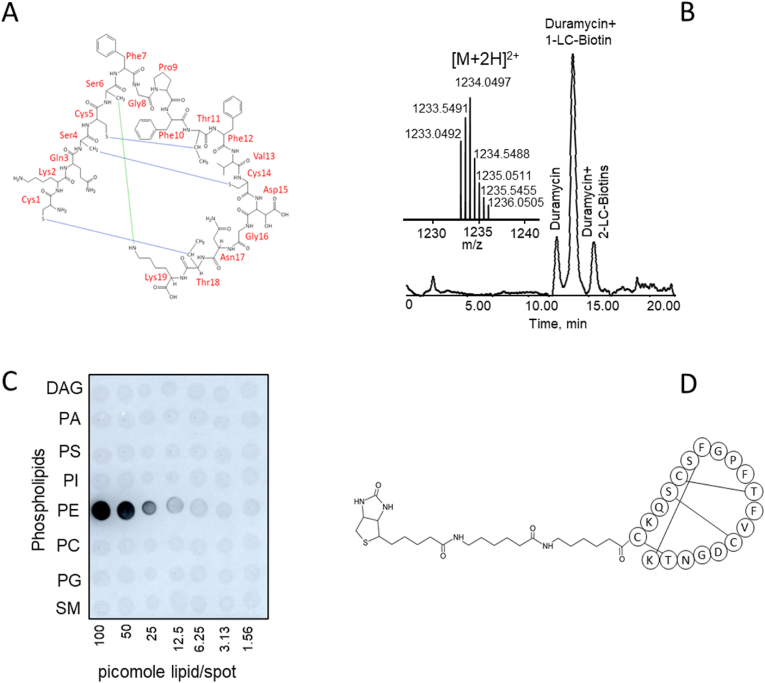
Oxidation of lipids can occur via enzymatic or non-enzymatic means. The primary oxidation products, hydroperoxy-phospholipids, can lead to truncation of the parent lipid, generating reactive aldehydes that either reside with the short leaving fragments, such as HNE, HHE etc., or reactive aldehydes that remain with the parent truncated lipid (see Scheme 1). The fact that PEs are specifically targeted for oxidation during ferroptosis and that truncated PE products have been identified strongly suggests that oxidatively-truncated PE-modified proteins are also direct consequences of ferroptosis. These are different from the alternative short reactive electrophilic lipid fragments which can be generated from any class of polyunsaturated phospholipids. In addition, non-enzymatic decomposition of hydroperoxy-phospholipids is not dependent on the structure of the polar headgroup. This prompted us to develop a new technology that takes advantage of the lantibiotic, duramycin, as an innovative specific affinity probe for the enrichment, isolation, and identification of truncated PE-protein adducts generated during ferroptosis. Proteins that are modified by a truncated reactive PE via one of its reactive acyl chains, will generate a PE-protein adduct possessing a free PE headgroup. Duramycin is a 19 amino acid type B lantibiotic produced by Streptoverticillium cinnamoneus, which binds PE in a 1:1 ratio [54,55]. It has a specific 3-dimensional structure due to three lanthionine bonds providing extreme stability [Fig. 5A]. Duramycin specifically recognizes the headgroup of PE with high affinity (low nM range) as compared to other phospholipids [54,55]. Its residues 5 through 14 are responsible for the PE headgroup recognition [54,55]. The free N-terminal cysteine and the free epsilon amino group of the second lysine in duramycin provide convenient locations for site-directed conjugations. We used redox lipidomics and redox proteo-lipidomics developed in our lab to identify the truncated PE-protein adducts.
Fig. 5.
Duramycin as an affinity probe to identify PE-lipoxidated proteins. 5A: duramycin structure. 5B: Purification of the duramycin-biotin conjugate by C8 chromatography and MS analysis of the mono-conjugated product. 5C: Specificity testing of the duramycin-biotin probe using a lipid array. 5D: Simplified structure of the duramycin-biotin conjugate.
Conjugation of the lantibiotic duramycin with a long chain (LC-LC) biotin containing a 30.5 Å spacer arm via a N-hydroxysuccinimide (NHS) active ester yielded 3 products: unreacted duramycin, and duramycin containing mono- and di-conjugated long-chain biotins. These were separated by C8 reverse phase chromatography [Fig. 5B]. The mono-conjugated duramycin was isolated and purified for further use. Its doubly-charged ion was confirmed by high resolution LC-MS analysis [Fig. 5B] The result indicated that the measured mass (average, 1234.0497 as an [M+2H]2+ ion) was within 4.05 ppm of the theoretical mass (1234.0547 as an [M+2H]2+ ion) for the mono-conjugate. Specificity testing of the probe was performed utilizing a lipid array consisting of DAG, PA, PS, PI, PE, PC, PG and SM [Fig. 5C]. Detection was assessed by enhanced chemiluminescence using a Streptavidin-HRP conjugated second step to the conjugated duramycin [Fig. 5C and D]. The duramycin-LC-biotin reacted only with PE, indicating that the high degree of specificity was retained after modification, where signals as low as 3–6 pmol/spot could be detected.
We next sought to determine if ethanolamine would be a suitable compound for displacing PE from the PE-duramycin interaction. To this end, duramycin agarose affinity columns were prepared using an NHS-activated resin. Two types of small unilamellar vesicles (SUVs) were prepared: DOPC only and DOPC/SAPE. SUVs containing DOPC only or DOPC/SAPE (2 mg of total lipid) were incubated with individual columns of the duramycin affinity resin with a capacity for twice that of the lipid load for 1 h at room temperature. The columns were washed in 20 mM HEPES containing 0.1 M NaCl/10 mM KCl and 1.0 ml fractions were collected and monitored by ESI-MS in positive mode in a m/z range from 400 to 1800 for DOPC (m/z 786, [M+H]+). The PC only SUVs were unable to bind and eluted in the loading step [Supplementary Fig. 5]. The PC/PE containing SUVs were bound to the resin and were not detected in the load or wash fractions and failed to elute until 0.2 M ethanolamine was added. This indicated that ethanolamine was a suitable eluting agent for the duramycin-PE affinity applications. We further tested this elution strategy with the duramycin-biotin conjugate [Fig. 5D]. To this end, 50 μl of a 1.5 h reaction mixture containing SAPE (1 mM) + Fe/Ascorbate in 20 mM ammonium bicarbonate buffer was added to 60 μl (0.19 μmole) of the lantibiotic probe and the solution was adjusted to 1.0 ml with ammonium bicarbonate buffer. The incubation was allowed to proceed with mixing for 2 h at room temperature. 100 μl of washed Streptavidin magnetic beads was then added and incubated with mixing for an additional 2 h at room temperature. The beads were loaded onto an empty column and 1.0 ml of the load flow-through was collected [Supplementary Fig. 6A]. The beads were washed with an additional 1.0 ml of bicarbonate buffer [Supplementary Fig. 6B]. The bound material was eluted with 1.0 ml of bicarbonate buffer containing 0.2 M ethanolamine (Supplementary Fig. 6C). Assessment was by C8 LC-ESI-MS analysis. The results indicated that the duramycin-biotin conjugate was able to capture intact SAPE and its oxidized and truncated products, including lyso-PE. All of the aforementioned PE-headgroup containing species were eluted in the presence of 0.2 M ethanolamine [Supplementary Fig. 6C].
