Abstract
Background
Heat shock protein beta-1 (HSPB1) is a crucial biomarker for pathological processes in various cancers. However, the clinical value and function of HSPB1 in breast cancer has not been extensively explored. Therefore, we adopted a systematic and comprehensive approach to investigate the correlation between HSPB1 expression and clinicopathological features of breast cancer, as well as determine its prognostic value. We also examined the effects of HSPB1 on cell proliferation, invasion, apoptosis, and metastasis.
Methods
We investigated the expression of HSPB1 in patients with breast cancer using The Cancer Genome Atlas and immunohistochemistry. Chi-squared test and Wilcoxon signed-rank test were used to examine the relationship between HSPB1 expression and clinicopathological characteristics.
Results
We observed that HSPB1 expression was significantly correlated with the stage N, pathologic stages, as well as estrogen and progesterone receptors. Furthermore, high HSPB1 expression resulted in a poor prognosis for overall survival, relapse-free survival, and distant metastasis-free survival. Multivariable analysis showed that patients with poor survival outcomes had higher tumor, node, metastasis, and pathologic stages. Pathway analysis of HSPB1 and the altered neighboring genes suggested that HSPB1 is involved in the epithelial-to-mesenchymal transition. Functional analysis revealed showed that transient knockdown of HSPB1 inhibited the cell migration/invasion ability and promoted apoptosis.
Conclusions
HSPB1 may be involved in breast cancer metastasis. Collectively, our study demonstrated that HSPB1 has prognostic value for clinical outcomes and may serve as a therapeutic biomarker for breast cancer.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12885-023-10983-3.
Keywords: HSPB1, Breast cancer, Prognostic biomarker, Cell proliferation, Metastasis
Background
Breast cancer is one of the most common malignant tumors in women. The incidence rate is rising each year, which has a significant negative impact on women's health and quality of life [1–3]. Despite advancements in current treatment techniques, including surgery and chemotherapy, the outcome of breast cancer remains unsatisfactory [4]. Therefore, finding novel molecular indicators for breast cancer are urgently required.
Heat shock protein 27, also referred to as heat shock protein beta-1 (HSPB1), belongs to the small HSP family. Its purpose is to stop or prevent cellular proteins from denaturing or unfolding in response to stress or elevated temperatures [5]. HSPB1 regulates many pathological processes in cancer, including drug resistance, apoptosis, and metastasis [6–8]. HSPB1 is considered an important molecular target for tumor growth inhibition and apoptosis induction [9] and is vital in the regulation of tumorigenesis and the development of some cancers [10–12]. For example, esophageal squamous cell carcinoma with overexpression of HSPB1 has a worse prognosis [13]. Upregulation of HSPB1 is related to poor overall survival in hepatocellular carcinoma and promotes tumorigenesis [14]. Further research has revealed that HSPB1 knockout led to a decrease in insulin levels and expression of growth factor-like binding protein 2, which may promote the proliferation and metastasis of hepatocellular carcinoma [15]. Moreover, HSPB1 expression is related to the epithelial-mesenchymal transition (EMT). Through the upregulation of Snail1 and PRRX1, HSPB1 overexpression promotes EMT and drives the migration and invasion of salivary adenoid cystic carcinoma cells [16]. Additionally, HSPB1 interferes with bone metastasis in breast cancer [17] and modulates the PTEN levels in human breast cancer cells [18]. The potential relevance of HSPB1 and its underlying mechanisms in the development of breast cancer have yet to be elucidated.
While the relationship between HSPB1 and tumorigenesis has been demonstrated, limited evidence has illustrated the clinical significance and function of HSPB1 in breast cancer. In this study, we used a systematic and comprehensive approach to assess the relationship between HSPB1 expression and clinicopathological characteristics in patients with breast cancer. We also determined the prognostic value of HSPB1. Additionally, we examined the effects of HSPB1 on cell proliferation, invasion, apoptosis, and metastasis. We believe that this study has identified HSPB1 as a therapeutic target for breast cancer.
Methods
Patient tissue specimens
A total of 18 cancerous tissues (n = 15) and normal adjacent tissue (n = 3) were collected from patients undergoing surgery for breast cancer at the First Affiliated Hospital of Shenzhen University from April 2019 to February 2021. These samples were taken from tissues removed surgically. In addition, we collected 20 samples of in situ breast cancer and lung metastases. The tissues were quickly stored at − 80 °C until use, according to the Tumor Bank protocol [19]. Written informed consent was provided by all participants.
HSPB1 expression and univariate/multivariate regression analysis
We obtained data with the relevant clinical characteristics from The Cancer Genome Atlas (TCGA). TCGA is an open-access resource, that includes 33 types of cancer from approximately 20,000 patients [20]. These data were used for HSPB1 expression analysis as well as preliminary analyses of univariate logistic regression and multivariate Cox regression to examine prognostic factors and clinical outcomes, including overall survival (OS), disease-specific survival (DSS), and progress free interval (PFI). Patients with breast cancer were examined to determine whether HSPB1 expression was correlated with clinicopathological variables to better understand the prognostic value of HSPB1 based on status (tumor or normal), patient age (≤ 60 or > 60 years of age), tumor (T) stage (T1, T2, T3, or T4), node (N) stage (N0, N1, N2, or N3), metastasis (M) stage (M0 or M1), pathological stage (stage I, stage II, or stage III), estrogen receptors (ER; negative or positive), progesterone receptors (PR; negative or positive), and human epidermal growth factor receptor 2 (HER2; negative or positive). We examined the association between HSPB1 expression and clinical characteristics using Chi-square and Wilcoxon signed-rank tests.
Cell culture, cell transfection and quantitative real-time PCR
We obtained six cell lines from the Chinese Academy of Sciences, including human normal mammary epithelial cells: MCF10A, and all breast cancer cell line types: luminal A: T47D, luminal B: BT474, HER-2 positive: SK-BR-3, Triple-negative A: MDA-MB-453, Triple-negative B: MDA-MB-231. A mix of 10% fetal bovine serum (FBS) and RPMI Medium 1640 (DMEM; Gibco, Carlsbad, CA, USA) was used to culture each cell line. Two sequences targeting HSPB1 siRNAs were synthesized by Gene Pharma (Suzhou, China): si-1: 5′-GCCAUUAUUAGAGACCUCATT-3′ and si-2: 5′-UCACCAUCCCAGUCACCUUTT-3′. Cells were transfected using Lipofectamine 2000 (Invitrogen, United States). The Pure Link RNA Mini Kit (Invitrogen, United States) was used to extract total RNA. Quantitative real-time PCR (RT-PCR) was performed under the following conditions: 95 °C for 5 min, and 40 cycles of 95 °C for 30 s, 60 °C for 45 s and 72 °C for 30 s. Primers were purchased from Gene Pharma (Suzhou, China) using the following sequences: HSPB1, 5′-CTCTGAAGGGTCCGAAGTGAT-3′ and 5′-ATTCCTGTGGTGGTCCAAAAC-3′; Actin: 5′-CACCATTGGCAATGAGCGGTTC-3′ and 5′-AGGTCTTTGCGGATGTCCACGT-3′.
Western blot and immunohistochemical analysis
Samples were tested for total protein concentration using the bicinchoninic acid method. A 12% SDS-PAGE gel was used to separate 20 µg of each protein sample. We then transferred the separated proteins onto polyvinylidene fluoride membranes for primary antibody detection. The membranes were then incubated for 1 h at room temperature with alkaline phosphatase-conjugated secondary antibodies (Roche, Switzerland), followed by three washes with Tris-buffered saline with Tween 20 for 15 min. Primary antibodies against HSPB1 (1: 1000 dilution), glyceraldehyde-3-phosphate dehydrogenase (1: 5,000 dilution), vimentin (1: 1000 dilution), N-cadherin (1: 1000 dilution) and E-cadherin (1: 1000 dilution) were used. All the antibodies used in this study were purchased from CST (USA). The labeled proteins were visualized via chemiluminescent imaging.
Immunohistochemical assays were performed on human breast cancer and adjacent tissues. All fresh tissues were cryopreserved before being processed into histology blocks for sectioning. Briefly, after deparaffinization and rehydration, antigen retrieval was performed by heating 5μm -thick sections at 95 °C for 15 min in 10 mM citrate buffer (pH 6.0). After incubation with the primary antibody against HSPB1 (1: 1000 dilution) for 12 h, the sections were counterstained with hematoxylin to label the nuclei. Two pathologists blindly evaluated and scored all stained sections to determine the degree of immunostaining.
