Abstract
Engineered living materials (ELMs) fabricated by encapsulating microbes in hydrogels have great potential as bioreactors for sustained bioproduction. While long-term metabolic activity has been demonstrated in these systems, the capacity and dynamics of gene expression over time is not well understood. Thus, we investigate the long-term gene expression dynamics in microbial ELMs constructed using different microbes and hydrogel matrices. Through direct gene expression measurements of engineered E. coli in F127-bisurethane methacrylate (F127-BUM) hydrogels, we show that inducible, input-responsive genetic programs in ELMs can be activated multiple times and maintained for multiple weeks. Interestingly, the encapsulated bacteria sustain inducible gene expression almost 10 times longer than free-floating, planktonic cells. These ELMs exhibit dynamic responsiveness to repeated induction cycles, with up to 97% of the initial gene expression capacity retained following a subsequent induction event. We demonstrate multi-week bioproduction cycling by implementing inducible CRISPR transcriptional activation (CRISPRa) programs that regulate the expression of enzymes in a pteridine biosynthesis pathway. ELMs fabricated from engineered S. cerevisiae in bovine serum albumin (BSA) - polyethylene glycol diacrylate (PEGDA) hydrogels were programmed to express two different proteins, each under the control of a different chemical inducer. We observed scheduled bioproduction switching between betaxanthin pigment molecules and proteinase A in S. cerevisiae ELMs over the course of 27 days under continuous cultivation. Overall, these results suggest that the capacity for long-term genetic expression may be a general property of microbial ELMs. This work establishes approaches for implementing dynamic, input-responsive genetic programs to tailor ELM functions for a wide range of advanced applications.
Keywords: Hydrogels, Input-responsive engineered living materials (ELMs), ELM bioproduction, CRISPR gene Activation (CRISPRa), Programmable bioproduction
Graphical abstract
Highlights
-
•
Engineered Living Materials (ELMs) extend the inducible-response in bacteria by 2.5 more weeks than liquid culture.
-
•
Input-responsive ELMs can respond to multiple induction cycles.
-
•
Bioproduction can be repeatedly initiated with CRISPR gene activation programs.
-
•
Product switching with multi-input genetic programs demonstrated in yeast ELMs.
-
•
Sustained, multi-week inducible expression may be a general feature of microbial ELMs.
1. Introduction
Bioproduction with engineered microbes plays an increasingly important role in the industrial synthesis of drugs, chemicals, food and peptides [1]. Microbes can be encapsulated in 3D biocompatible polymeric scaffolds to create engineered living materials (ELMs) with changes in metabolism [2] that offer significant advantages compared to cells cultured in traditional liquid suspension [[2], [3], [4], [5], [6]]. ELMs fabricated by encapsulating microbes in hydrogels have long-term, sustained metabolic functions that can be harnessed for bioproduction cycles lasting up to one year [3]. Microbe encapsulation in biomaterials has long been known to give rise not only to changes in metabolic capacity, but also in growth rate and colony morphology [[7], [8], [9], [10], [11]]. At present, much less is known about how encapsulation affects the capacity and dynamics of long-term microbial gene expression.
Synthetic biologists have developed a number of strategies, including implementing inducible gene expression and dynamic gene regulatory networks, to program bioproduction functions by controlling the timing and expression levels of biosynthetic genes [[12], [13], [14], [15], [16], [17]]. Microbial ELMs responsive to external stimuli have been successfully implemented through the use of inducible gene expression systems [6,[18], [19], [20], [21], [22], [23]]. Tunable differences in biosynthetic output have been achieved in stimuli-responsive microbial ELMs by varying the number of light-inducer pulses [6]. Input-responsive ELMs have exhibited programmable genetic responses to small-molecule inducers following multiple cycles of preservation and cold-storage [4,18]. Collectively, these results are consistent with the idea that microbes encapsulated in hydrogels can respond to inducers and express heterologous genes under long-term continuous culture conditions. If encapsulated microbes retain the ability to express high levels of heterologous genes, then it may be possible to develop input-responsive ELMs [24] with bioproduction functions that can be cycled ON and OFF, or switched between multiple products. These dynamically-programmable functions could dramatically increase the versatility and reliability of on-demand bioproduction, without the process complexities or cold storage typically needed to achieve such capabilities with free-floating, planktonic cells [3,25].
We sought to investigate how hydrogel encapsulation affects the capacity and dynamics of microbial gene expression. We fabricated two different microbial ELM systems. We encapsulated and casted engineered Eschericia coli in Pluronic F127-bisurethane methacrylate (F127-BUM) hydrogels. E. coli is a gram-negative, generally-recognized-as-safe (GRAS) bacterium that can utilize glucose efficiently to make a wide array of bioproducts [26]. Additionally, we encapsulated engineered Saccharomyces cerevisiae in bovine serum albumin (BSA) - polyethylene glycol diacrylate (PEGDA) hydrogels via stereolithographic apparatus (SLA) 3D printing [27,28]. The eukaryotic yeast S. cerevisiae has a long history as a bioproduction host [29]. We created an approach to measure gene expression directly in hydrogels and quantified the impact of encapsulation on the timing and levels of heterologous E. coli gene expression. Interestingly, the ability to generate high levels of heterologous gene expression was retained up to ten times longer in encapsulated E. coli than in free-floating, planktonic cells. The successful construction of input-responsive E. coli ELMs incorporating CRISPR gene activation (CRISPRa) programs [14,15] that cycle pteridine biosynthesis [15,30] ON and OFF over the course of multiple weeks illustrates that complex bioproduction functions can be implemented in these systems. We show that input-responsive bioproduct switching can be achieved in dynamically-programmed S. cerevisiae ELMs over the course of 27 days of continuous culture. Taken together, this work demonstrates that encapsulated microbes can express high levels of heterologous genes and suggests that the capacity for long-term gene expression may be a general feature of microbial ELMs.
2. Materials and methods
2.1. E. coli ELM materials and methods
2.1.1. E. coli plasmid construction and preparation
All plasmids used in this study are listed in Supplementary Table S1. Plasmids were designed using the Benchling sequence designer. PCR fragments for assembly were amplified using Phusion DNA Polymerase (Thermo Fisher Scientific) for cloning using 5X In-Fusion HD kits (Takara Bio). The assembled plasmids were transformed into chemically competent E. coli NEB Turbo cells (New England Biolabs) in Luria-Bertani (LB) medium or agar plates supplemented with antibiotics (100 μg/mL carbenicillin and/or 25 μg/mL chloramphenicol). Plasmids were purified using QIAprep Spin Miniprep Kits (Qiagen 27,104) according to the manufacturer's protocol. Plasmid concentrations were quantified via spectrophotometry (Nanodrop 2000c, Cat. ND-2000C). Constructs were confirmed by Sanger sequencing (Genewiz-Azenta), and sequencing results were analyzed using Benchling. For E. coli ELM experiments, E. coli MG1655 cells were grown overnight at 37 °C with orbital shaking at 220 RPM in MOPS-based EZ-Rich Defined Medium (EZ-RDM, Teknova M2105) with 0.2% glucose, unless otherwise specified, and appropriate antibiotics (100 μg/mL carbenicillin and/or 25 μg/mL chloramphenicol).
2.1.2. F127-BUM synthesis
F127-BUM was synthesized according to the protocol described in Millik et al. [31]. Briefly, Pluronic F127 (60 g, 4.8 mmol) was dried under vacuum at room temperature (RT) in a round bottom flask, and anhydrous CH2Cl2 (550 mL) was charged to the flask under a N2 atmosphere. The mixture was stirred until F127 was completely dissolved before adding dibutyltin dilaurate. 2-Isocyanatoethyl methacrylate (3.5 mL, 24.8 mmol) was diluted in anhydrous CH2Cl2 (50 mL) and added to the reaction mixture dropwise. The reaction was allowed to run for 2 days. F127-BUM was condensed by evaporating CH2Cl2, precipitated in Et2O (2000 mL) overnight and decanted. The precipitate was washed twice in Et2O before drying overnight under vacuum (RT). The final product was stored in the dark at 4 °C before use.
2.1.3. E. coli ELMs fabrication
E.coli laden F127-BUM hydrogels were prepared using a method previously developed by Johnston & Yuan et al. [3] with the following modifications: starter culture of E. coli MG1655 cells was grown in 3 mL EZ-RDM with appropriate antibiotics and incubated overnight at 37 °C with 220 RPM orbital shaking. Hydrogels were seeded with 1.5 × 108 cells/g of hydrogel. The microbial hydrogel mixture was transferred to a syringe while kept cold (sol-state, 5 °C) and then warmed to room temperature (∼21 °C) to allow the sol-to-gel transition. The shear-responsive hydrogel was then extruded into a cylindrical silicone mold (diameter = 4 mm, height = 2 mm) placed between two glass slides. The hydrogel was allowed to sit for 15 min in the mold to assure a proper shape conformation. The entire mold assembly was photocured for 3 min on each side under a UV 365 nm lamp (UVP 95-0005-05) at 18.6 mW/cm2 (Supplementary Fig. S1). The fully-cured hydrogels were rinsed with 100 μL EZ-RDM and incubated overnight at 37 °C in 2 mL of EZ-RDM with appropriate antibiotics. For hand-extruded (uncast) hydrogels, 100 μL of microbial gel mixture was extruded directly from the syringe onto a glass slide and UV photocured according to the steps described above. The photocured hydrogels were cut into four equal parts of ∼25 μL before incubating in EZ-RDM.
2.1.4. Direct-gel fluorescence measurement for cast hydrogels
End-point fluorescence measurements for both hydrogels and the surrounding liquid media were collected using a microplate reader (Biotek Synergy HTX). The following settings were used for the different constitutively-expressed fluorescent proteins: sfGFP (excitation: 485 nm, emission: 528 nm, gain: 35) and mRFP1 (excitation: 540 nm, emission: 600 nm, gain: 35). For directly measuring fluorescence from hydrogels, the hydrogels were rinsed once with 100 μL EZ-RDM and transferred into a 96-well clear flat-bottom plate (Corning 3916) with wells pre-filled with 100 μL of EZ-RDM. To ensure proper fluorescence readings, the cylindrical cast hydrogels were positioned at the center of the well with the flat circular surface facing the bottom of the plate. For all experiments, three to five technical replicates per treatment were used. 100 μL of the surrounding liquid media containing planktonic cells were also measured for fluorescence comparison purposes. Data were plotted using Prism (GraphPad).
2.1.5. Fluorescence microscopy of cast hydrogels
Cast F127-BUM hydrogels containing E. coli MG1655 with CRISPRa-directed sfGFP expression (pCK389.306 and pPC003) were continuously cultured for two days before imaging. The hydrogel was removed from the spent media and washed with 100 μL of EZ-RDM media before it was placed in a chamber slide (Ibidi μ-slide 8 well, Cat. 80,826) pre-filled with 100 μL of EZ-RDM media. Hydrogel images were taken using an EVOS FL auto microscope without any image manipulation (Thermo Fisher).