Assessment of oxidatively-truncated PE-lipoxidated proteins in ferroptotic cells using a duramycin affinity probe.
We then used the lantibiotic probe to identify the truncated PE-lipoxidated proteins in a variety of cell types undergoing ferroptosis, using the ferroptosis inducer, RSL3. Upon treatment with RSL3, HT-22 cells, MLE cells, M2 macrophages and H9c2 cells [Fig. 4 and Supplementary Fig. 7] underwent ferroptosis that could be rescued by the addition of Ferrostatin-1. Cell death was verified by LDH release. We identified a total of 48 truncated PE-lipoxidated proteins that increased in HT-22 cells upon RSL3 treatment and decreased upon treatment with Ferrostatin-1 [Supplementary Table 3A]. Likewise, the number of proteins that we identified which increased in the presence of RSL3 and decreased with Ferrostatin-1 treatment were as follows: 50 PE-lipoxidated proteins in MLE cells, 35 PE-lipoxidated proteins in H9c2 cells and 20 PE-lipoxidated proteins in M2 macrophages [Supplementary Tables 3A and 3B].
The subcellular components/locations of the identified PE-lipoxidated proteins in ferroptotic HT-22 cells, as reported by the Proteome Discoverer software, indicated that these proteins generally fell into four subcellular categories: nuclear proteins, cytoskeletal proteins, mitochondrial proteins, and plasma membrane proteins [see Supplementary Tables 3A and B and excel spreadsheets for each cell type]. The majority of the HT-22 proteins were localized in the nucleus (∼54%), followed by cytoskeletal and mitochondrial proteins (∼15 and 17%, respectively), and finally the plasma membrane (∼8%). Likewise, most PE-lipoxidated proteins in M2 macrophages were of nuclear origin (∼50%) followed by cytoskeletal and plasma membrane proteins (both ∼20%) and mitochondrial proteins (∼5%). In contrast, H9c2 cells displayed their highest proportion of PE-lipoxidated proteins during ferroptosis (46%) in the cytoskeletal compartment, followed by the nuclear compartment (∼20%), plasma membrane (∼11%) and mitochondria (∼6%). MLE cells displayed their highest percentage of PE-lipoxidated proteins in the nucleus (∼32%) followed by the plasma membrane (∼18%), cytoskeleton (∼12%) and mitochondria (∼8%). Proteins from other subcellular compartments were also identified. Interestingly, among the PE-lipoxidated proteins identified in HT-22 cells was the sterol carrier protein-2 (SCP-2), a non-specific lipid binding protein which is involved in the transfer of oxidized phospholipids including oxidized PE [56,57]. An earlier study had implicated SCP-2 in ferroptosis [31]. This study indicated that inhibitors of SCP-2 induced anti-ferroptotic activity, but the effect was transient and only served to slow down the ferroptotic kinetics [31].
Our data reported in this manuscript strongly suggests that the lipidation of proteins during ferroptosis is a non-specific event that depends upon the protein and the PE levels in the cells. The degree of PE-lipoxidation may vary with cell type, choice of ferroptotic stimulus, and its time of incubation. In this study, we used four different cell types from two different mammalian species. These cells displayed differential sensitivity to the ferroptotic stimulus used in this study (RSL3) as shown in Supplementary Fig. 7. Therefore, we do not expect that the same proteins will be captured in these cell lines. One protein, elongation factor 1-alpha 1, was identified in all three mouse cell lines (MLE, HT-22, and M2 macrophages). Three proteins, ubiquitin-60 S ribosomal protein L40, heat shock cognate 71 kDa protein, and prelamin-A/C, were commonly captured in MLE and M2 macrophages. Similarly, cofilin-1 and peptidyl-prolyl cis-trans isomerase A were the common captures in HT-22 and M2 macrophages. Two proteins, histone H4 and myosin, were common between M2 macrophages and H9C2 [see Supplementary Fig. 8]. In HT-22 cells, over 95% of the proteins were identified in four or more RSL3 and RSL3+Fer 1 experimental replicate samples (total 6 samples), over 85% of all identified proteins were identified in five of six experimental samples, and over 70% of the proteins were identified in all six experimental samples. These data clearly demonstrate the reproducibility of the lantibiotic probe. Thus, our results indicate that multiple PE-lipoxidated proteins are formed during the course of ferroptosis. The degree of PE-lipoxidation of proteins may also depend on the choice of ferroptotic stimulator and its concentration and time of incubation. The cell type chosen also adds another degree of variability.
Modeling/docking studies of oxidatively-truncated PE species to the PE-lipoxidated proteins determined from the lantibiotic screen.
We have identified a number of oxidatively-truncated PE species that are formed during ferroptosis. In conjunction with our lantibiotic probe, we have identified several dozens of PE-lipoxidated proteins in different cell types undergoing ferroptosis [Supplementary Tables 3A and B]. We sought to determine if there were any differences in binding energies among oxidatively-truncated PE species as compared to the parent SAPE to the various proteins identified in the lantibiotic screen. For these studies, ten proteins for which either X-ray structures or homology models were available were randomly selected from the pool of PE-lipoxidated proteins identified in the lantibiotic screening [Supplementary Table 4]. The binding characteristics of two of our identified truncated PE species, PE(18:0/C5-oxo-anoyl) and PE(18:0/C9-oxo-anoyl), onto those proteins were investigated by docking simulations. The selection of proteins was representative of both the different types of cell lines and the observed subcellular localizations.
Our Scheme 1 indicates that oxidatively-truncated PE-protein adducts can potentially be formed with nucleophilic amino acids such as lysine (Lys), histidine (His) and cysteine (Cys) reacting with the electrophilic terminal oxygen of the truncated PE species. Interestingly, in our list of randomly selected proteins from our screen, lysine residues far outnumbered the combined histidines and cysteines [Supplementary Table 4]. The average binding energies per protein/truncated-PE species from the docking simulations [Supplementary Table 5] as well as the lowest binding energies (most favorable conformation) obtained for each protein/truncated-PE complex [Supplementary Table 6], indicated that the truncated-PE species have binding energies at least as good as the parent SAPE molecule. In particular, the ensemble-averaged results presented in Supplementary Table 5 suggest that the truncated species may potentially have even higher affinities than their non-truncated parent molecules to bind proteins. A histogram plot of distances (<4.0 Å) of the terminal oxygen of the truncated-PE species from either Lys, His or Cys residues found in the 10 randomly selected proteins [Supplementary Fig. 9A] indicated that among the three nucleophilic amino acids, lysines were significantly more likely to be targeted by truncated PEs for adduct formation, compared to histidines and cysteines. However, this is based on structural considerations as lysines are typically more abundant in proteins than histidines or cysteines. From a reactivity point of view, Cys » His > Lys > Arg [58]. Thus, in a model peptide containing Cys, His and Lys residues, Cys will be preferentially targeted in terms of reactivity. Supplementary Fig. 9B illustrates an example of a docking pose of a truncated reactive PE(18:0/C5-oxo-ane) positioned ideally (distance <4.0 Å) to form an adduct with cytochrome c amino acids Lys 80 or Cys 18. The binding sites in the docking simulations were selected based on their binding energy values which are determined using scoring functions defined in the commonly used docking software (in our case SMINA which is a fork of Autodock Vina). The best binding site (as shown in Supplementary Fig. 9B), is typically the one that has the lowest binding energy amongst all the possible binding sites. The multiple docking simulations of at least 10 randomly selected proteins showed the preferential binding of the truncated PEs compared to the non-truncated PE, as seen by their lower binding energy values (Supplementary Tables 5 and 6), in support of the observation that oxidatively-truncated species favor/promote the formation of PEox-protein adducts. Interestingly, when average binding energies for SAPE or the truncated species TrPE_C9 and TrPE_C5 were plotted against the number of nucleophilic amino acids (K, H or C) in the proteins, there was a general positive correlation (0.7–0.8) indicating that the binding becomes stronger with the increasing number of nucleophilic amino acids in general [Supplementary Fig. 10].