Kaplan–Meier plotter and PrognoScan database analysis
We analyzed the correlation between HSPB1 transcription levels and OS, relapse-free survival (RFS), and distant metastasis-free survival (DMFS) in patients with breast cancer using the Kaplan–Meier plotter database (http://www.kmplot.com/) [21]. For all tests, the hazard ratios (HR) with 95% confidence intervals were defined as significant at p < 0.05. The sample sizes of the high- and low-HSPB1 groups were 939 and 940 for OS, 2,465 and 2,464 for RFS, and 1,383 and 1,382 for DMFS, respectively.
The PrognoScan database (http://dna00.bio.kyutech.ac.jp/PrognoScan/index.html) [22] was used to analyze the correlation between HSPB1 expression and prognosis in patients with breast cancer, including OS, RFS, and DMFS (these data from the GSE1456-GPL96 and GSE2990 cohorts). Samples were divided into groups of high or low HSPB1 expression levels, with the lowest 50% and the top 50% considered low and high expression, respectively.
Analysis of HSPB1-interacting genes and proteins
The gene–gene interaction network and protein–protein interaction network of HSPB1 were constructed using GeneMANIA (http://www.genemania.org) and STRING online (https://string-db.org/) [23, 24]. To verify the correlation between HSPB1 and the altered neighboring genes, a breast cancer cohort from the TCGA database was analyzed using bc-GenExMinerv 4.8 (http://bcgenex.ico.unicancer.fr/BC-GEM/GEM-Accueil.php?js=1) [25] and RNA-seq data (N = 4,712). Using the GSCALite database (http://bioinfo.life.hust.edu.cn/web/GSCALite/) [26], pathways were analyzed for the altered neighboring genes.
Cell viability and colony formation assay
We tested the viability of cells using a Cell Counting Kit-8 (CCK8; Dojindo, Kumamoto, Japan) in accordance with the manufacturer's protocol. In 96-well plates, culture media (100μL) and 3,000 cells were plated. After culturing for 0 to 72 h at 37 °C in a humidified incubator with 5% CO2, each well was incubated for 2 h with 10 μL of CCK-8 reagent. Measurement of absorbance at 450 nm and assessment of proliferation ability were conducted using a spectrophotometer (Bio-Rad Laboratories, CA, USA). The colony-forming potential of all cancer cells was assessed by seeding them onto six-well plates at a density of 300 cells/well. After 10–14 days of culture, cells were removed from the medium, fixed with 4% paraformaldehyde, stained with 0.1% crystal violet, and washed thrice with phosphate buffered saline (PBS) before imaging.
Wound healing and Transwell® invasion assays
Transfected SK-BR-3 and MDA-MB-231 cells (si-NC and siHSPB1) were seeded and cultured in an FBS containing medium at 37 °C until 100% confluence. A straight scratch was made through each culture using a 200 μL pipette tip. After wounding, cells were washed three times with PBS and then replenished with fresh serum-free media. The wound area was photographed immediately (t = 0 h) and after 72 h (t = 72 h) using an inverted microscope.
Transwell® chambers coated with Matrigel were filled with a 200 μL cell suspension in serum-free medium. The lower chamber was filled with 600 μL of complete medium. Subsequently, the plates were incubated for 24 h. By optical microscopy, we analyzed the migrated cells by fixing them with 4% paraformaldehyde and staining them with 0.1% crystal violet.
Analyses of cell apoptosis
A culture plate was seeded with cells and grown to 70% confluence. For the cell apoptosis assay, SK-BR-3 and MDA-MB-231 cells were stained with 5 µL Annexin V-fluorescein isothiocyanate and 5 µL propidium iodide. A FACS-can flow cytometer and Cell Quest software were used for the follow-up analysis (Becton Dickinson, USA).
Statistical analysis
All statistical analyses were conducted using R version 3.6.3. The Wilcoxon rank sum test was used to analyze the difference in HSPB1 expression between normal (n = 113) and tumor tissues (n = 1,119). We examined the relationship between HSPB1 expression and clinicopathological characteristics using the Chi-squared test and Wilcoxon signed-rank test. We used Kaplan–Meier to evaluate the prognostic value of HSPB1 expression. All tests were defined as significant at p < 0.05.
Results
HSPB1 expression increased in patients with breast cancer
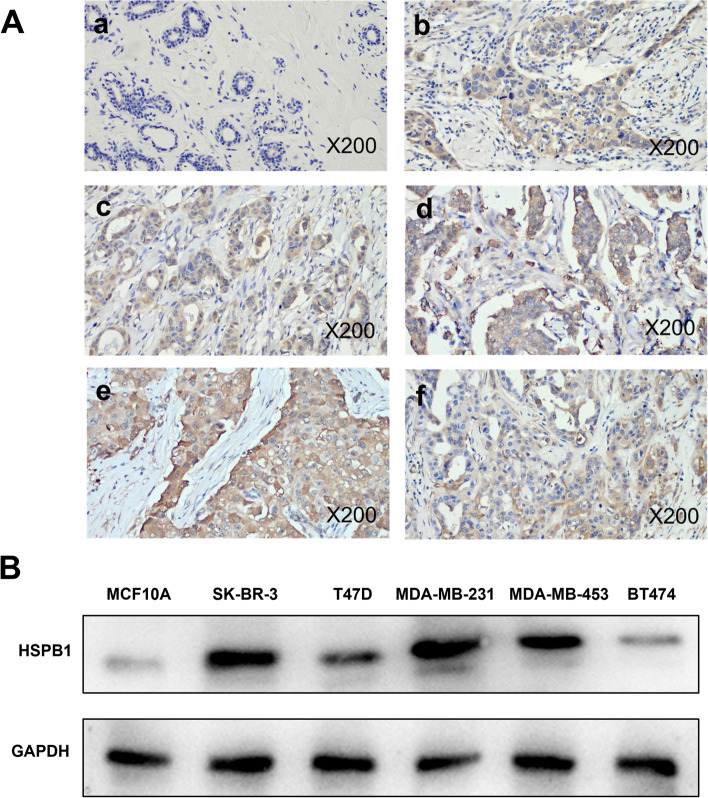
A higher expression of HSPB1 in breast cancer tissue when comparing the immunohistochemical analysis in of 18 cancerous and non-cancerous sample pairs (Fig. 1A). In addition, HSPB1 expression was increased in the breast cancer cell lines SK-BR-3, T47D, MDA-MB-231, MDA-MB-453, and BT474, especially in SK-BR-3 and MDA-MB-231 cells, compared with that in normal breast MCF10A cells (Fig. 1B). The findings collectively support the hypothesis that HSPB1 is highly expressed in patients with breast cancer.
Fig. 1.
HSPB1 expression in breast cancer tissues and cells. A Immunohistochemical analysis of HSPB1 expression in breast tumor tissues (b-f) and normal tissues (a) (200 × magnification,). B Western blots showing the expression of HSPB1 protein in six breast cancer cell lines. The blots were cut prior to hybridisation with antibodies during blotting
HSPB1 expression and clinical variables of patients with breast cancer
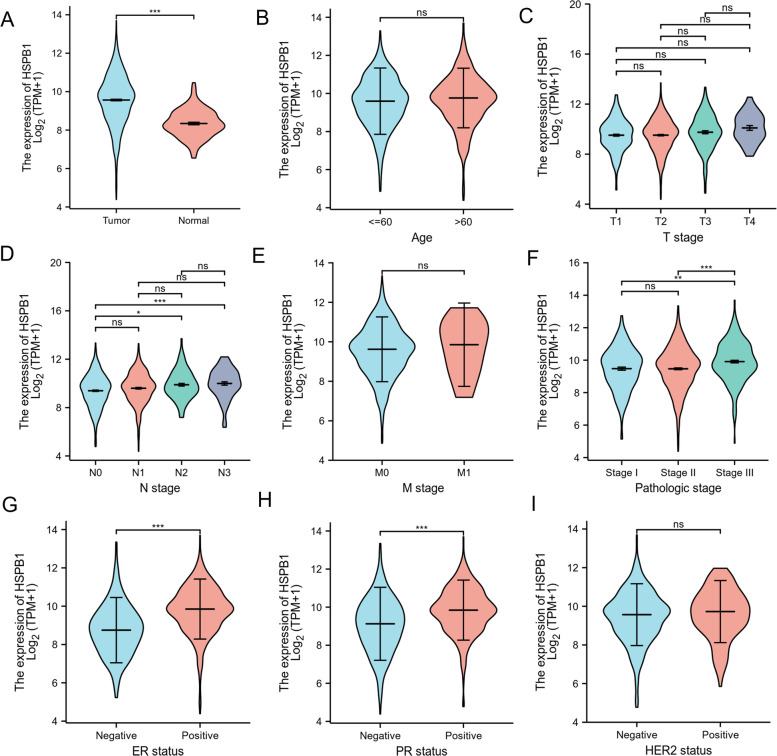
To better understand the relevance and underlying mechanisms of HSPB1 expression in breast cancer, we summarized the distribution of clinicopathological information of patients in HSPB1 high expression group and HSPB1 low expression group (Table 1). The data showed that the distribution of breast cancer patients in the high expression group and the low expression group was significantly different in N stage (p = 0.003), pathologic stage (p = 0.032), ER status (p < 0.001), and PR status (p < 0.001), but there was no significant difference between HSPB1 expression and age, T stage, M stage, and HER2 status (All p > 0.05). We further examined the relationship between HSPB1 expression and clinical characteristics, including the patient status (tumor or normal) (Fig. 2A), age (≤ 60 and > 60) (Fig. 2B), T stage (T1, T2, T3, or T4) (Fig. 2C), N stage (N0, N1, N2, or N3) (Fig. 2 D), M stage (M0 or M1) (Fig. 2E), pathologic stage (stage I, stage II, or stage III) (Fig. 2F), ER (negative or positive) (Fig. 2G), PR (negative or positive) (Fig. 2H). HER2 (negativeo or psitive) (Fig. 2I). The results showed the analysis of the pathologic stages showed that HSPB1 expression significantly increased in stages II and III compared with stage I (p < 0.001). Additionally, based on ER, PR, and HER2 expression, we observed that HSPB1 expression was significantly higher in receptor-positive samples than in receptor-negative samples (ER: p < 0.001; PR: p < 0.001).