2.1.6. Inducible expression in E.coli liquid continuous cultures
A starter culture of E. coli MG1655 containing an aTc-inducible CRISPRa plasmid (pCK389.306) and an sfGFP reporter plasmid (pPC003) was grown overnight at 37 °C with 220 RPM orbital shaking in 3 mL EZ-RDM with 0.2% glucose and appropriate antibiotics. An OD600 measurement was taken the next day using a spectrophotometer to calculate the cell concentrations for controlling the seeding density of 2 mL liquid cultures in 14-mL tubes (Fisher Scientific, Cat. 149569C) at 1.5 × 108 cells/ml media. Anhydrotetracycline (aTc) inducer (final concentration of 200 nM) was added into the media to induce gene expression in planktonic microbes. Fluorescence from 100 μL of culture was measured every 24 h using a plate reader. For continuous culture, the cells were pelleted by centrifugation at 7000 RPM for 5 min. The spent media was removed and the cells were resuspended in fresh EZ-RDM.
2.1.7. Dynamic inducible expression in E. coli ELMs
A starter culture of E. coli MG1655 containing an aTc-inducible CRISPRa plasmid (pCK389.306) and an sfGFP reporter plasmid (pPC003) was grown overnight at 37 °C with 220 RPM orbital shaking in 3 mL of EZ-RDM with 0.2% glucose and antibiotics for selection. Microbial hydrogels were prepared and incubated according to the ELM fabrication protocol described above. aTc inducer (final concentration of 200 nM) was added into the surrounding EZ-RDM to induce gene expression of the hydrogel-encapsulated microbes. Fluorescence of both the hydrogels and the surrounding liquid media was measured every 24 h using the direct-gel measurement method described above. For dynamic induction experiments, the hydrogels were rinsed once with 100 μL EZ-RDM before measurement and serially diluted every 24 h into fresh EZ-RDM either with or without inducers depending on the induction scheme/cycle.
2.1.8. Dynamic inducible CRISPRa program for bioproduction in E. coli ELMs
A starter culture of E. coli MG1655 containing an aTc-inducible CRISPRa plasmid (pCK389.306) and a pteridine biosynthetic pathway plasmid (pCK014) was grown overnight at 37 °C with 220 RPM orbital shaking in 3 mL EZ-RDM with 0.2% glucose and appropriate selection antibiotics (100 μg/mL carbenicillin and/or 25 μg/mL chloramphenicol). Microbial hydrogels were prepared and incubated according to the ELM fabrication protocol above. aTc inducer (final concentration of 200 nM) was added into the surrounding EZ-RDM to induce gene expression in the hydrogel-encapsulated microbes. Pteridine fluorescence for both the hydrogels and the surrounding liquid media was measured every 24 h using the pteridine measurement method described below. For the dynamic induction experiments, the hydrogels in the cultures were rinsed once with 100 μL EZ-RDM and serially diluted every 24 h into fresh EZ-RDM with or without inducers, depending on the induction scheme.
2.1.9. Pteridine fluorescence measurement
End-point fluorescence measurements for both the hydrogels and the surrounding liquid media were made using a microplate reader equipped with a monochromator (TECAN infinite M1000). The following settings were used to measure the fluorescence of pteridines in hydrogels or from surrounding liquid media: excitation 340 nm ± 5 nm, emission 440 nm ± 5 nm, gain of 150. For directly measuring the fluorescence from hydrogels, the hydrogels were transferred into a 96-well clear flat-bottom plate (Corning 3916) with wells pre-filled with 100 μL EZ-RDM. To ensure uniform fluorescence readings, the cylindrical hydrogels were positioned at the center of the well with the flat circular surface facing the bottom of the plate. Two to four technical replicates per treatment were measured. Data were plotted using Prism (GraphPad). Logistic fits for expression rate determination were performed in Python (ver 3.8.5).
2.1.10. LC-MS for pteridine detection
The LC-MS analysis of culture supernatants was adapted from a prior method described in Kiattisewee et al. [15]. Culture supernatants were collected by centrifugation of the spent cultures (without the hydrogels) at 7000 RPM for 5 min. LC−MS analysis was completed using an Agilent UPLC 1290 system equipped with a QTOF-MS 6530 and a ZORBAX RRHT Extend-C18, 80 Å, 2.1 × 50 mm, 1.8 μm column and an electrospray ion source. LC conditions: solvent A—water with 0.1% formic acid; solvent B—methanol with 0.1% formic acid. Gradient: 2 min at 95%:5%:0.2 (A:B:flow rate in mL/min), 4 min ramp from 95%:5%:0.2 (A:B:flow rate in mL/min) to 70%:30%:0.2, 1 min ramp back to 95%:5%:0.2 (A:B:flow rate in mL/min), and 2 min post time. The MS acquisition (positive ion mode) scan covered m/z 80−3000. Analysis of pyruvoyl tetrahydropterin was performed with extracted m/z (M + H) 238.0935. Because an analytical standard for pyruvoyl tetrahydropterin was not commercially available, we report relative production levels as ion counts. We showed that the retention time of pyruvoyl tetrahydropterin (PT, 2.6 min) is significantly different than that of dihydrobiopterin (BH2, 1.2 min), previously reported in Kiattisewee et al., which shared the same chemical formula (C9H13N5O3) and molecular ion (M + H: m/z = 238.0935) (Supplementary Fig. S7a). The retention time of BH2. was determined using a commercially available standard (Cayman Chemical). Data were plotted using Prism (GraphPad).
2.2. S. cerevisiae ELM materials and methods
2.2.1. S. cerevisiae plasmid construction and preparation
All plasmids used in this study are listed in Supplementary Table S1. PCR fragments for assembly were amplified using Q5 High-Fidelity DNA Polymerase (New England BioLabs) for cloning using Q5 High-Fidelity 2X Master Mix (New England BioLabs). The assembled plasmids were transformed into electrocompetent E. coli DH10B cells (Thermo Fisher Scientific) and selected on LB agar plates supplemented with antibiotics (100 μg/mL Ampicillin or 50 μg/mL Kanamycin). Plasmids were purified using a GeneJET Plasmid Miniprep Kit (Thermo Fisher K0502) according to the manufacturer's protocol. Plasmid concentrations were quantified via spectrophotometry (Nanodrop 2000c, Cat. ND-2000C). Constructs were confirmed by Sanger sequencing, and the sequencing results were analyzed using Benchling. For proteinase A production, the plasmid PlTy3-GAL1-ScPEP4 was digested with XhoI for integration into the delta region and transformed into S. cerevisiae BY4741 cells via the Frozen EZ Yeast Transformation II Kit (Zymo Research). Successful transformants were selected on YPD agar plates supplemented with Geneticin G418 (200 μg/mL) and confirmed via colony PCR. For the production of betaxanthins, the plasmid p415-UraInt-pCup1-MjDOD-CYP76AD5 was linearized using NotI for integration into the Ura locus and transformed into protease-producing S. cerevisiae BY4741 cells via the Frozen EZ Yeast Transformation II Kit (Zymo Research) to yield strain spk05. Successful transformants were selected on SC agar plates lacking Leucine and confirmed via colony PCR as well as by the presence of fluorescence.
2.2.2. Conjugation of PEGDA to BSA
BSA-PEGDA conjugates were formulated according to a protocol described in Sanchez-Rexach et al. and optimized for SLA 3D printing [27,28]. To make 20 g of resin, 10 wt% PEGDA (Mn = 700) (Sigma Aldrich) was dissolved in 11.2 mL of deionized water. Then, 30 wt% BSA in powder form (Nova Biologics) was added slowly to the PEGDA solution to form BSA-PEGDA conjugates via an aza-Michael addition reaction. The resin mixture was stored overnight at 4 °C before use.
2.2.3. S. cerevisiae ELMs fabrication
A starter culture of S. cerevisiae spk05 cells was grown in 4 mL YPD media (10 g/L yeast extract, 20 g/L peptone, 20 g/L glucose) with 50 μg/mL of geneticin (G418) for counterselection against bacteria and incubated overnight at 30 °C with 220 RPM orbital shaking. OD600 measurements were taken the next day using a spectrophotometer to calculate the cell concentrations for controlling seeding density. For 20 g of BSA-PEGDA resin, 1 × 109 cells/mL culture was introduced to the formulation. Then, 0.075 wt% Ru(bpy)3Cl2 (Sigma Aldrich) and 0.24 wt% sodium persulfate (Sigma Aldrich) were sequentially added into the resin-microbial mixture as photoinitiators. ELM constructs were printed using a SLA 3D printer (Formlabs Form 2) in Open Mode using a layer height of 100 μm and photocured using a 405 nm violet laser (250 mW) with 140 μm laser spot size.
2.2.4. Dynamically-inducible bioproduction in S. cerevisiae ELMs
S. cerevisiae-laden BSA-PEGDA hydrogels were prepared according to the ELM fabrication protocol given above. The ELM constructs were placed in 50 mL culture tubes and incubated in 20 mL of YPD media at 30 °C with 220 RPM orbital shaking. The production of betaxanthins was induced by adding 0.5 mM Cu2SO4.5H2O inducer into the YPD media, while proteinase A production was induced by the addition of 1 wt% galactose to the YPD media. For the dynamic induction experiment, the culture media was replaced every 48 h with fresh YPD with one inducer depending on the induction scheme. Culture supernatants were collected for fluorescence measurements by centrifugation of the spent cultures (without the hydrogels) at 4400 RPM for 10 min.
2.2.5. Measurements of betaxanthins and proteinase A
Fluorescence measurements of culture supernatants were made using a microplate reader (Fluoroskan Ascent FL - Thermo Labsystems). The following settings were used for both the betaxanthins and the proteinase A fluorescence measurements: excitation at 485 nm and emission at 520 nm. For the betaxanthins measurements, 300 μL of supernatant were transferred to a 96-well clear flat-bottom microplate (Greiner Bio-One, UV-Star®). For the proteinase A measurements, an Amplite® Universal Fluorimetric Protease Activity Assay Kit (AAT Bioquest) was used to quantify the amount of functional proteinase A in 100 μL of supernatant. Three replicates were used. Data were plotted using Prism (GraphPad).
