Abstract
The re-emergence of monkeypox (MPX), in the era of COVID-19 pandemic is a new global menace. Regardless of its leniency, there are chances of MPX expediting severe health deterioration. The role of envelope protein, F13 as a critical component for production of extracellular viral particles makes it a crucial drug target. Polyphenols, exhibiting antiviral properties have been acclaimed as an effective alternative to the traditional treatment methods for management of viral diseases. To facilitate the development of potent MPX specific therapeutics, herein, we have employed state-of-the-art machine learning techniques to predict a highly accurate 3-dimensional structure of F13 as well as identify binding hotspots on the protein surface. Additionally, we have effectuated high-throughput virtual screening methodology on 57 potent natural polyphenols having antiviral activities followed by all-atoms molecular dynamics (MD) simulations, to substantiate the mode of interaction of F13 protein and polyphenol complexes. The structure-based virtual screening based on Glide SP, XP and MM/GBSA scores enables the selection of six potent polyphenols having higher binding affinity towards F13. Non-bonded contact analysis, of pre- and post- MD complexes propound the critical role of Glu143, Asp134, Asn345, Ser321 and Tyr320 residues in polyphenol recognition, which is well supported by per-residue decomposition analysis. Close-observation of the structural ensembles from MD suggests that the binding groove of F13 is mostly hydrophobic in nature. Taken together, this structure-based analysis from our study provides a lead on Myricetin, and Demethoxycurcumin, which may act as potent inhibitors of F13. In conclusion, our study provides new insights into the molecular recognition and dynamics of F13-polyphenol bound states, offering new promises for development of antivirals to combat monkeypox. However, further in vitro and in vivo experiments are necessary to validate these results.
Keywords: Monkeypox virus, Myricetin, Demethoxycurcumin, Curcumin, Ellagic acid, Molecular dynamics simulation
1. Introduction
The resurgence of monkeypox (MPX), a zoonotic disease, with an unknown animal reservoir is caused by a member of the Poxviridae family i.e. monkeypox virus (MPXV) and has put the public in distress [1]. While it was originally found in African rainforests, the recent outbreak (since May 2022) deals with the spread of this viral infection in other non-endemic countries which has the potential to cause a global epidemic [2]. This viral transmission through bodily fluids and respiratory droplets, exhibits symptoms similar to small pox such as fever, swollen lymph nodes and a body rash that lasts for a few weeks [[3], [4], [5]]. However, severe medical complications such as lymphadenopathy, which distinguishes MPX from chickenpox can be a leading cause of 3–5% deaths [6]. The skin lesions in the eruptive phase, progresses to macules, papules, vesicles and pustules, which later develops crusts leading to hyper-pigmentation [7]. Most severe cases of MPX follows pitted facial scars, corneal ulceration, vision loss, cutaneous bacterial infections, bronchopneumonia, respiratory distress etc. Earlier studies also suggests the frequent occurrence of chickenpox and MPX co-infection [[8], [9], [10]].
In this scenario, due to lack of MPX specific therapeutics, three available drugs tecovirimat ST-246, brincidofovir and cidofovir are put to use as an emergency. However, as a consequence of limited effectiveness of these available drugs in human use and with the emergence of new viral variants, they are not considered as viable frontline treatment options. Evidences from literature demonstrate the inhibitory role of these drugs towards a viral structural protein F13. This palmitylated structural F13 protein, found in most of the orthopoxvirus is highly conserved in nature. F13 have been vividly studied to play a critical role in viral replication and transcription by producing extracellular virions (EEV) and attaching or releasing enveloped virions (EV) from the cell membrane [11]. The addition of 16-carbon fatty acid tails during palmitylation is indispensable for membrane association, proper localization and functioning of F13 protein. For that reason, F13 protein is considered as a promising drug target for discovery and development of anti-MPXV therapies. To effectively treat individuals infected with the virus, it may be necessary to use a combination of therapies. Owing to this solution, a recent study on Resveratrol, a natural polyphenol has been found to inhibit viral replication including MPX but its mechanism of action is poorly understood and needs further investigation [12].
The prevalence of plant-based polyphenols as potent antiviral agents is rising due to their potential health benefits with the ability of inhibiting diverse stages of viral replication, wide availability, inexpensive production as well as low side-effects [11,13,14]. Depending on the nature of the virus (DNA or RNA), different polyphenols interacts with the viral particle [15,16]. Recent studies have provided evidences of natural polyphenols as important targets for development of effective therapeutics against various viral diseases. The effectiveness of plant-derived polyphenols observed against cardiovascular diseases, neurodegenerative diseases, diabetes, and cancer, among other things, makes them competent enough to be used as a functional food or in drug preparations [[17], [18], [19]]. Previous state-of-the-art studies demonstrate the multi-mechanistic antiviral nature of polyphenols that might be facilitated due to their antioxidant activities, inhibition of viral replication, etc [20].
Given that the antiviral activities of these natural polyphenols needs more comprehensive study, herein, we have made an attempt to identify potent lead molecules exhibiting inhibitory pursuits against F13 as well as their plausible binding mode [21]. However, the lack of structural information for F13 protein hinders the development procedure of potent therapeutics. The current study utilizes the state-of-the-art machine learning techniques implemented in AlphaFold2 for prediction of a high-quality protein structure. All-atoms MD simulations of the F13-polyphenol complexes, following high-throughput virtual screening, apprehends the function and behaviour of the protein as well as the bound polyphenols at an atomic level. Our research establishes a baseline for investigating the potential of machine learning approaches in structure prediction and provides direction for identifying conceivable molecular recognition mechanisms for ligands. As a whole, our study shed light on the dynamics of polyphenols-bound F13 conformations, paving the way for the development of novel antivirals against a range of pharmacological targets.
2. Materials and methods
2.1. Nearly accurate structure of F13 protein by dint of state-of-the-art techniques
Initially, the total reference proteome from three genomes of MPXV (Congo virus strain (1996), ID: NC_003310.1, West African strain (2018), ID: MT903345.1, and West African strain (2022), ID: ON563414.3) was obtained from NCBI protein database. Following collection of the F13 protein from three proteomes, we compared the similarities and differences of those sequences at amino acid level using MultAlin tool [22]. In the initial period of acquiring the 3-dimensional (3D) information of the F13 protein, the Congo viral F13 sequence, being the ancient strain (ID: NC_003310.1) was subjected to BLASTp [23] and PSI-Blast [24] search against PDB to identify potent structural homologs for protein modelling. Due to lack of suitable homologs in PDB, the state-of-the-art machine learning structure prediction protocol implemented in AlphaFold2, was used to generate high-quality protein models. Further validation of the predicted model from AlphaFold2 was carried out using the highest predicted local distance difference test (pLDDT) score, which indicates the confidence of the model [25]. In addition, we also deployed several standard protein model evaluation servers such as PROCHECK [26] and ERRAT [27] programs available in SAVEs server, ProSA [28], ProQ [29], and MolProbity [30] to assess the stereo-chemical qualities of the modelled F13.
Post-F13 modelling using AlphFold2, the protein was subjected to all-atoms MD simulation to evaluate its structural stability and dynamics in apo state. In this study, a TIP3 water layer was added around the protein to create a fully hydrated and neutralized system with 0.15 M NaCl. Using GROMACS v2021.4, a MD simulation of 200 ns was performed in this neutralized system [31] (as explained in section 2.3 below) in replicates. The simulation protocol was adapted from our earlier studies [32,33]. Subsequently, we used GROMOS-based clustering approach with a cut-off of 0.2 nm to select a representative snapshot from the top ranked cluster for further exercises.
2.2. Prediction of F13 binding cavity and virtual screening of antiviral polyphenols
To optimize the hydrogen bonds and ensure proper charged states for the ionizable amino acid residues, we prepared the modelled F13 protein (obtained from the top ranked cluster post-MD) using the Protein Preparation Wizard of Maestro [34] with a pH of 7.0, which was determined using the PROPKA module [35].
The deep PointSite model [36] was further availed to annotate the binding site residues of the F13 protein, where residues having values equivalent to 1 are more likely to be involved in the binding activity [37]. Following active site identification, the receptor grid was generated by specifying active site residues using Maestro 12.8 (Schrödinger, LLC, New York, 2021-2). We screened 57 natural polyphenols having reported antiviral activity virtually, focusing on five residues (His115, Lys117, Asn132, His325, and Lys327) in the binding groove of the F13 protein. To further validate our binding site residue prediction, we utilized another machine learning approach, PrankWeb [38].
To evaluate the efficacy of natural products, we selected 57 polyphenols (Table S1) from the scientific literature with available structures in the public domain [39] and retained their structures in sdf format from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/). To undergo the process of molecular recognition, the ligands were prepared with the modelled F13 protein using the LigPrep tool available in Maestro 12.8 (Schrödinger, LLC, New York, 2021-2). The default parameters (pH 7.0) were selected for generating tautomers as well as possible states of the molecules, where the pKa values of these states was calculated using the Epik module [35]. The final structures of the ligands were optimized using the OPLS4 force field to generate low-energy 3D ligand conformers [40].
Following the preparation of the F13 protein and ligands, glide docking programme implemented in the Schrodinger suite (Schrödinger, LLC, New York, 2021–22) was used for studying the molecular interactions between F13 and polyphenols. The docking calculations were performed in SP (standard precision) mode followed by XP (extra precision) mode [41]. The docked F13-polyphenol complexes were visualised using PyMOL (http://www.pymol.org/pymol) and LigPlot+ [42].