Table 1.
The relationship between the high and low expression of HSPB1 and different clinical indicators in patients with breast cancer
| Characteristic | Low expression of HSPB1 | High expression of HSPB1 | p |
|---|---|---|---|
| n | 541 | 542 | |
| Age, n (%) | 0.133 | ||
| < = 60 | 313 (28.9%) | 288 (26.6%) | |
| > 60 | 228 (21.1%) | 254 (23.5%) | |
| T stage, n (%) | 0.409 | ||
| T1 | 143 (13.2%) | 134 (12.4%) | |
| T2 | 321 (29.7%) | 308 (28.5%) | |
| T3 | 62 (5.7%) | 77 (7.1%) | |
| T4 | 15 (1.4%) | 20 (1.9%) | |
| N stage, n (%) | 0.003 | ||
| N0 | 282 (26.5%) | 232 (21.8%) | |
| N1 | 175 (16.4%) | 183 (17.2%) | |
| N2 | 52 (4.9%) | 64 (6%) | |
| N3 | 26 (2.4%) | 50 (4.7%) | |
| M stage, n (%) | 0.403 | ||
| M0 | 469 (50.9%) | 433 (47%) | |
| M1 | 8 (0.9%) | 12 (1.3%) | |
| Pathologic stage, n (%) | 0.032 | ||
| Stage I | 91 (8.6%) | 90 (8.5%) | |
| Stage II | 327 (30.8%) | 292 (27.5%) | |
| Stage III | 102 (9.6%) | 140 (13.2%) | |
| Stage IV | 7 (0.7%) | 11 (1%) | |
| ER status, n (%) | < 0.001 | ||
| Negative | 187 (18.1%) | 53 (5.1%) | |
| Positive | 330 (31.9%) | 463 (44.7%) | |
| PR status, n (%) | < 0.001 | ||
| Negative | 225 (21.8%) | 117 (11.3%) | |
| Positive | 290 (28%) | 398 (38.5%) | |
| HER2 status, n (%) | 0.337 | ||
| Negative | 295 (40.6%) | 263 (36.2%) | |
| Positive | 78 (10.7%) | 79 (10.9%) |
The p-values indicate significant differences between the low and the high expression of HSPB1 in clinical variables. For age, median (interquartile range), and other clinical variables, the Wilcoxon rank sum test and the chi-square test were used to calculate the p-values, respectively
TMN stage was according to the seventh edition of the Guidelines for the American Journal of Critical Care. ER estrogen receptor, PR progesterone receptor, HER2 human epidermal growth factor receptor. Bold values indicate that p < 0.05
Fig. 2.
Association between the HSPB1 expression levels and clinical characteristics in patients with breast cancer. The relationship between HSPB1 expression and clinical characteristics, including the patient A status (tumor or normal), B age (≤ 60 and > 60), C T stage (T1, T2, T3, or T4), D N stage (N0, N1, N2, or N3), E M stage (M0 or M1), F pathologic stage (stage I, stage II, or stage III), G estrogen receptor (ER; negative or positive), H progesterone receptor (PR; negative or positive). I human epidermal growth factor receptor 2 (HER2; negative or positive)
Validation of the prognostic value of HSPB1 in patients with breast cancer
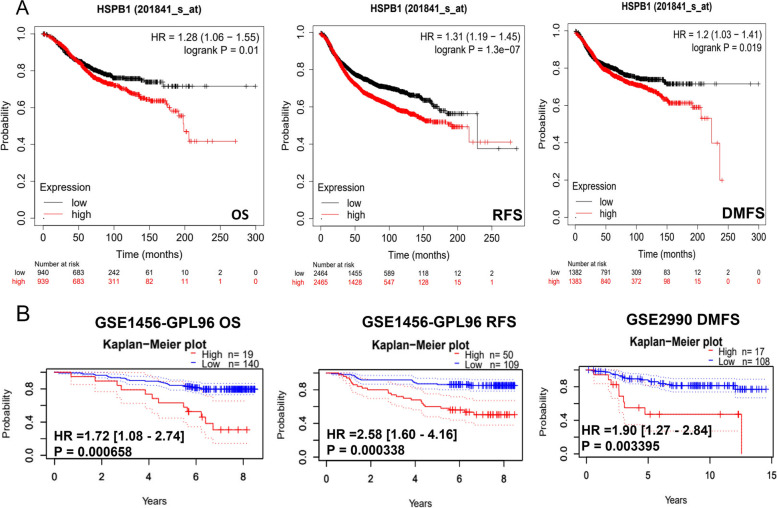
As HSPB1 expression levels are intimately related to breast cancer progression, Kaplan–Meier survival curves were used to compare the expression levels of HSPB1 with prognosis (Fig. 3A). The expression of HSPB1 is significantly correlated with poor prognosis (OS: HR = 1.28, 95% CI: 1.06–1.55, p = 0.01; RFS: HR = 1.31, 95% CI: 1.19–1.45, p = 1.3e-07; DMFS: HR = 1.2, 95% CI: 1.03–1.41, p = 0.019). A prognostic database, PrognoScan, was employed to test the clinical outcome effect of HSPB1. As shown in Fig. 3B, in the GSE1456-GPL96 and GSE2990 cohorts, high-HSPB1 were significantly worse than low-HSPB1 on OS, RFS, and DMFS.
Fig. 3.
An analysis of the prognostic value of HSPB1. A Survival curves for overall survival (OS), relapse Free Survival (RFS), and distant metastasis free survival (DMFS) using the Kaplan –Meier plotter. B Survival curves for OS, RFS, and DMFS using the PrognoScan database
Logistic regression analysis (Table 2) showed a strong relationship between HSPB1 expression and N stage (N1&N2&N3 vs. N0; OR = 1.427, 95% CI: 1.121–1.818, p = 0.004), pathologic stage (Stage III &Stage IV vs. Stage I & Stage II; OR = 1.516, 95% CI: 1.144–2.014, p = 0.004), ER status (Positive vs. Negative; OR = 4.950, 95% CI: 3.561–6.982, p < 0.001), and PR status (Positive vs. Negative; OR = 2.639, 95% CI: 2.019–3.464, p < 0.001), but there was no significant difference between HSPB1 expression and age, T stage, M stage, HER2 status, and radiation_therapy (All p > 0.05).
Table 2.
Clinical pathological features related with HSPB1 expression according to logistic regression analysis
| Characteristics | Total(N) | Odds Ratio(OR) | P value |
|---|---|---|---|
| Age (> 60 vs. < = 60) | 1,083 | 1.211 (0.953–1.540) | 0.118 |
| T stage (T3&T4 vs. T1&T2) | 1,080 | 1.322 (0.955–1.836) | 0.093 |
| N stage (N1&N2&N3 vs. N0) | 1,064 | 1.427 (1.121–1.818) | 0.004 |
| M stage (M1 vs. M0) | 922 | 1.625 (0.666–4.182) | 0.293 |
| Pathologic stage (Stage III&Stage IV vs. Stage I&Stage II) | 1,060 | 1.516 (1.144–2.014) | 0.004 |
| ER status (Positive vs. Negative) | 1,033 | 4.950 (3.561–6.982) | < 0.001 |
| PR status (Positive vs. Negative) | 1,030 | 2.639 (2.019–3.464) | < 0.001 |
| HER2 status (Positive vs. Negative) | 715 | 1.136 (0.797–1.620) | 0.480 |
| radiation_therapy (Yes vs. No) | 987 | 1.171 (0.910–1.506) | 0.219 |
TMN stage was according to the seventh edition of the Guidelines for the American Journal of Critical Care. ER estrogen receptor, PR progesterone receptor, HER2 human epidermal growth factor receptor. Bold values indicate that p < 0.05
We conducted univariate and multivariate Cox regression analyses of OS, DSS, PFI, RFS, and DMFS to investigate the relationship between prognostic factors and clinical outcomes (Tables 3, 4, 5, 6, and 7 respectively). As summarized in Table 3, in univariate Cox regression analysis we found that age (> 60 years), advanced T, N, M, and advanced pathological stages were all significantly associated with poor OS. However, patients who received radiation_therapy were significantly associated with better OS (p = 0.004). In multivariable Cox regression analysis, age and M stage were independent predictors of OS. Significant difference in HSPB1 expression observed in univariate Cox regression (p = 0.037), but there was no significant difference in multivariate analysis of factors potentially predictive of OS.