2-mercaptoethanol as a potent nucleophile to prevent the formation of PE-lipoxidated protein adducts.
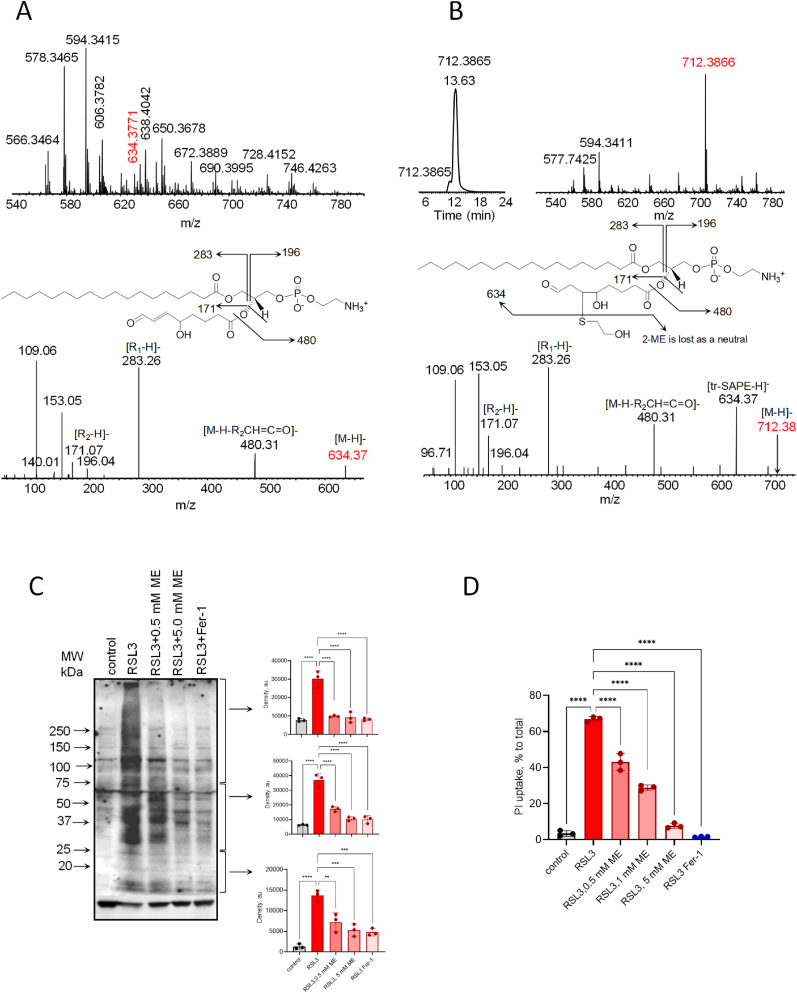
Our non-enzymatic and enzymatic model system data with SAPE and the peptide IHYKYMCNS suggested that the Cys residue was the most susceptible to attack and that the truncated species (PE(26:2)+2 oxygens) was one of the most reactive electrophiles. Given these results we reasoned that an SH containing nucleophile, such as 2-mercaptoethanol, could effectively compete with the PE-peptide and PE-protein adduct formation. Negative mode LCMS analysis [Fig. 6A] indicated that truncated PE species are formed in 30 min in the peptide/SAPE/iron/ascorbate system, similar to that seen in Fig. 2B. The PE(26:2)+2 oxygens species (m/z 634.3771) is noted in red along with its MS/MS fragmentation pattern. The addition of 5 mM 2-mercaptoethanol to the system indicates that a new truncated species is formed (m/z 712.3866) which now dominates this region of the spectrum [Fig. 6B]. This species was identified as a 2-mercaptoethanol adduct of the reactive PE(26:2)+2 oxygen species [Fig. 6B] with a mass increase of +78 Da. Its fragmentation pattern results in the regeneration of the m/z 634.37 species with the loss of the 2-mercaptoethanol as a neutral. Given these results, we reasoned that 2-mercaptoethanol could inhibit PE-protein adduct formation in HT-22 cells undergoing ferroptosis. To this end, cells were induced to undergo ferroptosis with RSL3 (0.5 μM, 4 h) in the absence or presence of 2-mercaptoethanol (0, 0.5 or 5 mM) or ferrostatin (0.4 μM). Proteins were extracted and processed for SDS gel analysis followed by transfer onto nitrocellulose membranes. Membranes were incubated with the duramycin-biotin probe followed by incubation with Streptavidin-HRP using chemiluminescence as a readout. Our results indicate that cells treated with RSL3 plus 2-mercaptoethanol (5 mM) significantly inhibited the PE-lipoxidation of proteins and exhibited chemiluminescence staining similar to controls and ferrostatin-treated cells [Fig. 6C]. Similar to the model system gels, the RSL3 treated lane indicated diffuse protein bands, most likely due to the hydrophilic/hydrophobic and potential variable nature of the individual proteins upon PE-lipoxidation. This is most notable in the higher molecular weight portion of the gel. Densitometric scans of the low, medium and high molecular weight ranges of the gels indicated a significant inhibition of PE-lipoxidation at all concentrations of 2-mercaptoethanol tested [Fig. 6C]. Inhibition of cell death was confirmed as assessed by PI uptake of ferroptotic HT-22 cells in the absence or presence of 2-mercaptoethanol or ferrostatin [Fig. 6D]. Taken together, these results indicate that PE-lipoxidated proteins are participants in ferroptosis and may contribute to a point of no return in the cell death process.
Fig. 6.