3. Results
3.1. Direct measurement of gene expression in ELMs
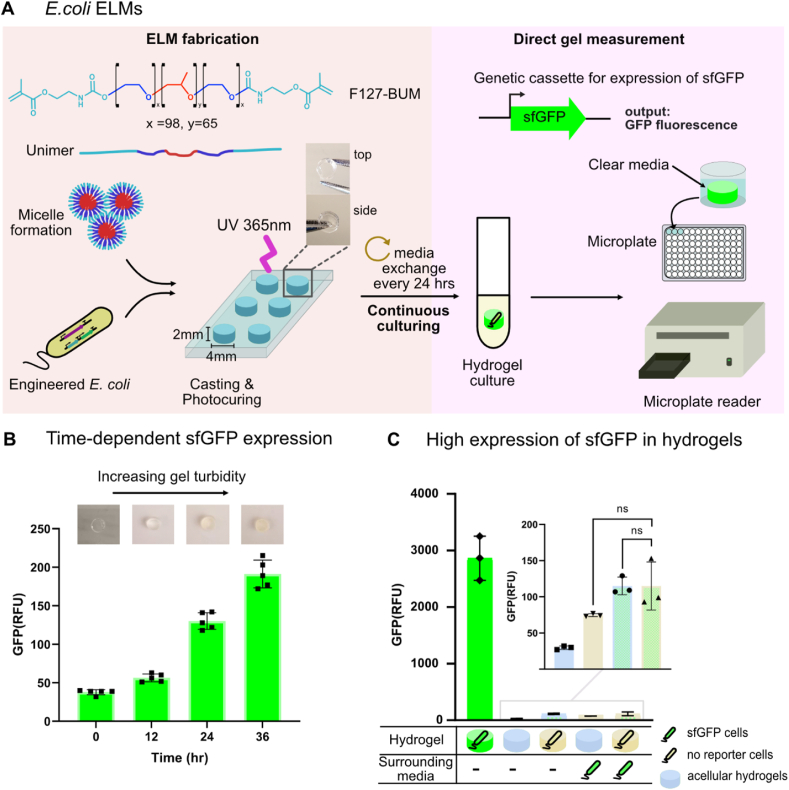
Fluorescent reporter protein expression has been an invaluable tool for quantifying the timing and strength of microbial gene expression [32,33]. For microbes encapsulated in hydrogels, fluorescent reporter gene expression can be monitored with microscopy [34] using relatively laborious multi-step sample preparation and image analysis workflows [35]. We developed a method for ‘direct gel measurement’ that uses a plate reader (Fig. 1a) to quantify gene expression, allowing a much larger number of hydrogel samples to be easily analyzed than with microscopy. Once we standardized the process of hydrogel casting and photocuring, we found that reporter protein expression could be measured simply by removing the hydrogels from the culture conditions, rinsing them once to remove cells that had leaked from the hydrogels [3], and then placing them into microplate wells.
Fig. 1.
E.coli ELMs for engineered on-demand bioproduction. a) Schematic of E. coli ELMs for expression and bioproduction in continuous culture. ELMs are fabricated by seeding Pluronic F127-BUM hydrogels with engineered E. coli cells. The mixture is transferred via syringe into a cylindrical silicone mold (diameter = 4 mm and height = 2 mm). The mold is placed in between glass slides and photocured with UV light (365 nm). Cast hydrogels are continuously cultured in EZ-RDM media. Fluorescent reporter gene expression from cells encapsulated in hydrogels is quantified using the direct-gel measurement method: cultured hydrogels are placed at the center of a microplate well pre-filled with fresh media for quantification using a microplate reader (see methods for additional details). b) sfGFP reporter expression from the pBT001-J2:RR2.CM.J23118 plasmid was measured at the time points indicated using the direct-gel measurement method. A gradual increase in ELM turbidity was observed with progressing culturing time (top inset). Bars represent the mean ± standard deviation from n = 5 technical replicates. c) High expression of sfGFP from encapsulated E. coli harboring pWS028.J3.J23106.sfGFP plasmid was measured using the direct-gel measurement method shown in a. Cast hydrogels and surrounding media were independently seeded with or without cells: acellular (light blue), no reporter (beige) or sfGFP-expressing (green) cells. Bars represent the mean ± standard deviation from n = 3 technical replicates. Statistical significance was assessed using a two-tailed unpaired Student's t-test (p > 0.05). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Initially, we applied our direct gel measurement workflow to E. coli ELMs fabricated from F127-BUM hydrogels using a hand-extrusion method [3]. F127-BUM is a polymer material with a temperature-dependent sol-gel transition at ∼17 °C, compatible with growth of microbial cells [[2], [3], [4],36]. This hydrogel is shear-thinning, making it easily extrudable [3]. F127-BUM hydrogels are also optically transparent, which permits the use of fluorescent proteins to monitor changes in gene expression of encapsulated microbes (Fig. 1a and b top inset). We observed poor consistency in measured reporter protein fluorescence levels, which we attributed to the highly irregular shapes of the hand-extruded hydrogels (Supplementary Fig. S2). By casting the hydrogels in silicone molds, we were able to enforce uniform shapes and dimensions (Fig. 1a). We measured constitutive (always ON) green fluorescent protein (sfGFP) expression across sets of E. coli ELMs cast in a mold with 2 mm thickness and two different diameters, 3 mm and 4 mm (Supplementary Fig. S3 and Fig. 1b).
We reasoned that maximizing coverage of the microplate well with the cast hydrogel would give more reliable fluorescence measurements. Consistent with this view, the larger 4 mm diameter hydrogels generated higher sfGFP signals (130 ± 15 versus 76 ± 11 relative fluorescence units, RFUs) and had a coefficient of variation that was 57% smaller than the 3 mm diameter hydrogels (Supplementary Table S2). Increasing the hydrogel diameter to 10 mm appeared to qualitatively reduce sfGFP fluorescence compared to the 4 mm diameter hydrogels, except when there was a corresponding increase in the volume of culture media (Supplementary Fig. S4). The 4 mm diameter hydrogels have a larger surface area-to-volume ratio than the 10 mm diameter hydrogels, which could improve mass transfer into the smaller diameter ELMs and improve gene expression [[37], [38], [39]]. Based on the results in this section, the rest of the E. coli ELMs experiments were conducted with the 4 mm diameter hydrogels.
As a final optimization of our direct gel measurement method, we evaluated the relative performance of sfGFP, as above, with a commonly employed red fluorescent protein variant, mRFP1. For this experiment, E. coli constitutively expressing one of the two fluorescent proteins were cast into 4 mm diameter hydrogels and placed into liquid media (Fig. 1b, Supplementary Fig. S3, and Supplementary Table S2). We observed a steady increase in fluorescence for hydrogels seeded with sfGFP-expressing E. coli, saturating over the course of 36 h (Fig. 1b). Across all time points, the mean coefficient of variation for sfGFP fluorescence was only 8.9 ± 0.4%, while the mRFP1 counterparts yielded a much larger mean coefficient of variation (35.0 ± 28.7%, Supplementary Table S2). The background fluorescence contributions from blank hydrogels (1.0% of the measured value for hydrogel-encapsulated E. coli expressing sfGFP), hydrogel-encapsulated cells with no reporter (1.6%), and free-floating planktonic cells (1.4%), are all very low (Fig. 1c). Thus, we conclude that the direct gel measurement method provides a reliable approach for quantifying gene expression in hydrogels, and that most of the measured fluorescence is generated by hydrogel-encapsulated cells expressing the reporter protein.
3.2. Persistence of genetic activity in ELM continuous culture
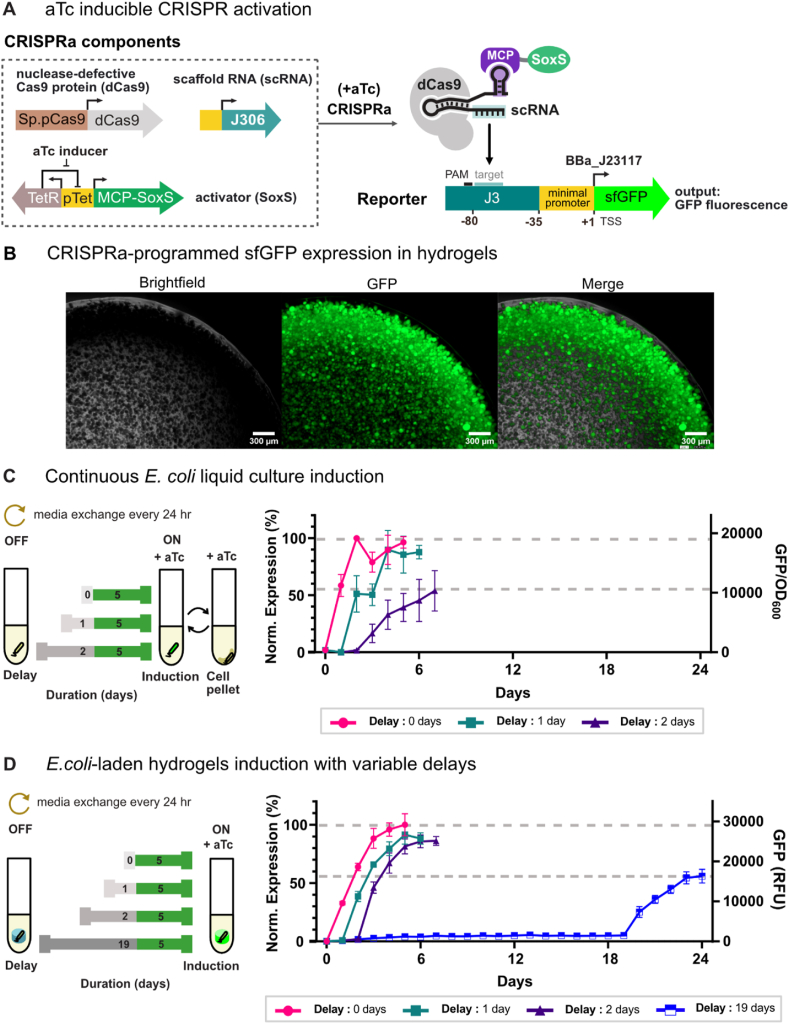
To evaluate the potential for input-responsive ELMs to function in long-term bioproduction applications, we first investigated whether encapsulated microbes can respond to external stimuli under continuous culture conditions. For these experiments, we implemented an inducible system for CRISPR-based transcriptional activation (CRISPRa) [[14], [15], [16],40] (Fig. 2a). In this system, the addition of anhydrous tetracycline (aTc) to the culture media induces heterologous expression of the biochemical machinery for CRISPRa: a nuclease defective Cas9 protein (dCas9), guide RNAs modified to contain a protein recruitment domain (scRNAs), and a SoxS transcriptional activator protein. The CRISPRa machinery generates scRNA-directed gene expression by recruiting the SoxS transcriptional activator to targeted promoters specified by the scRNA spacer sequence.
Fig. 2.