Table 3.
Univariate and Multivariable analysis of factors potentially predictive of overall survival
| Characteristics | Total(N) | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|---|
| Hazard ratio (95% CI) | P value | Hazard ratio (95% CI) | P value | ||
| Age | 1082 | ||||
| < = 60 | 601 | Reference | |||
| > 60 | 481 | 2.020 (1.465–2.784) | < 0.001 | 3.430 (1.821–6.460) | < 0.001 |
| T stage | 1079 | ||||
| T1 | 276 | Reference | |||
| T2&T3&T4 | 803 | 1.482 (1.007–2.182) | 0.046 | 1.014 (0.371–2.776) | 0.978 |
| N stage | 1063 | ||||
| N0 | 514 | Reference | |||
| N1&N2&N3 | 549 | 2.239 (1.567–3.199) | < 0.001 | 1.878 (0.885–3.985) | 0.101 |
| M stage | 922 | ||||
| M0 | 902 | Reference | |||
| M1 | 20 | 4.254 (2.468–7.334) | < 0.001 | 5.195 (1.718–15.708) | 0.004 |
| Pathologic stage | 1059 | ||||
| Stage I | 180 | Reference | |||
| Stage II&Stage III&Stage IV | 879 | 2.210 (1.313–3.721) | 0.003 | 2.120 (0.473–9.507) | 0.326 |
| ER status | 1032 | ||||
| Negative | 240 | Reference | |||
| Positive | 792 | 0.712 (0.495–1.023) | 0.066 | 0.572 (0.229–1.425) | 0.230 |
| PR status | 1029 | ||||
| Negative | 342 | Reference | |||
| Positive | 687 | 0.732 (0.523–1.024) | 0.068 | 0.728 (0.309–1.716) | 0.468 |
| HER2 status | 715 | ||||
| Negative | 558 | Reference | |||
| Positive | 157 | 1.593 (0.973–2.609) | 0.064 | 0.678 (0.317–1.450) | 0.316 |
| radiation_therapy | 986 | ||||
| No | 434 | Reference | |||
| Yes | 552 | 0.576 (0.394–0.841) | 0.004 | 0.653 (0.354–1.203) | 0.172 |
| HSPB1 | 1082 | ||||
| Low | 541 | Reference | |||
| High | 541 | 1.208 (1.074–1.455) | 0.037 | ||
TMN stage was according to the seventh edition of the Guidelines for the American Journal of Critical Care. ER estrogen receptor, PR progesterone receptor, HER2 human epidermal growth factor receptor. Bold values indicate that p < 0.05
Table 4.
Univariate and Multivariable analysis of factors potentially predictive of disease specific survival
| Characteristics | Total(N) | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|---|
| Hazard ratio (95% CI) | P value | Hazard ratio (95% CI) | P value | ||
| Age | 1062 | ||||
| < = 60 | 590 | Reference | |||
| > 60 | 472 | 1.445 (0.941–2.219) | 0.093 | 1.280 (0.751–2.180) | 0.364 |
| T stage | 1059 | ||||
| T1 | 274 | Reference | |||
| T2&T3&T4 | 785 | 1.781 (1.033–3.071) | 0.038 | 1.271 (0.525–3.080) | 0.595 |
| N stage | 1044 | ||||
| N0 | 511 | Reference | |||
| N1&N2&N3 | 533 | 3.797 (2.222–6.489) | < 0.001 | 2.840 (1.451–5.559) | 0.002 |
| M stage | 903 | ||||
| M0 | 884 | Reference | |||
| M1 | 19 | 7.454 (3.988–13.931) | < 0.001 | 6.663 (3.139–14.142) | < 0.001 |
| Pathologic stage | 1041 | ||||
| Stage I | 178 | Reference | |||
| Stage II&Stage III&Stage IV | 863 | 3.396 (1.478–7.803) | 0.004 | 1.118 (0.300–4.165) | 0.869 |
| ER status | 1013 | ||||
| Negative | 232 | Reference | |||
| Positive | 781 | 0.559 (0.351–0.891) | 0.015 | 0.435 (0.193–0.978) | 0.044 |
| PR status | 1010 | ||||
| Negative | 334 | Reference | |||
| Positive | 676 | 0.519 (0.334–0.807) | 0.004 | 0.707 (0.321–1.555) | 0.388 |
| HER2 status | 704 | ||||
| Negative | 550 | Reference | |||
| Positive | 154 | 1.477 (0.740–2.948) | 0.269 | ||
| radiation_therapy | 977 | ||||
| No | 430 | Reference | |||
| Yes | 547 | 0.791 (0.483–1.295) | 0.351 | ||
| HSPB1 | 1062 | ||||
| Low | 528 | Reference | |||
| High | 534 | 1.515 (1.239–1.837) | 0.029 | ||
TMN stage was according to the seventh edition of the Guidelines for the American Journal of Critical Care. ER estrogen receptor, PR progesterone receptor, HER2 human epidermal growth factor receptor. Bold values indicate that p < 0.05
Table 5.
Univariate and Multivariable analysis of factors potentially predictive of progress free interval
| Characteristics | Total(N) | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|---|
| Hazard ratio (95% CI) | P value | Hazard ratio (95% CI) | P value | ||
| Age | 1082 | ||||
| < = 60 | 601 | Reference | |||
| > 60 | 481 | 1.253 (0.904–1.738) | 0.175 | ||
| T stage | 1079 | ||||
| T1 | 276 | Reference | |||
| T2&T3&T4 | 803 | 1.886 (1.241–2.867) | 0.003 | 1.609 (0.731–3.542) | 0.237 |
| N stage | 1063 | ||||
| N0 | 514 | Reference | |||
| N1&N2&N3 | 549 | 2.333 (1.621–3.357) | < 0.001 | 1.771 (1.123–2.794) | 0.014 |
| M stage | 922 | ||||
| M0 | 902 | Reference | |||
| M1 | 20 | 8.315 (4.829–14.315) | < 0.001 | 6.005 (3.115–11.573) | < 0.001 |
| Pathologic stage | 1059 | ||||
| Stage I | 180 | Reference | |||
| Stage II&Stage III&Stage IV | 879 | 2.268 (1.325–3.880) | 0.003 | 0.867 (0.316–2.381) | 0.782 |
| ER status | 1032 | ||||
| Negative | 240 | Reference | |||
| Positive | 792 | 0.622 (0.436–0.887) | 0.009 | 0.700 (0.394–1.244) | 0.224 |
| PR status | 1029 | ||||
| Negative | 342 | Reference | |||
| Positive | 687 | 0.558 (0.400–0.779) | < 0.001 | 0.593 (0.343–1.025) | 0.061 |
| HER2 status | 715 | ||||
| Negative | 558 | Reference | |||
| Positive | 157 | 1.228 (0.712–2.119) | 0.461 | ||
| radiation_therapy | 986 | ||||
| No | 434 | Reference | |||
| Yes | 552 | 0.899 (0.631–1.281) | 0.555 | ||
| HSPB1 | 1082 | ||||
| Low | 541 | Reference | |||
| High | 541 | 1.425 (1.137–1.750) | 0.041 | ||
TMN stage was according to the seventh edition of the Guidelines for the American Journal of Critical Care. ER estrogen receptor, PR progesterone receptor, HER2 human epidermal growth factor receptor. Bold values indicate that p < 0.05
Table 6.