2-mercaptoethanol prevents the formation of PE-lipoxidated proteins during ferroptosis. 6A: Truncated PE region from the peptide/SAPE/iron/ascorbate model system in the absence of 2-mercaptoethanol and MS/MS of the m/z 634.37 truncated species. 6B: Addition of 5 mM 2-mercaptoethanol to the model system showing the truncated PE-2-mercaptoethanol adduct (m/z 712.38) and its fragmentation. 6C: HT-22 cells were induced to undergo ferroptosis with RSL3 (0.5 μM, 4 h) in the absence or presence of 2-mercaptoethanol (ME, 0, 0.5 or 5 mM) or ferrostatin-1 (Fer-1, 0.4 μM). Proteins were extracted and processed for SDS gel analysis followed by transfer onto nitrocellulose membranes. Membranes were incubated with the duramycin-biotin probe followed by incubation with Streptavidin-HRP and assessed by
chemiluminescence and densitometric scanning of the high, medium and low molecular weight region of the gel (arrows). 6D: PI uptake of HT-22 ferroptotic cell death in the absence or presence of 2-mercaptoethanol (ME). *p < 0.05; **p < 0.01; ***p < 0.005; ****p < 0.001.
4. Discussion
A number of links and pathogenic associations have been made between ferroptosis and several major disease conditions and acute injuries such as degenerative and neo-plastic diseases, ischemic injury in the liver, kidney, brain (due to stroke or intracerebral hemorrhage) and heart, pulmonary diseases including asthma [33] as well as traumatic brain injury [[24], [25], [26], [27], [28], [29], [30]]. A critical open question with regard to ferroptosis is how, specifically, the accumulation of lipid hydroperoxides translates into ferroptotic cell death? Our redox lipidomics work has established that lipid hydroperoxides, including 15-OOH-AA-PE, are present in very low steady-state concentrations and are unstable due to their ability to generate new peroxyl- and alkoxyl-radicals and readily decompose to secondary products, particularly in the presence of iron [29,30]. Indeed, decomposition of the hydroperoxy-intermediates into secondary products may be the cause of their low abundance in cells [59], supported by our identification of oxidatively-truncated PE species with truncated chain lengths ranging from 5 to 9 carbons. Our model systems with SAPE/iron/H2O2, SAPE/iron/ascorbate and SAPE/15LOX, the latter two systems using a model peptide, indicated that in addition to the numerous truncated species that can be formed, a variety of peptide-truncated-PE adducts and peptide-intact-PEox adducts were formed in the model systems lending to the extensive diversity of potential modified proteins that may occur during ferroptosis. Electrophoretic migration of the lipid-modified model peptide resulted in diffuse, non-distinct bands owing to its hydrophilic/hydrophobic properties and potential variations in the types of adducts formed. Our model systems also suggested that Cys was the preferred nucleophilic residue that was modified. Interestingly, one of the major reactive truncated PE species that was produced was PE(26:2) +2 oxygens (m/z 634.3771). A recently published paper by Tyurina et al. [60] detected the same PE species (PE(26:2 + 2 oxygens or PE-(18:0/hydroxy-8-oxo-oct-6-enoic acid (HOOA)) as well as a 16:0 truncated PE species (PE-(16:0/hydroxy-8-oxo-oct-6-enoic acid (HOOA)) and an 18:0 truncated PC species (PC-(18:0/hydroxy-8-oxo-oct-6-enoic acid (HOOA)) in ferroptotic A375 melanoma cells, all with the same 8 carbon truncated sn-2 chain, that positively correlated with cell death in vitro and in vivo, indicating the potential importance of this particular PE truncation.
Another critical open question that remains is why are PEs specifically targeted for oxidation during ferroptosis? The predominance of PUFA chains in the PE class of lipids and the fact that PE is the second most abundant phospholipid in eukaryotic cells may, in part, answer this question [35]. However, the detailed mechanisms of ferroptosis, in general, remain largely unknown and the specific roles of oxygenated PEs (PEox) is only beginning to emerge [[20], [21], [22], [23],26,27,[31], [32], [33]]. In this study, we have developed a new technology that takes advantage of the lantibiotic duramycin, as a specific affinity probe for the enrichment, isolation and identification of the truncated and intact PE-protein adducts generated during ferroptosis. This new technology allows the specific enrichment of PE-protein adducts which retain a free PE headgroup. This is applicable to ferroptotic death mechanisms since PEs have been identified as the selective targets of pro-ferroptotic lipid peroxidation. Our results indicate that, in addition to the protein adducts resulting from the small reactive lipid fragments such as HNE, HHE, etc., PE-lipoxidated adducts are also formed during ferroptosis. In fact, when binding energies were compared between the SAPE parent and two of the identified oxidatively-truncated PE species, our results suggested that the truncated products bound equally as well to the proteins identified in our lantibiotic screen. The higher affinity potential of the truncated species for proteins would only serve to favor/promote the formation of truncated PE-protein adducts.
Nucleophilic amino acids are known to make significant contributions to protein function such as in catalysis, serving as sites for post-translational modification, making them ideal targets of electrophilic groups. A chemical proteomic platform for the global and quantitative analysis of lysine residues in native biological systems in a previous study resulted in the identification of several hundred lysine residues with heightened reactivity towards electrophiles [61]. The study further emphasized a broad potential and diverse functional consequences of liganding lysine residues within the human proteome. Similar to the study by Hacker et al. [61], our modeling studies showed that a stronger binding of the truncated PE species to proteins correlated with the number of electrophilic amino acids in general (K, H and C). In terms of chemical reactivity, however, Cys residues are far more reactive than His, Lys or Arg residues [58]. Along these lines, our peptide model system data suggested that Cys was the preferred residue for adduction with the highly reactive PE(26:2)+2 oxygens or PE-(18:0/hydroxy-8-oxo-oct-6-enoic acid (HOOA)) truncated species. Because of cysteine's heightened reactivity, we demonstrated that upon the addition of a suitable SH competing nucleophile, 2-mercaptoethanol, we were able to inhibit adduct formation to a large degree in our model systems as well as in cellular proteins from ferroptotic HT-22 cells, culminating in the inhibition of HT-22 ferroptotic death. Inhibition of ferroptosis by 2-mercaptoethanol was originally reported in 2012 [62]. Its effect on erastin-induced ferroptosis was ascribed to its ability to circumvent the inhibition of system Xc- by promoting Cys uptake through an alternative pathway [62]. In addition to this possibility and the possibility that reduction of other cellular proteins by 2-mercaptoethanol could contribute to the overall inhibition of ferroptosis, our newly acquired data indicates that the inhibition of PE-lipoxidation of proteins may also play an important part.
Despite all of the above, the MS-based identification of the adducted truncated PE species to specific amino acid residues still remains elusive, most likely due to several reasons. First, the lantibiotic isolation of the PE-modified proteins was performed on denatured proteins after which tryptic digestion was performed. The resulting PE-lipoxidated fragments will, more than likely, display both hydrophilic/hydrophobic characteristics as shown in our peptide model system and will either fail to elute from the C18 reverse phase column or may exhibit an extremely broad retention time window at best. Second, the adducted portion of the protein may be somewhat resistant to tryptic digestion due to the nucleophilic amino acid modification which may lead to steric hindrance, leaving only the non-modified portion of the protein accessible to digestion and MS-based identification. Third, there may be multiple modifications of any particular protein by several truncated PEs, by several short chain leaving fragments (departing electrophiles) or any combination of these, which may occur within a given digested fragment. This would result in resistance to enzymatic digestion and would severely compromise chromatographic behavior as noted above. Fourth, there may be highly oligomeric protein aggregates formed during ferroptosis that were unable to be assessed, also due to their resistance to denaturation/digestion. The extent of the denaturation/digestion process as well as the degree of lipidation during ferroptosis cannot be expected to occur to the same degree from sample to sample in a biological system, especially since adduction is a highly variable process [63]. Finally, our truncated database that was constructed for SAPE parent molecules containing up to 3 oxygens in the arachidonic fatty acyl chain generated over 300 potential truncated species. The heterogeneity of these species, based on their truncated chain length and reactivity of the oxygen containing groups, can generate a multiplicity of lipoxidated protein adducts that are formed during ferroptotic death, each in extremely low abundance which may be below the threshold level of detection.