Persistence of genetic activity in E. coli ELMs. a) Schematic of an inducible CRISPR activation (CRISPRa) program for sfGFP expression. CRISPRa-directed expression of sfGFP uses nuclease-defective Cas 9 (dCas9) and a scaffold RNA (scRNA) that specifies a target site upstream of the engineered J3 promoter. scRNA (J306) is a modified guide RNA that includes a 3′ MS2 hairpin to recruit a transcriptional activator (SoxS) fused to the MS2 coat protein (MCP). The CRISPRa-programmed expression of sfGFP is induced upon the addition of anhydrotetracycline (aTc), which expresses MCP-SoxS from the pTet promoter. b) CRISPRa-programmed sfGFP expression in hydrogel-encapsulated E. coli. Microscopy images were taken from a two-day old continuously-cultured ELM (scale bar = 300 μm). c) Gene expression dynamics were evaluated in bacterial liquid continuous culture following variable delays of 0, 1, and 2 days (gray) before CRISPRa-programmed sfGFP expression was initiated and carried out for 5 days (green) by the addition of aTc inducer. Media was continuously removed and replenished every 24 h throughout the entire experiment, keeping the culture volume and composition constant via cell pelleting. sfGFP expression was tracked over time following continuous culture induction delays. Values represent the mean ± standard deviation from n = 3 replicates. d) Gene expression dynamics in ELMs were evaluated following variable delays of 0, 1, 2 and 19 days (gray) of continuous culture before initiation of CRISPRa-programmed sfGFP expression for 5 days (green). Media was replenished every 24 h throughout the entire experiment. The impact of induction delay duration on ELM gene expression levels was quantified by tracking sfGFP expression. Values represent the mean ± standard deviation from n = 5 technical replicates, except for 19-day delay samples, where n = 3 technical replicates. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
To track the capacity and dynamics of input-responsive gene expression over time, the sfGFP reporter was placed under the control of the inducible CRISPRa machinery (Fig. 2a). We could then compare the relative levels of sfGFP fluorescence when aTc induction was initiated at the beginning of the experiment (i.e., 0-day delay) with sfGFP outputs produced when induction was delayed for multiple days under continuous cultivation. We also fabricated input-responsive E. coli ELMs with inducible CRISPRa-programmed sfGFP expression and monitored the gene expression dynamics over multiple weeks of continuous cultivation using the method described in Section 3.1. Microscopy images confirmed that the inducible CRISPRa program was active in ELMs, as indicated by a large abundance of well-distributed, sfGFP-expressing microcolonies (Fig. 2b).
Free-floating, planktonic E. coli induced at the beginning of the experiment without delay reached about 50% of their maximum sfGFP expression after one day, and their highest levels of expression after two days (Fig. 2c). When induction was delayed for one day (1-day delay), no significant difference in expression capacity compared to the 0-day delay samples was observed. However, when the delay period was increased to two days (2-day delay), the expression capacity of the free-floating planktonic cells dropped significantly, and was only 57% of the maximum expression achieved by the 0-day delay samples (Fig. 2c).
Across the set of E. coli ELM samples, about 60% of the final endpoint expression level was reached two days after induction, with the highest levels of expression occurring three days after induction (Fig. 2d). E. coli ELMs had no discernable differences in the capacity or dynamics of sfGFP expression following 1 or 2 days of delayed induction compared to the 0-day delay sample (Fig. 2d, Supplementary Figs. S5a and S6). Even when the induction delay was extended to 19 days of continuous culture, the ELMs retained the capacity to express 56% of the maximum level achieved with the 0-day delay samples. By Day 2 of aTc induction, the instantaneous rate of sfGFP expression in the 19-day delayed induction samples was reduced 59% compared to the 0-day delay samples (Supplementary Fig. S6). Thus, we do see that long-term continuous cultivation affects the capacity and dynamics of ELM gene expression. Nonetheless, these results show that input-responsive genetic programs in E. coli ELMs can be activated for multiple weeks and that encapsulation greatly increases the capacity for long-term, inducible gene expression compared to cells grown in liquid suspension culture.
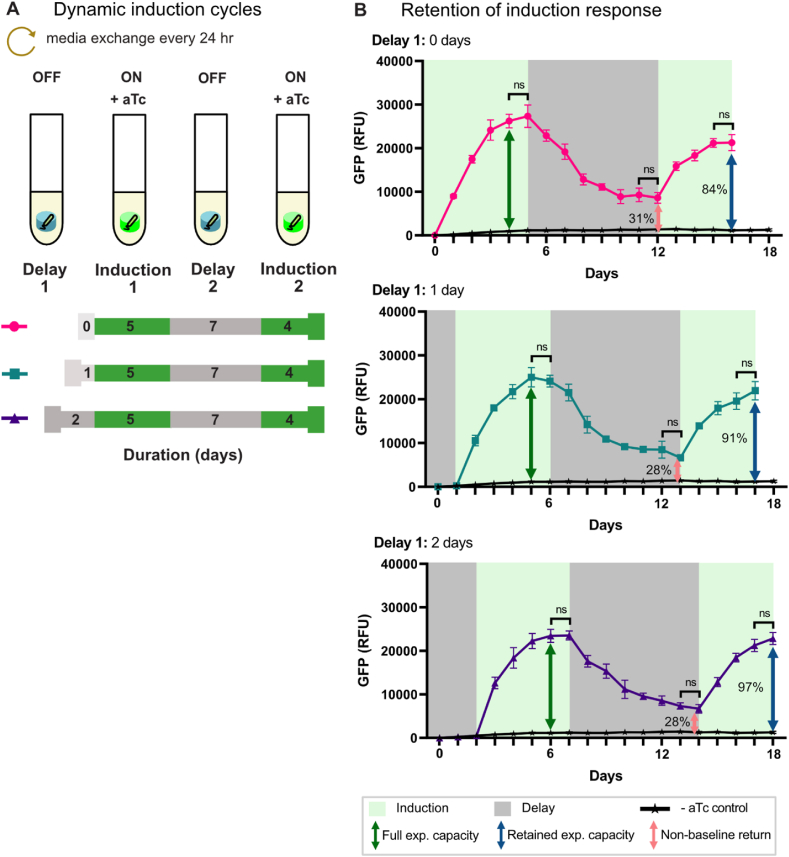
3.3. Gene expression capacity retained in multiple-cycles of induction
To function as inducible bioproduction platforms, ELMs must be able to generate heterologous gene expression levels sufficient for biosynthesis [4]. In the previous section, we showed that heterologous gene expression can be induced after multiple weeks of continuous culture. In this section, we investigated the expression dynamics of repeated heterologous gene expression induction events. The goal was to quantify the fraction of inducible gene expression capacity retained after multiple-cycles of induction [3,5,6]. These experiments consisted of two Delay periods (expression OFF), where ELMs were cultured in media alone, and two Induction periods (expression ON), where aTc was added to induce sfGFP gene expression from the CRISPRa program (Fig. 2, Fig. 3a). ELMs were cultured 0, 1 and 2 days without aTc during Delay period 1 (Delay 1). Induction period 1 (Induction 1) was continued until sfGFP gene expression reached a maximum plateau. The ELMs were then cultured in media without aTc until sfGFP fluorescence decreased and plateaued again at a relatively low level (Delay 2). Heterologous gene expression was re-initiated during Induction period 2 (Induction 2) by re-adding aTc to the media, and the cells were then cultured until saturating sfGFP fluorescence was obtained a second time.
Fig. 3.
Retention of induction response in E. coli ELMs. a) Schematic of dynamic induction cycles. Alternating delay (gray) and induction (green) and cycles were implemented by placing the ELMs into fresh media every 24 h. Varying delay lengths were applied to the first delay cycle (Delay 1). b) Retention of induction response following a subsequent induction cycle. Subplots (top to bottom): Delay 1 = no-delay, 1 day, 2-days, with uninduced, no aTc controls (Supplementary Fig. S5b) replotted on each subplot. Full expression capacity is defined as the maximum expression level achieved during Induction 1. Retained expression capacity and non-baseline returns for each Delay 1 length were calculated relative to the full expression capacity. Plotted values represent the mean ± standard deviation from n = 5 technical replicates, except for uninduced∗∗ samples (∗∗n = 2 technical replicates). Green and gray shades denote induction and delay periods, respectively. Statistical significance in sfGFP fluorescence at two consecutive time points was evaluated using a two-tailed unpaired Student's t-test (p > 0.05). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
For encapsulated cells induced without an initial delay in the first round (Delay 1 = 0 days), sfGFP expression increased from the initial baseline and reached maximum expression after four days of continuous culture (Fig. 3b, top). sfGFP fluorescence began decreasing as soon as aTc was withdrawn at the start of Delay 2. sfGFP fluorescence continued to decrease exponentially and plateaued after 6 days, reaching a baseline that was about 70% lower than the Induction 1 maximum. Increases in sfGFP expression were observed immediately upon aTc re-addition at the start of Induction 2. Within three days, sfGFP fluorescence reached a second maximum, equivalent to 84% of the Induction 1 maximum. No further increases in sfGFP expression were observed and the experiment was stopped at Day 16.
Samples cultured 1 and 2 days without aTc during Delay 1 exhibit expression dynamics very similar to the 0-day Delay 1 samples (Fig. 3b, middle and bottom). The Delay 1 = 1 day and Delay 1 = 2 day samples reach their first maxima within four days after initiating Induction 1. Exponential decreases in sfGFP fluorescence began when aTc was withdrawn at the start of Delay 2, and continued for 6–7 days, until second baselines about 70% lower than the respective Induction 1 maxima were reached. In both cases, inducing gene expression a second time (i.e., Induction 2) yields sfGFP maximum expression levels in 3–4 days that are within 10% of the Induction 1 maxima. Collectively, the data in this section show that encapsulated bacteria can generate high levels of heterologous gene expression following a second induction event and provide an initial characterization of inducible gene expression dynamics in this ELM system.