Then, Molecular Mechanics-Generalized Born Surface Area (MM/GBSA) method implemented in Prime (Schrödinger, LLC, New York, 2021-2) was used to predict the binding free energy (ΔGbind) of the docked ligands at the ligand binding site of F13 protein [43]. The methodology for calculating MM/GBSA was adapted from Genheden et al. (2015) [44]– [45]. Finally, we screened the top ranked complexes from these generated poses based on their SP, XP, and ΔGbind scores (high) from MM/GBSA analysis. For further validation of our docking protocol employed in Glide, we docked the modelled F13 and polyphenols using AutoDock Vina v1.5.7 [46].
2.3. Understanding the atomistic behaviour of the modelled F13 protein and screened polyphenols using all-atoms MD simulation
To explore the intrinsic stability and conformational flexibility of the six (screened from molecular docking studies) docked F13-polyphenol complexes, we employed all-atoms MD simulations using a structure based balanced force field, CHARMM36m [47] in TIP3P [48] water model in GROMACS. Ligand topologies were generated using CHARMM General Force Field (CGenFF) [49]. To neutralise each system, 0.15M NaCl was added to each system. Following electro-neutralization, each system was subjected to energy minimization using steepest descent method for 5000 steps. The energy minimized systems were then equilibrated using NVT and NPT ensemble with periodic boundary conditions using a Berendsen thermostat [50] and Nose Hoover thermostat [51,52]. A production MD of 200 ns was run with constant pressure (1 bar) and temperature (303K) using the Parrinello-Rahman barostat algorithm [53].
2.4. MD trajectory analysis
GROMACS utility toolkits were used to analyse the quality of the trajectories through determining their conformational stability and flexibility. To determine the dynamics and stability, various properties of the system including backbone root mean squared deviation (RMSD), Cα-root mean squared fluctuations (RMSF), radius of gyration (Rg), solvent accessible surface area (SASA) and intermolecular hydrogen bonds were computed using gmx rms, gmx rmsf, gmx gyrate, gmx sasa and gmx hbond respectively. The time dependent structural changes in the secondary structural elements of each system during MD simulations were mapped using VMD (in a timescale of 200 ns). XmGrace (https://plasma-gate.weizmann.ac.il/Grace/), PyMOL and BIOVIA Discovery Studio Visualizer (BIOVIA, San Diego, CA, USA) were used for visualization of 2D graphs and structures respectively and the plots were generated in OriginPro and Adobe Illustrator 2.8.
RMSD-based clustering analysis of all the systems was carried out by using gmx cluster with a cut-off of 0.2 nm using the last 100 ns MD trajectories. Principal component analysis (PCA) separates the collective motions from the local dynamics to a small subset comprised of principal components (PCs) that define the collective motion. To observe these collective motions within the F13-polyphenol complexes, PCA was performed using gmx covar and gmx anaeig built-in modules of GROMACS using the last 100 ns MD trajectories.
2.5. Estimation of binding free energy by molecular mechanics/Poisson-Boltzmann surface area (MM/PBSA) approach
The binding free energy of the F13-polyphenol complexes involved in their binding process, was calculated using the g_mmpbsa tool [54], which uses molecular-mechanics based Poisson–Boltzmann Surface Area (MM/PBSA) calculations [55]. A total of 200 snapshots were extracted from the last 100 ns MD trajectories at equal interval of time for determining the binding energy (ΔGbind). To better understand the contribution of individual amino acids to the binding free energy and per-residue decomposition analysis was performed.
3. Results and discussion
The rapid emergence of different MPXV variants and upsurge in the number of cases led to the growing demand for more effective treatments, concerning the limited number of current treatment options. F13 is a crucial drug target for the development of potent antiviral drugs or inhibitors that lacks close human homologs. Studies have shown that targeting antiviral plant-based polyphenols against envelope proteins for designing effective inhibitors can trigger the development of effective therapeutics [[56], [57], [58], [59], [60]]. Although polyphenols have been found to have various biological functions, their specific mechanism of action against viruses is not well-defined [61].
A recent research study has shed light on how polyphenols can inhibit viral entry, reproduction and DNA inhibition [21], which provides valuable insights into their mechanism of exhibiting antiviral activity against viruses. In particular, herein, we highlight the importance of investigating the antiviral properties of polyphenols against MPXV and aims to identify potent antivirals against an AI-based model of F13 using state-of-the-art computational methods.
3.1. Optimization and structural dynamics of AlphaFold modelled F13
To avail improved performances in molecular docking studies, 3D structures of the target protein plays a vital role. Mostly, the availability of experimentally solved structures in PDB provides adequate structural information related to the protein target. However, there still exists an abundance of non-redundant protein sequences, which are poorly characterized. Our target protein, F13, is a non-redundant protein, which lacks experimental structures. Therefore, to access its structural information as well as viability in different physiological conditions, we tried to obtain its 3D structure. Initially, we performed multiple sequence alignment of F13 protein encoded by the three available MPX genomes of different strains (as illustrated in Fig. S2A). The BLAST search for structural homologs yielded a structure of Serratia plymuthica phospholipase D (PDB ID: 7E0M) with an identity of 24.28% and 88% query coverage, which did not meet the 30% cut off value, required for obtaining a precise model. Therefore, we implemented the DeepMind's AlphaFold2 [[62], [63], [64]] to predict a protein structure with higher accuracy with a backbone and side chain coordinates consistent with the expected structures in presence of ions. The precise AlphaFold2 predicted model of F13 protein acquired a high pLDDT score of 92.50% (Fig. S1A), suggesting its higher confidence score. The generated model from AlphaFold2 was further subjected to various model evaluation servers to examine its prediction accuracy and stereo-chemical qualities. The overall-quality of our modelled F13 was further supported by the ERRAT score of 90.12% which indicates acceptable protein environment and evaluated the non-bonded interactions between different atom types. The PROCHECK [65] analysis was used to assess the amino acid residues' placement in the Ramachandran plot [26]. The distributions of φ (phi) and ψ (psi) angles of the amino acid residues obtained by Ramachandran plots showed that 88.10% were in most favoured regions, 11.7% in additional allowed region and 0.3% in generously allowed region with no residues in disallowed region (Table S2).
Furthermore, ProSA [28], ProQ [29], and MolProbity [30] servers were also used for stereochemical calculations. The ProQ analysis of the F13 model revealed an LG score of 7.17 (where, LG-score>4 signifies extremely good model). The ProSA-web tool was used for refinement and validation of the modelled protein by comparing the model's energy with the potential mean force derived from a large set of known protein structures to check for the native protein folding energy. Energy profile of the proposed model and the Z-score value (a measure of model quality as it measures the total energy of the structures) calculates the interaction energy per residue using a distance-based pair potential. The ProSA analysis of the model F13 achieves a Z-score of −9.15 kcal/mol (where the negative ProSA energy reflects reliability of the model) reflecting the quality of the model. The MolProbity web server provided a detailed atomic contact analysis to identify any steric problems or dihedral-angle diagnostics. Furthermore, analysis of the modelled F13 in MolProbity server showed no bad bonds and angles within the amino acid residues which additionally confirmed the reliability of our model (Fig. S2B). Altogether, these consistent results from our analysis exhibits the better performance of our AlphaFold2 modelled F13 protein.
3.2. Exploring the structural reliability of the AlphaFold2 modelled F13 protein
As suggested from previous studies, the reliability of AlphaFold2 modelled protein structures is non speculative without performing further post-modelling refinement techniques [66]. Hence, to further gauge the suitability of the AlphaFold2 modelled F13 in high-throughput drug discovery in a real-world scenario, refinement of the structure was done through all-atoms MD simulations in triplicates (Fig. 1 ) [66]. The modelled F13 and its close homolog phospholipase D from Serratia plymuthica shares a highly conserved HKD motifs including catalytic residues histidine (His115, His325) and lysine (Lys118 and Lys327), which is accountable for self-dimerization (Figs. S3A and B) [67]. The utilization of PointSite and PrankWeb (Fig. S4) tools identified key binding site residues His115, Lys117, Asn132, His325, and Lys327 which is in correlation with other studies [68]. Upon structural superimposition of the model structure with the reference structure (phospholipase D) (Fig. S1B) it was observed that replicate 1 and 3 had a lower RMSD of 1.0 Å than replicates 2 (1.15 Å).
Fig. 1.
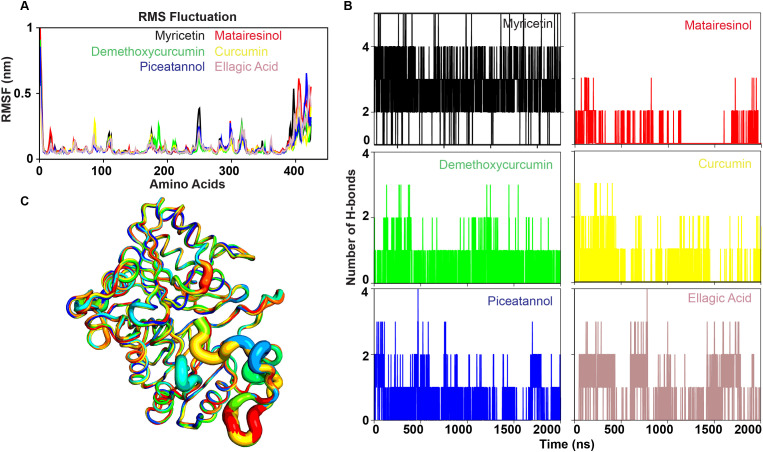
F13 protein modelled with AlphaFold2 and embedded in the TIP3P water model and phosphate groups for all-atoms MD simulations. And dynamics of protein simulation plotted in box and whisker method using Origin Pro. (A) Backbone RMSD of MD simulated replicates of F13 in its Apo state (B) Root mean square fluctuations of the Cα-atoms of the triplicates. (C) Rg plot representing the compactness of the triplicates. (D) The SASA plot as a function of time for triplicates.
To assess the stability of MD simulated structures, we computed RMSD, RMSF, Rg and SASA profile for the triplicates. Based on Fig. 1A, it can be observed that replicate 1, 2 and 3 of the Apo state exhibited a consistent trend with an average RMSD value of 0.23 nm ± 0.03 nm, 0.19 ± 0.02 nm and 0.20 ± 0.03 nm until 200 ns. This value is below the threshold of 0.25 nm, which is considered as a reliable indicator of a structure's close resemblance to the reference structure according to the literature [69]. Analysis of the last 100 ns MD trajectories of each system, records the behavioural changes of F13 protein at amino acid level including the RMSF of Cα-atoms. The findings revealed that the N-terminal domain exhibited less fluctuation on contrary to the C-terminal domain in all replicates (Fig. 1B).
The information concerning the system's equilibrium measured with Rg, states the system compactness depending upon increased or decreased perturbation [70]. The mean Rg of the replicate 1, 2 and 3 were recorded as 2.15 nm, 2.15 nm and 2.17 nm respectively (Fig. 1C). The lowest Rg value exhibited by the replicate 1 and 2 was maintained till 200 ns. The average SASA values for the replicates 1, 2 and 3 were calculated as 192 nm2, 193 nm2 and 198 nm2 respectively (Fig. 1D).
On the top of mentioning that all the replicates yield overall good results, we concluded that replicate 1 showed the best structural dynamics, hence was selected for further exercises.
3.3. Molecular recognition of the antiviral polyphenols by optimized AlphaFold2 F13 protein
Molecular docking analysis is a useful method for predicting protein-ligand binding activities and can be used to virtually screen large databases of compounds [71]. In this study, screening of 57 polyphenols was conducted, resulting in the identification of 128 docked ligand conformations. These conformations were then screened based on their Glide XP and MM/GBSA scores, and the top 6 conformations were selected for further analysis of their intermolecular interactions, as detailed in Table 1 and Table S1.
Table 1.
List of polyphenols used in this study with their docking scores, MM/GBSA binding free energy and amino acids forming important non-bonded contacts with AlphaFold2 modelled F13.
| Complex | SP score (kcal/mol) | XP score (kcal/mol) | MM/GBSA (kcal/mol) | Interaction |
|
|---|---|---|---|---|---|
| Hydrogen Bonds | Hydrophobic Interactions | ||||
| Myricetin | −6.71 | −8.40 | −36.56 | Trp280, Glu424, Asn341, Asp134 | Met239, Leu423 |
| Matairesinol | −5.71 | −7.76 | −29.98 | Asn240, Asn341 | Arg136, Val141, Trp280, Leu423 |
| Demethoxy-curcumin | −6.26 | −7.31 | −43.91 | Ser137, Asn240, Trp280, Asn341, Ser130 | His325, Arg282, Leu423, Trp280, Val141 |
| Piceatannol | −6.00 | −7.30 | −38.82 | Asn132, Asn240, Trp280, Asn341 | Leu423 |
| Curcumin | −5.47 | −5.92 | −31.89 | Lys117, Asn240, Asn341 | Tyr48, Leu423, His325 |
| Ellagic Acid | −5.06 | −5.24 | −36.57 |
Trp280, Glu424, Asp134 |
Trp280, His325, Val141 |
Binding free energy of Matairesinol, Curcumin, Ellagic Acid, Demethoxycurcumin, Piceatannol, and Myricetin attained as −29.98 kcal/mol, −31.89 kcal/mol, −36.57 kcal/mol, −43.91 kcal/mol, −38.82 kcal/mol, and −36.56 kcal/mol, XP scores −7.76 kcal/mol, −5.92 kcal/mol, −5.24 kcal/mol, −7.31 kcal/mol, −7.30 kcal/mol, and −8.40 kcal/mol, respectively. These screened polyphenols have previously been reported to have antiviral activity by targeting viral particles directly [21].
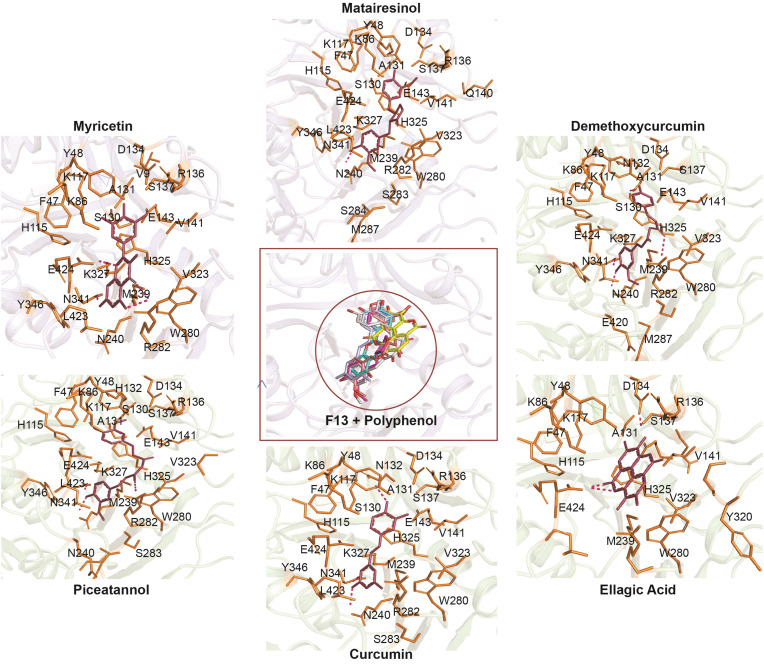
Moreover, we identified specific amino acid residues, such as Asn240, Trp280, Asn341, Val141, His325, and Leu423 that contributed towards the intermolecular interactions between the polyphenols and modelled F13 protein. Strong hydrogen bonds were observed in all the six complexes, including certain dominating hydrophobic contacts. A 2-dimensional representation of the non-bonded interactions between F13 and polyphenols is depicted in Fig. S5, while a superimposed view of the top six docked complexes displaying the binding pocket of the F13 protein and non-bonded interactions of the six highest-ranking complexes are shown in Fig. 2 . Further validation of our docking protocol through AutoDock Vina also reveals that the binding mode of the screened ligands is similar to ones generated using Glide scoring functions. Fig. S6 is the comparative illustration of the orientation of polyphenol binding to the modelled F13. The binding affinities of the screened polyphenols as generated by AutoDock Vina have been enlisted in Table S3.
Fig. 2.
Illustration of the superimposed view of top-ranked docked poses of polyphenols and the binding poses of polyphenols within the binding site of modelled F13.3-D interaction between F13 and the top six polyphenols (F13-Myricetin, F13-Matairesinol, F13-Demethoxycurcumin, F13-Piceatannol, F13-Curcumin, and F13-Ellagic Acid).
Myricetin, also known as 3,5,7-trihydroxy-2-(3′,4′,5′-trihydroxyphenyl)-4H-chromen-4-one, belongs to the group of flavonoids, found in diverse groups of plants such as bayberry, vine tea, grape and pomegranate in the form of glycosides [72]. Earlier studies confirm the antiviral, antitumor [73], antioxidant [57], antibacterial [58] as well as anti-inflammatory effects of Myricetin for other viruses such as SARS-CoV-2 [[74], [75], [76]]. It is noteworthy to mention that Myricetin prevents viral infection by stimulating epithelial Cl− secretion [77]. The presence of 3′-OH, 4′-OH and 5′-OH on the B-ring of Myricetin makes them more effective in nature [78]. Studies have shown that the solubility rate of Myricetin in organic solvents is less in room temperature [79]. Most importantly, it is moderately membrane permeable with an apparent permeability coefficient, Papp = 1.7 × 10−6 cm/s [80].
Demethoxycurcumin and Curcumin belongs to the curcuminoid group of polyphenols. Curcumin, also known as diferuloylmethan are extracted from ground rhizomes of the plant Curcuma longa L. (Zingiberaceae) [81]. The antiviral activity of Curcumin against many enveloped viruses has attracted a considerable group of researchers. Meanwhile, a study based on SARS-CoV-2 also highlights the inhibitory activity of Demethoxycurcumin [82]. Piceatannol and Matairesinol belong to stilbenes and lignans groups of polyphenols respectively. Though the antiviral properties of these groups of polyphenols including curcuminoids have not been thoroughly explored, however there are evidences on use of these phytocompounds for inhibition of viruses such as herpes simplex, HIV, influenza and human papilloma, etc. [59,60]. Ellagic Acid, being a member of phenolic acid group possesses high antioxidant and medicinal properties including anti-viral, anti-bacterial, anti-carcinogenic and anti-inflammatory activities [56]. Understanding the structural basis of the antiviral activity exhibited by these polyphenols will provide new insights into their mode of action.
3.4. Essential dynamics of F13-polyphenol complexes
To understand the atomistic movement of each residue in each F13-polyphenol system governed by inter-atomic interactions, we further calculated the RMSD, RMSF, Rg, and SASA profiles of the entire MD simulated systems. The average RMSD values for Myricetin, Matairesinol, Demethoxycurcumin, Piceatannol, Curcumin, and Ellagic Acid-F13 complexes were calculated as 0.2 nm, 0.26 nm, 0.17 nm, 0.18 nm, 0.23 nm and 0.16 nm respectively (Fig. 3 A). All F13-polyphenol complexes exhibited a distribution RMSD value of 0.2 nm, except for Piceatannol, which showed a RMSD of 0.1 nm over the 200 ns (Fig. S7A). These RMSD analyses showed that the complexes formed by Myricetin, Matairesinol, Demethoxycurcumin, Piceatannol, Curcumin and Ellagic Acid endured stability with minimal fluctuation over the 200 ns simulation time. However, we observed maximum stability in Ellagic Acid followed by Demethoxycurcumin, Piceatannol and Myricetin.
Fig. 3.
Box-whisker plot displaying backbone RMSD, Cα-RMSF profile, Rg and SASA of top six polyphenols bound with F13 over the time scale of 200 ns. (A) RMSD of backbone atoms of F13. (B) RMSF profiles of residues of F13 in complex with all top six polyphenols. (C) The compactness of top six F13-polyphenols bound states assessed by plotting the radius of gyration profile. (D) SASA plot for top six polyphenols bound with F13 during 200 ns MD.
The average ligand RMSD analysis showed a consistent trend with minimal fluctuation in Ellagic acid, indicating a stable complex with no significant movement or structural changes observed during the 200 ns simulation period. However, the ligand RMSD profiles of Myricetin, Matairesinol, Demethoxycurcumin, Piceatannol, and Curcumin, exhibited little higher RMSD over the simulation period (Fig. S7B). The increased RMSD of the ligands may be due to their change in orientation in the binding site cavity of the modelled F13 protein.
The average RMSF profile of Cα-atoms of each system provides crucial insight into the degree of residual fluctuations in F13 upon ligand binding [33]. Our study displayed that the first five residues in the N-terminal region and residues Cys394 to Glu424 in the C-terminal region shows higher mobility in all six systems. The loops connecting α-β-α units at the N-terminal domain and the loops connecting helices and strands at C-terminal region displayed high degree mobility, which might be resulted due to the low pLDDT score in the AlphaFold model, whereas, helices and strands had minimal fluctuation. Remarkably, we observed mixed loop/β-strand conformations around 244–254 amino acids in F13-polyphenol complexes (Fig. 3B).
Compactness and folding mechanism of the F13-polyphenol complexes was assessed from their Rg [83]. The average Rg values for the above-mentioned systems were calculated as 2.14 nm, 2.18 nm, 2.16 nm, 2.17 nm, 2.16 nm and 2.15 nm respectively. The F13-polyphenol complexes showed a minor deviation in Rg values, indicating towards their stability and compactness maintained over the time scale of 200 ns MD simulation (Fig. 3C). Also, the calculated solvent accessible surface area of these systems exhibited stable SASA (∼193 nm2–197 nm2) throughout the MD simulation (Fig. 3D). Altogether, we can conclude that, among all the analyses performed on the F13-polyphenol complexes so far, Myricetin, Demethoxycurcumin and Ellagic Acid have maintained stable trend throughout the simulation which also compliment with the findings of docking analysis.
3.5. All-atoms MD simulation in assessing mobility and flexibility of polyphenol bound F13 systems
3.5.1. Flexibility/plasticity among the F13-polyphenol complexes
As mentioned earlier, the RMSF profiles of the Cα-atoms in each system estimates the stability and plasticity of each MD simulated system as well as the role of each fluctuating residue in protein flexibility upon ligand binding. The average RMSF values for Myricetin, Matairesinol, Demethoxycurcumin, Piceatannol, Curcumin, and Ellagic Acid-bound F13 complexes are 0.09 nm, 0.10 nm, 0.08 nm, 0.09 nm, 0.08 nm and 0.09 nm respectively. We also observed a higher statistical residual fluctuation of residues Cys394-Glu424 towards the C-terminal end with minimal residual fluctuations in the loop region from residue Ile244-Phe254 (Fig. 4 A).
Fig. 4.
The average B-factor/thermal-fluctuation profile of MD simulation and dynamics of intermolecular hydrogen bonds of the F13-ligand complexes over a timescale of 200 ns. (A) The average RMSF profile of F13-polyphenol complexes. (B) The superimposed F13 bounded top six polyphenols complex B-factor/thermal-fluctuation profile shown in putty format (thicker region with high degree of mobility/flexibility). (C) Intermolecular H-bond dynamics in top six F13-polyphenol complexes during 200 ns MD simulation.
Hydrogen bonds and hydrophobic contacts are important for protein-ligand complexes [84] and increasing the number of hydrophobic atoms can enhance the biological activity of a lead drug [85]. Intermolecular contact analysis can reveal differences between docked complexes and MD simulated systems, which may be due to structural rearrangements or ligand reorientation within the binding pocket of the protein. Table S4 enlists the key residues involved in forming important hydrophobic contacts as well as hydrogen bonds. Glu143, Asp134, Asn345, Ser321, Lys86, Lys117 and Tyr320 were identified as residues crucial for ligand recognition. The various non-bonded contacts including H-bonds formed between F13 and the screened plant-based polyphenols have been illustrated in Fig. 4B. Myricetin and Piceatannol strongly binds to F13 in the binding pocket by forming an average of 4 hydrogen bonds per frame followed by Ellagic Acid, Demethoxycurcumin, Curcumin and Matairesinol (∼2–3 hydrogen bonds per frame). The hydrogen bond occupancies of donor and acceptor atoms of F13 and polyphenols are listed in Table S5. We observed that Glu143 possesses the highest hydrogen bond occupancy of 97% in Myricetin complex followed by Glu424 (28.07%) in Piceatannol and Asp134 (25.52) and Glu417 (12.74%) in Ellagic Acid. On the other hand, we noticed least percentage of hydrogen-bond occupancies in Demethoxycurcumin, Curcumin and Matairesinol complexes.
The B-factor/thermal fluctuation analysis of the top six polyphenols bound F13 complexes is consistent with the RMSF profile of F13-ligand complexes, showing high degree of fluctuation at the C-terminal region (Fig. 4C).
3.5.2. Understanding the global motion of F13-polyphenol complexes
The global motion of ligand-bound states can be captured by using the principal components (PCs) from the MD trajectories by diagonalizing the covariance matrices of the main-chain atoms of the protein through the eigenvectors (EVs) or, the PCs and their eigenvalues [86,87]. A scatter plot was generated using the first two principal components (PC1 and PC2) obtained from PCA for each system, including Myricetin, Matairesinol, Demethoxycurcumin, Piceatannol, Curcumin and Ellagic Acid. The covariance matrices of the Cα-atoms revealed that trace values 24.58 nm2, 37.94 nm2, 21.76 nm2, 25.73 nm2, 18.74 nm2, and 26.62 nm2 for each system respectively. Curcumin, with least trace value followed by Demethoxycurcumin and Myricetin had fewer fluctuations in Cα-atoms in comparison with other systems, suggesting towards their more defined and stable conformation in the phase space as represented by PCA plot. The 2D projection of the first two eigenvectors in phase space supported the compactness of all polyphenols (Figs. S8A and B Supporting text).
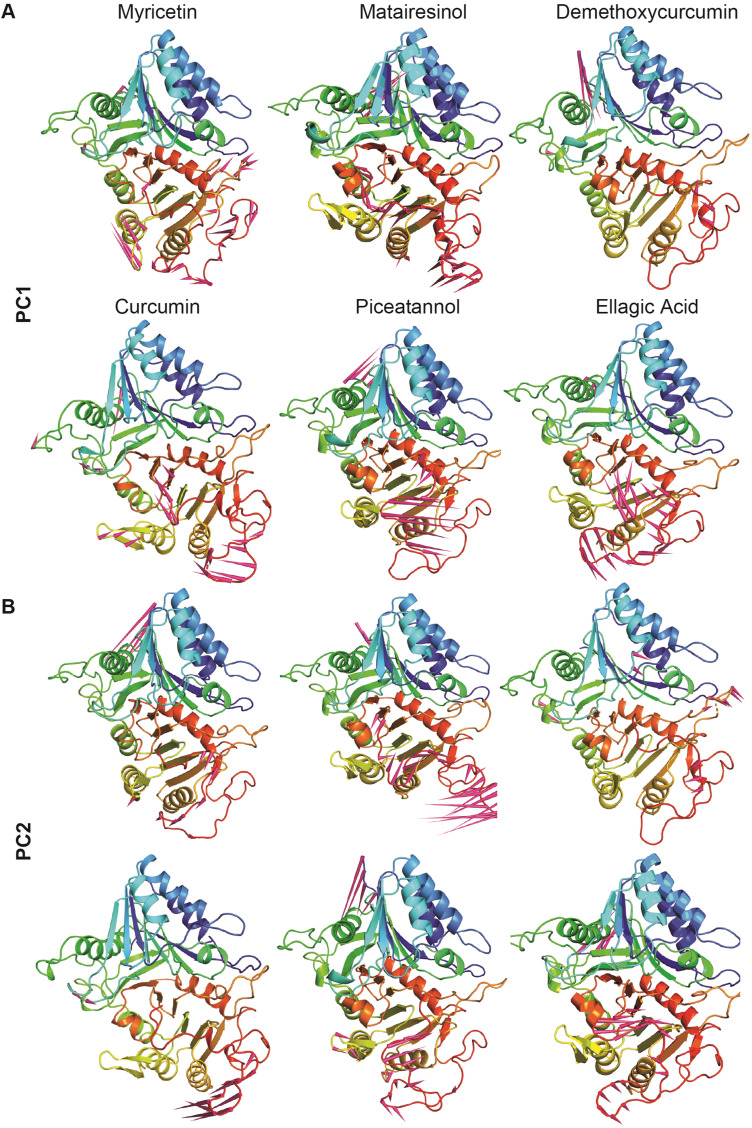
To understand the direction of movement captured by the eigenvectors, porcupine plots were generated using the extreme projections on both PC1 and PC2 (Fig. 5 ). The direction of the arrow in main chain atom represents the direction of motion, while the length of the arrow characterizes the movement strength (Supporting Text). The N-terminal end and the larger loop in the C-terminal domain was found to contribute to the major movements observed in Matairesinol, Piceatannol, Curcumin, and Ellagic Acid as indicated by larger cones in a cone plot. These results are consistent with the results of the RMSF analysis. Taking into consideration all the above-mentioned analyses, we can conclude that Myricetin and Demethoxycurcumin forms the most stable and compact complex with F13 possessing least fluctuation at the amino acid level.
Fig. 5.
Porcupine plot displaying the movement of main-chain atoms of each system corresponds to PC1 and PC2 obtained by performing PCA on MD trajectories. (A) Porcupine plot displaying the motion of top six polyphenol complex with modelled protein F13 obtained from PC1. (B) Porcupine plot displaying the motion of top six polyphenol complex with modelled protein F13 obtained from PC2.
3.5.3. Structural heterogeneity among the F13-polyphenol complexes
To understand the structural heterogeneity in the MD simulated ensembles, we implemented RMSD-based gmx cluster tool with a cut-off of 0.2 nm over the last 100 ns trajectories to generate structural ensembles. Fig. 6 A demonstrates the electrostatic surface potential map of the MD simulated F13-polyphenol complexes. The distribution of charges observed in the surface of the MD simulated F13 reveals that the polyphenols are buried inside a hydrophobic cavity formed by negatively charged residues. The structural superposition of the docked complexes with their respective cluster representatives obtained from clustering analysis have been displayed in Fig. 6 (B). The Cα-RMSD values for the protein-ligand complexes Myricetin, Matairesinol, Demethoxycurcumin, Piceatannol, Curcumin and Ellagic Acid are found to be 0.9 Å, 0.7 Å, 0.8 Å, 0.9 Å, 0.8 Å and 1.10 Å respectively. MD simulations revealed that the ligand binding orientation in the top cluster was slightly different from the docked state, but the overall ligand binding orientation remained relatively stable. We also observed the formation of β-strands in Tyr381-Tyr383 region in Myricetin and Ellagic Acid complexes, with formation of small 3–10 α-helices in regions Tyr107-Leu110 in Ellagic Acid and Gln209-Leu211 in Myricetin, Arg395-Leu397 in Myricetin, Matairesinol, Curcumin and Ellagic Acid complexes. The formation of β-strands can be observed in the loop in C-terminal region which has been observed to be a major contributor towards flexibility of the F13 protein structure (supported from the results of RMSF and PCA). These findings suggest that the F13-polyphenol complexes are stable during MD simulations and the binding orientation of the ligand is maintained till 200 ns.
Fig. 6.
Structural superimposition of the top ranked cluster representatives of F13 bound polyphenol complexes obtained from the clustering analysis and their electrostatic surface potential map. (A) Distribution of charges noticed in the electrostatic surface potential map of the superposed MD simulated F13-polyphenol complexes. (B) Structural heterogeneity observed in the superposed structures of the docked as well as MD simulated F13-polyphenol complexes.
VMD's timeline viewer tool was used to analyse time-dependent changes in secondary structure elements of each F13 polyphenol complex system throughout the 200 ns MD simulation. Our analysis suggests that ligand interactions can alter conformational and structural changes in the protein (Fig. S9, supporting text).
3.6. Binding free energy of F13-polyphenol complexes
The quantitative understanding of the strength of any bio-molecular interaction involved in ligand recognition (in our case) or any catalytic process involves the use of diverse computational approaches such as free energy perturbation (FEP), thermodynamics integration (TI), linear interaction energies (LIE), molecular mechanics Poisson-Boltzmann surface area (MM/PBSA) and molecular mechanics Generalized Born surface area (MM/GBSA) approaches [55,[88], [89], [90], [91], [92], [93], [94]]. However, the use of simple scoring functions often ignores the contribution of important energy terms such as free energy of solvation. Therefore, the MM/PBSA approach has been used to compute the interaction energies and study biomolecular complexes. The binding free energy of each protein-ligand systems from the MD trajectories are computed using 200 snapshots through molecular mechanics based MM/PBSA method. The energetic terms contributing to the total binding free energy is summarized in (Table 2 ).
Table 2.
MM/PBSA binding free energy of the F13-polyphenol complexes computed using 200 snapshots extracted at equal interval of time from the last 100 ns trajectories of each system.
| Complex | Vander Waal energy (KJ/mol) | Electrostatic energy (KJ/mol) | Polar solvation energy (KJ/mol) | SASA energy (KJ/mol) | Binding energy (KJ/mol) |
|---|---|---|---|---|---|
| Myricetin | −109.01 ± 0.96 | −103.50 ± 1.75 | 191.29 ± 2.41 | −14.29 ± 0.07 | −35.50 ± 1.18 |
| Matairesinol | −24.84 ± 2.11 | −4.36 ± 0.69 | 22.17 ± 4.05 | −3.32 ± 0.27 | −10.23 ± 4.02 |
| Demethoxycurcumin | −146.01 ± 2.13 | −54.17 ± 1.27 | 153.88 ± 2.72 | −18.51 ± 0.24 | −64.86 ± 1.30 |
| Piceatannol | −121.55 ± 0.78 | −36.46 ± 1.60 | 123.10 ± 2.13 | −14.63 ± 0.06 | −49.44 ± 1.42 |
| Curcumin | −88.32 ± 2.58 | −20.86 ± 1.30 | 72.79 ± 3.49 | −12.11 ± 0.34 | −48.53 ± 1.80 |
| Ellagic Acid | −79.53 ± 1.62 | −35.96 ± 2.68 | 97.47 ± 3.15 | −10.03 ± 0.12 | −28.25 ± 1.60 |
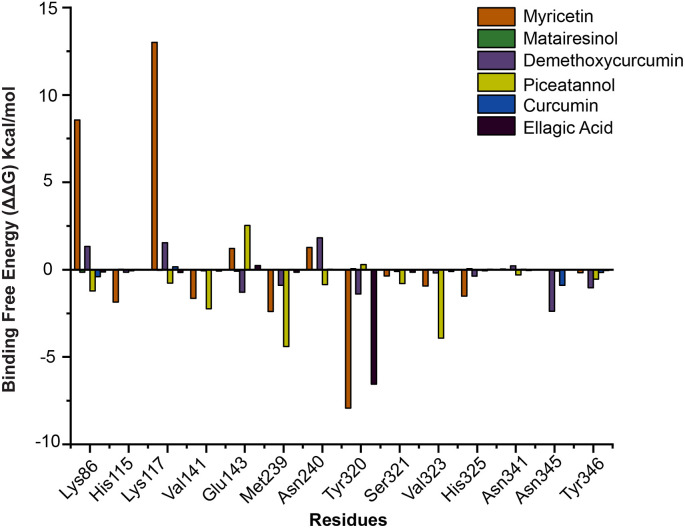
It was observed that Curcumin, Demethoxycurcumin, Piceatannol and Myricetin complexes displayed higher binding free energy of −48.53 ± 1.80 kJ/mol, −64.86 ± 1.30 kJ/mol, −49.44 ± 1.42 kJ/mol and −35.50 ± 1.18 kJ/mol respectively. Among the energy components, van der Waals energy contributed mostly towards the binding process of polyphenols towards F13. The polar and apolar energetic contributions of these compounds towards binding free energy from the last 100 ns of equilibrated trajectories are depicted in Fig. S10. However, as binding free energy calculation does not reveal about the true entropic changes [95], per-residue decomposition analysis is conducted to understand the contributions of the critical or, ligand binding residues towards the binding process (Fig. 7 ). In all the ligand bound complexes Lys117, Glu143, Ser321 and Asn345 formed H-bonds, while, Trp280, Met239, His325, Val141 and Tyr346 contributed through pi-alkyl contacts. Most of the residues participating in ligand recognition contributed towards negative free energy of the F13-ligand bound complexes.
Fig. 7.
Per-residue decomposition analysis displaying the energetic contribution of important amino acids towards binding free energy of F13-polyphenol complexes.
In this study, we performed a structure-based high-throughput virtual screening on 57 naturally available antiviral polyphenols that were published in the literature, against the AlphaFold2 modelled F13 protein. The utility of state-of-the-art machine learning approach implemented in AlphaFold2 as well as all-atoms MD simulation elucidates the differential conformational states and mode of interaction of the F13-polyphenol complexes. Previous studies have shown the accuracy of protein backbone and side chain prediction of AlphaFold2 in active and inactive states, which could be a major limitation while studying the structural behaviour of the protein in any alternative conformation [[96], [97], [98], [99]]. The antiviral activities of the diverse groups of polyphenols attract researches for their extensive study. Considering their stability, reactivity, synergism as well as bioavailability during their recovery, processing, storage and market applications are some of the important aspects that needs to be taken care of [100]. These polyphenols exert antiviral activities based on the nature of their interacting virus (DNA or RNA virus). In our study, we performed state-of-the-art all-atoms MD simulations on the top six screened polyphenols to get a clear vision on the mechanism of ligand recognition. Though we are confident about our results from the study, the use of 57 polyphenols with known antiviral activity for virtual screening have limited our study to a small chemical space. The significance of experimental research and the valuable insights it can yield in the field of biological science, is well known. However, due to various constraints and limitations of resources, our study is solely based on computational approaches. The accuracy and efficacy of our screened polyphenols against F13 protein of MPXV can be further achieved through in vivo and in vitro experiments.
4. Conclusion
The major envelop protein F13 plays a crucial role in viral entry and formation of EEV, making it a promising target for drugs to treat poxvirus infections. To understand the dynamics and differential modes of interaction of plant-based antiviral polyphenols and F13, the implementation of the state-of-the-art machine learning based AlphaFold2 in structure prediction succeeds in modelling a high precision F13 model in its apo state. Keen observation of the modelled protein reveals its resemblance with to the members of Phospholipase-D family, with a highly conserved HxKxxxxD (HKD) motif responsible for self-dimerization. The predicted structure of F13 adopts β-α-β-α-β sandwich fold, which is composed of two distinct domains connected by a flexible loop.
The intermolecular contact analysis based on molecular docking and all-atoms MD simulations reveals that, Myricetin and Demethoxycurcumin are the top two plant-based antiviral polyphenols that consistently interacts with the binding site residues of modelled F13 by forming strong hydrogen bonds as well as hydrophobic contacts. The above-mentioned findings perfectly corroborate with binding free energy calculation.
In the MD simulated polyphenol-F13 complexes, we have observed that minor structural changes in the protein structure have resulted from the high degree of fluctuation observed in the loop of the C-terminal domain. However, this fluctuation might have occurred due to the low pLDDT score acquired by the region during model prediction.
Based on intermolecular contact analyses and molecular mechanics based binding free energy calculations, we have identified Glu143, Asp134, Asn345, Ser321, Lys86, Lys117 and Tyr320 as putative active site residues involved in ligand recognition and binding process. Our docking and MD simulations suggest that Myricetin and Demethoxycurcumin could be the promising polyphenols for developing antivirals against MPXV by inhibiting F13.
However, further in vitro and in vivo investigations are needed to confirm these results. Our study is the first attempt to understand the dynamics of F13 and plant-based polyphenols that exhibit antiviral properties, and further experimental validation is required to develop effective therapeutics against MPXV.
Authors contributions
Madhusmita Rout: Methodology, Software Investigation, Data curation, Visualization, Writing – original draft. Sarbani Mishra: Methodology, Software Investigation, Data curation, Visualization, Writing – original draft. Suchanda Dey: Writing – review & editing. Mahender Kumar Singh: Data analysis, Budheswar Dehury: Conceptualization, Methodology, review & editing, and Supervision, Sanghamitra Pati: Writing – review & editing, and funding acquisition.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Acknowledgements
The authors are thankful to the Department of Health Research, Ministry of Health and Family Welfare, Govt. of India for providing financial support under the DHR-Young Scientist grant (No. R.12014/41/2021-HR/E-Office:8116317) to BD. The authors also acknowledge Indian Council of Medical Research, New Delhi for providing financial support for the study through intramural funding.
Biographies
Ms. Madhusmita Rout (M.Sc.) is a Research Intern in the Bioinformatics Division of ICMR-Regional Medical Research Centre, Bhubaneswar, Odisha. Her research interests focus on application of machine learning in structure-based drug design.
Ms. Sarbani Mishra (M.Sc.) is a Research Intern in the Bioinformatics Division of ICMR-Regional Medical Research Centre, Bhubaneswar, Odisha. Her research focuses on viral genomics, immunoinformatics and antiviral drug development.
Mrs. Suchanda Dey (M.Sc.) is a final year Ph.D. scholar of Biomics and Biodiversity Lab, Siksha ‘O’ Anusandhan (deemed to be) University, Bhubaneswar. Her research interests include microbial comparative genomics and antimicrobial drug resistance.
Dr. Mahender Kumar Singh (Ph.D.) is a data scientist by profession and hard-core computational biologist. His research focuses on development of algorithms, data mining and machine learning.
Dr. Budheswar Dehury (Ph.D.) a Young Scientist in the Bioinformatics division of ICMR-Regional Medical Research Centre, Bhubaneswar, Odisha. His research focuses on structural biology of membrane proteins, protein stability and molecular dynamics.
Dr. Sanghamitra Pati (MBBS, MD and MPH) is the Director of ICMR- Regional Medical Research Centre, Bhubaneswar, Odisha and is an expert in the field of public health, meta-analysis, virology and multimorbidity.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.compbiomed.2023.107116.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Petersen B.W., Damon I.K. Orthopoxviruses: vaccinia (smallpox vaccine), Variola (smallpox), monkeypox, and cowpox. Mand. Douglas, Bennett’s Princ. Pract. Infect. Dis. 2014;2:1694–1702.e3. doi: 10.1016/B978-1-4557-4801-3.00135-1. [DOI] [Google Scholar]
- 2.Mileto D., et al. New challenges in human monkeypox outside Africa: a review and case report from Italy. Trav. Med. Infect. Dis. 2022;49 doi: 10.1016/j.tmaid.2022.102386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Venkatesan P. Monkeypox transmission—what we know so far. Lancet Respir. Med. 2022 doi: 10.1016/s2213-2600(22)00386-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kmiec D., Kirchhoff F. Monkeypox: a new threat? Int. J. Mol. Sci. 2022;23(14) doi: 10.3390/ijms23147866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.The Lancet, “Monkeypox: a global wake-up call,”. Lancet. 2022;400(10349):337. doi: 10.1016/S0140-6736(22)01422-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Release P. 2022. “Multi-country Monkeypox Outbreak: Situation Update (10 June 2022),”; pp. 1–9.https://www.who.int/emergencies/disease-outbreak-news/item/2022-DON392 June. [Online]. Available: [Google Scholar]
- 7.Monkeypox.”.
- 8.Rimoin Aw K.-I., Kisalu N., et al. Emerg Infect Dis; 2001– 2004. Endemic Human Monkeypox, Democratic Republic of Congo; p. 934. –7, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hutin Y.J., Williams R.J., Malfait P. Outbreak of human monkeypox, democratic republic of Congo 1996 to 1997. Emerg. Infect. Dis. 2001;7:434–438. doi: 10.3201/eid0703.010311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jezek Z., Szczeniowski M., Paluku K.M., Mutombo M. Human monkeypox: confusion with chickenpox. Acta Trop. 1988;45(4):297–307. [PubMed] [Google Scholar]
- 11.Smith G.L., Vanderplasschen A., Law M. The formation and function of extracellular enveloped vaccinia virus. J. Gen. Virol. 2002;83(12):2915–2931. doi: 10.1099/0022-1317-83-12-2915. [DOI] [PubMed] [Google Scholar]
- 12.Cao S., Realegeno S., Pant A., Satheshkumar P.S., Yang Z. Suppression of poxvirus replication by resveratrol. Front. Microbiol. 2017;8(NOV):1–10. doi: 10.3389/fmicb.2017.02196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar N., Goel N. Phenolic acids: natural versatile molecules with promising therapeutic applications. Biotechnol. Reports. 2019;24 doi: 10.1016/j.btre.2019.e00370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.El-Toumy S.A., Salib J.Y., El-Kashak W.A., Marty C., Bedoux G., Bourgougnon N. Antiviral effect of polyphenol rich plant extracts on herpes simplex virus type 1. Food Sci. Hum. Wellness. 2018;7(1):91–101. doi: 10.1016/j.fshw.2018.01.001. [DOI] [Google Scholar]
- 15.Sundararajan A., et al. Influenza virus variation in susceptibility to inactivation by pomegranate polyphenols is determined by envelope glycoproteins. Antivir. Res. 2010;88(1):1–9. doi: 10.1016/j.antiviral.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu G., et al. Antiviral activity and possible mechanisms of action of pentagalloylglucose (PGG) against influenza A virus. Arch. Virol. 2011;156(8):1359–1369. doi: 10.1007/s00705-011-0989-9. [DOI] [PubMed] [Google Scholar]
- 17.Cory H., Passarelli S., Szeto J., Tamez M., Mattei J. The role of polyphenols in human health and food systems: a mini-review. Front. Nutr. 2018;5(September):1–9. doi: 10.3389/fnut.2018.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shahidi F., Ambigaipalan P. Phenolics and polyphenolics in foods, beverages and spices: antioxidant activity and health effects – a review. J. Funct.Foods. 2015;18:820–897. doi: 10.1016/j.jff.2015.06.018. [DOI] [Google Scholar]
- 19.Cheynier V. Phenolic compounds: from plants to foods. Phytochemistry Rev. 2012;11(2–3):153–177. doi: 10.1007/s11101-012-9242-8. [DOI] [Google Scholar]
- 20.Montenegro-landívar M.F., et al. Polyphenols and their potential role to fi ght viral diseases : an overview. Sci. Total Environ. 2021;801 doi: 10.1016/j.scitotenv.2021.149719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Montenegro-Landívar M.F., et al. Polyphenols and their potential role to fight viral diseases: an overview. Sci. Total Environ. 2021;801 doi: 10.1016/j.scitotenv.2021.149719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16(22):10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu J.‐H. Basic local alignment search tool. Catal. Today Z. 2020 doi: 10.1002/9783527809080.cataz02180. [DOI] [Google Scholar]
- 24.Altschul S.F., et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jumper J., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596(7873):583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramachandran G.N., Ramakrishnan C., Sasisekharan V. Stereochemistry of polypeptide chain configurations. J. Mol. Biol. 1963;7(1):95–99. doi: 10.1016/S0022-2836(63)80023-6. [DOI] [PubMed] [Google Scholar]
- 27.Colovos C., Yeates T.O. Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. 1993;2(9):1511–1519. doi: 10.1002/pro.5560020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wiederstein M., Sippl M.J. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35(SUPPL.2) doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wallner B., Elofsson A. Can correct protein models be identified? Protein Sci. 2003;12(5):1073–1086. doi: 10.1110/ps.0236803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davis I.W., et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35(SUPPL.2) doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bekker H., et al. Gromacs: a parallel computer for molecular dynamics simulations. Phys. Comput. 1993;92:252–256. January. [Google Scholar]
- 32.Dehury B., et al. Structural analysis and molecular dynamics simulations of novel δ-endotoxin Cry1Id from Bacillus thuringiensis to pave the way for development of novel fusion proteins against insect pests of crops. J. Mol. Model. 2013;19(12):5301–5316. doi: 10.1007/s00894-013-2010-x. [DOI] [PubMed] [Google Scholar]
- 33.Dehury B., Tang N., Blundell T.L., Kepp K.P. Structure and dynamics of γ-secretase with presenilin 2 compared to presenilin 1. RSC Adv. 2019;9(36):20901–20916. doi: 10.1039/c9ra02623a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Madhavi Sastry G., Adzhigirey M., Day T., Annabhimoju R., Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 2013;27(3):221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- 35.Shelley J.C., Cholleti A., Frye L.L., Greenwood J.R., Timlin M.R., Uchimaya M. Epik: a software program for pKa prediction and protonation state generation for drug-like molecules. J. Comput. Aided Mol. Des. 2007;21(12):681–691. doi: 10.1007/s10822-007-9133-z. [DOI] [PubMed] [Google Scholar]
- 36.Yan X., et al. PointSite: a point cloud segmentation tool for identification of protein ligand binding atoms. J. Chem. Inf. Model. 2022;62(11):2835–2845. doi: 10.1021/acs.jcim.1c01512. [DOI] [PubMed] [Google Scholar]
- 37.Grosenbach D.W., et al. Oral tecovirimat for the treatment of smallpox. N. Engl. J. Med. 2018;379(1):44–53. doi: 10.1056/nejmoa1705688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jendele L., Krivak R., Skoda P., Novotny M., Hoksza D. PrankWeb: a web server for ligand binding site prediction and visualization. Nucleic Acids Res. 2019;47(W1):W345. doi: 10.1093/nar/gkz424. –W349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ben-Shabat S., Yarmolinsky L., Porat D., Dahan A. Antiviral effect of phytochemicals from medicinal plants: applications and drug delivery strategies. Drug Deliv. Transl. Res. 2020;10(2):354–367. doi: 10.1007/s13346-019-00691-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu C., et al. OPLS4: improving force field accuracy on challenging regimes of chemical space. J. Chem. Theor. Comput. 2021;17(7):4291–4300. doi: 10.1021/acs.jctc.1c00302. [DOI] [PubMed] [Google Scholar]
- 41.Friesner R.A., et al. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 2006;49(21):6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 42.Singh A.K., Kushwaha P.P., Prajapati K.S., Shuaib M., Gupta S., Kumar S. Identification of FDA approved drugs and nucleoside analogues as potential SARS-CoV-2 A1pp domain inhibitor: an in silico study. Comput. Biol. Med. 2021;130 doi: 10.1016/j.compbiomed.2020.104185. October 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jacobson M.P., et al. A hierarchical approach to all-atom protein loop prediction. Proteins Struct. Funct. Genet. 2004;55(2):351–367. doi: 10.1002/prot.10613. [DOI] [PubMed] [Google Scholar]
- 44.S G., U R. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expet Opin. Drug Discov. 2015;10(5):449–461. doi: 10.1517/17460441.2015.1032936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jawarkar R.D., et al. QSAR based virtual screening derived identification of a novel hit as a SARS CoV-229E 3CLpro Inhibitor: GA-MLR QSAR modeling supported by molecular Docking, molecular dynamics simulation and MMGBSA calculation approaches. Arab. J. Chem. 2022;15(1) doi: 10.1016/j.arabjc.2021.103499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trott O., Olson A.J., Vina AutoDock. Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading,”. J. Comput. Chem. 2009 doi: 10.1002/jcc.21334. NA-NA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang J., Mackerell A.D. CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J. Comput. Chem. 2013;34(25):2135–2145. doi: 10.1002/jcc.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79(2):926–935. doi: 10.1063/1.445869. [DOI] [Google Scholar]
- 49.Vanommeslaeghe K., MacKerell A.D. Automation of the CHARMM general force field (CGenFF) I: bond perception and atom typing. J. Chem. Inf. Model. 2012;52(12):3144–3154. doi: 10.1021/ci300363c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Victor R. Berendsen and nose-hoover thermostats temperature in MD MD at constant temperature - NVT ensemble. Unknown. 2007:1–4. [Google Scholar]
- 51.Hoover W.G. Canonical dynamics: equilibrium phase-space distributions. Phys. Rev. 1985;31(3):1695–1697. doi: 10.1103/PhysRevA.31.1695. [DOI] [PubMed] [Google Scholar]
- 52.Nosé S. A molecular dynamics method for simulations in the canonical ensemble. Mol. Phys. 2002;100:191–198. [Google Scholar]
- 53.Parrinello M., Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 1981;52(12):7182–7190. doi: 10.1063/1.328693. [DOI] [Google Scholar]
- 54.Kumari R., Kumar R., Lynn A. G-mmpbsa -A GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model. 2014;54(7):1951–1962. doi: 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
- 55.Kollman P.A., et al. Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc. Chem. Res. 2000;33(12):889–897. doi: 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- 56.Oroian M., Escriche I. Antioxidants: characterization, natural sources, extraction and analysis. Food Res. Int. 2015;74:10–36. doi: 10.1016/j.foodres.2015.04.018. [DOI] [PubMed] [Google Scholar]
- 57.Bertin R., et al. Activity of myricetin and other plant-derived polyhydroxyl compounds in human LDL and human vascular endothelial cells against oxidative stress. Biomed. Pharmacother. 2016;82:472–478. doi: 10.1016/j.biopha.2016.05.019. [DOI] [PubMed] [Google Scholar]
- 58.Arita-Morioka K.I., Yamanaka K., Mizunoe Y., Tanaka Y., Ogura T., Sugimoto S. Inhibitory effects of Myricetin derivatives on curli-dependent biofilm formation in Escherichia coli. Sci. Rep. 2018;8(1) doi: 10.1038/s41598-018-26748-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abba Y., Hassim H., Hamzah H., Noordin M.M. Antiviral activity of resveratrol against human and animal viruses. Adv. Virol. 2015;2015 doi: 10.1155/2015/184241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Naithani R., et al. Antiviral activity of phytochemicals: a comprehensive review. Mini-Rev. Med. Chem. 2008;8(11):1106–1133. doi: 10.2174/138955708785909943. [DOI] [PubMed] [Google Scholar]
- 61.Mehany T., Khalifa I., Barakat H., Althwab S.A., Alharbi Y.M., El-Sohaimy S. Polyphenols as promising biologically active substances for preventing SARS-CoV-2: a review with research evidence and underlying mechanisms. Food Biosci. 2021;40 doi: 10.1016/j.fbio.2021.100891. January. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jumper J., et al. Applying and improving AlphaFold at CASP14. Proteins: Struct., Funct., Bioinf. 2021;89(12):1711–1721. doi: 10.1002/prot.26257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Laskowski R.A., Thornton J.M. PDBsum extras: SARS-CoV-2 and AlphaFold models. Protein Sci. 2022;31(1):283–289. doi: 10.1002/pro.4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jones D.T., Thornton J.M. The impact of AlphaFold2 one year on. Nat. Methods. 2022;19(1):15–20. doi: 10.1038/s41592-021-01365-3. [DOI] [PubMed] [Google Scholar]
- 65.Laskowski R.A., MacArthur M.W., Thornton J.M. 2012. PROCHECK : Validation of Protein-Structure Coordinates; pp. 684–687. [DOI] [Google Scholar]
- 66.Cavasotto C.N., Aucar M.G., Adler N.S. Computational chemistry in drug lead discovery and design. Int. J. Quant. Chem. 2019;119(2) doi: 10.1002/qua.25678. [DOI] [Google Scholar]
- 67.Wang F., Liu S., Mao X., Cui R., Yang B., Wang Y. Crystal structure of a phospholipase d from the plant-associated bacteria serratia plymuthica strain as9 reveals a unique arrangement of catalytic pocket. Int. J. Mol. Sci. 2021;22(6):1–17. doi: 10.3390/ijms22063219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Scardino V., Di Filippo J.I., Cavasotto C.N. How good are AlphaFold models for docking-based virtual screening? iScience. 2023;26 doi: 10.1016/j.isci.2022.105920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bolhuis P.G. Sampling kinetic protein folding pathways using all-atom models. Lect. Notes Phys. 2006;703:393–433. doi: 10.1007/3-540-35273-2_11. [DOI] [Google Scholar]
- 70.Sahoo C.R., Paidesetty S.K., Dehury B., Padhy R.N. Molecular dynamics and computational study of Mannich-based coumarin derivatives: potent tyrosine kinase inhibitor. J. Biomol. Struct. Dyn. 2020;38(18):5419–5428. doi: 10.1080/07391102.2019.1701554. [DOI] [PubMed] [Google Scholar]
- 71.Patel M., et al. Novel phytochemical inhibitors targeting monkeypox virus thymidine and serine/threonine kinase: integrating computational modeling and molecular dynamics simulation. J. Biomol. Struct. Dyn. 2023 doi: 10.1080/07391102.2023.2179547. [DOI] [PubMed] [Google Scholar]
- 72.Jiang S., et al. Design, synthesis and antibacterial activities against Xanthomonas oryzae pv. oryzae, Xanthomonas axonopodis pv. Citri and Ralstonia solanacearum of novel myricetin derivatives containing sulfonamide moiety. Pest Manag. Sci. 2020;76(3):853–860. doi: 10.1002/ps.5587. [DOI] [PubMed] [Google Scholar]
- 73.Yan T., et al. Preparation, characterization and evaluation of the antioxidant capacity and antitumor activity of myricetin microparticles formated by supercritical antisolvent technology. J. Supercrit. Fluids. 2021;175 doi: 10.1016/j.supflu.2021.105290. [DOI] [Google Scholar]
- 74.Pasetto S., Pardi V., Murata R.M. Anti-HIV-1 activity of flavonoid myricetin on HIV-1 infection in a dual-chamber in vitro model. PLoS One. 2014;9(12) doi: 10.1371/journal.pone.0115323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cho J.K., et al. Geranylated flavonoids displaying SARS-CoV papain-like protease inhibition from the fruits of Paulownia tomentosa. Bioorg. Med. Chem. 2013;21(11):3051–3057. doi: 10.1016/j.bmc.2013.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xiao T., et al. Myricetin inhibits SARS-CoV-2 viral replication by targeting mpro and ameliorates pulmonary inflammation. Front. Pharmacol. 2021;12 doi: 10.3389/fphar.2021.669642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sun H., Niisato N., Nishio K., Hamilton K.L., Marunaka Y. Distinct action of flavonoids, myricetin and quercetin, on epithelial Cl- Secretion: useful tools as regulators of Cl- secretion. BioMed Res. Int. 2014;2014 doi: 10.1155/2014/902735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kaul R., Paul P., Kumar S., Büsselberg D., Dwivedi V.D., Chaari A. Promising antiviral activities of natural flavonoids against sars-cov-2 targets: systematic review. Int. J. Mol. Sci. 2021;22(20) doi: 10.3390/ijms222011069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xue C.F., Guo J.M., Qian D.W., Duan J.A., Shu Y. Absorption of flavonoids from Abelmoschus manihot extract by in situ intestinal perfusion. Yaoxue Xuebao. 2011;46(4):454–459. [PubMed] [Google Scholar]
- 80.Yee Shiyin. In vitro permeability across caco-2 cells (colonic) can predict in vivo (small intestinal) absorption in man—fact or myth. Pharm. Res. (N. Y.) 1997;14:763–766. doi: 10.1023/a:1012102522787. [DOI] [PubMed] [Google Scholar]
- 81.Borra S.K., Mahendra J., Gurumurthy P., Jayamathi, Iqbal S.S., Mahendra L. Effect of curcumin against oxidation of biomolecules by hydroxyl radicals. J. Clin. Diagn. Res. 2014;8(10) doi: 10.7860/JCDR/2014/8517.4967. CC01–CC05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Khaerunnisa S., Kurniawan H., Awaluddin R., Suhartati S. Potential inhibitor of COVID-19 main protease (M pro) from several medicinal plant compounds by molecular docking study. Preprints. 2020:1–14. March. [Google Scholar]
- 83.Dehury B., Tang N., Mehra R., Blundell T.L., Kepp K.P. Side-by-side comparison of Notch- and C83 binding to γ-secretase in a complete membrane model at physiological temperature. RSC Adv. 2020;10(52):31215–31232. doi: 10.1039/d0ra04683c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Panigrahi S.K. Strong and weak hydrogen bonds in protein-ligand complexes of kinases: a comparative study. Amino Acids. 2008;34(4):617–633. doi: 10.1007/s00726-007-0015-4. [DOI] [PubMed] [Google Scholar]
- 85.Varma A.K., Patil R., Das S., Stanley A., Yadav L., Sudhakar A. Optimized hydrophobic interactions and hydrogen bonding at the target-ligand interface leads the pathways of Drug-Designing. PLoS One. 2010;5(8) doi: 10.1371/journal.pone.0012029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Girdhar K., et al. Novel insights into the dynamics behavior of glucagon-like peptide-1 receptor with its small molecule agonists. J. Biomol. Struct. Dyn. 2019;37(15):3976–3986. doi: 10.1080/07391102.2018.1532818. [DOI] [PubMed] [Google Scholar]
- 87.Dehury B., Behera S.K., Mahapatra N. Structural dynamics of Casein Kinase I (CKI) from malarial parasite Plasmodium falciparum (Isolate 3D7): insights from theoretical modelling and molecular simulations. J. Mol. Graph. Model. 2017;71:154–166. doi: 10.1016/j.jmgm.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 88.Parenti M.D., Rastelli G. Advances and applications of binding affinity prediction methods in drug discovery. Biotechnol. Adv. 2012;30(1):244–250. doi: 10.1016/j.biotechadv.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 89.Meirovitch H. Recent developments in methodologies for calculating the entropy and free energy of biological systems by computer simulation. Curr. Opin. Struct. Biol. 2007;17(2):181–186. doi: 10.1016/j.sbi.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 90.Kollman P. Free energy calculations: applications to chemical and biochemical phenomena. Chem. Rev. 1993;93(7):2395–2417. doi: 10.1021/cr00023a004. [DOI] [Google Scholar]
- 91.Ytreberg F.M., Swendsen R.H., Zuckerman D.M. Comparison of free energy methods for molecular systems. J. Chem. Phys. 2006;125(18) doi: 10.1063/1.2378907. [DOI] [PubMed] [Google Scholar]
- 92.Foloppe N., Hubbard R. Towards predictive ligand design with free-energy based computational methods? Curr. Med. Chem. 2006;13(29):3583–3608. doi: 10.2174/092986706779026165. [DOI] [PubMed] [Google Scholar]
- 93.Homeyer N., Gohlke H. Free energy calculations by the molecular mechanics Poisson-Boltzmann surface area method. Mol. Inform. 2012;31(2):114–122. doi: 10.1002/minf.201100135. [DOI] [PubMed] [Google Scholar]
- 94.Gohlke H., Kiel C., Case D.A. Insights into protein-protein binding by binding free energy calculation and free energy decomposition for the Ras-Raf and Ras-RalGDS complexes. J. Mol. Biol. 2003;330(4):891–913. doi: 10.1016/S0022-2836(03)00610-7. [DOI] [PubMed] [Google Scholar]
- 95.Lai B., Oostenbrink C. Binding free energy, energy and entropy calculations using simple model systems. Theor. Chem. Acc. 2012;131(10):1–13. doi: 10.1007/s00214-012-1272-1. [DOI] [Google Scholar]
- 96.Ramanathan A., Savol A., Burger V., Chennubhotla C.S., Agarwal P.K. Protein conformational populations and functionally relevant substates. Acc. Chem. Res. 2014;47(1):149–156. doi: 10.1021/ar400084s. [DOI] [PubMed] [Google Scholar]
- 97.Modi V., Dunbrack R.L. Defining a new nomenclature for the structures of active and inactive kinases. Proc. Natl. Acad. Sci. U.S.A. 2019;116(14):6818–6827. doi: 10.1073/pnas.1814279116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Weis W.I., Kobilka B.K. The molecular basis of G protein-coupled receptor activation. Annu. Rev. Biochem. 2018;87:897–919. doi: 10.1146/annurev-biochem-060614-033910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Xie T., Saleh T., Rossi P., Kalodimos C.G. Conformational states dynamically populated by a kinase determine its function. Science. 2020;370(6513) doi: 10.1126/science.abc2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Galanakis C.M. Polyphenols: properties, recovery, and applications. Polyphenols Prop. Recover. Appl. 2018:1–438. doi: 10.1016/C2016-0-05057-X. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.