We have confirmed the existence of PE-lipoxidated proteins in ferroptotic cells by the lantibiotic method. The degree of PE-lipoxidation may vary with cell type, choice of ferroptotic stimulus and its time of incubation. It is possible that proteins in different cellular compartments may undergo a “progressive” degree of PE-lipidation over time. The PE-protein modifications in various cellular compartments may serve to inhibit/alter protein function and/or induce protein aggregation which may be resistant to degradation and accumulate as unfolded/damaged proteins. Indeed, many neurodegenerative diseases exhibit aggregates that exhibit proteolytic resistance. Of course, the formation of any adduct will depend upon the concentration of the target protein, its subcellular location, and the rate at which the adduct is formed in conjunction with any ongoing repair mechanisms and oxidant-scavenging reactions [39,64].
Several unanswered questions remain regarding the PE-lipoxidation of proteins during ferroptosis. Is this an inconsequential consequence of the ferroptotic process or do these PE-protein adducts play a significant role in the cell's demise? A recently published article indicated that the ER is one of the initial key sites of lipid peroxidation and suggested an ordered progression of membrane peroxidation [65]. In this respect, protein lipidation by truncated PE-protein adducts may also follow a similar progression. The current study suggests that different compartments in different cell types may be differentially PE-lipoxidated and this may depend upon the presence and density of PE in subcellular structures. While oxidatively-truncated PE-lipoxidated proteins may inhibit/alter protein function, they may also serve as anchors to various membranes via their unmodified fatty acyl chain. This special type of lipidation by the truncated PE species may alter the protein's location via tethering to a variety of cellular/subcellular membranes, especially in cases where multiple truncated PE adducts are formed. Along these lines, the amino group of the “free” PE headgroup may also serve as a crosslinking site with other proteins within the cell or cellular membranes. Notably, dialdehyde products of lipid peroxidation such as malonyl-dialdehyde, acroleyl-dialdehyde [37,38], may also contribute to the cross-linking and oligomerization of PE-lipoxidated proteins.
Overall, in line with the robust and diversified lipid peroxidation typically seen in ferroptosis, the execution of this cell death program is also associated with the generation of oxidatively-truncated PE species and the accumulation of multiple oxidatively modified proteins, including a variety of PE-lipoxidated proteins for each cell type as determined by our lantibiotic affinity technology. This is in sharp contrast to several other death programs such as necroptosis and pyroptosis, where the engagement of single proteins (MLKL and gasdermine, respectively), has been documented that are critical to these cell death processes [[9], [10], [11], [12], [13]]. It is unlikely that protein-lipidation during ferroptosis has a regulatory role since these types of modifications are normally considered irreversible. However, in other pathophysiological conditions, PE-protein lipidation may alter protein structure, redox state, catalytic function and/or signaling in response to environmental/metabolic changes. With this in mind, our lantibiotic technology would be directly applicable to address these questions. Whether proteins exist that are more prone to PE-lipoxidation remains a matter of investigation. In the case of aging, some proteins, such as heat shock proteins and elongation factors associated with protein synthesis, have been shown to be carbonylated/lipidated consistently from bacteria to humans [37]. Whether a similar situation exists during ferroptotic death or other pathophysiological conditions or whether PE-lipoxidation has a place in normal cellular physiology, remains to be determined. However, the significance of the identification of PE-lipoxidation adducts of proteins is dual, as they may contribute to a “point of no return” in ferroptosis and may potentially be used as biomarkers of ferroptosis.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the National Institutes of Health grant numbers: AI156924, AI156923, AI145406, HL114453, NS061817, NS076511, CA165065, R01GM139297 and Polish National Science Centre no. 2019/35/D/ST4/02203.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2023.102758.
Contributor Information
A.A. Amoscato, Email: amoscatoaa@upmc.edu.
V.E. Kagan, Email: kagan@pitt.edu.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
Data availability
Data will be made available on request.
References
- 1.Savill J., Dransfield I., Gregory C., Haslett C. A blast from the past: clearance of apoptotic cells regulates immune responses. Nat. Rev. Immunol. 2002;2:965–975. doi: 10.1038/nri957. [DOI] [PubMed] [Google Scholar]
- 2.Medina C.B., Ravichandran K.S. Do not let death do us part: ‘find-me’ signals in communication between dying cells and the phagocytes. Cell Death Differ. 2016;23:979–989. doi: 10.1038/cdd.2016.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wolf Y., Boura-Halfon S., Cortese N., Haimon Z., Sar Shalom H., Kuperman Y., Kalchenko V., Brandis A., David E., Segal-hayoun Y., Chappell-Maor L., Yaron A., Jung S. Brown-adipose-tissue macrophages control tissue innervation and homeostatic energy expenditure. Nat. Immunol. 2017;18:665–674. doi: 10.1038/ni.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kong Y., Janssen B.J., Malinauskas T., Vangoor V.R., Coles C.H., Kaufmann R., Ni T., Gilbert R.J.C., Padilla-Parra S., Pasterkamp R.J., Jones E.Y. Structural basis for plexin activation and regulation. Neuron. 2016;91:548–560. doi: 10.1016/j.neuron.2016.06.018. 10.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weiskopf K., Schnorr P.J., Pang W.W., Chao M.P., Chhabra A., Seita J., Feng M., Weissman I.L. Myeloid cell origins, differentiation, and clinical implications. Microbiol. Spectr. 2016;4:1–28. doi: 10.1128/microbiolspec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oldenborg P.A. CD47: a cell surface glycoprotein which regulates multiple functions of hematopoietic cells in health and disease. ISRN Hematol. 2013;2013 doi: 10.1155/2013/614619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kagan V.E., Tyurina Y.Y., Tyurin V.A., Mohammadyani D., Friedman Angeli J.P., Baranov S.V., Klein-Seetharaman J., Friedlander R.M., Mallampalli R.K., Conrad M., Bayir H. Cardiolipin signaling mechanisms: collapse of asymmetry and oxidation. Antioxidants Redox Signal. 2015;22(18):1667–1680. doi: 10.1089/ars.2014.6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ward T.H., Cummings J., Dean E., Greystoke A., Hou J.M., Backen A., Ranson M., Dive C. Biomarkers of apoptosis. Br. J. Cancer. 2008;99:841–846. doi: 10.1038/sj.bjc.6604519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen X., He W.T., Hu L., Li J., Fang Y., Wang X., Xu X., Wang Z., Huang K., Han J. Pyroptosis is driven by non-selective gasdermin-D pore and its morphology is different from MLKL channel-mediated necroptosis. Cell Res. Sep. 2016;26(9):1007–1020. doi: 10.1038/cr.2016.100. Epub 2016 Aug 30. PMID: 27573174; PMCID: PMC5034106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen J., Kos R., Garssen J., Redegeld F. Molecular insights into the mechanism of necroptosis: the necrosome as a potential therapeutic target. Cells. 2019;8(12):1486. doi: 10.3390/cells8121486.PMID:31766571. PMCID: PMC6952807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parisi L.R., Sowlati-Hashjin S., Berhane I.A., Galster S.L., Carter K.A., Lovell J.F., Chemler S.R., Karttunen M., Atilla-Gokcumen G.E. Membrane disruption by very long chain fatty acids during necroptosis. ACS Chem. Biol. 2019;14(10):2286–2294. doi: 10.1021/acschembio.9b00616. Epub 2019 Sep 20. PMID: 31490656; PMCID: PMC6800604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ding J., Wang K., Liu W., She Y., Sun Q., Shi J., Sun H., Wang D.C., Shao F. Pore-forming activity and structural autoinhibition of the gasdermin family. Nature. 2016;535(7610):111–116. doi: 10.1038/nature18590. Epub 2016 Jun 8. Erratum in: Nature. 2016 Dec 1;540(7631):150. PMID: 27281216. [DOI] [PubMed] [Google Scholar]
- 13.Broz P., Pelegrín P., Shao F. The gasdermins, a protein family executing cell death and inflammation. Nat. Rev. Immunol. 2020;20(3):143–157. doi: 10.1038/s41577-019-0228-2. Epub 2019 Nov 5. PMID: 31690840. [DOI] [PubMed] [Google Scholar]
- 14.Lin X., Lu X.X., Luo G., Xiang H H. Progress in PD-1/PD-L1 pathway inhibitors: from biomacromolecules to small molecules. Eur. J. Med. Chem. 2020;186 doi: 10.1016/j.ejmech.2019.111876. Epub 2019 Nov 15. PMID: 31761384. [DOI] [PubMed] [Google Scholar]
- 15.Johnson D.C., Taabazuing C.Y., Okondo M.C., Chui A.J., Rao S.D., Brown F.C., Reed C., Peguero E., de Stanchina E., Kentsis A., Bachovchin D.A. DPP8/DPP9 inhibitor-induced pyroptosis for treatment of acute myeloid leukemia. Nat. Med. 2018;24(8):1151–1156. doi: 10.1038/s41591-018-0082-y. Epub Jul 2. PMID: 29967349; PMCID: PMC6082709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu H., Zhang S., Wu J., Chen M., Cai M.C., Fu Y., Li W., Wang J., Zhao X., Yu Z., Ma P., Zhuang G. Molecular targeted therapies elicit concurrent apoptotic and GSDME-dependent pyroptotic tumor cell death. Clin. Cancer Res. 2018;24(23):6066–6077. doi: 10.1158/1078-0432.CCR-18-1478. Epub 2018 Jul 30. PMID: 30061362. [DOI] [PubMed] [Google Scholar]
- 17.Fu Y., Hong L., Xu J., Zhong G., Gu Q., Gu Q., Guan Y., Zheng X., Dai Q., Luo X., Liu C., Huang Z., Yin X.M., Liu P., Li M. Discovery of a small molecule targeting autophagy via ATG4B inhibition and cell death of colorectal cancer cells in vitro and in vivo. Autophagy. 2019;15(2):295–311. doi: 10.1080/15548627.2018.1517073.Epub2018Sep20. PMID: 30176161; PMCID: PMC6333450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li K., Wu D., Chen X., Zhang T., Zhang L., Yi Y., Miao Z., Jin N., Bi X., Wang H., Xu J., Wang D. Current and emerging biomarkers of cell death in human disease. BioMed Res. Int. 2014;2014(2014) doi: 10.1155/2014/690103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang M., Antoine D.J., Weemhoff J.L., Jenkins R.E., Farhood A., Park B.K., Jaeschke H. Biomarkers distinguish apoptotic and necrotic cell death during hepatic ischemia-reperfusion injury in mice. Liver Transplant. 2014;20(11):1372–1382. doi: 10.1002/lt.23958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stockwell B.R., Jiang X. The chemistry and biology of ferroptosis. Cell. Chem. Biol. 2020;27(4):365–375. doi: 10.1016/j.chembiol.2020.03.013. PMID: 32294465; PMCID: PMC7204503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao J.Y., Dixon S.J. Mechanisms of ferroptosis. Cell. Mol. Life Sci. 2016;73(11–12):2195–2209. doi: 10.1007/s00018-016-2194-1. Epub 2016 Apr 5. PMID: 27048822; PMCID: PMC4887533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seibt T.M., Proneth B., Conrad M. Role of GPX4 in ferroptosis and its pharmacological implication. Free Radic. Biol. Med. 2019;133:144–152. doi: 10.1016/j.freeradbiomed.2018.09.014. Epub 2018 Sep 13. PMID: 30219704. [DOI] [PubMed] [Google Scholar]
- 23.Rwigema J.C., Beck B., Wang W., Doemling A., Epperly M.W., Shields D., Goff J.P., Franicola D., Dixon T., Frantz M.C., Wipf P., Tyurina Y., Kagan V.E., Wang H., Greenberger J.S. Two strategies for the development of mitochondrion-targeted small molecule radiation damage mitigators. Int. J. Radiat. Oncol. Biol. Phys. 2011;80(3):860–868. doi: 10.1016/j.ijrobp.2011.01.059. Epub 2011 Apr 13. PMID: 21493014; PMCID: PMC3104115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang W.S., SriRamaratnam R., Welsch M.E., Shimada K., Skouta R., Viswanathan V.S., Cheah J.H., Clemons P.A., Shamji A.F., Clish C.B., Brown L.M., Girotti A.W., Cornish V.W., Schreiber S.L., Stockwell B.R. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156:317–331. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li W., Feng G., Gauthier J.M., Lokshina I., Higashikubo R., Evans S., Liu X., Hassan A., Tanaka S., Cicka M., Hsiao H.M., Ruiz-Perez D., Bredemeyer A., Gross R.W., Mann D.L., Tyurina Y.Y., Gelman A.E., Kagan V.E., Linkermann A., Lavine K.J., Kreisel D. Ferroptotic cell death and TLR4/Trif signaling initiate neutrophil recruitment after heart transplantation. J. Clin. Invest. 2019;129(6):2293–2304. doi: 10.1172/JCI126428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kenny E.M., Fidan E., Yang Q., Anthonymuthu T.S., New L.A., Meyer E.A., Wang H., Kochanek P.M., Dixon C.E., Kagan V.E., Bayir H. Ferroptosis contributes to neuronal death and functional outcome after traumatic brain injury. Crit. Care Med. 2019;47(3):410–418. doi: 10.1097/CCM.0000000000003555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anthonymuthu T.S., Kenny E.M., Lamade A.M., Kagan V.E., Bayır H H. Oxidized phospholipid signaling in traumatic brain injury. Free Radic. Biol. Med. 2018;124:493–503. doi: 10.1016/j.freeradbiomed.2018.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Magtanong L., Dixon S.J. Ferroptosis and brain injury. Dev. Neurosci. 