Univariate and Multivariable analysis of factors potentially predictive of relapse-free survival
| Characteristics | Total(N) | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|---|
| Hazard ratio (95% CI) | P value | Hazard ratio (95% CI) | P value | ||
| Age | 4929 | ||||
| < = 60 | 2464 | Reference | |||
| > 60 | 2465 | 1.125 (0.832–2.056) | 0.127 | 1.245(0.796–2.214) | 0.414 |
| T stage | 4920 | ||||
| T1 | 1274 | Reference | |||
| T2&T3&T4 | 3646 | 1.315 (1.007–2.731) | 0.052 | 1.373 (0.826–3.134) | 0.125 |
| N stage | 4894 | ||||
| N0 | 2143 | Reference | |||
| N1&N2&N3 | 2751 | 2.215 (2.222–5.194) | < 0.001 | 2.853 (1.321–5.317) | 0.013 |
| M stage | 4801 | ||||
| M0 | 4637 | Reference | |||
| M1 | 164 | 6.153 (3.064–12.612) | < 0.001 | 5.132 (3.428–13.241) | < 0.001 |
| Pathologic stage | 4887 | ||||
| Stage I | 1035 | Reference | |||
| Stage II&Stage III&Stage IV | 3852 | 3.491 (1.613–7.423) | 0.007 | 1.307 (0.403–3.459) | 0.749 |
| ER status | 4872 | ||||
| Negative | 938 | Reference | |||
| Positive | 3934 | 0.459 (0.313–0.725) | 0.003 | 0.413 (0.213–0.728) | 0.045 |
| PR status | 4860 | ||||
| Negative | 1104 | Reference | |||
| Positive | 3756 | 0.617 (0.414–0.876) | 0.017 | 0.834 (0.414–1.625) | 0.407 |
| HER2 status | 4507 | ||||
| Negative | 3754 | Reference | |||
| Positive | 753 | 1.603 (0.629–2.714) | 0.315 | ||
| radiation_therapy | 4736 | ||||
| No | 1975 | Reference | |||
| Yes | 2761 | 0.691 (0.315–1.176) | 0.453 | ||
| HSPB1 | 4929 | ||||
| Low | 2464 | Reference | |||
| High | 2465 | 1.479 (1.124–1.893) | 0.038 | ||
TMN stage was according to the seventh edition of the Guidelines for the American Journal of Critical Care. ER estrogen receptor, PR progesterone receptor, HER2 human epidermal growth factor receptor. Bold values indicate that p < 0.05
Table 7.
Univariate and Multivariable analysis of factors potentially predictive of distant metastasis-free survival
| Characteristics | Total(N) | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|---|
| Hazard ratio (95% CI) | P value | Hazard ratio (95% CI) | P value | ||
| Age | 2765 | ||||
| < = 60 | 1382 | Reference | |||
| > 60 | 1383 | 1.152 (0.831–1.625) | 0.273 | ||
| T stage | 2759 | ||||
| T1 | 573 | Reference | |||
| T2&T3&T4 | 2186 | 1.974 (1.164–2.977) | 0.004 | 1.527 (0.822–2.312) | 0.313 |
| N stage | 2753 | ||||
| N0 | 1374 | Reference | |||
| N1&N2&N3 | 1378 | 2.712 (1.742–3.529) | < 0.001 | 1.631 (1.042–2.636) | 0.034 |
| M stage | 2649 | ||||
| M0 | 1913 | Reference | |||
| M1 | 736 | 7.137 (3.785–12.465) | < 0.001 | 5.024 (3.607–10.143) | < 0.001 |
| Pathologic stage | 2746 | ||||
| Stage I | 381 | Reference | |||
| Stage II&Stage III&Stage IV | 2365 | 3.539 (1.613–5.746) | 0.013 | 1.463 (0.607–2.594) | 0.815 |
| ER status | 2731 | ||||
| Negative | 643 | Reference | |||
| Positive | 2088 | 0.703 (0.342–1.327) | 0.019 | 0.820 (0.492–1.393) | 0.314 |
| PR status | 2716 | ||||
| Negative | 721 | Reference | |||
| Positive | 1995 | 0.524 (0.311–0.816) | < 0.001 | 0.573 (0.249–1.214) | 0.073 |
| HER2 status | 1843 | ||||
| Negative | 1449 | Reference | |||
| Positive | 394 | 1.304 (0.801–2.024) | 0.526 | ||
| radiation_therapy | 2679 | ||||
| No | 1163 | Reference | |||
| Yes | 1516 | 0.649 (0.434–1.407) | 0.434 | ||
| HSPB1 | 2765 | ||||
| Low | 1382 | Reference | |||
| High | 1383 | 1.614 (1.307–1.942) | 0.045 | ||
TMN stage was according to the seventh edition of the Guidelines for the American Journal of Critical Care. ER estrogen receptor, PR progesterone receptor, HER2 human epidermal growth factor receptor. Bold values indicate that p < 0.05
More advanced T, N, M, and pathologic stages were associated with worse DSS, and ER or PR positive statuses were associated with better DSS. The advanced N and M stages were independent predictors of DSS (Table 4), PFI (Table 5), RFS (Table 6), and DMFS (Table 7). Significant difference in HSPB1 expression observed in univariate Cox regression (DSS: p = 0.029; PFI: p = 0.041; RFS: p = 0.038; DMFS: p = 0.045), but there was no significant difference in multivariate analysis of factors potentially predictive of DSS, PFI, RFS, and DMFS.
HSPB1 expression differences observed in the univariate Cox regression and multivariate analyses were not significant.
These data suggest that patients with breast cancer with high HSPB1 expression have a poor prognosis. HSPB1 is not an independent marker for OS, DSS, PFI, RFS, or DMFS.
Identification of HSPB1-interacting genes and proteins and pathway analysis
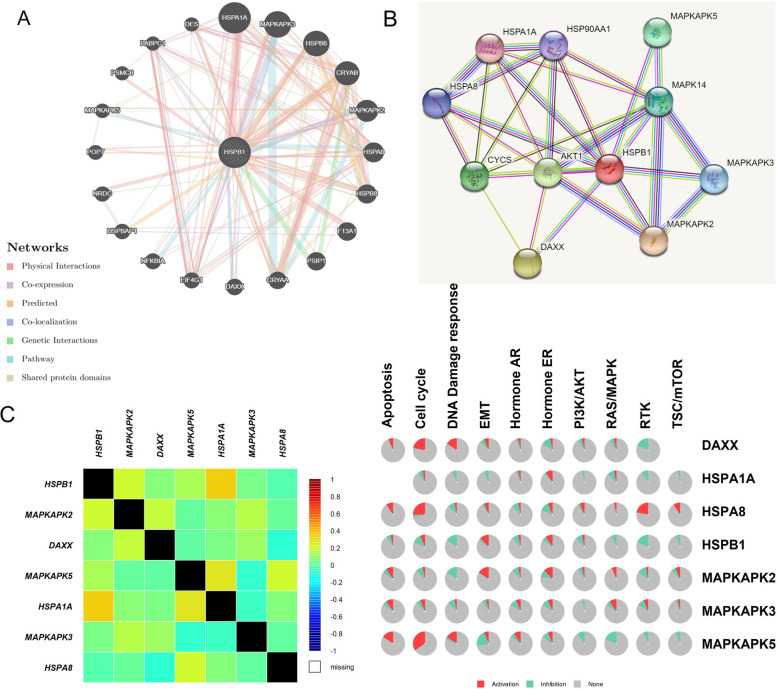
We used GeneMANIA to create the gene–gene interaction network for HSPB1 and the altered neighboring genes (Fig. 4A). We observed that the 20 most frequently altered genes were remarkably associated with HSPB1 expression. The proteins expressed by these six genes (mitogen-activated protein kinase activated protein kinase 2 (MAPKAPK2), death domain-associated protein (DAXX), MAPKAPK5, heat shock protein family A member 1A (HSPA1A), MAPKAPK3, and heat shock protein family A member 8 (HSPA8)) were found to interact with HSPB1 in the STRING database, with correlation scores of 0.998, 0.996, 0.992, 0.964, 0.981, and 0.972, respectively (Fig. 4B). We then used bc-GenExMinerv 4.8 to confirm the relationship between HSPB1 and the six genes (Fig. 4C). HSPB1 expression was positively associated with HSPA1A (r = 0.35, p < 0.0001), and moderate associated with MAPKAPK2 (r = 0.200, p < 0.0001). Our study used the GSCALite database to analyze pathways of altered neighboring genes (Fig. 4D). These results indicated that HSPB1 expression activated the EMT. Moreover, we observed that DAXX, HSPA8, and MAPKAPK5 mainly activate the cell cycle. However, the expression of HSPB1 mainly inhibited DNA damage response, and the Ras/MAPK, and RTK. These findings suggested that HSPB1 was associated with the occurrence, progression, and metastasis of breast cancer.
Fig. 4.