3.4. Dynamic inducible CRISPRa programs for bioproduction in ELMs
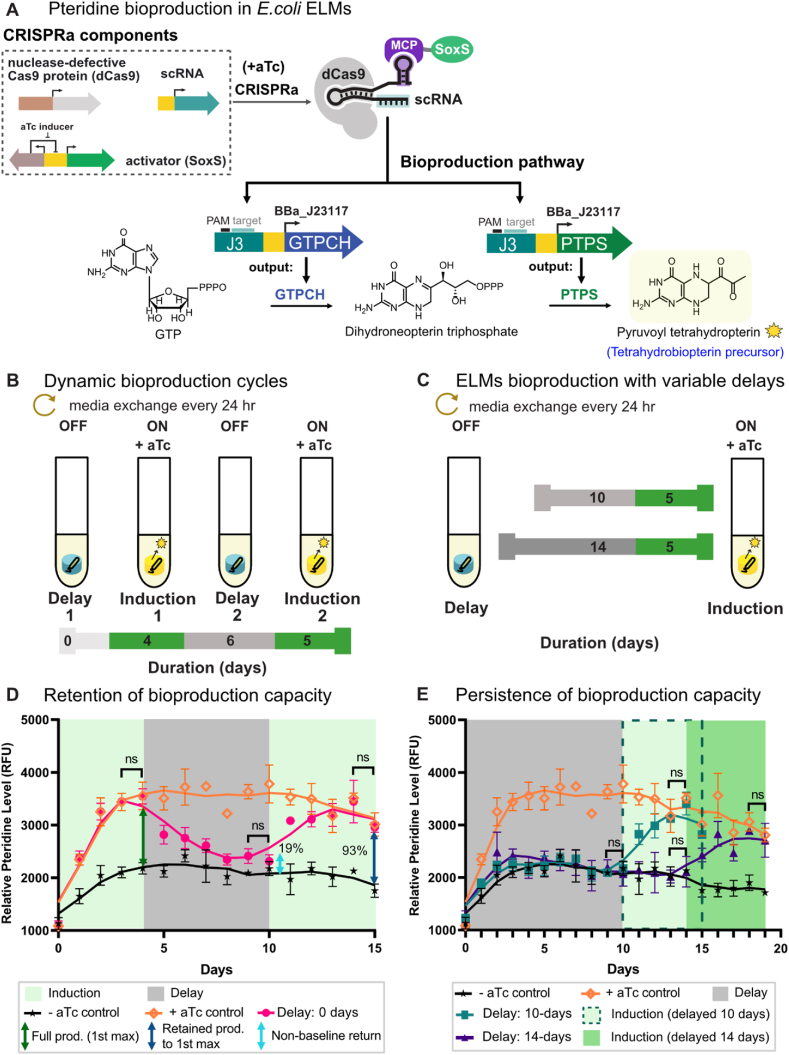
Motivated by our findings that ELMs have long-term, dynamically-responsive genetic activity, we examined whether the metabolic activity of encapsulated microbes can be dynamically controlled with inducible CRISPRa programs. We constructed an aTc-inducible CRISPRa program to activate expression of a two-gene metabolic pathway producing a pteridine derivative, pyruvoyl tetrahydropterin (PT) [30]. Pteridines constitute a large class of compounds with therapeutic potential as drugs for metabolic deficiencies, cancer, inflammation, and more [41]. In our system, aTc-induction activates expression of GTPCH from E. coli MG1655 and PTPS from M. alpina, which together catalyze the conversion of guanosine triphosphate (GTP) into PT (Fig. 4a). PT is fluorescent, which allowed us to monitor ELM production of PT by adapting an established pteridine detection assay [15,30], for use with our direct gel measurement method (Fig. 4 d,e and Supplementary Fig. S7b). Although the lack of readily-available commercial standards prevented quantification of PT production, the presence of PT produced by the ELMs and secreted into the media could be confirmed using LC-MS (Supplementary Fig. S7c).
Fig. 4.
Pyruvoyl tetrahydropterin synthesis in E. coli ELMs. a) Schematic of aTc-inducible CRISPRa programs controlling the expression of two enzymes (GTPCH and PTPS) catalyzing the conversion of guanosine triphosphate (GTP) into the pteridine derivative pyruvoyl tetrahydropterin (PT). b) Dynamic induction cycles on E. coli ELM for PT bioproduction. Bioproduction cycles alternating between periods of induction (production ON, green), and delay (production OFF, gray), were implemented under continuous culture conditions for 15 days. Media was replenished every 24 h. c) PT bioproduction capacity retained following a subsequent induction cycle. PT fluorescence from ELM constructs was measured every 24 h, with uninduced (no aTc) and induced (+aTc) controls included in the plot. Full production capacity is defined as the maximum pteridine level achieved in Induction 1. Retained production capacity and non-baseline returns were calculated relative to the full production capacity. Values represent the mean ± standard deviation from n = 3–4 technical replicates. Green and gray shades denote induction and delay periods, respectively. Statistical significance in pteridine fluorescence at two consecutive time points was evaluated using a two-tailed unpaired Student's t-test (p > 0.05). d) PT bioproduction in ELMs was measured following variable delays of 10 or 14 days (gray) of continuous culture before CRISPRa-programmed enzyme expressions were initiated by the addition of aTc inducer and carried out for 5 days (green). e) Persistence of bioproduction in ELMs was characterized as measured fluorescence from pyruvoyl tetrahydropterin every 24 h, with uninduced (no aTc) and induced (+aTc) controls included in the plot. Values represent the mean ± standard deviation from n = 3–4 technical replicates. Statistical significance in pteridine fluorescence at two consecutive time points was evaluated using a two-tailed unpaired Student's t-test (p > 0.05). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
We measured PT bioproduction across two programmed-bioproduction cycles that alternated between induced (production ON) and non-induced (production OFF) conditions. In the first instance, induction was initiated at the start of the experiment (Delay 1 = 0 days) and carried out for four days (Fig. 4b). PT production increased immediately upon aTc addition, reaching the first maximum production level on Day 3 (Fig. 4d). Turning production OFF by withdrawing aTc during Delay 2 caused an immediate decrease in PT production, as indicated by the immediate reduction in measured fluorescence. Six days after aTc withdrawal, PT fluorescence plateaued at 19% of the maximum fluorescence measured in Induction 1. aTc was added back to the culture media during Induction 2, and PT production increased again, reaching 93% of the first maximum within three days (Fig. 4d). The first and second maxima were indistinguishable from the fluorescence generated by always-ON ELMs constitutively-expressing the PT pathway (+aTc control), showing that dynamically-programmed cycles can access relatively high PT bioproduction levels.
We evaluated whether the inducible bioproduction phenotype was impacted by the length of time spent under continuous culture conditions by extending Delay 1 from 0 to 10 or 14 days (Fig. 4c). No significant PT fluorescence was observed until aTc was added to the culture media (Fig. 4e). At saturation, PT production levels for ELMs with Delay 1 = 10 or 14 days were similar to the Delay 1 = 0 days and always-ON (+ aTc control) samples. Overall, the inducible dynamics of the bioproduction cycles resemble the sfGFP gene expression dynamics presented in Section 3.3, indicated by immediate increase in the induced response, reaching a maximum after 3–4 days and returning to the baseline within 5 days upon inducer withdrawal. The results in this section show that E. coli ELMs are genetically and metabolically active under long-term culture conditions, and that the metabolic activities of encapsulated cells can be dynamically controlled with inducible CRISPRa programs.
3.5. Multi-input genetic programs in S. cerevisiae ELMs
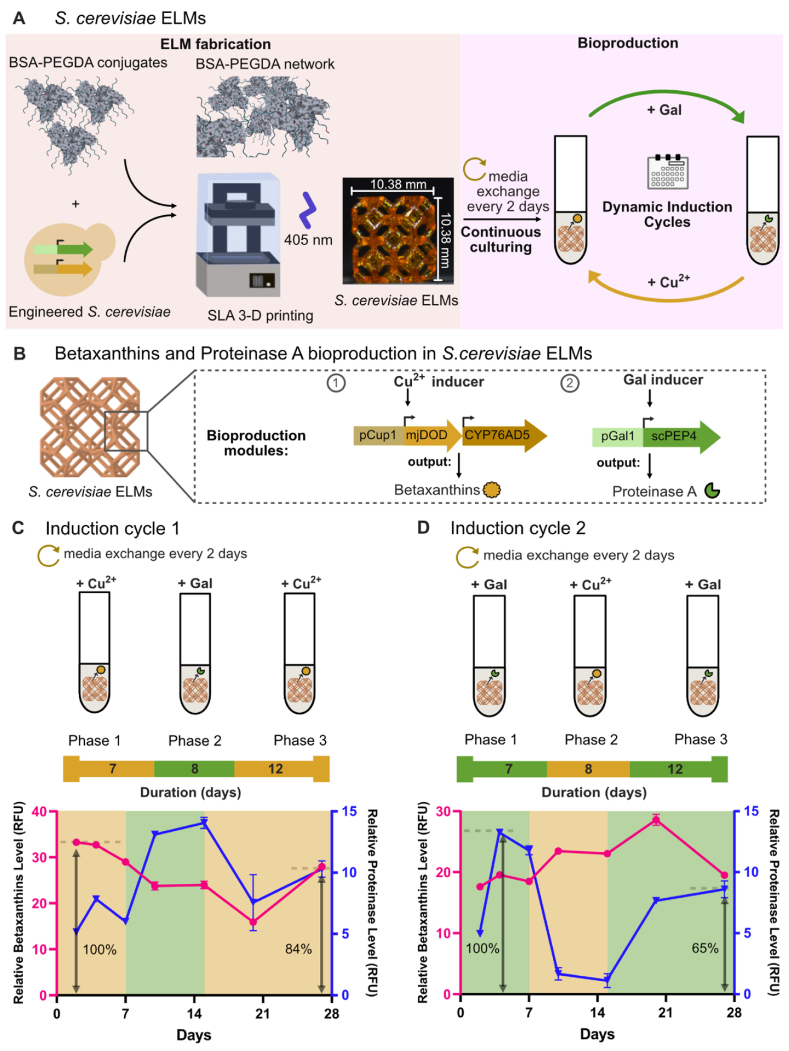
We investigated whether S. cerevisiae ELMs exhibit a similar capacity for long-term, dynamically-programmable gene expression as we observed for E. coli ELMs. The F127-BUM hydrogels employed in the sections above have been used to encapsulate yeast for sustained bioproduction [[2], [3], [4],36]. Here, we used a second type of acrylate-based hydrogel conjugated with bovine serum albumin (BSA) protein [27,28]. Recent applications of BSA-PEGDA have employed vat photopolymerization to fabricate microbial ELMs [28]. This fabrication method yields mechanically robust protein-based hydrogel networks that have good mechanical properties (moduli and toughness) and are also enzymatically degradable [27,28,42]. We fabricated yeast hydrogel-based ELMs by encapsulating S.cerevisiae spk05 cells in BSA - polyethylene glycol diacrylate (PEGDA) bioconjugates (Fig. 5a). The ELMs were printed using a stereolithographic apparatus (SLA) 3D printer and photocured as described in the methods [27,28].
Fig. 5.