2018;40(5–6):382–395. doi: 10.1159/000496922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang L.L., Liao X.H., Sun H., Jiang X., Liu Q., Zhang L. Augmenter of liver regeneration protects the kidney from ischaemia-reperfusion injury in ferroptosis. J. Cell Mol. Med. 2019;23(6):4153–4164. doi: 10.1111/jcmm.14302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Del Re D.P., Amgalan D., Linkermann A., Liu Q., Kitsis R.N. Fundamental mechanisms of regulated cell death and implications for heart disease. Physiol. Rev. 2019;99(4):1765–1817. doi: 10.1152/physrev.00022.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Friedmann Angeli J.P., Schneider M., Proneth B., Tyurina Y.Y., Tyurin V.A., Hammond V.J., Herbach N., Aichler M., Walch A., Eggenhofer E., Basavarajappa D., Rådmark O., Kobayashi S., Seibt T., Beck H., Neff F., Esposito I., Wanke R., Förster H., Yefremova O., Heinrichmeyer M., Bornkamm G.W., Geissler E.K., Thomas S.B., Stockwell B.R., O'Donnell V.B., Kagan V.E., Schick J.A., Conrad M. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat. Cell Biol. 2014;16(12):1180–1191. doi: 10.1038/ncb3064.Epub2014Nov17. PMID: 25402683; PMCID: PMC4894846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kagan V.E., Mao G., Qu F., Angeli J.P., Doll S., Croix C.M. St, Dar H.H., Liu B., Tyurin V.A., Ritov V.B., Kapralov A.A., Amoscato A.A., Jiang J., Anthonymuthu T., Mohammadyani D., Yang Q., Proneth B., Klein-Seetharaman J., Watkins S., Bahar I., Greenberger J., Mallampalli R.K., Stockwell B.R., Tyurina Y.Y., Conrad M., Bayır H. Oxidized arachidonic and adrenic PEs navigate cells to ferroptosis. Nat. Chem. Biol. 2017;13(1):81–90. doi: 10.1038/nchembio.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wenzel S.E., Tyurina Y.Y., Zhao J., St Croix C.M., Dar H.H., Mao G., Tyurin V.A., Anthonymuthu T.S., Kapralov A.A., Amoscato A.A., Mikulska-Ruminska K., Shrivastava I.H., Kenny E.M., Yang Q., Rosenbaum J.C., Sparvero L.J., Emlet D.R., Wen X., Minami Y., Qu F., Watkins S.C., Holman T.R., VanDemark A.P., Kellum J.A., Bahar I., Bayır H., Kagan V.E. PEBP1 wardens ferroptosis by enabling lipoxygenase generation of lipid death signals. Cell. 2017;171(3) doi: 10.1016/j.cell.2017.09.044. (2017) 628-641.e26. doi: 10.1016/j.cell.2017.09.044. PMID: 29053969; PMCID: PMC5683852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zarkovic N., Cipak A., Jaganjac M., Borovic S., Zarkovic K. Pathophysiological relevance of aldehydic protein modifications. J. Proteonomics. 2013;92:239–247. doi: 10.1016/j.jprot.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 35.Calzada C., Onguka O., Claypool S.M. Phosphatidylethanolamine metabolism in health and disease. Int. Rev. Cell Mol. Biol. 2016;321:29–88. doi: 10.1016/bs.ircmb.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wiernicki B., Dubois H., Tyurina Y.Y., Hassannia B., Bayir H., Kagan V.E., Vandenabeele P., Wullaert A., Vanden Berghe T. Excessive phospholipid peroxidation distinguishes ferroptosis from other cell death modes including pyroptosis. Cell Death Dis. 2020;11:922–932. doi: 10.1038/s41419-020-03118-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Demasi M., Augusto O., Bechara E.J.H., Bicev R.N., Cerqueira F.M., da Cunha F.M., Denicola A., Gomes F., Miyamoto S., Netto L.E.S., Randall L.M., Stevani C.V., Thomson L. Oxidative modification of proteins: from damage to catalysis, signaling, and beyond. Antioxidants Redox Signal. 2021;35(12):1016–1079. doi: 10.1089/ars.2020.8176. 2021. [DOI] [PubMed] [Google Scholar]
- 38.Barrera G., Pizzimenti S., Ciamporcero E.S., Daga M., Ullio C., Arcaro A., Cetrangolo G.P., Ferretti C., Dianzani C., Lepore A., Gentile F. Role of 4-hydroxynonenal-protein adducts in human diseases. Antioxidants Redox Signal. 2014;22(18):1681–1702. doi: 10.1089/ars.2014.6166. 2014. [DOI] [PubMed] [Google Scholar]
- 39.Chen Y., Liu Y., Lan T., Qin W., Zhu Y., Qin K., Gao J., Wang H., Hou X., Chen N., Friedmann Angeli J.P., Conrad M., Wang C. Quantitative profiling of protein carbonylations in ferroptosis by an aniline-derived probe. J. Appl. Comput. Sci. 2018;140(13):4712–4720. doi: 10.1021/jacs.8b01462. [DOI] [PubMed] [Google Scholar]
- 40.Ichimura Y., Kirisako T., Takao T., Satomi Y., Shimonishi Y., Ishihara N., Mizushima N., Tanida I., Kominami E., Ohsumi M., Noda T., Ohsumi Y. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 41.Kapralov A.A., Yang Q., Dar H.H., Tyurina Y.Y., Anthonymuthu T.S., Kim R., St Croix C.M., Mikulska-Ruminska K., Liu B., Shrivastava I.H., Tyurin V.A., Ting H.C., Wu Y.L., Gao Y., Shurin G.V., Artyukhova M.A., Ponomareva L.A., Timashev P.S., Domingues R.M., Stoyanovsky D.A., Greenberger J.S., Mallampalli R.K., Bahar I., Gabrilovich D.I., Bayır H., Kagan V.E. Redox lipid reprogramming commands susceptibility of macrophages and microglia to ferroptotic death. Nat. Chem. Biol. Mar. 2020;16(3):278–290. doi: 10.1038/s41589-019-0462-8. Epub 2020 Feb 17. PMID: 32080625; PMCID: PMC7233108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dhe-Paganon S., Werner E.D., Chi Y.-I., Shoelso S. Structure of the globular tail of nuclear lamin. J. Biol. Chem. 2002;277(20):17381–17384. doi: 10.1074/jbc.C200038200. [DOI] [PubMed] [Google Scholar]
- 43.Kiss B., Duelli A., Radnai L., Kékesi K.A., Katona G., Nyitray L. Crystal structure of the S100A4–nonmuscle myosin IIA tail fragment complex reveals an asymmetric target binding mechanism. Proc. Natl. Acad. Sci. USA. 2012;109(16):6048–6053. doi: 10.1073/pnas.1114732109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Evrard C., Capron A., Marchand C., Clippe A., Wattiez R., Soumillion P., Knoops B., Declercq J.-P. Crystal structure of a dimeric oxidized form of human peroxiredoxin 5. J. Mol. Biol. 2004;337(5):1079–1090. doi: 10.1016/j.jmb.2004.02.017. [DOI] [PubMed] [Google Scholar]
- 45.Maximciuc A.A., Putkey J.A., Shamoo Y., MacKenzie K.R. Complex of calmodulin with a ryanodine receptor target reveals a novel, flexible binding mode. Structure. 2006;14(10):1547–1556. doi: 10.1016/j.str.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 46.Xu Z., Page R.C., Gomes M.M., Kohli E., Nix J.C., Herr A.B., Patterson C., Misra S. Structural basis of nucleotide exchange and client binding by the Hsp 70 cochaperone Bag 2. Nat. Struct. Mol. Biol. 2008;15(12):1309–1317. doi: 10.1038/nsmb.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ueda J., Harada A., Urahama T., Machida S., Maehara K., Hada M., Makino Y., Nogami J., Horikoshi N., Osakabe A. Testis-specific histone variant H3t gene is essential for entry into spermatogenesis. Cell Rep. 2017;18(3):593–600. doi: 10.1016/j.celrep.2016.12.065. [DOI] [PubMed] [Google Scholar]
- 48.S. Release, 1: Maestro Schrödinger, LLC: New York, NY, USA 2018.