Identification of HSPB1-interacting genes and proteins and pathway analysis. The gene–gene interaction network and protein–protein interaction network of HSPB1 were constructed using GeneMANIA A and STRING B. C The correlation between HSPB1 and the altered neighboring genes using bc-GenExMiner v 4.8. D The GSCALite protocol was used to analyze the pathway activity (activation and inhibition)
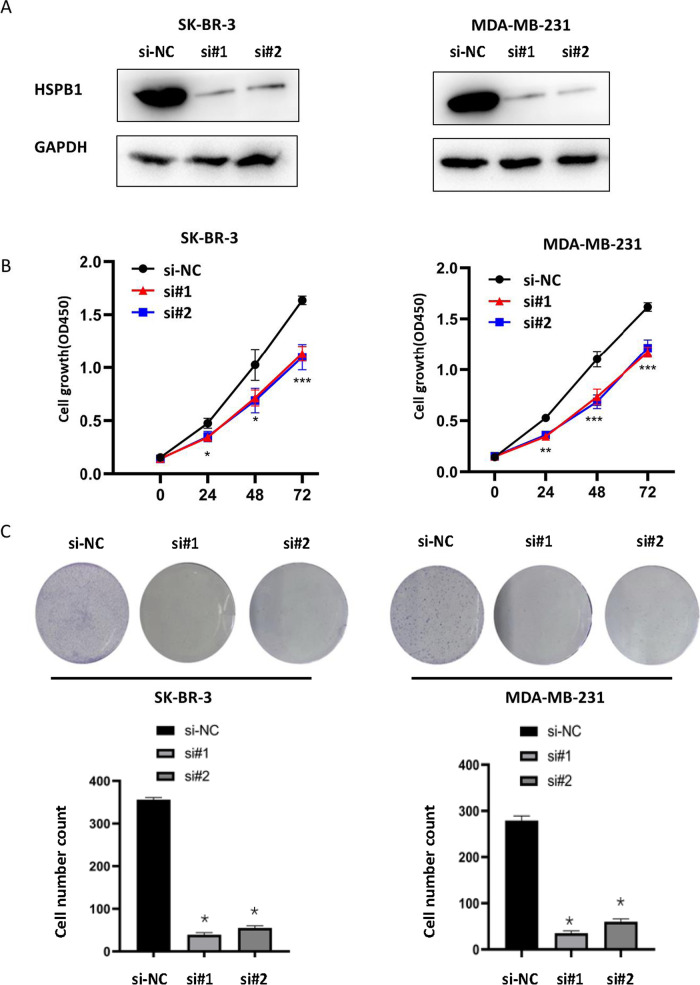
Transient knockdown of HSPB1 inhibited proliferation in breast cancer cells
In this study, SK-BR-3 and MDA-MB-231 breast cancer cells with the high protein expression of HSPB1 were selected as research subjects. Two siRNAs targeting HSPB1 were transfected into SK-BR-3 and MDA-MB-231 cells to knock down HSPB1. These results suggested that HSPB1 was successfully knocked down (Fig. 5A). Additionally, we tested the ability of HSPB1 to promote cell growth by using CCK8 and colony formation assays (Fig. 5B and C). Cell proliferation was significantly decreased in SK-BR-3 and MDA-MB-231 cells after transient HSPB1 knockdown compared with that in the si-NC group. Moreover, the transient knockdown lines exhibited significantly inhibited cell colony-forming numbers compared to the si-NC group.
Fig. 5.
Transient knockdown of HSPB1 affects breast cancer cell proliferation. A Verification of HSPB1 expression in SK-BR-3 and MDA-MB-231 cell lines via western blot. Growth curves B and Colony-forming efficiency C in SK-BR-3 and MDA-MB-231 cells before and after HSPB1 transient knockdown. The quantification of each analysis is shown in the following figure. All assays were performed in triplicate. The data are presented as means ± SEM. (*p < 0.05; **p < 0.01; *** p < 0.001). The blots were cut prior to hybridisation with antibodies during blotting
HSPB1 transient knockdown inhibited cell migration/invasion and promoted cell apoptosis in breast cancer cells
Cell invasion ability of SK-BR-3 and MDA-MB-231 cells was assessed at 0 and 72 h after injury using a wound-healing assay. Our data showed that in the scratch experiment, the wound healing rate of HSPB1 knockdown treated cells was significantly lower than that of control cells, and their migration ability was significantly reduced (Fig. 6A). Additionally, Transwell® inserts were was used to evaluate cell migration (Fig. 6B). The number of migrated SK-BR-3 and MDA-MB-231 cells after HSPB1 transient knockdown was lower than that of the si-NC group. Next, we studied the effect of HSPB1 on breast cancer cell apoptosis using flow cytometry. HSPB1 transient knockdown promoted apoptosis in SK-BR-3 and MDA-MB-231 cells (Fig. 6C). These findings support the hypothesis that HSPB1 affects the migration, invasion, and apoptosis in breast cancer.
Fig. 6.
Transient knockdown of HSPB1 affects breast cancer cell migration, invasion, and apoptosis. A The effect of transient knockdown of HSPB1 expression on the cell migration was determined by using the wound healing assay. The quantification of each analysis is shown in the right figure. The experiments were carried out in triplicate (**p < 0.01). The scratch area was calculated using Image J software. Cell scratch area (0 h) minus cell scratch area (72 h) to get the cell migration area, the percentage of cell migration area to cell scratch area (0 h) is the cell migration index. B Transwell® assay was used to determine the cell invasion after HSPB1 transient knockdown. The quantification of each analysis is shown in the right figure. The experiments were carried out in triplicate (***p < 0.001). C The impact of transient knockdown HSPB1 expression on cellular apoptosis as determined via flow cytometry. All assays were performed in triplicate. SK-BR-3 and MDA-MB-231 breast cancer cell lines were used in this study. The right picture shows the percentage of cell apoptosis. The experiments were carried out in triplicate (***p < 0.001)
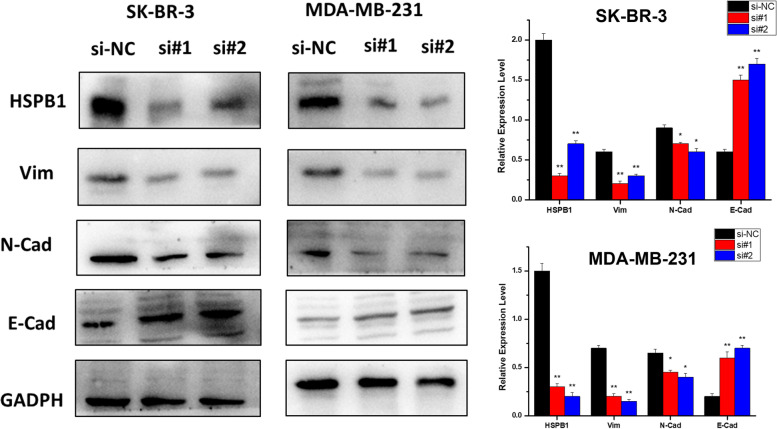
HSPB1 might be involved in the metastasis of breast cancer
To evaluate the effect of HSPB1 on EMT in breast cancer cells. The expression of E-cadherin, N-cadherin, and vimentin were analyzed after HSPB1 transient knockdown. We discovered that HSPB1 knockdown significantly reduced the expression of vimentin and N-cadherin, but E-cadherin expression was significantly increased (Fig. 7). We hypothesized that HSPB1 is involved in breast cancer metastasis. To explore this hypothesis, HSPB1 expression was first analyzed in the metastasis and in situ groups of with breast cancer. HSPB1 was highly expressed in patients with metastatic breast cancer, but low in those with carcinoma in situ (Fig. 8A). We then used RT-PCR to detect the expression of HSPB1 in triple-negative breast cancer cells (MDA-MB-468, MDA-MB-157, and MDA-MB-231) and non-triple-negative breast cancer cells (MCF-7, MDA-MB-453, and BT474). Concordantly, HSPB1 expression was significantly increased in triple-negative breast cancer cells (p < 0.01) (Fig. 8B). Overall, HSPB1 may be involved in breast cancer metastasis.
Fig. 7.
Transient knockdown of HSPB1 affects the cell epithelial-mesenchymal transition (EMT) process. A Vimentin, N-cadherin, and E-cadherin expression levels after siHSPB1-treatment as measured using western blotting. All assays were performed in triplicate. The blots were cut prior to hybridisation with antibodies during blotting. The quantification of each analysis is shown in the right figure. The experiments were carried out in triplicate (*p < 0.05; **p < 0.01)
Fig. 8.
A Immunohistochemical staining showing the expression of HSPB1 in situ and in breast cancer metastasis. B The expression of HSPB1 in three triple-negative breast cancer cell lines (MDA-MB-468, MDA-MB-157, and MDA-MB-231) and three other breast cancer cell lines (MCF-7, MDA-MB-453, and BT474) were measured through quantitative real-time PCR (** p < 0.01)
Discussion
Despite continuous improvements in breast cancer research and treatment methods, the incidence rate continues to increase. Therefore, there is a pressing need to explore the mechanisms leading to breast cancer metastasis. HSPB1 is widely expressed in various tumors [7, 10, 27] and may contribute to tumor proliferation, migration, and drug resistance [28, 29]. Our study evaluated the expression levels, clinicopathological associations, clinical significance, and influence on metastasis of HSPB1 in breast cancer.