Dynamic inducible bioproduction in S. cerevisiae ELMs. a) Schematic of bioproducing S.cerevisiae ELMs. These ELMs are fabricated by seeding BSA-PEGDA hydrogels with engineered S. cerevisiae. The ELMs are printed using a SLA 3-D printer and continuously cultured with media exchange every 2 days. For scheduled bioproduction, ELMs were dynamically induced to control expression of native and heterologous enzymes. b) Multi-input genetic programs in S. cerevisiae ELMs for multi-product biosynthesis: 1) copper (II)-inducible expression of dioxygenase enzyme (mjDOD gene product) with constitutive expression of P450 enzyme (CYP76AD5 gene product) for betaxanthins production, and 2) galactose-inducible expression of secreted proteinase A enzyme (scPEP4 gene). c) Bioproduction Induction cycle 1 consists of 3 phases (Phase 1 = 7 days, Phase 2 = 8 days, Phase 3 = 12 days). Top: Alternating induction by selective addition of copper (II) inducer (orange) in Phases 1 and 3, and galactose inducer (green) in Phase 2. Bottom: Time-dependent betaxanthins and proteinase A production levels. Values represent the mean ± standard deviation from n = 3 technical replicates. d) Bioproduction Induction cycle 2 with 3 phases (phase lengths are identical to c). Top: Alternating induction by selective addition of galactose inducer (green) in Phases 1 and 3, and copper (II) inducer in Phase 2. Bottom: Time-dependent betaxanthins and proteinase A production levels. Values represent the mean ± standard deviation from n = 3 technical replicates. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
The S. cerevisiae cells were programmed to express two different proteins, each under the control of a different inducible promoter. Expression of secreted proteinase A enzyme from the scPEP4 gene was placed under the control of a galactose-inducible promoter. Copper (II)-inducible expression of a dioxygenase enzyme (mjDOD gene) combined with constitutively-expressed P450 enzyme (CYP76AD5 gene) catalyzes the production of secreted betaxanthin pigment molecules [3] (Fig. 5b). With this design, inducers can be selectively added or withdrawn from the media to schedule increases or decreases, respectively, in production of the target molecules (Fig. 5c and d - top). In the experiments that follow, the relative production levels of proteinase A and betaxanthins were monitored over the course of 27 days under continuous cultivation (Fig. 5c and d - bottom). Specifically, we employed a fluorometric enzyme activity assay to measure relative proteinase A expression levels. Betaxanthins are formed from the spontaneous reactions of betalamic acid and heterogeneous primary amines within the cell [43]. We quantified the relative production of the resulting mixtures of betaxanthins by measuring the fluorescence of yellow pigment betaxanthin molecules [44].
We performed two different dynamic induction cycles, each consisting of three phases: Phase 1 = 7 days, Phase 2 = 8 days, and Phase 3 = 12 days. In the first dynamic induction cycle, copper (II) was added in the first phase, withdrawn and replaced with galactose in the second phase, and added again in the third phase while galactose was withdrawn (Fig. 5c). The second dynamic induction cycle was carried out in a similar manner, where galactose was added in the first phase, replaced with copper (II) in the second phase, and then re-added to the third phase where copper (II) was withdrawn (Fig. 5d).
In the first induction cycle, galactose-induced proteinase A production in Phase 2 was 1.5–2.1 fold higher than the levels measured in the absence of galactose (Phases 1 and 3) (Fig. 5c, bottom). The production of betaxanthins in Phase 3, induced with the addition of copper (II) on the 15th day of the experiment, reached 84% of the maximum level of induced betaxanthins production observed in Phase 1. In the second induction cycle, relatively large differences in proteinase A production were observed, and 65% of the maximum Phase 1 expression was also achieved by Day 27 in Phase 3 (Fig. 5d, bottom). The transitions between the phases for betaxanthins production were less distinct in the second induction cycle compared to the first induction cycle. Nonetheless, clear variations in the levels of measured betaxanthins could be seen at the endpoints of the phases. Thus, as with the E. coli ELMs, the S. cerevisiae ELMs retain the capacity to express multiple genes in multi-week continuous cultures. Moreover, these results constitute a proof-of-concept demonstration that multi-input, multi-output gene expression programs can be implemented in ELMs to permit switching between multiple bioproducts.
4. Discussion and conclusions
Bioproduction using microbes engineered with synthetic biology provides a promising approach for manufacturing unnatural and natural chemicals [45]. ELMs fabricated by encapsulating microbes in biocompatible hydrogels can produce a wide range of value-added biochemicals. Recent demonstrations have included E. coli ELMs secreting >150 mg/L of l-DOPA after only 22 h and S. cerevisiae ELMs secreting 1.5–1.7 g/L of 2,3-butanediol after only 48 h of culturing [3]. ELMs can also exhibit bioproduction phenotypes that are difficult to achieve with free-floating, planktonic cells. Planktonic cells in continuous culture tend to suffer from genetic instability, which generally prevents their use for long-term bioproduction [46]. Previous work has shown that ELMs have long-term metabolic activity that can be sustained over multiple rounds of culturing and preservation, permitting long-term, high-yield bioproduction [[2], [3], [4],10,11,47]. We found that encapsulated microbes have persistent genetic activity that can be induced multiple weeks longer than in planktonic cells (Fig. 2, Fig. 3). These results have immediate implications for the further development of ELMs as platforms for on-demand bioproduction and for understanding how microbes interact with materials to generate novel phenotypes.
We successfully applied our dynamic CRISPR-based expression programs to a two-gene heterologous pathway in E. coli ELMs for PT bioproduction, a direct precursor of tetrahydropterin (BH4) for phenylketonuria treatment [15,30,48]. PT production from E. coli input-responsive ELMs can be repeatedly cycled ON/OFF and delayed by multiple weeks of culturing before turning the production ON via induction. We obtained a bioproduction profile similar to that of the fluorescent reporter gene expression, showing both persistence and retention of bioproduction activity upon production cycling and delay. In S. cerevisiae ELMs, induction of multiple genetic programs could be scheduled one at a time, permitting the switch between bioproduction of betaxanthins and proteinase A over multiple weeks of continuous cultures. The successful extension of long-term programmable gene expression from bacterial to yeast ELMs suggests that long-term, multi-week genetic activity may be a general property of microbe-laden ELMs.
ELMs deployed as portable bioreactors for chemical bioproduction could provide localized, on-demand access to commodity and specialty chemicals needed for health, biodefense and consumer goods manufacturing [3,4,49]. This capability would be especially impactful when unpredicted market fluctuations, such as during a pandemic, create acute supply shortages, or when supply chain access is limited, such as during military operations or space exploration. With current technologies, product cycling, or the process of stopping and then re-starting production, requires either discarding spent culture and re-inoculating with fresh cells, or large-volume cold-storage that can preserve cellular bioproduction capacity [50]. While both approaches are easy to achieve at a laboratory scale, the cost becomes expensive at larger production scales of dozens or even thousands of liters [51]. By controlling induction timing in input-responsive ELMs, cost-effective product cycling could be implemented. The ability to switch between multiple products simply by inducing a separate genetic program within the encapsulated cells adds entirely new flexibility to ELM bioproduction.
At present, the exact physiological state of hydrogel-encapsulated microbes remains uncertain [52]. It is well-understood that planktonic microbial cells grown in liquid culture typically enter stationary phase after two days and suppress translation for survival, resulting in the decline of protein synthesis levels [53]. By comparison, we found that continuously-cultured ELMs can express heterologous gene products at high levels for at least 2.5 weeks. We observed increasing turbidity of continuously-cultured ELMs over time, indicative of growth and colonization within the hydrogel microstructures (Fig. 1b - top inset, Supplementary Fig. S8) [54]. At this point, we cannot quantify how much of the observed increases in gene expression arise from increases in the number of living cells within the ELM. Nonetheless, taken together, these results are consistent with the idea that at least some portion of the microbes within these materials are not in a dormant state akin to the stationary or death phase [55].
Hydrogel formulation is known to influence microbial cellular phenotypes in ELM cultures [7,52]. The hydrogel matrix asserts mechanical forces [56] to the encapsulated microbes, resulting in growth rate changes relative to planktonic cells in liquid cultures [57,58]. Variations in oxygen transport into the hydrogel matrix can affect the size of the encapsulated colony [8], the expression and maturation of chromophores [23,59], and microbial metabolism [2]. Compared to planktonic cells, immobilized bacteria trapped in biofilms [60] and in rigid 3D extracellular matrix [61] experience lower fluxes through the tricarboxylic acid (TCA) cycle. Because the TCA cycle is used for energy generation in oxygen-rich conditions, lower TCA cycle fluxes could lead to decreased immobilized cell growth rates [58,62]. Similarly, immobilized yeast are known to have less flux through the glycolytic pathway than planktonic cells and experience higher rates of internal carbohydrate reserve utilization in starvation conditions [63]. Future work to link hydrogel environments and the resulting functional properties could enable rational design of engineered microenvironments to obtain the desired cellular responses for enhanced bioproduction phenotypes.
On-going developments continue to expand the synthetic biology toolset for implementing dynamic gene regulation in microbes [64]. CRISPR-Cas transcriptional control has emerged as a modular and programmable route for implementing genetic circuitry in diverse systems, including prokaryotes, cell-free systems, and here in ELMs [[14], [15], [16]]. Functional CRISPRa tools require expression of multiple components — dCas9, activation domain, and guide RNAs — to regulate downstream genetic circuitry (Fig. 2, Fig. 4a). Our demonstration that these tools can be applied in ELMs suggests that more complex, multi-gene programs combining CRISPR activation and inhibition (CRISPRa/i) can be active in ELM systems. These next-generation tools would allow us to build complex regulatory programs by simultaneously activating or repressing multiple genes within engineered pathways and the host genome [16]. We expect that developing programmable ELMs capable of coordinating dynamic control over complex multi-gene programs will result in entirely new capabilities for high-yield bioproduction [65], including rapid product switching and cycling, product diversification, and in situ process monitoring [[66], [67], [68]]. Creating dynamically-programmable, input-responsive ELMs will also be valuable for studying how encapsulation affects cellular physiology and for synthesizing ELMs as smart biosensors [69], as devices for the in-situ delivery of biomedicines [6,70], or as environmental sentinels [18].
Credit author statement
Widianti Sugianto: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing - original draft, and Writing - review & editing. Gokce Altin-Yavuzarslan: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Validation, and Visualization. Benjamin I. Tickman: Conceptualization, Methodology, Validation, and Writing - original draft. Cholpisit Kiattisewee: Data curation, Formal analysis, Methodology, Visualization, and Writing - review & editing. Shuo-Fu Yuan, Sierra M. Brooks, and Jitkanya Wong: Methodology and Resources. Hal S. Alper: Conceptualization, Funding acquisition, and Supervision. Alshakim Nelson: Conceptualization, Funding acquisition, and Supervision. James M. Carothers: Conceptualization, Funding acquisition, Project administration, Supervision, Writing - original draft, and Writing - review & editing.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: James M. Carothers is an advisor to Wayfinder Biosciences.