- 49.Koes D.R., Baumgartner M.P., Camacho C.J. Lessons learned in empirical scoring with smina from the CSAR 2011 benchmarking exercise. J. Chem. Inf. Model. 2013;53(8):1893–1904. doi: 10.1021/ci300604z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang S., Krieger J.M., Zhang Y., Kaya C., Kaynak B., Mikulska-Ruminska K., Doruker P., Li H., Bahar I. ProDy 2.0: increased scale and scope after 10 Years of protein dynamics modelling with Python. Bioinformatics. 2021;37(20):3657–3659. doi: 10.1093/bioinformatics/btab187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bakan A., Meireles L.M., ProDy I. Bahar. Protein dynamics inferred from theory and experiments. Bioinformatics. 2011;27(11):1575–1577. doi: 10.1093/bioinformatics/btr168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pi J., Wu X., Feng Y. Fragmentation patterns of five types of phospholipids by ultra-high-performance liquid chromatography electrospray ionization quadrupole time-of-flight tandem mass spectrometry. Anal. Methods. 2016;8:1319–1332. [Google Scholar]
- 53.Wakita C., Maeshima T., Yamazaki A., Shibata T., Ito S., Akagawa M., Ojika M., Yodoi J., Uchida K. Stereochemical configuration of 4-hydroxy-2-nonenal-cysteine adducts and their steroselective formation in a redox-regulated protein. J. Biol. Chem. 2009;284:28810–28822. doi: 10.1074/jbc.M109.019927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McAuliffe O., Ross R.P., Hill C. Lantibiotics: structure, biosynthesis and mode of action. FEMS Microbiol. Rev. 2001;25:285–308. doi: 10.1111/j.1574-6976.2001.tb00579.x. [DOI] [PubMed] [Google Scholar]
- 55.Zhao M. Lantibiotics as probes for phosphatidylethanolamine. Amino Acids. 2011;41:1071–1079. doi: 10.1007/s00726-009-0386-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gallegos A.M., Atshaves B., Storey S.M., Starodub O., Petrescu A.D., Huang H., McIntosh A.L., Martin G.G., Chao H., Kier A.B., Schroeder F. Gene structure, intracellular localization and functional roles of sterol carrier protein-2. Prog. Lipid Res. 2001;40:498–563. doi: 10.1016/s0163-7827(01)00015-7. [DOI] [PubMed] [Google Scholar]
- 57.Burgardt N.I., Gianotti A.R., Ferreyra R.G., Ermacora M.R. A structural appraisal of sterol carrier protein 2. Biochim. Biophys. Acta. 2017;1865:565–577. doi: 10.1016/j.bbapap.2017.03.002. [DOI] [PubMed] [Google Scholar]
- 58.Milkovic L., Zarkovic N., Marusic Z., Zarkovic K., Jaganjac M. The 4-hydroxynonenal-protein adducts and their biological relevance: are some proteins preferred targets? Antioxidants. 2023;12(856):1–18. doi: 10.3390/antiox12040856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ayala A., Muñoz M.F., Argüelles S. Lipid peroxidation: production, metabolism, and signaling mechanisms of malondialdehyde and 4-hydroxy-2-nonenal. Oxid. Med. Cell. Longev. 2014 doi: 10.1155/2014/360438. 2014 (2014) 1-31 article ID 360438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tyurina Y.Y., Kapralov A.A., Tyurin V.A., Shurin G., Amoscato A.A., Rajasundaram D., Tian H., Bunimovich Y.L., Nefedova Y., Herrick W.G., Parchment R.E., Doroshow J.H., Bayir H., Srivastava A.K., Kagan V.E. Redox phospholipidomics discovers pro-ferroptotic death signals in A375 melanoma cells in vitro and in vivo. Redox Biol. 2023;61 doi: 10.1016/j.redox.2023.102650.Epub2023Feb28. PMID: 36870109; PMCID: PMC9996109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hacker S., Backus K., Lazear M., Forli S., Correia B.E., Cravatt B.F. Global profiling of lysine reactivity and ligandability in the human proteome. Nat. Chem. 2017;9:1181–1190. doi: 10.1038/nchem.2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dixon S.J., Lemberg K.M., Lamprecht M.R., Skouta R., Zaitsev E.M., Gleason C.E., Patel D.N., Bauer A.J., Cantley A.M., Yang W.S., Morrison B., Ferroptosis B.R. Stockwell. Iron Dependent Form Nonapoptotic Cell Death Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tzeng S.-C., Maier C.S. Label-free proteomics assisted by affinity enrichment for elucidating the chemical reactivity of the liver mitochondrial proteome toward adduction by the lipid electrophile 4-hydroxy-2-nonenal (HNE) Front. Chem. 2016;4(2):1–17. doi: 10.3389/fchem.2016.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dalle-Donne I., Aldini G., Carini M., Colombo R., Rossi R., Milzani A. Protein carbonylation, cellular dysfunction, and disease progression. J. Cell Mol. Med. 2006;10(2):389–406. doi: 10.1111/j.1582-4934.2006.tb00407.x.PMID:16796807. PMCID: PMC3933129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.von Krusenstiern N., Robson R.N., Qian N., Qui B., Hu F., Reznik E., Smith N., Zandkarimi F., Ester V.M., Dupont M., Hirschhorn T., Shchepinov M.S., Min W., Woerpel K.A., Stockwell B.R. Identification of essential sites of lipid peroxidation in ferroptosis. Nat. Chem. Biol. 2023 doi: 10.1038/s41589-022-01249-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.