Many studies have investigated HSPB1 in relation to various cancer types. Huang et al. (2010) reported that HSPB1 was overexpressed in gastric adenocarcinoma tissue and that serum levels of HSPB1 were increased in patients with gastric adenocarcinoma, which may indicate gastric malignancy and thus its detection may be helpful for screening gastric adenocarcinoma [30]. Another study suggested that the expression of HSPB1 may be used to predict poor prognosis and transfer tendency in prostate cancer and demonstrated that HSPB1 was promoted in an insulin-like growth factor 1-dependent manner [31]. They also reported that the phosphorylation of extracellular signal-regulated kinase 1 and Akt stabilizes the BAD /14–3-3 protein complex, reducing the rate of prostate cancer cell apoptosis. In addition, HSPB1 expression in prostate cancer cells is significantly increased after androgen deprivation and chemotherapy and acts as a molecular chaperone for cell protection, making cells resistant to drugs [32]. These results indicated that HSPB1 could be used as a target for radiotherapy sensitization in prostate cancer. Furthermore, HSPB1 antibody levels are elevated in patients with breast cancer [33]. Despite these observations, a comprehensive study of HSPB1 in breast cancer has not yet been conducted, and it remains unclear how HSPB1 affects breast cancer occurrence and development [34]. According to our findings, the expression of HSPB1 was considerably upregulated in breast cancer tissues, which is consistent with a previous report [35]. Notably, we also confirmed that HSPB1 expression was associated with the clinical features of patients with breast cancer. Higher HSPB1 expression was closely correlated with pathologic stage, ER, and PR. The increased expression of HSPB1 observed in late-stage malignancies suggests that HSPB1 may contribute to cancer development. Therefore, we propose HSPB1 as a marker of poor survival in patients with breast cancer.
To evaluate the prognostic potential of HSPB1 in breast cancer, we used the Kaplan–Meier survival curve to analyze the effect of HSPB1 expression level on survival. Patients with higher HSPB1 level had remarkably worse OS, RFS, and DMFS. Consistent results were obtained using the online tool PrognoScan. However, HSPB1 expression differences were not significant in univariate logistic regression and multivariate analyses for OS, DSS, PFI, RFS, and DMFS. We confirmed that HSPB1 is not an independent marker for OS, DSS, PFI, RFS, or DMFS. According to these findings, HSPB1 may be a prognostic biomarker for breast cancer and may facilitate the development of targeted precision oncology.
Apoptosis plays a vital role in cancer [36]. HSPB1 directly inhibits the activation of caspases to inhibit cell apoptosis [37], prevent multiple apoptotic effects from inducing cell death, and regulate apoptosis signaling pathway, [38]. In this study, a transient knockdown of HSPB1 inhibited cell proliferation and migration/invasion activity and promoted apoptosis of breast cancer cells. EMT is the most crucial pathway for tumor cell invasion and metastasis [39–41]. During this process, cells gain the ability to move, invade, and separate from the epithelial membrane. EMT is associated with tumorigenesis, metastasis, and drug resistance [42]. Similar to Yun et al. [43], we confirmed that the transient knockdown of HSPB1 significantly reduced the expression of vimentin, and N-cadherin, but upregulated E-cadherin expression. As an oncogene, HSPB1 promotes the activity of breast cancer cells by regulating the EMT process, thus establishing HSPB1 as a promising biomarker for the diagnosis and treatment of breast cancer; however, the potential mechanism by which HSPB1 affects cell proliferation and EMT requires further investigation.
Conclusions
Overall, high HSPB1 expression predicted poor clinical outcomes, meaning that it holds potential as a novel prognostic biomarker for breast cancer. HSPB1 knockdown inhibited the proliferation, migration, invasion, and apoptosis of breast cancer cells. This study advances our current understanding of the role of HSPB1 as a prognostic marker for breast cancer treatment.
Supplementary Information
Acknowledgements
We kindly thank the TCGA database (https://portal.gdc.cancer.gov/), the Kaplan-Meier plotter database (http://www.kmplot.com/), The PrognoScan database (http://dna00.bio.kyutech.ac.jp/PrognoScan/index.html), the GeneMANIA (http://www.genemania.org) and STRING online (https://string-db.org/), the bc-GenExMinerv 4.8 (http://bcgenex.ico.unicancer.fr/BC-GEM/GEM-Accueil.php?js=1) and the GSCALite database (http://bioinfo.life.hust.edu.cn/web/GSCALite/).
Statement
This study was approved by Shenzhen Second People's Hospital for experiments, including any relevant details. All authors confirm that all methods were carried out in accordance with relevant guidelines and regulations
Abbreviations
- CCK8
Cell counting kit 8
- DAXX
Death domain associated protein
- DMFS
Distant metastasis-free survival
- DSS
Disease-specific survival
- EMT
Epithelial-mesenchymal transition
- ER
Estrogen receptor; FBS, fetal bovine serum
- HER2
Human epidermal growth factor receptor 2
- HR
Hazard ratio
- HSPA1A
Heat shock protein family A member 1A
- HSPA8
Heat shock protein family A member 8
- HSPB1
Heat shock protein beta-1
- MAPKAPK
Mitogen-activated protein kinase activated protein kinase
- OS
Overall survival
- PBS
Phosphate buffered saline
- PFI
Progression-free interval
- PR
Progesterone receptor
- RFS
Relapse-free survival
- RT-PCR
Quantitative real-time PCR
- TCGA
The Cancer Genome Atlas
Authors’ contributions
QH: study design and original draft writing; QH and JW: analysis and interpretation of data; JW: project investigation; NX: reviewed. All authors approved the submitted version. QH and JW have contributed equally to this work.
Funding
This project was supported by the National Natural Science Foundation of China, China (No. 82172356, No. 81972003), the Natural Science Foundation of Guangdong, China (No. 2021A1515012144).
Availability of data and materials
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials.
Declarations
Ethics approval and consent to participate
This study was approved by The Institutional Research Ethics Committee of Shenzhen Second People’s Hospital. The patients provided written informed consent to participate in this study. All authors confirm that all methods were carried out in accordance with relevant guidelines and regulations.
Consent for publication
Not applicable.
Competing interests
The authors have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Qin Huo and Juan Wang contributed equally to this work.