Acknowledgements
This work was supported by US National Science Foundation (NSF) award EFMA-2029249 AM01 (to J.M.C., A.N., and H.S.A.) and NSF award CBET 1844152 (to J.M.C.).
We thank members of the Carothers, Nelson, Alper, and Marchand groups at UW and UT-Austin for materials, technical assistance, and comments on the manuscripts.
Part of this work (microscopy) was conducted at the Molecular Analysis Facility, a National Nanotechnology Coordinated Infrastructure site at the University of Washington which is supported in part by the National Science Foundation (grant NNCI-1542101), the University of Washington, the Molecular Engineering & Sciences Institute, the Clean Energy Institute, and the National Institutes of Health.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mtbio.2023.100677.
Contributor Information
Alshakim Nelson, Email: alshakim@uw.edu.
James M. Carothers, Email: jcaroth@uw.edu.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Liu Y., Nielsen J. Recent trends in metabolic engineering of microbial chemical factories. Curr. Opin. Biotechnol. 2019;60:188–197. doi: 10.1016/j.copbio.2019.05.010. [DOI] [PubMed] [Google Scholar]
- 2.Butelmann T., Priks H., Parent Z., Johnston T.G., Tamm T., Nelson A., Lahtvee P.-J., Kumar R. Metabolism control in 3D-printed living materials improves fermentation. ACS Appl. Bio Mater. 2021;4:7195–7203. doi: 10.1021/acsabm.1c00754. [DOI] [PubMed] [Google Scholar]
- 3.Johnston T.G., Yuan S.-F., Wagner J.M., Yi X., Saha A., Smith P., Nelson A., Alper H.S. Compartmentalized microbes and co-cultures in hydrogels for on-demand bioproduction and preservation. Nat. Commun. 2020;11:563. doi: 10.1038/s41467-020-14371-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yuan S.-F., Brooks S.M., Nguyen A.W., Lin W.-L., Johnston T.G., Maynard J.A., Nelson A., Alper H.S. Bioproduced proteins on demand (Bio-POD) in hydrogels using Pichia pastoris. Bioact. Mater. 2021;6:2390–2399. doi: 10.1016/j.bioactmat.2021.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saha A., Johnston T.G., Shafranek R.T., Goodman C.J., Zalatan J.G., Storti D.W., Ganter M.A., Nelson A. Additive manufacturing of catalytically active living materials. ACS Appl. Mater. Interfaces. 2018;10:13373–13380. doi: 10.1021/acsami.8b02719. [DOI] [PubMed] [Google Scholar]
- 6.Sankaran S., Becker J., Wittmann C., del Campo A. Optoregulated drug release from an engineered living material: self-replenishing drug depots for long-term, light-regulated delivery. Small. 2019;15 doi: 10.1002/smll.201804717. [DOI] [PubMed] [Google Scholar]
- 7.Bhusari S., Sankaran S., del Campo A. 2022. Regulating Bacterial Behavior within Hydrogels of Tunable Viscoelasticity. 2022.01.06.475183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.B. Pabst, B. Pitts, E. Lauchnor, P.S. Stewart, Gel-Entrapped Staphylococcus aureus Bacteria as Models of Biofilm Infection Exhibit Growth in Dense Aggregates, Oxygen Limitation, Antibiotic Tolerance, and Heterogeneous Gene Expression , Antimicrob. Agents Chemother.. 60 (n.d.) 6294–6301. 10.1128/AAC.01336-16. [DOI] [PMC free article] [PubMed]
- 9.Martínez-Calvo A., Bhattacharjee T., Bay R.K., Luu H.N., Hancock A.M., Wingreen N.S., Datta S.S. Morphological instability and roughening of growing 3D bacterial colonies. Proc. Natl. Acad. Sci. USA. 2022;119 doi: 10.1073/pnas.2208019119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rathore S., Desai P.M., Liew C.V., Chan L.W., Heng P.W.S. Microencapsulation of microbial cells. J. Food Eng. 2013;116:369–381. doi: 10.1016/j.jfoodeng.2012.12.022. [DOI] [Google Scholar]
- 11.John R.P., Tyagi R.D., Brar S.K., Surampalli R.Y., Prévost D. Bio-encapsulation of microbial cells for targeted agricultural delivery. Crit. Rev. Biotechnol. 2011;31:211–226. doi: 10.3109/07388551.2010.513327. [DOI] [PubMed] [Google Scholar]
- 12.Hartline C.J., Schmitz A.C., Han Y., Zhang F. Dynamic control in metabolic engineering: theories, tools, and applications. Metab. Eng. 2021;63:126–140. doi: 10.1016/j.ymben.2020.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dinh C.V., Prather K.L. Layered and multi-input autonomous dynamic control strategies for metabolic engineering. Curr. Opin. Biotechnol. 2020;65:156–162. doi: 10.1016/j.copbio.2020.02.015. [DOI] [PubMed] [Google Scholar]
- 14.Fontana J., Dong C., Kiattisewee C., Chavali V.P., Tickman B.I., Carothers J.M., Zalatan J.G. Effective CRISPRa-mediated control of gene expression in bacteria must overcome strict target site requirements. Nat. Commun. 2020;11:1618. doi: 10.1038/s41467-020-15454-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kiattisewee C., Dong C., Fontana J., Sugianto W., Peralta-Yahya P., Carothers J.M., Zalatan J.G. Portable bacterial CRISPR transcriptional activation enables metabolic engineering in Pseudomonas putida. Metab. Eng. 2021;66:283–295. doi: 10.1016/j.ymben.2021.04.002. [DOI] [PubMed] [Google Scholar]
- 16.Tickman B.I., Burbano D.A., Chavali V.P., Kiattisewee C., Fontana J., Khakimzhan A., Noireaux V., Zalatan J.G., Carothers J.M. Multi-layer CRISPRa/i circuits for dynamic genetic programs in cell-free and bacterial systems. Cell Syst. 2022;13:215–229.e8. doi: 10.1016/j.cels.2021.10.008. [DOI] [PubMed] [Google Scholar]
- 17.Fontana J., Sparkman-Yager D., Zalatan J.G., Carothers J.M. Challenges and opportunities with CRISPR activation in bacteria for data-driven metabolic engineering. Curr. Opin. Biotechnol. 2020;64:190–198. doi: 10.1016/j.copbio.2020.04.005. [DOI] [PubMed] [Google Scholar]
- 18.Tang T.-C., Tham E., Liu X., Yehl K., Rovner A.J., Yuk H., de la Fuente-Nunez C., Isaacs F.J., Zhao X., Lu T.K. Hydrogel-based biocontainment of bacteria for continuous sensing and computation. Nat. Chem. Biol. 2021;17:724–731. doi: 10.1038/s41589-021-00779-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi W.S., Kim M., Park S., Lee S.K., Kim T. Patterning and transferring hydrogel-encapsulated bacterial cells for quantitative analysis of synthetically engineered genetic circuits. Biomaterials. 2012;33:624–633. doi: 10.1016/j.biomaterials.2011.09.069. [DOI] [PubMed] [Google Scholar]
- 20.Liu X., Tang T.-C., Tham E., Yuk H., Lin S., Lu T.K., Zhao X. Stretchable living materials and devices with hydrogel–elastomer hybrids hosting programmed cells. Proc. Natl. Acad. Sci. USA. 2017;114:2200–2205. doi: 10.1073/pnas.1618307114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Graham A.J., Dundas C.M., Partipilo G., Mahfoud Ie M., FitzSimons T., Rinehart R., Chiu D., Tyndall A.E., Rosales A.M., Keitz B.K. 2021. Transcriptional Regulation of Synthetic Polymer Networks. [DOI] [Google Scholar]
- 22.González L.M., Mukhitov N., Voigt C.A. Resilient living materials built by printing bacterial spores. Nat. Chem. Biol. 2020;16:126–133. doi: 10.1038/s41589-019-0412-5. [DOI] [PubMed] [Google Scholar]
- 23.Sankaran S., del Campo A. Optoregulated protein release from an engineered living material. Adv. Biosyst. 2019;3 doi: 10.1002/adbi.201800312. [DOI] [PubMed] [Google Scholar]
- 24.Rivera-Tarazona L.K., Campbell Z.T., Ware T.H. Stimuli-responsive engineered living materials. Soft Matter. 2021;17:785–809. doi: 10.1039/D0SM01905D. [DOI] [PubMed] [Google Scholar]
- 25.Llorente B., Williams T.C., Goold H.D., Pretorius I.S., Paulsen I.T. Harnessing bioengineered microbes as a versatile platform for space nutrition. Nat. Commun. 2022;13:6177. doi: 10.1038/s41467-022-33974-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Selas Castiñeiras T., Williams S.G., Hitchcock A.G., Smith D.C. E. coli strain engineering for the production of advanced biopharmaceutical products. FEMS (Fed. Eur. Microbiol. Soc.) Microbiol. Lett. 2018;365:fny162. doi: 10.1093/femsle/fny162. [DOI] [PubMed] [Google Scholar]
- 27.Sanchez-Rexach E., Smith P.T., Gomez-Lopez A., Fernandez M., Cortajarena A.L., Sardon H., Nelson A. 3D-Printed bioplastics with shape-memory behavior based on native bovine serum albumin. ACS Appl. Mater. Interfaces. 2021;13:19193–19199. doi: 10.1021/acsami.0c22377. [DOI] [PubMed] [Google Scholar]
- 28.G. Altin-Yavuzarslan, S.M. Brooks, S.-F. Yuan, J.O. Park, H.S. Alper, A. Nelson, Additive Manufacturing of Engineered Living Materials with Bio-Augmented Mechanical Properties and Resistance to Degradation , Adv. Funct. Mater.. n/a (n.d.) 2300332. 10.1002/adfm.202300332. [DOI] [PMC free article] [PubMed]
- 29.Nielsen J., Keasling J.D. Engineering cellular metabolism. Cell. 2016;164:1185–1197. doi: 10.1016/j.cell.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 30.Ehrenworth A.M., Sarria S., Peralta-Yahya P. Pterin-dependent mono-oxidation for the microbial synthesis of a modified monoterpene indole alkaloid. ACS Synth. Biol. 2015;4:1295–1307. doi: 10.1021/acssynbio.5b00025. [DOI] [PubMed] [Google Scholar]
- 31.Millik S.C., Dostie A.M., Karis D.G., Smith P.T., McKenna M., Chan N., Curtis C.D., Nance E., Theberge A.B., Nelson A. 3D printed coaxial nozzles for the extrusion of hydrogel tubes toward modeling vascular endothelium. Biofabrication. 2019;11 doi: 10.1088/1758-5090/ab2b4d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meyerovich M., Mamou G., Ben-Yehuda S. Visualizing high error levels during gene expression in living bacterial cells. Proc. Natl. Acad. Sci. USA. 2010;107:11543–11548. doi: 10.1073/pnas.0912989107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andersen J.B., Sternberg C., Poulsen L.K., Bjørn S.P., Givskov M., Molin S. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl. Environ. Microbiol. 1998;64:2240–2246. doi: 10.1128/AEM.64.6.2240-2246.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnston T.G., Fillman J.P., Priks H., Butelmann T., Tamm T., Kumar R., Lahtvee P.-J., Nelson A. Cell-laden hydrogels for multikingdom 3D printing. Macromol. Biosci. 2020;20 doi: 10.1002/mabi.202000121. [DOI] [PubMed] [Google Scholar]
- 35.Donius A.E., Bougoin S.V., Taboas J.M. FRET imaging in three-dimensional hydrogels. JoVE. 2016 doi: 10.3791/54135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.S.M. Brooks, K.B. Reed, S.-F. Yuan, G. Altin-Yavuzarslan, R. Shafranek, A. Nelson, H.S. Alper, Enhancing long-term storage and stability of engineered living materials through desiccant storage and trehalose treatment , Biotechnol. Bioeng.. n/a (n.d.). 10.1002/bit.28271. [DOI] [PubMed]
- 37.Ogbonna J.C., Matsumura M., Kataoka H. Effective oxygenation of immobilized cells through reduction in bead diameters: a review. Process Biochem. 1991;26:109–121. doi: 10.1016/0032-9592(91)80025-K. [DOI] [Google Scholar]
- 38.Liu Y., Xia X., Liu Z., Dong M. The next frontier of 3D bioprinting: bioactive materials functionalized by bacteria. Small. 2023;19 doi: 10.1002/smll.202205949. [DOI] [PubMed] [Google Scholar]
- 39.Cui Z., Feng Y., Liu F., Jiang L., Yue J. 3D bioprinting of living materials for structure-dependent production of hyaluronic acid. ACS Macro Lett. 2022;11:452–459. doi: 10.1021/acsmacrolett.2c00037. [DOI] [PubMed] [Google Scholar]
- 40.Dong C., Fontana J., Patel A., Carothers J.M., Zalatan J.G. Synthetic CRISPR-Cas gene activators for transcriptional reprogramming in bacteria. Nat. Commun. 2018;9:2489. doi: 10.1038/s41467-018-04901-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carmona-Martínez V., Ruiz-Alcaraz A.J., Vera M., Guirado A., Martínez-Esparza M., García-Peñarrubia P. Therapeutic potential of pteridine derivatives: a comprehensive review. Med. Res. Rev. 2019;39:461–516. doi: 10.1002/med.21529. [DOI] [PubMed] [Google Scholar]
- 42.Narupai B., Smith P.T., Nelson A. 4D printing of multi-stimuli responsive protein-based hydrogels for autonomous shape transformations. Adv. Funct. Mater. 2021;31 doi: 10.1002/adfm.202011012. [DOI] [Google Scholar]
- 43.Grewal P.S., Modavi C., Russ Z.N., Harris N.C., Dueber J.E. Bioproduction of a betalain color palette in Saccharomyces cerevisiae. Metab. Eng. 2018;45:180–188. doi: 10.1016/j.ymben.2017.12.008. [DOI] [PubMed] [Google Scholar]
- 44.Khan M.I., Giridhar P. Plant betalains: chemistry and biochemistry. Phytochemistry. 2015;117:267–295. doi: 10.1016/j.phytochem.2015.06.008. [DOI] [PubMed] [Google Scholar]
- 45.Lee S.Y., Kim H.U., Chae T.U., Cho J.S., Kim J.W., Shin J.H., Kim D.I., Ko Y.-S., Jang W.D., Jang Y.-S. A comprehensive metabolic map for production of bio-based chemicals. Nat Catal. 2019;2:18–33. doi: 10.1038/s41929-018-0212-4. [DOI] [Google Scholar]
- 46.Xie D. Continuous biomanufacturing with microbes — upstream progresses and challenges. Curr. Opin. Biotechnol. 2022;78 doi: 10.1016/j.copbio.2022.102793. [DOI] [PubMed] [Google Scholar]
- 47.Liu X., Chung Y.-K., Yang S.-T., Yousef A.E. Continuous nisin production in laboratory media and whey permeate by immobilized Lactococcus lactis. Process Biochem. 2005;40:13–24. doi: 10.1016/j.procbio.2003.11.032. [DOI] [Google Scholar]
- 48.Michals-Matalon K., Bhatia G., Guttler F., Tyring S.K., Matalon R. Response of phenylketonuria to tetrahydrobiopterin. J. Nutr. 2007;137:1564S–1567S. doi: 10.1093/jn/137.6.1564S. [DOI] [PubMed] [Google Scholar]
- 49.Tsolas S.D., Hasan M.M.F. Survivability-aware design and optimization of distributed supply chain networks in the post COVID-19 era. J. Adv. Manufact. Process. 2021;3 doi: 10.1002/amp2.10098. [DOI] [Google Scholar]
- 50.Prakash O., Nimonkar Y., Shouche Y.S. Practice and prospects of microbial preservation. FEMS (Fed. Eur. Microbiol. Soc.) Microbiol. Lett. 2013;339:1–9. doi: 10.1111/1574-6968.12034. [DOI] [PubMed] [Google Scholar]
- 51.Katoh S., Horiuchi J., Yoshida F. second ed. Wiley; 2015. Biochemical Engineering: A Textbook for Engineers, Chemists and Biologists. [Google Scholar]
- 52.Priks H., Butelmann T., Illarionov A., Johnston T.G., Fellin C., Tamm T., Nelson A., Kumar R., Lahtvee P.-J. Physical confinement impacts cellular phenotypes within living materials. ACS Appl. Bio Mater. 2020;3:4273–4281. doi: 10.1021/acsabm.0c00335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pletnev P., Osterman I., Sergiev P., Bogdanov A., Dontsova O. Survival guide: Escherichia coli in the stationary phase. Acta Nat. 2015;7:22–33. [PMC free article] [PubMed] [Google Scholar]
- 54.Jeong Y., Kong W., Lu T., Irudayaraj J. Soft hydrogel-shell confinement systems as bacteria-based bioactuators and biosensors. Biosens. Bioelectron. 2023;219 doi: 10.1016/j.bios.2022.114809. [DOI] [PubMed] [Google Scholar]
- 55.Jeong Y., Irudayaraj J. Hierarchical encapsulation of bacteria in functional hydrogel beads for inter- and intra- species communication. Acta Biomater. 2023 doi: 10.1016/j.actbio.2023.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dufrêne Y.F., Persat A. Mechanomicrobiology: how bacteria sense and respond to forces. Nat. Rev. Microbiol. 2020;18:227–240. doi: 10.1038/s41579-019-0314-2. [DOI] [PubMed] [Google Scholar]
- 57.Kandemir N., Vollmer W., Jakubovics N.S., Chen J. Mechanical interactions between bacteria and hydrogels. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-29269-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shao X., Mugler A., Kim J., Jeong H.J., Levin B.R., Nemenman I. Growth of bacteria in 3-d colonies. PLoS Comput. Biol. 2017;13 doi: 10.1371/journal.pcbi.1005679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Müller J., Jäkel A.C., Richter J., Eder M., Falgenhauer E., Simmel F.C. Bacterial growth, communication, and guided chemotaxis in 3D-bioprinted hydrogel environments. ACS Appl. Mater. Interfaces. 2022 doi: 10.1021/acsami.1c20836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wan N., Wang H., Ng C.K., Mukherjee M., Ren D., Cao B., Tang Y.J. Bacterial metabolism during biofilm growth investigated by 13C tracing. Front. Microbiol. 2018;9 doi: 10.3389/fmicb.2018.02657. https://www.frontiersin.org/articles/10.3389/fmicb.2018.02657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Han Y., Jiang N., Xu H., Yuan Z., Xiu J., Mao S., Liu X., Huang J. Extracellular matrix rigidities regulate the tricarboxylic acid cycle and antibiotic resistance of three-dimensionally confined bacterial microcolonies. Adv. Sci. 2023;10 doi: 10.1002/advs.202206153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smet C., Van Derlinden E., Mertens L., Noriega E., Van Impe J.F. Effect of cell immobilization on the growth dynamics of Salmonella Typhimurium and Escherichia coli at suboptimal temperatures. Int. J. Food Microbiol. 2015;208:75–83. doi: 10.1016/j.ijfoodmicro.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 63.Doran P.M., Bailey J.E. Effects of immobilization on the nature of glycolytic oscillations in yeast. Biotechnol. Bioeng. 1987;29:892–897. doi: 10.1002/bit.260290711. [DOI] [PubMed] [Google Scholar]
- 64.Jang S., Lim H.G., Yang J., Seo S.W., Jung G.Y. In: Current Developments in Biotechnology and Bioengineering. Singh S.P., Pandey A., Du G., Kumar S., editors. Elsevier; 2019. Chapter 5 - synthetic regulatory tools to engineer microbial cell factories for chemical production; pp. 115–141. [DOI] [Google Scholar]
- 65.Fontana J., Voje W.E., Zalatan J.G., Carothers J.M. Prospects for engineering dynamic CRISPR–Cas transcriptional circuits to improve bioproduction. J. Ind. Microbiol. Biotechnol. 2018;45:481–490. doi: 10.1007/s10295-018-2039-z. [DOI] [PubMed] [Google Scholar]
- 66.Silver E.A., Pyke D.F., Thomas D.J. fourth ed. CRC Press; Boca Raton: 2016. Inventory and Production Management in Supply Chains. [DOI] [Google Scholar]
- 67.Rodrigo-Navarro A., Sankaran S., Dalby M.J., del Campo A., Salmeron-Sanchez M. Engineered living biomaterials. Nat. Rev. Mater. 2021;6:1175–1190. doi: 10.1038/s41578-021-00350-8. [DOI] [Google Scholar]
- 68.X. Liu, M.E. Inda, Y. Lai, T.K. Lu, X. Zhao, Engineered Living Hydrogels , Adv. Mater.. n/a (n.d.) 2201326. 10.1002/adma.202201326. [DOI] [PMC free article] [PubMed]
- 69.Liu X., Yuk H., Lin S., Parada G.A., Tang T.-C., Tham E., de la Fuente-Nunez C., Lu T.K., Zhao X. 3D printing of living responsive materials and devices. Adv. Mater. 2018;30 doi: 10.1002/adma.201704821. [DOI] [PubMed] [Google Scholar]
- 70.Zhu Y., Wang Z., Bai L., Deng J., Zhou Q. Biomaterial-based encapsulated probiotics for biomedical applications: current status and future perspectives. Mater. Des. 2021;210 doi: 10.1016/j.matdes.2021.110018. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.