Summary
The bulge of hair follicles harbors Nestin+ (neural crest like) stem cells, which exhibit the potential to generate various cell types including melanocytes. In this study, we aimed to determine the role of Sox9, an important regulator during neural crest development, in melanocytic differentiation of those adult Nestin+ cells. Immunohistochemical analysis after conditional Sox9 deletion in Nestin+ cells of adult mice revealed that Sox9 is crucial for melanocytic differentiation of these cells and that Sox9 acts as a fate determinant between melanocytic and glial fate. A deeper understanding of factors that regulate fate decision, proliferation and differentiation of these stem cells provides new aspects to melanoma research as melanoma cells share many similarities with neural crest cells. In summary, we here show the important role of Sox9 in melanocytic versus glial fate decision of Nestin+ stem cells in the skin of adult mice.
Subject areas: Molecular biology, Cell biology, Developmental biology
Graphical abstract
Highlights
-
•
Sox9 is expressed by developing melanocytic cells in adult hair follicles (HF)
-
•
Sox9 is essential for melanocytic differentiation of adult Nestin+ HF stem cells
-
•
Conditional deletion of Sox9 initiates glial differentiation of Nestin+ HF stem cells
-
•
Sox9 regulates proliferation and differentiation of Nestin+ HF stem cells
Molecular biology; Cell biology; Developmental biology
Introduction
Melanocytes, pigment-producing cells located in the epidermis and in hair follicles (HFs), supply surrounding keratinocytes with melanin. During embryonic development they originate from neural crest cells where they share a direct lineage with Schwann cells, the main glial cell type of the peripheral nervous system. The fate decision between those two cell types is regulated by a network of transcription factors. Afterward, cells that are determined to become melanocytes migrate as melanoblasts into the skin.1,2 The localisation of epidermal melanocytes in adult mice is restricted to sparsely haired areas such as the ear or the tail, while HF melanocytes reside in all skin regions covered by hair. In case of an injury as well as during the regular hair cycle, melanocytes are newly generated by melanocyte stem cells (McSCs).3 Dysfunction of McSCs not only results in hair greying but also initiates the formation of malignant melanoma – the deadliest type of skin cancer.4 McSCs reside in the HF.3 Hair follicle are skin appendages which protrude from the epidermis into the dermis. Each follicle is adjacent to a sebaceous gland and an arrector pili muscle and undergoes a repetitive cycling process, entering stages of growth (anagen), regression (catagen), and quiescence (telogen).5 Consequently, the pool of HF cells is continuously repopulated by HF stem cells such as keratinocyte stem cells, McSCs and other multipotent stem cells.6 All of them are slow cycling cells, which are mostly kept in the bulge – an HF stem cell niche located below the sebaceous gland. During anagen, the progeny of those stem cells migrates across the HF toward their final location, e.g. the hair bulb.7,8 The HF anatomy is complex and changes during the different stages.5 Therefore, in this study we focused on three areas: the bulge as a stem cell niche, the outer root sheath (ORS), which harbors migrating progenitor cells, and the bulb where mature melanocytes provide melanin to the hair shaft-forming keratinocytes.9 To maintain the cycling process of the HF, the homeostasis between stem cell self-renewal and specific differentiation must be strictly regulated.6
Besides various stem cell populations, a population of Nestin expressing cells (Nestin+ cells) resides in the bulge. In vitro and in vivo studies revealed their multipotency as they feature the potential to differentiate into neurons, blood vessels, glial cells, keratinocytes, and melanocytes.10 Some of those cells migrate out of the HF into the interfollicular region (IF).11 Nestin+ cells are also referred to as “adult neural crest like stem cells” in the literature.12 We recently revealed that these stem cells contribute to the melanocytic cell pool in vivo.13 A still outstanding question is which key factors regulate melanocytic differentiation of multipotent stem cells in the adult. As melanocytes emerge from neural crest cells during embryonic development, we focused in this study on key embryonic transcription factors.
The differentiation of (embryonic) Nestin+ neural crest stem cells is controlled by various transcription factors including the Sex-determining-region Y related HMG Box (Sox) family.14 Sox genes are key regulators of embryonic development and control stem cell behavior and cell fate determination.15 Sox9 represents a member of the SoxE family, a subgroup of the Sox proteins, and has been reported to regulate melanoglial differentiation of neural crest cells during development by inhibiting neuronal fate and instructing glial and melanocyte differentiation.16 Similar effects had been observed in the developing spinal cord as Sox9 predisposes cells toward gliogenesis.17 In neural progenitor cells and stem cells of the HF Sox9 is essential for stem cell maintenance throughout adulthood.18,19 Still, its role in adult McSCs and mature melanocytes in the HF is controversially discussed as two studies report contradictory findings regarding Sox9 expression in HF melanocytic cells.20,21 In this study, we aimed to determine the role of Sox9 in distinct developmental steps within the melanocytic lineage of adult Nestin+ HF cells. Therefore, we used a tamoxifen inducible Nestin-CreERT2 mouse line22 crossbred with mice carrying loxP-flanked Sox923 and the Cre-reporter GFP.24 Using this mouse line enabled us to timely control the deletion of Sox9 in adult Nestin+ cells and to follow the recombined cells over time by their expression of GFP as recently described.13 Recombination was induced starting at postnatal day 56 (P56) when HFs were still largely synchronized and in late telogen stage.25 Immunohistochemical analysis seven and 28 days post recombination revealed Sox9 as a regulator of fate decision in Nestin+ cells in adult HFs. More interestingly, it provides a new aspect to investigate melanoma initiation as recent studies described the potential of melanoma cells to dedifferentiate into neural-crest-like cells to initiate metastatic invasion.26
Results
Nestin+ stem cells and melanocytic cells of the hair follicle express Sox9 in vivo
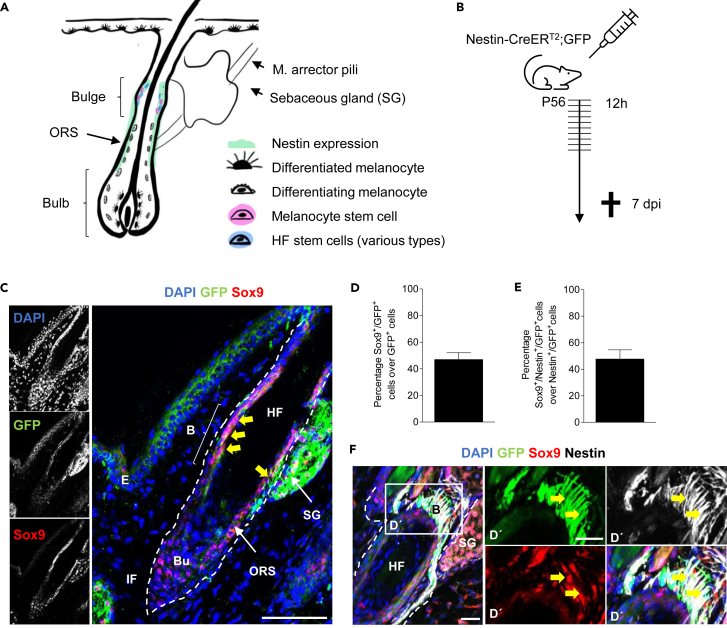
Nestin+ cells of the HF are multipotent and can differentiate into several cell types in vitro.10 Recently, we revealed that these cells generate melanocytes in vivo by labeling Nestin+ cells and tracking their cell descendants using tamoxifen inducible Nestin-CreERT2; GFP mice. Within this mouse model we determined a recombination rate of approximately 60%. The recombination rate represents the ratio of successfully recombined Nestin+ cells, marked by GFP expression, over all Nestin+ cells (for details, please see Stufchen et al.).13 Recombination efficiency around 60% was better than expected based on previously published results.22 Our previous study further revealed that Nestin expression was lost during melanocytic differentiation and that the localisation of Nestin+ cells differed from their progeny.13 Nestin expression was found mainly in cells in the bulge and ORS, regions that preferentially harbor stem and precursor cells, while cell descendants that had lost Nestin expression (Nestinneg/GFP+) were spread across the HF (Figure 1A), which is in line with previously published data.27
Figure 1.
Recombination in adult Nestin-CreERT2 transgenic mice targets Sox9-expressing hair follicle stem cells
(A) Schematic drawing of an adult hair follicle (HF) showing the localisation of Nestin-expressing cells (Nestin+) (green) and melanocytic cells in various differentiation stages; ORS = outer root sheath.
(B) Schematic drawing depicting the applied tamoxifen protocol to track Nestin+ cells and their progeny in vivo; Nestin-CreERT2; GFP mice (Ctrl mice) were intraperitoneally injected with tamoxifen every 12 h for 5 consecutive days starting at P56 and were killed 7 days post-injection (dpi).
(C) Representative immunohistochemical staining for GFP (green) and Sox9 (red) in tail skin of tamoxifen-treated Ctrl mice 7 dpi. Yellow arrows point toward recombined GFP+/Sox9+ cells in the bulge; dashed line marks the hair follicle; E = Epidermis, HF = Hair follicle, SG = Sebaceous gland, IF = Interfollicular area, B = Bulge, ORS = Outer root sheath, Bu = Bulb; DAPI is depicted in blue (scale bar = 50 μm).
(D) Immunohistochemical staining of GFP (green), Sox9 (red) and Nestin (white) in the hair follicle of Ctrl mice; Magnifications show signals in the bulge (D′); Yellow arrows point toward Sox9+/Nestin+/GFP+ cells; dashed line marks the hair follicle; HF = Hair follicle, SG = Sebaceous gland, B = Bulge; DAPI is depicted in blue (scale bar = 20 μm).
(E) The graphs show the quantification of Sox9+/GFP+ over total GFP+ cells per HF in tamoxifen-treated Ctrl mice 7 dpi; n = 4 mice, 4–10 HF per mice.
(F) Quantification of Sox9+/Nestin+/GFP+ over total Nestin+/GFP+ cells per HF in tamoxifen-treated Ctrl mice 7 dpi; n = 4 mice, 4–10 HF per mice. Data are presented as mean ± SEM.
To study the role of Sox9 in multipotent Nestin+ cells and their direct progeny in the skin, we first assessed Sox9 expression in the HF and epidermis in adult mice. For that we evaluated immunohistochemical expression of Sox9 and Nestin using Nestin-CreERT2; GFP mice which have received tamoxifen for 5 consecutive days and analyzed the tail skin 7 days postinjection (dpi; Figures 1B and S1). This tamoxifen protocol allowed us to label Nestin+ cells and their first progeny, which had been generated within one week. We observed that Sox9-expressing cells in HFs were mainly restricted to the ORS region and the bulge, in lower numbers also to the hair bulb (Figure 1C). Quantitative analysis of all recombined stem cells and their progeny (GFP+) revealed that Sox9 was expressed in 47.0% of the total GFP+ cell population (Figure 1D and Table S1). Here, co-expression was most prominent in the bulge and ORS, which indicates that Sox9 expression is primarily a feature of Nestin+ cells and early progenitors. However, Sox9 was only expressed in 46.7% of all Nestin+/GFP+ cells suggesting that Sox9 expression is either found in a specialized sub-population of Nestin+ cells (Figures 1E and 1F and Table S1) or that its expression oscillates.
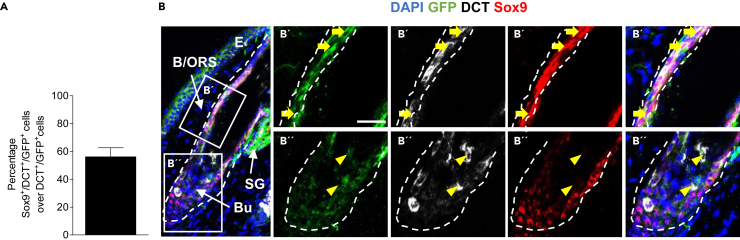
To address the question whether Sox9 is also expressed by cells of the melanocyte lineage generated by Nestin+ cells (hence GFP+ in recombined transgenic mice), we next performed Sox9 and dopachrome tautomerase (DCT, melanocyte lineage marker) immunohistochemical co-staining in the respective samples. Of interest, 56.2% of all DCT+/GFP+ cells expressed Sox9 (Figure 2A and Table S1) and their localization was also restricted primarily to the ORS, where amelanotic melanocytes and melanocytic progenitor cells reside (Figure 2B).28 These results indicate that Sox9 expression is not only a feature of Nestin+ stem cells but further attributed to developing melanocytic cells.
Figure 2.
Expression of Sox9 in ORS melanocytic cells
(A) Quantification of Sox9+/DCT+/GFP+ cells over total DCT+/GFP+ cells per HF in Ctrl mice; 7 dpi; n = 4 mice, 4–10 HF per mice; data are presented as mean ± SEM.
(B) Immunohistochemical staining of GFP (green), Sox9 (red) and DCT (white) in tamoxifen-treated Ctrl mice 7 dpi. Magnifications show signals in the ORS (B′) and hair bulb (B''); Yellow arrows point toward Sox9+/DCT+/GFP+ (upper panels, ORS) and yellow arrowheads point toward Sox9neg/DCT+/GFP+ cells (lower panels, Bulb); dashed line marks the hair follicle; E = Epidermis, HF = Hair follicle, B = Bulge, ORS = Outer root sheath, Bu = Bulb; DAPI is depicted in blue (scale bar = 20 μm).
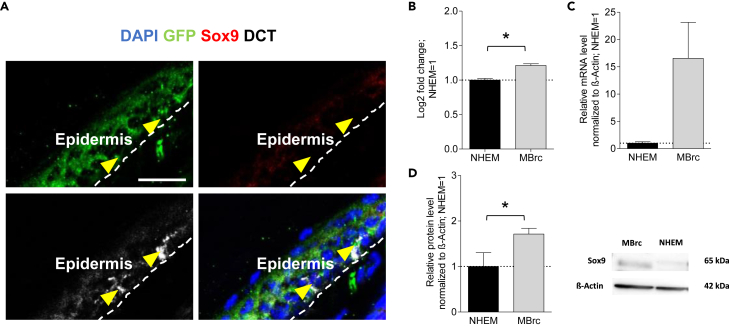
Sox9 expression is increased in developing melanocytic cells
Using tail skin of the transgenic mice allowed us to distinguish between HF and epidermal melanocytes as the latter are only found in restricted, sparsely haired areas in mice such as ear and tail skin.3 In contrast to the melanocyte pool in the HF, the epidermal melanocyte pool is not regularly repopulated under physiological conditions. Therefore, most of the epidermal melanocytes represent mature melanocytes.29 In contrast to our findings in the HF, we did not detect Sox9 expression in epidermal melanocytes in vivo (Figure 3A). In silico analysis from our group30 and in vitro analysis revealed low mRNA and protein expression of Sox9 in epidermal melanocytes and an upregulation in melanoblasts, the embryonic progenitor cells of melanocytes (Figures 3B–3D and Table S2). These findings indicate once more that Sox9 is expressed in early melanocytic cells.
Figure 3.
Increased expression of Sox9 in immature/dedifferentiated melanocytic cells
(A) Immunohistochemical staining of GFP (green), Sox9 (red) and DCT (white) in the epidermis of tamoxifen-treated Ctrl mice 7 dpi; n = 3 mice, 2–5 20-μm-thick sections of 1 cm long mice tail skin per mice. Yellow arrowheads point toward Sox9neg/DCT+ cells; dashed line marks the epidermis; E = Epidermis; DAPI is depicted in blue (scale bar = 25 μm).
(B and C) cDNA microarray (n = 3; Table S2, for details, please see Link-Paulus et al.30) and (C) qRT-PCR (n = 3; normalized to β-Actin) analyzing Sox9 expression in melanoblast-related-cells (MBrc) compared to its expression in normal human epidermal melanocytes (NHEM).
(D) Representative Western Blot image, lanes were cut out of a blot image including other, irrelevant lanes (n = 3; normalized to β-Actin); (B-D) students two-sided t-test was applied to determine statistical significance (∗p< 0.05). Data are presented as mean ± SEM.
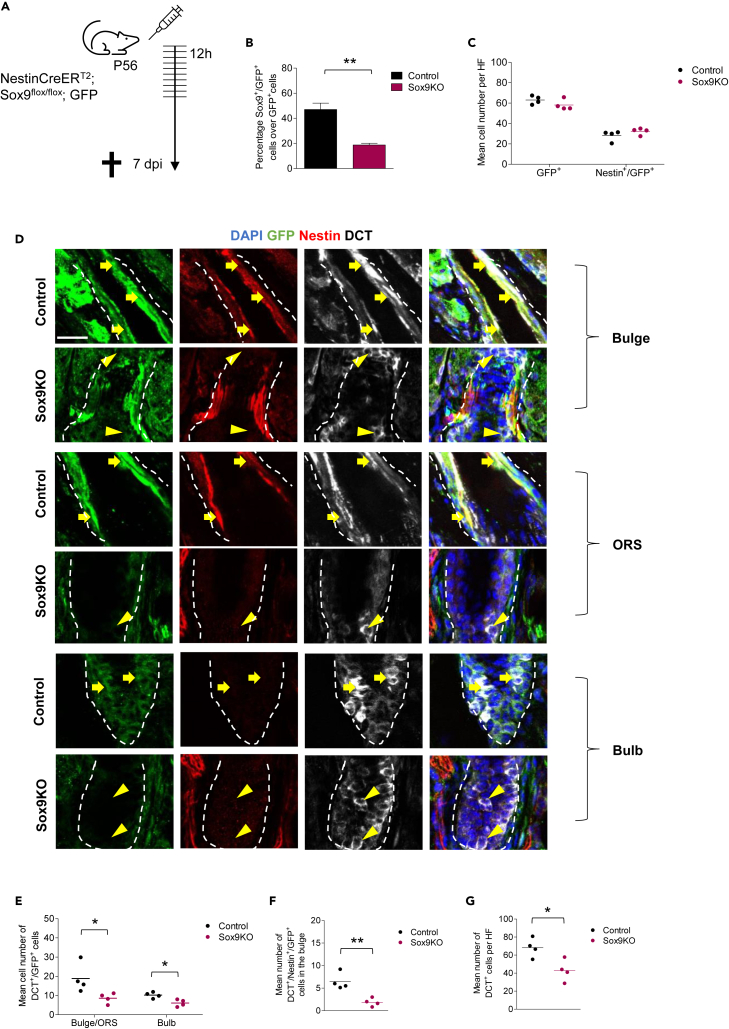
Sox9 is required for stem cells to differentiate into melanocytic cells
To address the function of Sox9 in melanocyte development, we conditionally ablated Sox9 in adult Nestin+ cells by crossing Nestin-CreERT2; GFP mice (mentioned above, now referred to as control “Ctrl”) with Sox9loxP/loxP mice (Nestin-CreERT2; Sox9fl/fl; GFP; from here on referred to as "Sox9KO"). Upon injection of tamoxifen at P56, Sox9 was specifically deleted in recombined Nestin+ cells and their progeny (GFP+ cells) in Sox9KO mice (Figure 4A). Since Cre-mediated recombination leads to a mosaic of recombined and non-recombined cells, we only assessed here the effects of Sox9 deletion in recombined cells. These effects are most likely cell-autonomous and not secondary. Immunohistochemical analysis for Sox9 seven days later confirmed successful knock out of Sox9 in Sox9KO mice as the ratio of Sox9+/GFP+ cells over total GFP+ cells per HF was significantly decreased by 58.5% compared to Ctrl mice (Figure 4B and Table S1). As only new transcription of Sox9 after knock out is affected, detection of previously translated Sox9 protein could account for the remaining 19% Sox9+ cells in the knock out mice.
Figure 4.
Sox9 ablation leads to a decrease in the number of DCT+(/Nestin+)/GFP+ cells in the HF
(A) Schematic demonstration of the applied tamoxifen protocol to specifically ablate Sox9 in Nestin+ cells in HF of adult mice in vivo; Either Nestin-CreERT2; GFP mice (Ctrl) or Nestin-CreERT2; GFP with floxed alleles of Sox9 (Sox9KO) were intraperitoneally injected with tamoxifen every 12 h for 5 consecutive days starting at P56 and sacrificed 7 dpi.
(B) Quantification of Sox9+/GFP+ cells over total GFP+ cells per HF in Sox9KO mice and Ctrl mice 7 dpi confirming successful knock out of Sox9; n = 4 mice per group; 4–10 HF per mice; data are presented as mean ± SEM.
(C) Quantification of the mean cell number of GFP+ or Nestin+/GFP+ cells per HF in Sox9KO mice and Ctrl mice 7 dpi; n = 4 mice per group; 12 HF per mice; data are presented as mean ± SEM.
(D) Immunohistochemical staining for GFP (green), Nestin (red) and DCT (white) in Sox9KO mice and Ctrl mice 7 dpi; dashed line marks the hair follicle; E = Epidermis, IF = Interfollicular area, B = Bulge, ORS = Outer root sheath, Bu = Bulb; Magnifications show signals in the bulge (upper panels), the ORS (middle panels) and the hair bulb (lower panels); DAPI is depicted in blue (scale bar = 20 μm); yellow arrows point toward DCT+/Nestin+/GFP+ cells (upper, middle panels) and DCT+/Nestinneg/GFP+ cells (middle, lower panels); yellow arrowheads point toward DCTneg/Nestinneg/GFP+ cells (upper panels) and DCT+/Nestinneg/GFPneg cells (middle panels (Sox9KO), lower panels).
(E) Quantification of the mean cell number of DCT+/GFP+ cells in the bulge/ORS and bulb in Sox9KO mice and Ctrl mice 7 dpi; n = 4 mice per group; 5–12 HF per mice; data are presented as mean ± SEM.
(F) Quantification of the mean cell number of DCT+/Nestin+/GFP+ cells in the bulge in Sox9KO mice and Ctrl mice 7 dpi; n = 4 mice per group; 5–12 HF per mice; data are presented as mean ± SEM.
(G) Quantification of the mean cell number of DCT+ cells per HF in Sox9KO mice and Ctrl mice 7 dpi; n = 4 mice per group; 12–15 HF per mice; data are represented as mean ± SEM. (B, E, F, G) students two-sided t-test was applied to determine statistical significance (∗: p < 0.05, ∗∗: p < 0.01).
Next, we analyzed the total cell number of GFP+ and Nestin+/GFP+ cells per HF and could not detect any significant changes comparing Ctrl and Sox9KO mice (Figure 4C and Table S3). This indicated that the total number of Nestin+ cells and their descendants had not been affected by Sox9 ablation 7 days post recombination. Nevertheless, we observed a change in the location of Nestin+/GFP+ cells in Sox9KO mice. Upon Sox9 deletion, the cells were mostly located in the bulge area, while in Ctrl mice about half of the Nestin+/GFP+ cells were also found in the ORS and in the bulb (Figures 4D and S3). To investigate whether Sox9 ablation affects melanocytic cells, we analyzed the numbers of DCT+/GFP+ cells (recombined, melanocytic cells), DCT+/Nestin+/GFP+ cells (recombined, melanocytic stem and progenitor cells) and DCT+ cells (melanocytic cells). Of interest, DCT+/GFP+ cells in Sox9KO mice were significantly reduced in the bulge/ORS by ∼55% and in the bulb by ∼60%, indicating a reduced number of melanocytic stem/progenitor cells and their descendants (Figure 4E and Table S3). In accordance, Sox9 depleted melanocytic stem and progenitor cells had also been significantly reduced in the bulge by ∼60% (Figure 4F and Table S3). This further resulted in a significant reduction of the total number of DCT+ cells in the HF of Sox9KO mice by ∼35% (Figure 4G and Table S3). As tamoxifen-treatment resulted in the deletion of Sox9 in at least 60% of GFP+ cells, a reduction of 50–60% of DCT+/(Nestin+/)GFP+ cells (Figure 4A) indicates that Sox9 is strongly involved and critical in the melanocytic differentiation process of Nestin+ cells. To confirm that the loss of Sox9 not only leads to the loss of mere DCT expression but indeed minimized the number of melanocytic cells, we performed siRNA mediated knock down of Sox9 in cultivated human melanocytes (NHEM). Here, DCT expression levels remained stable upon Sox9 inactivation (Figure S4).
Knock out of Sox9 leads to hyperproliferation of Nestin+ cells
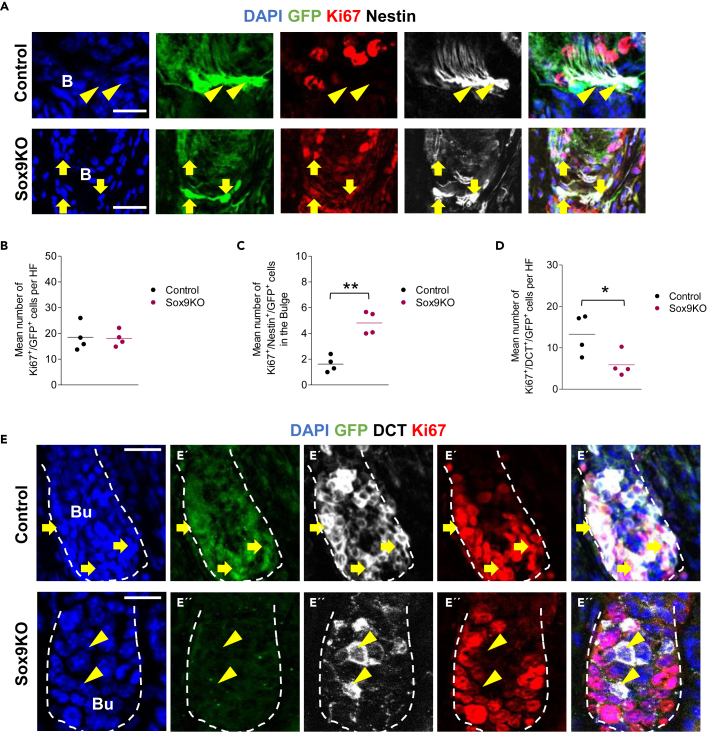
As we observed a reduced number of recombined melanocytic cells (DCT+/GFP+) and melanocytic stem and progenitor cells (DCT+/Nestin+/GFP+) upon Sox9 knock out 7 dpi, while the total number of recombined cells was unaffected (Figure 4C), we wondered whether the ablation of Sox9 led to an imbalance between differentiation and proliferation. To detect changes in the proliferation pattern of recombined cells and their progeny, we performed immunohistochemical staining against the proliferation marker Ki67. Here, we analyzed the absolute and relative number of Ki67+ cells in three different populations: GFP+ cells (recombined cells), DCT+/GFP+ cells (recombined, melanocytic cells), and Nestin+/GFP+ cells (recombined, stem and progenitor cells). In a quantitative analysis, the total number of proliferating GFP+ cells did not change comparing Ctrl and Sox9KO mice (Figures 5A, 5B, and Table S3). However, the Sox9 knock out led to a significant increase in the absolute number of proliferating stem and progenitor cells in the bulge (Figure 5C and Table S3). In contrast, we observed a reduction in the number of proliferating melanocytic cells by ∼45% in Sox9KO mice (Figures 5D, 5E, and Table S3).
Figure 5.
Sox9 ablation leads to a reduced number of proliferating melanocytic cells accompanied by a hyperproliferation of stem cells
(A) Immunohistochemical staining of GFP (green), Ki67 (red) and Nestin (white) in Ctrl mice and Sox9KO mice 7 dpi; magnifications show signals in the bulge in Ctrl mice (A′) and Sox9KO mice (A''); dashed line marks the hair follicle; B = Bulge; DAPI is depicted in blue (scale bar = 20 μm). Yellow arrows point toward triple positive cells (Ki67+/Nestin+/GFP+).
(B) Quantification of the mean number of Ki67+/GFP+ cells in Sox9KO and Ctrl mice 7 dpi; n = 4 mice per group; 6–12 HF per mice; data are presented as mean ± SEM.
(C) Quantification of the mean number of Ki67+/Nestin+/GFP+ cells in the bulge of Sox9KO and Ctrl mice 7 dpi; n = 4 mice per group; 4–12 HF per mice; data are presented as mean ± SEM.
(D) Quantification of the mean number of Ki67+/DCT+/GFP+ cells per HF in Sox9KO and Ctrl mice 7 dpi; n = 4 mice per group; 6–12 HF per mice; data are presented as mean ± SEM.
(E) Immunohistochemical staining of GFP (green), Ki67 (red) and DCT (white) cells in Ctrl mice and Sox9KO mice 7 dpi; magnifications show signals in the bulb of Ctrl mice (E′) and SoxKO mice (E''); dashed line marks the hair follicle; Bu = Bulb; DAPI is depicted in blue (scale bar = 20 μm); Yellow arrows point toward Ki67+/DCT+/GFP+ cells; yellow arrowheads point toward Ki67neg/DCT+/GFPneg cells (C) – (D) students two-sided t-test was applied to determine statistical significance (∗: p < 0.05, ∗∗: p < 0.01).
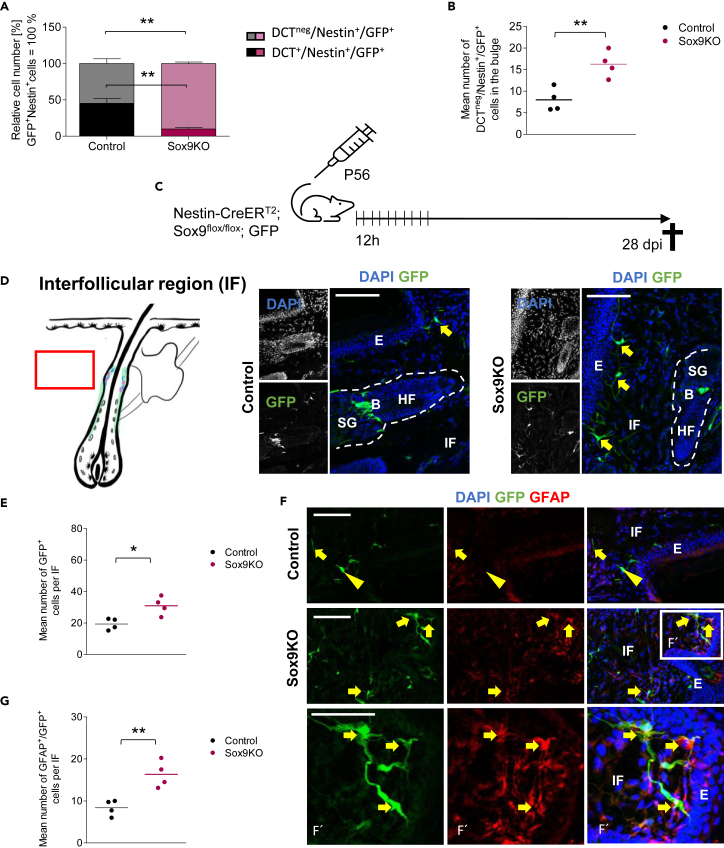
Fate switch of Nestin+ cells of the hair follicle upon Sox9 ablation
Sox9 ablation led to a decline exclusively in the number of DCT+ cells (Figure 4E–4G), while the total number of GFP+ cells and Nestin+/GFP+ cells did not noticeably change 7 dpi (Figure 4C). Therefore, we hypothesized that the loss of Sox9 did not result in mere cell death of melanocytic cells DCT+) but rather led to a switch in fate decision in recombined Nestin+ cells. To test this, we analyzed the stem cell population (Nestin+/GFP+) in detail and observed that the proportion of DCT+/Nestin+/GFP+ cells ("melanocyte stem and progenitor cells") in the Nestin+ population in the bulge was reduced from 45.2% in Ctrl to 10.2% in Sox9KO mice (Figure 6A, Table S1). Consequently, the number of DCTneg/Nestin+/GFP+ cells in the bulge was significantly increased and their proportion among all Nestin+ recombined cells was significantly higher in Sox9KO (90.0%) compared to Ctrl mice (54.8%; Figures 6A, 6B, Tables S1, and S3). As previous studies demonstrated the potential of HF Nestin+ stem cells to generate glial cells and neurons (Amoh et al., 2005a, 2017), we investigated whether the newly generated population of DCTneg/GFP+ cells differentiated into cells of the peripheral nervous system. These cells could have left the HF and migrated into the IF space, a region known to harbor Schwann cells and neurons.31 Therefore, we analyzed GFP+ cells in the HF and in the IF in Sox9KO and Ctrl mice 28 dpi (Figure 6C and S2). We chose this later time point of analysis to give newly generated cell populations enough time to differentiate, migrate, and acquire specific marker protein expression. Of interest, we observed that the total number of GFP+ cells in the IF was significantly increased 28 dpi in Sox9KO (Figures 6D, 6E, and Table S3). Further analysis were conducted by co-staining against neuronal maker Doublecortin (Dcx), Beta-III-Tubulin (β-III-Tub) and the glial marker Glial fibrillary acidic protein (GFAP). Although we found few cells co-expressing GFP with either Dcx or β-III-Tub in Ctrl and Sox9KO mice, we did not detect any changes in the absolute cell number of double-positive cells, neither in the HF nor in the IF (Figures S5 and S6). Therefore, we assumed that Sox9 knock out did not affect the number of recombined cells acquiring neuronal identity. In contrast, we observed an almost 2-fold increase in the number of DCTneg/GFP+ cells expressing the glial marker GFAP in the IF of Sox9KO mice compared to Ctrl (Figures 6F–6G, Table S3, and Figure S7). Hence, our datarevealed that Sox9-deficient cells rather differentiate into glial cells and migrate into the IF than remain as melanocytic cells in the HF. To confirm the glial cell type and to gain more mechanistic information, we stained for the Schwann cell lineage marker Sox10 and detected a significantly increased number of Sox10+/DCTneg/GFP+ cells in the IF (Figure S8). Those data indicate that Sox9 may regulate adult melanoglial fate decision coactively with other transcription factors like Sox10.
Figure 6.
Sox9 ablation leads to an increase of GFP+/GFAP+ cells in the interfollicular area
(A) Quantification of DCTneg/Nestin+/GFP+ and DCT+/Nestin+/GFP+ cells in Nestin+/GFP + cells in the bulge of Sox9KO mice and Ctrl mice 7 dpi; n = 4 mice per group; 5–12 HF per mice; data are presented as mean ± SEM.
(B) Quantification of the mean cell number of DCTneg/Nestin+/GFP+ cells in the bulge in Sox9KO mice and Ctrl mice 7 dpi; n = 4 mice per group; 5–11 HF per mice; data are presented as mean ± SEM.
(C) Schematic demonstration of the experimental mouse model used to specifically ablate Sox9 in Nestin+ stem cells in the hair follicle of adult mice in vivo; adult Nestin-CreERT2;GFP mice (Ctrl mice) with floxed alleles of Sox9 (Sox9KO mice) were intraperitoneally injected with Tamoxifen every 12 h for 5 consecutive days starting at P56 and killed 28 dpi.
(D) Immunohistochemical staining of GFP (green) in the IF of tail skin of Sox9KO mice and Ctrl mice 28 dpi; schematic drawing demonstrates the localisation of the region of interest; dashed line marks the hair follicle; E = Epidermis, IF = Interfollicular area, B = Bulge, SG = Sebaceous gland; yellow arrows point toward GFP+ cells; DAPI is depicted in blue (scale bar = 50 μm).
(E) Quantification of the mean cell number of GFP+ cells per IF in Sox9KO mice and Ctrl mice 28 dpi; n = 4 mice per group; 4 IF per mice; data are presented as mean ± SEM.
(F) Immunohistochemical staining of GFP (green) and GFAP (red) in the IF area of tail skin of Sox9KO mice and Ctrl mice 28 dpi; E = Epidermis, IF = Interfollicular area; yellow arrows point toward GFAP+/GFP+ cells; yellow arrowheads point toward GFAPneg/GFP+ cells; Magnification shows GFAP+/GFP+ cells (F′); DAPI is depicted in blue (scale bar = 50 μm).
(G) Quantification of the mean cell number of GFAP+/GFP+ cells per IF in Sox9KO mice and Ctrl mice 28 dpi; n = 4 mice per group; 4 IF per mice; data are presented as mean ± SEM. (A, B, E, H) students two-sided t-test was applied to determine statistical significance (∗: p < 0.05, ∗∗: p < 0.01).
Discussion
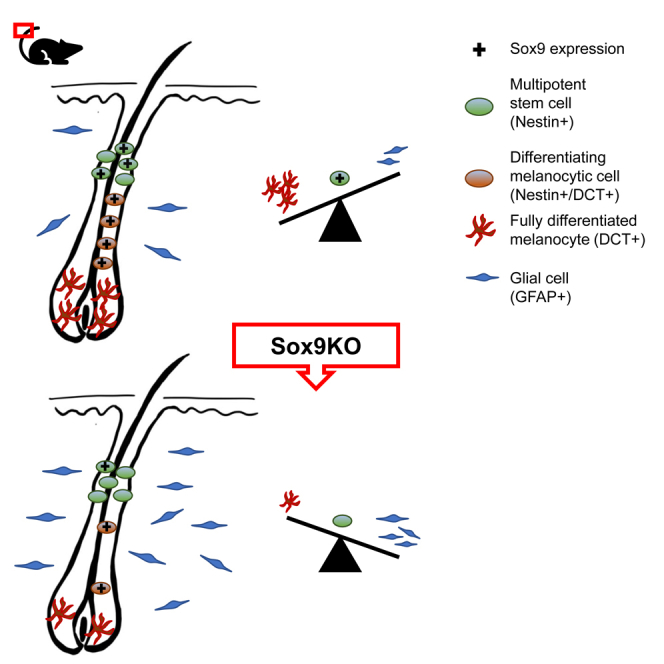
Melanocyte stem cells reside in the bulge of adult HF ensuring the repopulation of the epidermal and HF melanocyte pool after injuries or during the regular hair cycle.3 In addition to McSCs, multipotent Nestin+ cells located in the bulge of HFs contribute to the generation of melanocytes in vitro and in vivo.10,13 In this study, we revealed a key regulatory role of Sox9 in Nestin+ cells in the HF. We show that Sox9 ablation causes a decrease in the number of melanocytes generated by Nestin+ cells followed by an increased number of newly generated glial cells (GFAP+) in the interfollicular region indicating a pro-melanocytic role of Sox9 in the fate decision of Nestin+ HF stem cells.
Our study described the expression of Sox9 in melanocytic cells (DCT+) of the HF ORS, a region known to harbor Sox9+ cells and melanocyte progenitor cells.32 Until today, Sox9 expression in the melanocytic lineage was still under debate. While some studies claim a general lack of Sox9 in melanocytic cells,21 others support our finding in HF ORS melanocytes.20,33 Of interest, we did not observe any Sox9 expression in DCT+ epidermal melanocytes as opposed to a previous report.34 However, Sox9 expression and function was described to be region-dependent in the central nervous system35 and our findings support a differential Sox9 expression pattern in the adult skin. Still, we wondered why Sox9 was differentially expressed in epidermal and HF melanocytes. Although epidermal melanocytes appear as fully differentiated cells, the HF harbors melanocytes in diverse stages of differentiation depending on the HF stage.36 Our analysis revealed Sox9 expression mainly in melanocytic cells in the ORS of early anagen HF and therefore in melanocyte progenitor cells, which is supported by our analysis of Sox9 mRNA expression in melanoblasts and further by studies of other groups.37,38,39 The loss of Sox9 expression during differentiation in adult HFs could therefore explain controversial findings regarding Sox9 expression in melanocytes as other studies might not have analyzed the same timepoints and consequently not the exact same hair follicle stage in adult mice skin. We detected Sox9 expression in ORS melanocytic progenitor cells which originated from Nestin+ cells within one week (7 dpi) in early anagen, while Shakhova and colleagues clearly demonstrated the lack of Sox9 in melanocyte stem cells in the bulge and mature melanocytes in the HF bulb in a more advanced HF stage.21 Therefore, Sox9 expression seems to be strictly regulated and only found at a specific timepoint in immature melanocytic cells at early anagen. Of interest, a differentiation-dependent activity of Sox transcription factors in the HF has been reported for Sox10. Sox10, likewise Sox9 a member of the SoxE family, exhibits a dual role in the melanocytic lineage as its depletion in mature melanocytes leads to hair greying while its overexpression in immature melanocytes disrupts McSC maintenance and leads to hair greying as well.40 This indicates a dynamic, yet strictly regulated, expression of SoxE transcription factors in the melanocyte lineage, which may also apply to Sox9. The differential expression and role of Sox9 in McSCs is further supported as different types of McSCs have been described in the past years. Indeed, there are two different types of McSCs of adult HFs in telogen, one located in a structure called secondary hair germ, which is only apparent in telogen hair follicles, expressing markers of more mature melanocytes, the other located in the bulge, expressing CD34, Nestin and Sox9 and exhibiting a neural crest gene expression profile.33 A similar population of McCSs has been described as bipotent melanoglial stem cells. These cells express Nestin and Sox9 and can differentiate into glial cells and melanocytic cells in vitro.41 Both types share similarities with the targeted population (Nestin+) of our study. Accordingly, Sox9 would then be a specific feature of early melanocyte progenitors originated from multipotent Nestin+ cells from the bulge. Of interest, all Nestin+ described cell populations, including our target population, exhibit neural crest character. It has recently been shown that melanoma cells enter a neural crest stem cell like cell stage26 and that HF McSCs can give rise to malignant melanoma.4 Therefore, adult Nestin+/DCT+ cells may represent a source of malignant melanoma or at least a new tool to study melanoma cells in vivo.
The role of Sox9 in neural crest cells during embryonic development with “melanoglial” fate as well as its relevance in adult HF stem cells (K15+ keratinocyte stem cells, KcSCs) has been suggested previously,16,42,43 we are here the first to show its regulatory role in the differentiation process of adult neural crest like Nestin+ cells in the HF. Our results show that Sox9 is involved in the early melanocyte differentiation process of Nestin+ cells. Nestin+/K15neg stem cells and K15+ KcSCs both contribute to the KcSC pool.43 Because Sox9 is known for its relevance in K15+ KcSCs, we cannot fully exclude that the Sox9 knock out impacted the keratinocytic differentiation of Nestin+ stem cell-derived KcSCs.44 This might have secondary effects on melanocytic differentiation as cell-cell interaction is a common phenomenon in the HF.45 However, the role of Sox9 in keratinocyte differentiation of Nestin+ stem cells has not been investigated so far and the role of Sox9 in K15+ KcSCs does not automatically determine its role in Nestin+ stem cells, especially as Sox9 is known for its cell-type specific regulations.35,46
The role as important regulator of early melanocyte differentiation is furthersupported by the observed increase in DCTneg/GFAP+ glial cells 28 days upon Sox9KO. Consequently, the knock out of Sox9 induces a fate switch of Nestin+ cells and thereby an altered lineage progression. This also supports our findings of the immature stage of Sox9+ McSCs as they still exhibited the ability to change their cell fate. The tight relationship of glial cells and melanocytic cells has been reported in many studies.33,41,47 Common ancestor cells have not only been described in embryonic development,48,49 but also in adult HFs.41 Therefore, our findings are in line with several publications, which revealed the potential of melanocytic and glial cells to convert (transdifferentiate) into each other.2,48,49,50 Of interest, our study furthermore revealed increased Sox10 expression in the newly generated glial population in the IF. Besides its role in melanogenesis, Sox10 is indispensable in the embryonic and adult differentiation of Schwann cells.51 Our results indicate that Sox10 might physiologically be downregulated by Sox9 to supress gliogenesis. An inversely coupled expression and reciprocal regulation of both have previously been observed in glial52 and melanocytic cells.21 However, further experiments will be necessary to fully elucidate the transcriptional machinery, including downstream- and upstream regulators, controlling melanocyte versus glial differentiation in the adult HF.
In summary, we demonstrated the importance of Sox9 in inducing melanocyte fate determination of Nestin+ cells in the adult mouse HF in vivo. Sox9 ablation led to a decreased melanocytic differentiation accompanied by an increased number of GFAP+ glial cells in the IF area. Thereby, we identified the crucial role of Sox9 in a putative network with other transcription factors in fate decision of Nestin+ cells in the adult HF.
Limitation of the study
One limitation of the study is that we are not able to distinguish between a cell-autonomous function of Sox9 in melanocytic progenitor cells, or a secondary effect on melanocytic differentiation following the absence of Sox9 in all cell types descending from Nestin+ HF stem cells, e.g. keratinocytes. In addition, the HF cycle might also impact the cell numbers and analyzed HF are not fully synchronous, however, we focused on the analyses and comparison of HF in a similar size/volume. Another limitation as in all studies using cre-systems is that non-specific cre induction in other, Nestin negative cells cannot be completely ruled out.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Chicken polyclonal anti-Nestin | Aves | Cat#NES; RRID: AB_2194160 |
| Goat polyclonal anti-Sox9 | R&D Systems | Cat#AF3075; RRID: AB_477593 |
| Chicken polyclonal anti-GFP | Aves | Cat#GFP-1020; RRID: AB_10000240 |
| Goat polyclonal anti-GFP | Scigen | Cat#AB0066-200; RRID: AB_2333101 |
| Rat polyclonal anti-Ki67 | ThermoFisher | Cat#14-5698-82; RRID: AB_10854564 |
| Rabbit polyclonal anti-DCT | Abcam | Cat#ab74073; RRID: AB_1524517 |
| Chicken polyclonal anti-GFAP | Abcam | Cat# Ab4674; RRID: AB_304558 |
| Goat polyclonal anti-Dcx | Santa Cruz | Cat#sc8066; RRID:AB_2088494 |
| Rabbit polyclonal anti-Tubb3 | Abcam | Cat#ab18207; RRID:AB_444319 |
| Guinea pig polyclonal anti-Sox10 (AG Wegner) | Selfmade; 57 | N/A |
| Rabbit polyclonal anti-Sox9 | Merck | Cat# AB5535; RRID: AB_2239761 |
| Mouse polyclonal Anti-ß-Actin | Sigma-Aldrich | Klon AC-15/A5441 |
| Donkey polyclonal Biotin-conjugated anti-chicken IgY | Jackson ImmunoResearch | Cat#703-065-155; RRID: AB_2313596 |
| Donkey polyclonal Biotin-conjugated anti-goat IgG | Jackson ImmunoResearch | Cat#705-065-147; RRID: AB_2340397 |
| Alexa488-conjugated to Streptavidin | ThermoFisher | Cat# S11223; RRID: Not applicable |
| Donkey polyclonal Cy3-conjugated anti-chicken IgY | Jackson ImmunoResearch | Cat#703-166-155; RRID: AB_2340364 |
| Donkey polyclonal Cy3-conjugated anti-goat IgG | Jackson ImmunoResearch | Cat#705-165-147; RRID: AB_2307351 |
| Donkey polyclonal Cy3-conjugated anti-rabbit IgG | Jackson ImmunoResearch | Cat#711-165-152; RRID: AB_2307443 |
| Donkey polyclonal Cy3-conjugated anti-rat IgG | Jackson ImmunoResearch | Cat#712-165-153; RRID: AB_2340667 |
| Donkey polyclonal Cy5-conjugated anti-rabbit IgG | Jackson ImmunoResearch | Cat#711-175-152; RRID: AB_2340607 |
| Donkey polyclonal Alexa Fluor 647-conjugated anti-chicken IgY | Jackson ImmunoResearch | Cat#703-605-155; RRID: AB_2340379 |
| Donkey polyclonal Cy3-conjugated anti-guinea pig IgG | Jackson ImmunoResearch | Cat#706-165-148; RRID: AB_2340460 |
| HRP-coupled anti-rabbit | Chemicon | Cat#7074S; RRID: |
| HRP-coupled anti-mouse | Chemicon | Cat#7076S; RRID: |
| Critical commercial assays | ||
| E.Z.N.A. MicroElute Total RNA Kit | OmegaBiotech | Cat#R6831-02 |
| SuperScript II Reverse Transcriptase | Invitrogen | Cat#18064071 |
| LightCycler® 480 SYBR Green I Master | Roche Diagnostics | Cat#04887352001 |
| Pierce™ BCA Protein Assay Kit | ThermoFisher | Cat#23225 |
| Clarity™ Western ECL Substrate - Kit | Bio-Rad | Cat#1705061 |
| Experimental models: Cell lines | ||
| Human: Normal human epidermal melanocytes | Lonza | Cat#CC-2504; Lot# 0000493454 |
| Human: Normal human epidermal melanocytes | PromoCell | Cat#C-1240; |
| Melanoblast related cells (MBrc) | This Paper | 53 |
| Experimental models: Organisms/strains | ||
| CAG-CAT-eGFP mouse; FVB.B6-Tg(CAG-cat,-EGFP)1Rbns/KrnzJ |
24 | N/A |
| Nestin-GFP mouse; B6.Cg-Tg(Nes-EGFP)1Yamm | 55 | N/A |
| Sox9loxp/loxp:Sox9tm2Crm | 23 | N/A |
| Oligonucleotides | ||
| Primer Sox9 Forward: CGAACGCACATCAAGACGA | This paper | N/A |
| Primer Sox9 Reverse: AGGTGAAGGTGGAGTAGAGGC | This paper | N/A |
| Primer ß-Actin Forward: CTACGTCGCCCTGGACTTCGAGC | This paper | NA |
| Primer ß-Actin Reverse: GATGGAGCCGCCGATCCACACGG | This paper | N/A |
| Primer Dct Forward: TGGAGTGGTCCCTACATCCTA | This paper | N/A |
| Primer Dct Reverse: TCACTGGTGGTTTCTTCCG | This paper | N/A |
| Primer GFP Forward: GGTACATTGAGCAACTGACTG | This paper | N/A |
| Primer GFP Reverse:CTGCTAACCATGTTCATGCC | This paper | |
| siRNA targeting sequence: multiple (siPOOL – 30 siRNAs) | siTOOLs Biotech | NCBI Gene ID: 6662 |
| Softwareand algorithms | ||
| Fiji Imagej | N/A | https://imagej.net/ij/index.html |
| GraphPad Prism 5.0 | N/A | https://www.graphpad.com/scientific-software/prism/ |
| LabImage | N/A | https://www.kapelanbio.com/de/produkte/labimage-software.html |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact and is listed in the Author list, Anja Katrin Bosserhoff (anja.bosserhoff@fau.de)
Materials availability
All reagents generated in this study are available from the lead contact.
Materials availability statement
This study did not generate new unique reagents.
Experimental model and subject details
Cell lines
Normal human epidermal melanocytes (NHEM) were obtained from PromoCell (Heidelberg, Germany) or Lonza (Basel, Switzerland) and were derived from human neonatal foreskin tissue of Caucasian donors. Cell authentication was guaranteed by the manufacturer. The melanocytes were cultivated either in melanocyte serum-free M2 medium without PMA (phorbol myristate acetate) from PromoCell (Heidelberg, Germany) or in melanocyte serum-free medium with PMA from Lonza (Basel, Switzerland) at 37°C and 5% CO2. The de-differentiation procedure of melanocytes to MBrc was previously described.53 A special melanoblast growth medium contained MCBD 153 medium (Sigma-Aldrich, Steinheim, Germany) containing 8% chelated fetal bovine serum (FBS), 2% normal FBS (PAA Laboratories, Pasching, Austria), 2 mM glutamine, 1.66 ng/mL cholera toxin B, 10 ng/mL SCF (Sigma-Aldrich, Steinheim, Germany), 100 nM endothelin-3 and 2.5 ng/mL bFGF. Chelated FBS was prepared by mixing 1.2 g of Chelex-100 (Sigma-Aldrich, Steinheim, Germany) per 40 mL of FBS for 1.5hat 4°C with gentle stirring. Both cell lines were cultivated as previously described.54 Mycoplasma contamination was excluded on a regular basis by PCR.
Mice
All experiments were carried out in accordance with the European Communities Council Directive (86/609/EEC). Animal experiments were approved by the Governments of Upper Bavaria.
For all experiments, adult (8 to 12 weeks old) female and male mice were used. All animals were group-housed in standard cages, had ad libitum access to food and water and a 12 h light/dark cycle. We used Nestin-CreERT2 mice expressing Cre recombinase under the control of the Nestin promotor.55 These mice were crossbred to CAG CAT GFP reporter mice24 as recently described.13 Nuclear translocation of Cre by injection of tamoxifen leads to deletion of the STOP codon in front of the GFP, thereby inducing GFP expression in Nestin expressing cells as well as in their progeny. To delete Sox9 in adult Nestin+ HF cells, Nestin-CreERT2; GFP mice were crossbred to Sox9loxP/loxP mice, carrying loxP flanked exons 2 and 3 of the Sox9 gene.23 Upon recombination, Sox9 was specifically deleted in Nestin+ cells. Recombination was induced by injection of tamoxifen (50 μg/g body weight) twice per day for five consecutive days, starting at postnatal day 56 (P56). Experimental analyses were performed 7 days (∼P70) and 28 days (∼P90) after the tamoxifen injections. All animals have been described previously and all efforts were made to minimize the number of animals used and their suffering.
Method details
Transcriptome analysis with cDNA microarrays
To analyse the expression of Sox9 in melanoblast-related cells (MBrc) compared to normal human epidermal melanocytes (NHEM), we used datasets of our group generated with cDNA Microarrays (Affymetrix). Parts of those datasets have recently been published.30 The dataset contained two cell lines (NHEM, MBrc), each analyzed in three biological replicates. The raw data containing the log2signal were included in the supplementary file (Table S2).
Quantitative real-time PCR analysis
Isolation of total cellular RNA from cultured cells and generation of cDNAs by reverse transcription (RT) reaction was performed using E.Z.N.A. MicroElute Total RNA Kit (Omega, Norcross, GA, USA) and SuperScript II Reverse Transcriptase (Invitrogen, Groningen, Netherlands) following the manufacturer's protocols as described previously.54 Quantitative real-time PCR (qRT-PCR) analysis of gene expression was performed on a LightCycler® 480 II system (Roche Applied Science, Mannheim, Germany) with specific sets of oligonucleotide primers (β-Actin: 5’-CTACGTCGCCCTGGACTTCGAGC-3′, 5′-GATGGAGCCGCCGATCCACACGG-3′; Sox9: 5′-CGAACGCACATCAAGACGA-3′, 5′-AGGTGAAGGTGGAGTAGAGGC-3′; FoxD3: 5’-GACATGTTCGACAACGGCAG-3’, 5’-CTGTAAGCGCCGAAGCTCTG-3’; DCT: 5’- TGGAGTGGTCCCTACATCCTA-3’, 5’- TCACTGGTGGTTTCTTCCG-3’) as described previously.54 Primer sequences of all used primer sets are listed in the key resources table. Shortly, a volume of 1 μL complementary DNA template 0.5 μl (20 mM) of forward and reverse primers and 10 μL of SYBR Green I Master (Roche Applied Science, Mannheim, Germany) in a total of 20 μL reaction volume were applied to the following PCR program: 10 min 95°C (initial denaturation); 4.4°C/sec temperature transition rate up to 95°C for 10 sec, 10 sec 60°C, 20 sec 72°C, 87°C acquisition mode single, repeated for 45 times (amplification). Relative gene expression was expressed as a ratio of the expression level of the gene of interest to that of β-actin. The following cells were used: NHEM (normal human epidermal melanocytes); MBrc (Melanoblast-related cells).
Transfection of siRNA
For knockdown experiments, we seeded 80,000 NHEM (Lonza, Basel, Switzerland) per well of a six-well culture plate. On the following day, we transfected the cells with siRNA pool against human Sox9 (siTools Biotech GmbH, Planegg, Germany) or a control siPool (siCTR), respectively, using the Lipofectamine™ RNAiMAX transfection reagent (Life Technologies, Darmstadt, Germany). Total RNA was isolated after 96 hours of transfection and gene expressions were analysed as described in the section above.
Western blot
As previously described,56 3 x 106 cells were re-suspended in 200 μL RIPA buffer (Roche Applied Science, Mannheim, Germany) and lysed for 15minat 4°C. Insoluble fragments were removed by centrifugation at 13,000 r.p.m. for 10 min and the supernatant was stored at -20°C. In total, 30 μg of RIPA complete cell lysates was loaded per lane and separated on 10% SDS–PAGE gels and subsequently blotted onto a PVDF membrane. After blocking for 1 h with 5% non-fat dry milk/Tris-buffered saline with Tween20 (TBS-T) the membrane was incubated for 24 h with anti-Sox9 antibody (1:1,000 dilution; Abcam, Cambridge, UK) at 4°C or 1 h with anti-β-actin antibody (1:5,000 dilution; Sigma-Aldrich) at room temperature. After three washing steps with TBS-T, the membrane was incubated for 1 h with a horseradish peroxidase coupled secondary anti-rabbit (1:2,000 dilution in TBS-T) or anti-mouse (1:2,000 dilution in TBS-T) IgG antibody (Chemicon, Hofheim, Germany) and then washed again for three times in TBS-T. Finally, for the immunoreactions Clarity™ Western ECL Substrate (Bio-Rad) was added to the membranes for visualization with a Chemostar chemiluminescence imager (Intas, Goettingen, Germany). Protein levels were quantified relative to β-actin expression via computer-based densitometry of the scanned western blot images (using LabImage, Kapelan Bio-Imaging, Leipzig, Germany). The following cells were used: NHEM (normal human epidermal melanocytes); MBrc (Melanoblast-related cells).
Immunohistochemical staining of frozen murine tail skin slices
Mice were killed using CO2 and transcardially perfused with 50 mL phosphate-buffered saline (PBS, pH 7.4) followed by 50 mL 4% paraformaldehyde (PFA) at the rate of 20 mL/min. Skin samples of mice tail were subsequently transferred to a 30% sucrose solution. To produce cryosections, mice tail skin was embedded in TissueTec and frozen at -80°C. Longitudinal 20 μm tail sections were generated using a cryostat (Microm HM 500OM, Microm GmbH, Berlin, Germany). Slides were dried for 4hat room temperature and stored at -20°C until immunohistochemical analysis.
Immunofluorescent stainings were performed on 20 μm cryosections. Slices were washed three times with PBS and incubated with primary antibodies in PBS containing 0.5% Triton X-100 and 3% normal donkey serum (NDS) for 24-72hat 4°C. After incubation with the primary antibody, tissue was washed with PBS at room temperature and subsequently incubated with the secondary antibody in PBS containing 0.5% Triton X-100 and 3% NDS overnight at 4°C or for 2hat room temperature. After washing in PBS, nuclei were stained with DAPI (1:5000) and sections were mounted on coverslips using Aqua Poly Mount (Polysciences, Inc, Warrington, PA, USA). Negative control stainings were performed using secondary antibody only.
Z-stack images were taken using a Zeiss inverted Axio Observer 7 with ApoTome.2 equipped with an Axiocam 503, a Colibri 7 LED light source and 20x objective.
Antibodies
For immunohistochemical labeling of cells the following primary antibodies were used in this study: GFP (chicken polyclonal, 1:2,000, Aves, GFP-1020; RRID: AB_10000240), GFP (goat polyclonal, 1:500, Sicgen, AB0066-200; RRID: AB_2333101), Nestin (chicken polyclonal, 1:500, Aves, NES; RRID: AB_10805379), Sox9 (goat polyclonal, 1:200, R&D Systems, AF3075, RRDI: AB_2194160), Ki67 (rat polyclonal, 1:200, ThermoFisher, 14-5698-82, RRDI: AB_10854564), DCT (rabbit polyclonal, 1:500, Abcam, ab74073, RRID: AB_1524517), Dcx (goat polyclonal, 1:500, Santa Cruz, sc-8066; RRID: AB_2088494), GFAP (chicken polyclonal, 1:500, abcam, ab4674, RRDI: AB_304558), Tubb3 (rabbit polyclonal, 1:500, Abcam, ab18207, RRID: AB_444319) and Sox10 guinea pig polyclonal, selfmade.57
For visualization of primary antibody labeled proteins/cells the following secondary antibodies (all 1:400) were used: Biotin-conjugated anti-chicken IgY, and anti-goat IgG (donkey polyclonal, Jackson ImmunoResearch, Cambridgeshire, UK, 703-065-155, RRID: AB_2313596, and 705-065-147, RRID: AB_2340397) for amplification of the GFP reporter in combination with Alexa488-conjugated to Streptavidin (1:400; ThermoFisher, S11223); Cy3-conjugated anti-chicken IgY, anti-goat IgG, anti-rabbit IgG, and anti-rat IgG (donkey polyclonal, Jackson ImmunoResearch, 703-166-155, RRID: AB_2340364; 705-165-147, RRID: AB_2307351; 711-165-152, RRID: AB_2307443; 712-165-153, RRID: AB_2340667), Cy5-conjugated anti-rabbit IgG (donkey polyclonal, Jackson ImmunoResearch, 711-175-152, RRID: AB_2340607), and Alexa Fluor 647-conjugated anti-chicken IgY (donkey polyclonal, Jackson ImmunoResearch, 703-605-155, RRID: AB_2340379).
Software
Images were processed using Fiji ImageJ, Wayne Rasband, National Institute of Health, USA. Graphs were generated using GraphPad Prism 5.0 (GraphPad Software, Inc., San Diego, CA, USA).
Quantification and statistical analysis
Cell counting
To analyse the number of cells in the hair follicles, digital images of immunohistochemical-stained sections of mice tail skin (see above) were taken. The volume (area multiplied by the number of z-stacks) of analysed hair follicles or interfollicular regions was consistent between different slices and animals (Figures S1 and S2). We counted single/double/triple positive cells in the whole HF. We evaluated the number of signals in several HFs for n = 4 mice per group. The exact number of analysed HF per mice and per experiment is listed in Table S1.
Statistics
For statistical analysis significance levels were assessed using paired/unpaired Student’st-test with unequal variances. Differences were considered statistically significant at ∗p< 0.05, ∗∗p< 0.01, and ∗∗∗p< 0.001. All data are presented as mean ± SEM (standard error of the mean). All calculations were performed using the GraphPad Prism Software.
Acknowledgments
We thank Ryoichiro Kageyama for providing Nestin-CreERT2 mice. We thank Andreas Schedl and Michael Wegner for providing Sox9-floxed mice. We thank Ines Böhme for the help regarding in vitro experiments, Sneha Adhikarla and working group of Anja Bosserhoff and Ruth Beckervordersandforth for technical and methodical support.
The present work was performed in fulfilment of the requirements for obtaining the degree “Dr. med.” (IS).
Funding disclosure: This work was supported by grants from the IZKF (D31, D38, MD-Thesis scholarship for IS), the Wilhelm Sander Foundation (2018.113.2) and the DFG to AB (BO1573, CRC1540 (subproject C04), 460333672), RB (BE5136) and FB (BE 7623/1-1).
Author contributions
Conceptualization, I.S., F.B., M.F., R.B., and A.B.; Investigation, I.S., S.S., M.F., and S.F.; Formal analysis, I.S., S.S., M.F., and S.F.; Resources and Funding acquisition, R.B., F.B., M.F., and A.B.; Writing–Original draft, I.S. All authors contributed to manuscript revision, read, and approved the manuscript.
Declaration of interests
All authors declare that the submitted work was carried out in the presence of any personal, professional or financial relationships that could potentially be construed as a conflict of interest.
Inclusion and diversity
We worked to ensure sex balance in the selection of non-human subjects. While citing references scientifically relevant for this work, we also actively worked to promote gender balance in our reference list. We avoided “helicopter science” practices by including the participating local contributors from the region where we conducted the research as authors on the paper.
Published: May 19, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.106919.
Supplemental information
Data and code availability
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyse the data reported in this paper is available from the lead contact upon request.
References
- 1.Cichorek M., Wachulska M., Stasiewicz A., Tymińska A. Skin melanocytes: biology and development. Postepy Dermatol. Alergol. 2013;30:30–41. doi: 10.5114/pdia.2013.33376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Raamsdonk C.D., Deo M. Links between Schwann cells and melanocytes in development and disease. Pigment Cell Melanoma Res. 2013;26:634–645. doi: 10.1111/pcmr.12134. [DOI] [PubMed] [Google Scholar]
- 3.Gola M., Czajkowski R., Bajek A., Dura A., Drewa T. Melanocyte stem cells: biology and current aspects. Med. Sci. Mon. Int. Med. J. Exp. Clin. Res. 2012;18 doi: 10.12659/msm.883475. RA155-R159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun Q., Lee W., Mohri Y., Takeo M., Lim C.H., Xu X., Myung P., Atit R.P., Taketo M.M., Moubarak R.S., et al. A novel mouse model demonstrates that oncogenic melanocyte stem cells engender melanoma resembling human disease. Nat. Commun. 2019;10:5023. doi: 10.1038/s41467-019-12733-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Müller-Röver S., Handjiski B., van der Veen C., Eichmüller S., Foitzik K., McKay I.A., Stenn K.S., Paus R. A comprehensive guide for the accurate classification of murine hair follicles in distinct hair cycle stages. J. Invest. Dermatol. 2001;117:3–15. doi: 10.1046/j.0022-202x.2001.01377.x. [DOI] [PubMed] [Google Scholar]
- 6.Rompolas P., Greco V. Stem cell dynamics in the hair follicle niche. Semin. Cell Dev. Biol. 2014;25–26:34–42. doi: 10.1016/j.semcdb.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Waters J.M., Richardson G.D., Jahoda C.A.B. Hair follicle stem cells. Semin. Cell Dev. Biol. 2007;18:245–254. doi: 10.1016/j.semcdb.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 8.Oshima H., Rochat A., Kedzia C., Kobayashi K., Barrandon Y. Morphogenesis and renewal of hair follicles from adult multipotent stem cells. Cell. 2001;104:233–245. doi: 10.1016/s0092-8674(01)00208-2. [DOI] [PubMed] [Google Scholar]
- 9.Qiu W., Chuong C.M., Lei M. Regulation of melanocyte stem cells in the pigmentation of skin and its appendages: biological patterning and therapeutic potentials. Exp. Dermatol. 2019;28:395–405. doi: 10.1111/exd.13856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amoh Y., Li L., Katsuoka K., Hoffman R.M. Multipotent nestin-expressing hair follicle stem cells. J. Dermatol. 2009;36:1–9. doi: 10.1111/j.1346-8138.2008.00578.x. [DOI] [PubMed] [Google Scholar]
- 11.Uchugonova A., Duong J., Zhang N., König K., Hoffman R.M. The bulge area is the origin of nestin-expressing pluripotent stem cells of the hair follicle. J. Cell. Biochem. 2011;112:2046–2050. doi: 10.1002/jcb.23122. [DOI] [PubMed] [Google Scholar]
- 12.Sieber-Blum M., Grim M., Hu Y.F., Szeder V. Pluripotent neural crest stem cells in the adult hair follicle. Dev. Dynam. 2004;231:258–269. doi: 10.1002/dvdy.20129. [DOI] [PubMed] [Google Scholar]
- 13.Stufchen I., Beckervordersandforth R., Fischer S., Kappelmann-Fenzl M., Bosserhoff A.K., Beyer F. Two novel CreER(T2) transgenic mouse lines to study melanocytic cells in vivo. Pigment Cell Melanoma Res. 2022 doi: 10.1111/pcmr.13061. [DOI] [PubMed] [Google Scholar]
- 14.Haldin C.E., LaBonne C. SoxE factors as multifunctional neural crest regulatory factors. Int. J. Biochem. Cell Biol. 2010;42:441–444. doi: 10.1016/j.biocel.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamachi Y., Kondoh H. Sox proteins: regulators of cell fate specification and differentiation. Development. 2013;140:4129–4144. doi: 10.1242/dev.091793. [DOI] [PubMed] [Google Scholar]
- 16.Cheung M., Briscoe J. Neural crest development is regulated by the transcription factor Sox9. Development. 2003;130:5681–5693. doi: 10.1242/dev.00808. [DOI] [PubMed] [Google Scholar]
- 17.Stolt C.C., Lommes P., Sock E., Chaboissier M.C., Schedl A., Wegner M. The Sox9 transcription factor determines glial fate choice in the developing spinal cord. Genes Dev. 2003;17:1677–1689. doi: 10.1101/gad.259003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He N., Dong Z., Tai D., Liang H., Guo X., Cang M., Liu D. The role of Sox9 in maintaining the characteristics and pluripotency of Arbas Cashmere goat hair follicle stem cells. Cytotechnology. 2018;70:1155–1165. doi: 10.1007/s10616-018-0206-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jo A., Denduluri S., Zhang B., Wang Z., Yin L., Yan Z., Kang R., Shi L.L., Mok J., Lee M.J., Haydon R.C. The versatile functions of Sox9 in development, stem cells, and human diseases. Genes Dis. 2014;1:149–161. doi: 10.1016/j.gendis.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krahl D., Sellheyer K. Sox9, more than a marker of the outer root sheath: spatiotemporal expression pattern during human cutaneous embryogenesis. J. Cutan. Pathol. 2010;37:350–356. doi: 10.1111/j.1600-0560.2009.01369.x. [DOI] [PubMed] [Google Scholar]
- 21.Shakhova O., Cheng P., Mishra P.J., Zingg D., Schaefer S.M., Debbache J., Häusel J., Matter C., Guo T., Davis S., et al. Antagonistic cross-regulation between Sox9 and Sox10 controls an anti-tumorigenic program in melanoma. PLoS Genet. 2015;11:e1004877. doi: 10.1371/journal.pgen.1004877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Imayoshi I., Ohtsuka T., Metzger D., Chambon P., Kageyama R. Temporal regulation of Cre recombinase activity in neural stem cells. Genesis. 2006;44:233–238. doi: 10.1002/dvg.20212. [DOI] [PubMed] [Google Scholar]
- 23.Akiyama H., Chaboissier M.C., Martin J.F., Schedl A., de Crombrugghe B. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 2002;16:2813–2828. doi: 10.1101/gad.1017802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakamura T., Colbert M.C., Robbins J. Neural crest cells retain multipotential characteristics in the developing valves and label the cardiac conduction system. Circ. Res. 2006;98:1547–1554. doi: 10.1161/01.RES.0000227505.19472.69. [DOI] [PubMed] [Google Scholar]
- 25.Yi R. Concise review: mechanisms of quiescent hair follicle stem cell regulation. Stem Cell. 2017;35:2323–2330. doi: 10.1002/stem.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diener J., Sommer L. Reemergence of neural crest stem cell-like states in melanoma during disease progression and treatment. Stem Cells Transl. Med. 2021;10:522–533. doi: 10.1002/sctm.20-0351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li L., Mignone J., Yang M., Matic M., Penman S., Enikolopov G., Hoffman R.M. Nestin expression in hair follicle sheath progenitor cells. Proc. Natl. Acad. Sci. USA. 2003;100:9958–9961. doi: 10.1073/pnas.1733025100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishimura E.K. Melanocyte stem cells: a melanocyte reservoir in hair follicles for hair and skin pigmentation. Pigment Cell Melanoma Res. 2011;24:401–410. doi: 10.1111/j.1755-148X.2011.00855.x. [DOI] [PubMed] [Google Scholar]
- 29.Tobin D.J., Bystryn J.C. Different populations of melanocytes are present in hair follicles and epidermis. Pigm. Cell Res. 1996;9:304–310. doi: 10.1111/j.1600-0749.1996.tb00122.x. [DOI] [PubMed] [Google Scholar]
- 30.Linck-Paulus L., Lämmerhirt L., Völler D., Meyer K., Engelmann J.C., Spang R., Eichner N., Meister G., Kuphal S., Bosserhoff A.K. Learning from embryogenesis-A comparative expression analysis in melanoblast differentiation and tumorigenesis reveals miRNAs driving melanoma development. J. Clin. Med. 2021;10:2259. doi: 10.3390/jcm10112259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roosterman D., Goerge T., Schneider S.W., Bunnett N.W., Steinhoff M. Neuronal control of skin function: the skin as a neuroimmunoendocrine organ. Physiol. Rev. 2006;86:1309–1379. doi: 10.1152/physrev.00026.2005. [DOI] [PubMed] [Google Scholar]
- 32.Purba T.S., Haslam I.S., Shahmalak A., Bhogal R.K., Paus R. Mapping the expression of epithelial hair follicle stem cell-related transcription factors LHX2 and SOX9 in the human hair follicle. Exp. Dermatol. 2015;24:462–467. doi: 10.1111/exd.12700. [DOI] [PubMed] [Google Scholar]
- 33.Joshi S.S., Tandukar B., Pan L., Huang J.M., Livak F., Smith B.J., Hodges T., Mahurkar A.A., Hornyak T.J. CD34 defines melanocyte stem cell subpopulations with distinct regenerative properties. PLoS Genet. 2019;15:e1008034. doi: 10.1371/journal.pgen.1008034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Passeron T., Valencia J.C., Bertolotto C., Hoashi T., Le Pape E., Takahashi K., Ballotti R., Hearing V.J. SOX9 is a key player in ultraviolet B-induced melanocyte differentiation and pigmentation. Proc. Natl. Acad. Sci. USA. 2007;104:13984–13989. doi: 10.1073/pnas.0705117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vong K.I., Leung C.K.Y., Behringer R.R., Kwan K.M. Sox9 is critical for suppression of neurogenesis but not initiation of gliogenesis in the cerebellum. Mol. Brain. 2015;8:25. doi: 10.1186/s13041-015-0115-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Slominski A., Wortsman J., Plonka P.M., Schallreuter K.U., Paus R., Tobin D.J. Hair follicle pigmentation. J. Invest. Dermatol. 2005;124:13–21. doi: 10.1111/j.0022-202X.2004.23528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cook A.L., Smith A.G., Smit D.J., Leonard J.H., Sturm R.A. Co-expression of SOX9 and SOX10 during melanocytic differentiation in vitro. Exp. Cell Res. 2005;308:222–235. doi: 10.1016/j.yexcr.2005.04.019. [DOI] [PubMed] [Google Scholar]
- 38.Harris M.L., Baxter L.L., Loftus S.K., Pavan W.J. Sox proteins in melanocyte development and melanoma. Pigment Cell Melanoma Res. 2010;23:496–513. doi: 10.1111/j.1755-148X.2010.00711.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.D'Mello S.A.N., Finlay G.J., Baguley B.C., Askarian-Amiri M.E., Askarian-Amiri M.E. Signaling pathways in melanogenesis. Int. J. Mol. Sci. 2016;17:1144. doi: 10.3390/ijms17071144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harris M.L., Buac K., Shakhova O., Hakami R.M., Wegner M., Sommer L., Pavan W.J. A dual role for SOX10 in the maintenance of the postnatal melanocyte lineage and the differentiation of melanocyte stem cell progenitors. PLoS Genet. 2013;9:e1003644. doi: 10.1371/journal.pgen.1003644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Locher H., Saadah N., de Groot S., de Groot J.C.M.J., Frijns J.H.M., Huisman M.A. Hair follicle bulge cultures yield class III beta-tubulin-positive melanoglial cells. Histochem. Cell Biol. 2015;144:87–91. doi: 10.1007/s00418-015-1312-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsunogai Y., Miyadai M., Nagao Y., Sugiwaka K., Kelsh R.N., Hibi M., Hashimoto H. Contribution of sox9b to pigment cell formation in medaka fish. Dev. Growth Differ. 2021;63:516–522. doi: 10.1111/dgd.12760. [DOI] [PubMed] [Google Scholar]
- 43.Onishi S., Baba Y., Yokoi F., Ide K., Ohyama M., Nishifuji K. Progenitor cells expressing nestin, a neural crest stem cell marker, differentiate into outer root sheath keratinocytes. Vet. Dermatol. 2019;30:365–e107. doi: 10.1111/vde.12771. [DOI] [PubMed] [Google Scholar]
- 44.Vidal V.P.I., Chaboissier M.C., Lützkendorf S., Cotsarelis G., Mill P., Hui C.C., Ortonne N., Ortonne J.P., Schedl A. Sox9 is essential for outer root sheath differentiation and the formation of the hair stem cell compartment. Curr. Biol. 2005;15:1340–1351. doi: 10.1016/j.cub.2005.06.064. [DOI] [PubMed] [Google Scholar]
- 45.Nowak J.A., Polak L., Pasolli H.A., Fuchs E. Hair follicle stem cells are specified and function in early skin morphogenesis. Cell Stem Cell. 2008;3:33–43. doi: 10.1016/j.stem.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamashita S., Kataoka K., Yamamoto H., Kato T., Hara S., Yamaguchi K., Renard-Guillet C., Katou Y., Shirahige K., Ochi H., et al. Comparative analysis demonstrates cell type-specific conservation of SOX9 targets between mouse and chicken. Sci. Rep. 2019;9:12560. doi: 10.1038/s41598-019-48979-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Adameyko I., Lallemend F., Aquino J.B., Pereira J.A., Topilko P., Müller T., Fritz N., Beljajeva A., Mochii M., Liste I., et al. Schwann cell precursors from nerve innervation are a cellular origin of melanocytes in skin. Cell. 2009;139:366–379. doi: 10.1016/j.cell.2009.07.049. [DOI] [PubMed] [Google Scholar]
- 48.Graham A. Melanocyte production: dark side of the Schwann cell. Curr. Biol. 2009;19:R1116–R1117. doi: 10.1016/j.cub.2009.10.063. [DOI] [PubMed] [Google Scholar]
- 49.Mull A.N., Zolekar A., Wang Y.C. Understanding melanocyte stem cells for disease modeling and regenerative medicine applications. Int. J. Mol. Sci. 2015;16:30458–30469. doi: 10.3390/ijms161226207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Motohashi T., Yamanaka K., Chiba K., Aoki H., Kunisada T. Unexpected multipotency of melanoblasts isolated from murine skin. Stem Cell. 2009;27:888–897. doi: 10.1634/stemcells.2008-0678. [DOI] [PubMed] [Google Scholar]
- 51.Kuhlbrodt K., Herbarth B., Sock E., Hermans-Borgmeyer I., Wegner M. Sox10, a novel transcriptional modulator in glial cells. J. Neurosci. 1998;18:237–250. doi: 10.1523/JNEUROSCI.18-01-00237.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reiprich S., Cantone M., Weider M., Baroti T., Wittstatt J., Schmitt C., Küspert M., Vera J., Wegner M. Transcription factor Sox10 regulates oligodendroglial Sox9 levels via microRNAs. Glia. 2017;65:1089–1102. doi: 10.1002/glia.23146. [DOI] [PubMed] [Google Scholar]
- 53.Bosserhoff A.K., Ellmann L., Kuphal S. Melanoblasts in culture as an in vitro system to determine molecular changes in melanoma. Exp. Dermatol. 2011;20:435–440. doi: 10.1111/j.1600-0625.2011.01271.x. [DOI] [PubMed] [Google Scholar]
- 54.Bosserhoff A.K., Schneider N., Ellmann L., Heinzerling L., Kuphal S. The neurotrophin Neuritin1 (cpg15) is involved in melanoma migration, attachment independent growth, and vascular mimicry. Oncotarget. 2017;8:1117–1131. doi: 10.18632/oncotarget.13585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yamaguchi M., Saito H., Suzuki M., Mori K. Visualization of neurogenesis in the central nervous system using nestin promoter-GFP transgenic mice. Neuroreport. 2000;11:1991–1996. doi: 10.1097/00001756-200006260-00037. [DOI] [PubMed] [Google Scholar]
- 56.Feuerer L., Lamm S., Henz I., Kappelmann-Fenzl M., Haferkamp S., Meierjohann S., Hellerbrand C., Kuphal S., Bosserhoff A.K. Role of melanoma inhibitory activity in melanocyte senescence. Pigment Cell Melanoma Res. 2019;32:777–791. doi: 10.1111/pcmr.12801. [DOI] [PubMed] [Google Scholar]
- 57.Maka M., Stolt C.C., Wegner M. Identification of Sox8 as a modifier gene in a mouse model of Hirschsprung disease reveals underlying molecular defect. Dev. Biol. 2005;277:155–169. doi: 10.1016/j.ydbio.2004.09.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyse the data reported in this paper is available from the lead contact upon request.