References
- 1.Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries [J] CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Aggarwal S, Verma SS, Aggarwal S, et al. Drug repurposing for breast cancer therapy: old weapon for new battle[J] Semin Cancer Biol. 2021;68:8–20. doi: 10.1016/j.semcancer.2019.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liang Y, Zhang H, Song X, et al. Metastatic heterogeneity of breast cancer: molecular mechanism and potential therapeutic targets[J] Semin Cancer Biol. 2020;60:14–27. doi: 10.1016/j.semcancer.2019.08.012. [DOI] [PubMed] [Google Scholar]
- 4.Waks AG, Winer EP. Breast cancer treatment: a review[J] JAMA, J Am Med Assoc. 2019;321(3):288–300. doi: 10.1001/jama.2018.19323. [DOI] [PubMed] [Google Scholar]
- 5.Calderwood SK, Gong J. Heat shock proteins promote cancer: it's a protection racket[J] Trends Biochem Sci. 2016;41(4):311–323. doi: 10.1016/j.tibs.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu J, Liu T, Rios Z, et al. Heat Shock Proteins and Cancer.[J] Trends Pharmacol Sci. 2017;38(3):226–256. doi: 10.1016/j.tips.2016.11.009. [DOI] [PubMed] [Google Scholar]
- 7.Soleimani A, Jalili Nik M, Avan A, et al. The role of HSP27 in the development of drug resistance of gastrointestinal malignancies: current status and perspectives[J] J Cell Physiol. 2018;234(6):8241–8248. doi: 10.1002/jcp.27666. [DOI] [PubMed] [Google Scholar]
- 8.Choi SK, Kam H, Kim KY, et al. Targeting Heat Shock Protein 27 in Cancer: A Druggable Target for Cancer Treatment?[J] Cancers (Basel) 2019;11(8):1195. doi: 10.3390/cancers11081195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen X, Dong XS, Gao HY, et al. Suppression of HSP27 increases the antitumor effects of quercetin in human leukemia U937 cells[J] Mol Med Rep. 2016;13(1):689–696. doi: 10.3892/mmr.2015.4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okuno M, Adachi S, Kozawa O, et al. The clinical significance of phosphorylated heat shock protein 27 (HSPB1) in pancreatic cancer[J] Int J Mol Sci. 2016;17(1):137. doi: 10.3390/ijms17010137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hung CS, Huang CY, Hsu YW, et al. HSPB1 Rs2070804 polymorphism is associated with the depth of primary tumor[J] J Cell Biochem. 2020;121(1):63–69. doi: 10.1002/jcb.28266. [DOI] [PubMed] [Google Scholar]
- 12.Rajesh Y, Biswas A, Banik P, et al. Transcriptional regulation of HSPB1 by friend leukemia integration-1 factor modulates radiation and temozolomide resistance in glioblastoma[J] Oncotarget. 2020;11(13):1097–1108. doi: 10.18632/oncotarget.27425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, Feng Z, Wang W, et al. Expression of heat shock protein-27 (Hsp27) and P38MAPK in esophageal squamous cell carcinoma[J] Med Sci Monitor. 2017;23:5246–5253. doi: 10.12659/MSM.904912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y, Tao X, Jin G, et al. A targetable molecular chaperone Hsp27 confers aggressiveness in hepatocellular carcinoma[J] Theranostics. 2016;6(4):558–570. doi: 10.7150/thno.14693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hung C, Huang C, Lee C, et al. IGFBP2 plays an important role in heat shock protein 27-mediated cancer progression and metastasis[J] Oncotarget. 2017;8(33):54978–54992. doi: 10.18632/oncotarget.18989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen W, Ren X, Wu J, et al. HSP27 Associates with epithelial-mesenchymal transition, stemness and radioresistance of salivary adenoid cystic carcinoma[J] J Cell Mol Med. 2018;22(4):2283–2298. doi: 10.1111/jcmm.13510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gibert B, Eckel B, Gonin V, et al. Targeting heat shock protein 27 (HspB1) interferes with bone metastasis and tumour formation in vivo. Br J Cancer. 2012;107(1):63–70. doi: 10.1038/bjc.2012.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cayado-Gutiérrez N, Moncalero VL, Rosales EM, et al. Downregulation of Hsp27 (HSPB1) in MCF-7 human breast cancer cells induces upregulation of PTEN. Cell Stress Chaperones. 2013;18(2):243–249. doi: 10.1007/s12192-012-0367-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Florell SR, Coffin CM, Holden JA, et al. Preservation of RNA for functional genomic studies: a multidisciplinary tumor bank protocol[J] Modern Pathol. 2001;14(2):116–128. doi: 10.1038/modpathol.3880267. [DOI] [PubMed] [Google Scholar]
- 20.Liu J, Lichtenberg T, Hoadley KA, et al. An Integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018;173(2):400–416.e11. doi: 10.1016/j.cell.2018.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lanczky A, Gyorffy B. Web-based survival analysis tool tailored for medical research (KMplot): development and implementation. J Med Internet Res. 2021;23(7):e27633. doi: 10.2196/27633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mizuno H, Kitada K, Nakai K, et al. PrognoScan: a new database for meta-analysis of the prognostic value of genes. BMC Med Genomics. 2009;2:18. doi: 10.1186/1755-8794-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Franz M, Rodriguez H, Lopes C, et al. GeneMANIA update 2018. Nucleic Acids Res. 2018;46(W1):W60–W64. doi: 10.1093/nar/gky311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crosara KTB, Moffa EB, Xiao Y, et al. Merging in-silico and in vitro salivary protein complex partners using the STRING database: a tutorial. J Proteomics. 2018;171:87–94. doi: 10.1016/j.jprot.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 25.Jézéquel P, Campone M, Gouraud W, et al. bc-GenExMiner: an easy-to-use online platform for gene prognostic analyses in breast cancer. Breast Cancer Res Tr. 2012;131(3):765–775. doi: 10.1007/s10549-011-1457-7. [DOI] [PubMed] [Google Scholar]
- 26.Liu CJ, Hu FF, Xia MX, et al. GSCALite: a web server for gene set cancer analysis. Bioinformatics. 2018;34(21):3771–3772. doi: 10.1093/bioinformatics/bty411. [DOI] [PubMed] [Google Scholar]
- 27.Stope MB, Klinkmann G, Diesing K, et al. Heat shock protein HSP27 secretion by ovarian cancer cells is linked to intracellular expression levels, occurs independently of the endoplasmic reticulum pathway and HSP27’s phosphorylation status, and is mediated by exosome liberation[J] Dis Markers. 2017;2017:1–12. doi: 10.1155/2017/1575374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun X, Ou Z, Xie M, et al. HSPB1 as a novel regulator of ferroptotic cancer cell death[J] Oncogene. 2015;34(45):5617–5625. doi: 10.1038/onc.2015.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmitt E, Gehrmann M, Brunet M, et al. Intracellular and extracellular functions of heat shock proteins: repercussions in cancer therapy[J] J Leukocyte Biol. 2007;81(1):15–27. doi: 10.1189/jlb.0306167. [DOI] [PubMed] [Google Scholar]
- 30.Huang Q, Ye J, Huang Q, et al. Heat Shock Protein 27 is Over-Expressed in Tumor Tissues and Increased in Sera of Patients with Gastric Adenocarcinoma[J] Clin Chem Lab Med. 2010;48(2):263–9. doi: 10.1515/CCLM.2010.043. [DOI] [PubMed] [Google Scholar]
- 31.Zoubeidi A, Zardan A, Wiedmann RM, et al. Hsp27 promotes insulin-like growth factor-i survival signaling in prostate cancer via p90Rsk-dependent phosphorylation and inactivation of BAD[J] Cancer Res. 2010;70(6):2307–2317. doi: 10.1158/0008-5472.CAN-09-3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aloy M, Hadchity E, Bionda C, et al. Protective Role of Hsp27 Protein Against Gamma Radiation–Induced Apoptosis and Radiosensitization Effects of Hsp27 Gene Silencing in Different Human Tumor Cells[J]. International Journal of Radiation Oncology*Biology*Physics, 2008,70(2):543–553. [DOI] [PubMed]
- 33.Homaei-Shandiz F, Mehrad-Majd H, Tasbandi M, et al. Anti-heat shock protein-27 antibody levels in women with breast cancer: association with disease complications and two-year disease-free survival[J] Asian Pac J Cancer Prev. 2016;17(10):4655–4659. doi: 10.22034/APJCP.2016.17.10.4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu X, Feng C, Liu J, et al. Androgen Receptor and Heat Shock Protein 27 Co-Regulate the Malignant Potential of Molecular Apocrine Breast Cancer[J] J Exp Clin Canc Res. 2018;37(1):90. doi: 10.1186/s13046-018-0762-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang S, Zhang X, Wang H, et al. Heat shock protein 27 enhances SUMOylation of heat shock protein B8 to accelerate the progression of breast cancer[J] Am J Pathol. 2020;190(12):2464–2477. doi: 10.1016/j.ajpath.2020.04.012. [DOI] [PubMed] [Google Scholar]
- 36.Concannon CG, Gorman AM, Samali A. On the Role of Hsp27 in regulating apoptosis[J] Apoptosis (London) 2003;8(1):61–70. doi: 10.1023/A:1021601103096. [DOI] [PubMed] [Google Scholar]
- 37.Samali A, Robertson JD, Peterson E, et al. Hsp27 protects mitochondria of thermotolerant cells against apoptotic stimuli[J] Cell Stress Chaperones. 2001;6(1):49–58. doi: 10.1379/1466-1268(2001)006<0049:HPMOTC>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shan R, Liu N, Yan Y, et al. Apoptosis, autophagy and atherosclerosis: relationships and the role of Hsp27[J] Pharmacol Res. 2021;166:105169. doi: 10.1016/j.phrs.2020.105169. [DOI] [PubMed] [Google Scholar]
- 39.Derynck R, Weinberg RA. EMT and cancer: more than meets the eye[J] Dev Cell. 2019;49(3):313–316. doi: 10.1016/j.devcel.2019.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brabletz T, Kalluri R, Nieto MA, et al. EMT in Cancer[J] Nat Rev Cancer. 2018;18(2):128–134. doi: 10.1038/nrc.2017.118. [DOI] [PubMed] [Google Scholar]
- 41.Bakir B, Chiarella AM, Pitarresi JR, et al. EMT, MET, plasticity, and tumor metastasis[J] Trends Cell Biol. 2020;30(10):764–776. doi: 10.1016/j.tcb.2020.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pastushenko I, Blanpain C. EMT transition states during tumor progression and metastasis[J] Trends Cell Biol. 2019;29(3):212–226. doi: 10.1016/j.tcb.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 43.Yun CW, Kim HJ, Lim JH, et al. Heat Shock proteins: agents of cancer development and therapeutic targets in anti-cancer therapy[J] Cells (Basel, Switzerland) 2019;9(1):60. doi: 10.3390/cells9010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials.