Abstract
COMPEL is a database on composite regulatory elements, the basic structures of combinatorial regulation. Composite regulatory elements contain two closely situated binding sites for distinct transcription factors and represent minimal functional units providing combinatorial transcriptional regulation. Both specific factor–DNA and factor–factor interactions contribute to the function of composite elements (CEs). Information about the structure of known CEs and specific gene regulation achieved through such CEs appears to be extremely useful for promoter prediction, for gene function prediction and for applied gene engineering as well. The structure of the relational model of COMPEL is determined by the concept of molecular structure and regulatory role of CEs. Based on the set of a particular CE, a program has been developed for searching potential CEs in gene regulatory regions. WWW search and browse routines were developed for COMPEL release 3.0. The COMPEL database equipped with the search and browse tools is available at http://compel.bionet.nsc.ru/ . The program for prediction of potential CEs of NFAT type is available at http://compel.bionet.nsc.ru/FunSite.html and http://transfac.gbf.de/dbsearch/funsitep/s_comp.html
INTRODUCTION
During the last decade we have witnessed a tremendous progress in the experimental studies of transcriptional regulation. As a result, a large number of transcription factors has been cloned, the bulk of their target sites in gene regulatory regions has been discovered, and in many cases protein domains essential for direct factor–DNA and factor–factor interactions have been identified. These data should be collected and classified in specialized databases to make them applicable in both experimental and theoretical molecular genetic studies.
There are now a number of databases on transcriptional regulation available. A cluster of closely interrelated databases on protein and DNA sequences involved in transcriptional regulation comprises the following databases: EPD, TRANSFAC, TRRD and COMPEL. The Eukaryotic Promoter Database (EPD) contains general information about promoters, as they are defined by an experimentally proven transcription start site, and their tissue-specificity (1). The TRANSFAC database provides information on structure, function, sequence and classification of transcription factors, on their binding sites within genes as well as their DNA-binding profiles (2). An entry of one of the main TRANSFAC tables corresponds to a transcription factor or to an individual binding site. TRRD (Transcriptional Regulatory Region Database) collects information about the structure of whole regulatory regions of eukaryotic genes and about gene expression patterns as well (3,4). Each TRRD entry corresponds to an entire gene, and binding sites are considered as lowest level of hierarchy in gene transcriptional regulation (4,5).
In the last years it has become evident that the complex differential expression of genes in higher organisms is achieved through combinatorial regulation of transcription by specific combination of transcription factors binding to their target sites in the regulatory regions of these genes. We have developed the COMPEL database that emphasizes the key role of specific protein–protein interactions for gene regulation in a particular cellular content. In the COMPEL database we collect published information on composite regulatory elements. These are defined as pairs of closely situated binding sites, corresponding transcription factors, protein–protein interaction between them, and expression patterns provided by this combinatorial regulation (6,7). The databases TRRD, TRANSFAC and COMPEL contain cross-references to each other and a common table of genes (8,9). COMPEL has been developed in a joint effort of the Institute of Cytology and Genetics (Novosibirsk, Russia) and Gesellschaft für Bio-technologische Forschung mbH (Braunschweig, Germany) since 1995. The previous COMPEL releases have been previously described (2,6–8,10,11). Over the last year, the structure of the database has been improved considerably. One important new feature is the link to the EMBL databank (12). COMPEL is publicly available for non-commercial users and is distributed in three interlinked ASCII flat-files. We have also developed search and browse tools that are available via WWW at: http://compel.bionet.nsc.ru/
THE COMPOSITE REGULATORY ELEMENT CONCEPT
The term ‘composite element’ (CE) was introduced while studying the glucocorticoid response element in the mouse proliferin promoter where a glucocorticoid receptor binding site is adjacent to and functionally interacts with an AP-1 site (13). Further, this term was applied to quite different pairs of interacting sites and factors. Based on the known examples, we define a CE as a minimal functional unit where both protein–DNA and protein–protein interactions contribute to a highly specific pattern of gene transcriptional regulation (6,7).
There are two main types of CEs: synergistic and antagonistic. In synergistic CEs, simultaneous interactions of two factors with closely situated target sites result in a non-additive high level of transcriptional activation. Highly cooperative binding of factors to DNA and formation of a ternary complex protein–protein–DNA was experimentally proven in many cases. As a result of protein–protein interactions, a new protein surface may be formed which is characteristic for a certain factor pair. Interaction between two factors may be direct or mediated by a coactivator, for instance by p300/CBP. In some cases two factors independently bind to DNA but nevertheless synergistically activate transcription. The synergistic effect may then be accounted for by simultaneous interactions of activation domains of two factors with different components of the basal transcription complex or with specific co-activators, and/or direct factor–factor interactions may elicit conformational changes in activation domains. A number of factors are known to bend DNA and, thus, to facilitate binding of other factors.
Within an antagonistic CE two factors interfere with each other. In some cases competition for overlapping sites leads to mutually exclusive binding. There are other examples where factors can bind to DNA simultaneously, but binding of a repressing factor may mask an activation domain of an activator. A number of molecular mechanisms have been suggested for functioning of both synergistic and antagonistic CEs (7).
CE CLASSIFICATION
CEs can be classified by different criteria: (i) the cooperative effect of the transcription factors involved (synergism or antagonism); (ii) the structure of the transcription factors involved, namely the structure of DNA-binding domains; (iii) the specific function of a CE, for instance tissue-specific or inducible regulation.
To classify CEs in terms of DNA-binding domains we applied a previously developed transcription factor classification (2,14). The factors interacting at an individual CE mostly belong to different classes. Transcription factors of bZIP, REL and ETS classes play a very important role in CEs and ~50% of known CEs contain at least one binding site for one of these proteins. Generally, the transcription factors binding to the constituent sites of a CE recognize sequence motifs which clearly differ, one of them frequently being a highly purine-rich motif (on one strand), such as NF-κB, NF-ATp/c, ETS. Examples of the most frequent structural types of CEs collected in COMPEL are given in Table 1. They include 26 CEs of bZIP/REL type, 13 CEs of bZIP/ETS type, six CEs of REL/HMG type, five CEs of ETS/RUNT and three CEs of ETS/MADS type. Structurally similar elements are present in several different genes (Table 1), which apparently implies that such regulatory modules are functionally significant.
Table 1. Most frequent types of CEs according to the structure of DNA-binding domains of the transcription factors involved.
| Factor 1 | Factor 2 | Genes | COMPEL acc |
|---|---|---|---|
| CEs containing binding sites for ZIP and REL factors | |||
| AP-1 | NF-κB | E-selectin, human | C00097, C00102, C00103 |
| IFN-β, human | C00099 | ||
| IL-2, human | C00165 | ||
| AP-1 | NF-AT | GM-CSF, mouse | C00108 |
| GM-CSF, human | C00141, C00142, C00143, C00164 | ||
| IL-2, human | C00109 | ||
| IL-2, mouse | C00149, C00150, C00151, C00157 | ||
| IL-3, human | C00160 | ||
| IL-4, mouse | C00159 | ||
| IL-5, mouse | C00161 | ||
| C/EBP | NF-κB | IL-6, human | C00152 |
| IL-8, human | C00098 | ||
| SAA2, human | C00100 | ||
| SAA1, rat | C00101 | ||
| SAA, rabbit | C00148 | ||
| SAA3, mouse | C00153 | ||
| G-CSF, human | C00154 | ||
| ICAM-1, human | C00155 | ||
| CEs containing binding sites for ZIP and ETS factors | |||
| AP-1 | c-Ets-1, c-Ets-2 | Scavenger receptor, human | C00079, C00080 |
| GM-SCF, mouse | C00081 | ||
| Polyoma virus enhancer | C00082 | ||
| Collagenase, human | C00083 | ||
| uPA, human | C00084, C00085 | ||
| uPA, mouse | C00086 | ||
| JunB, mouse | C00087 | ||
| IgH, mouse | C00133 | ||
| TIMP-1, mouse | C00134 | ||
| IL-3, human | C00139 | ||
| C/EBPα | PU.1 | GM-CSF receptor α, human | C00186 |
| CEs containing binding sites for REL and HMG I factors | |||
| NF-κB | HMG I(Y) | E-selectin, human | C00056, C00058, C00059 |
| IFN-β, human | C00057 | ||
| GRO α, human | C00140 | ||
| NF-AT | HMG I(Y) | IL-4, mouse | C00167 |
| CEs containing binding sites for ETS and RUNT factors | |||
| ETS | AML1 | TCR β, human | C00020 |
| IgH µ, human | C00173 | ||
| TCR α, human | C00174 | ||
| Mo-MLV | C00184 | ||
| TCR β, mouse | C00185 | ||
| CEs containing binding sites for ETS and MADS factors | |||
| ETS | SRF | c-fos, human | C00022 |
| egr-1, mouse | C00126 | ||
| pip92, mouse | C00127 | ||
Since functional properties and tissue distribution of factors vary significantly within the same factor class, another criteria for classification is suggested based on combinatorial regulation provided by a CE (7). CEs are classified into five main groups as indicated in Table 2. CEs provide: (i) tissue-specific regulation, when one factor is tissue- or cell type-specific, and another is ubiquitous and constitutive (18 CEs); (ii) tissue-specific induction, when one factor is tissue-specific and another is inducible (22 CEs); (iii) inducible regulation, when one factor mediates a response to an extracellular signal and another is ubiquitous and constitutive (14 CEs); (iv) cross-coupling of signal transduction pathways, in the case of both factors being inducible through different pathways (64 CEs); (v) cell cycle-dependent regulation, when the activity of at least one factor is dependent on the cell cycle stage (three CEs). The majority of CEs (102) contain at least one site for an inducible factor (Table 2, blue cells).
Table 2. Classification of CEs according to the specific function they provide.
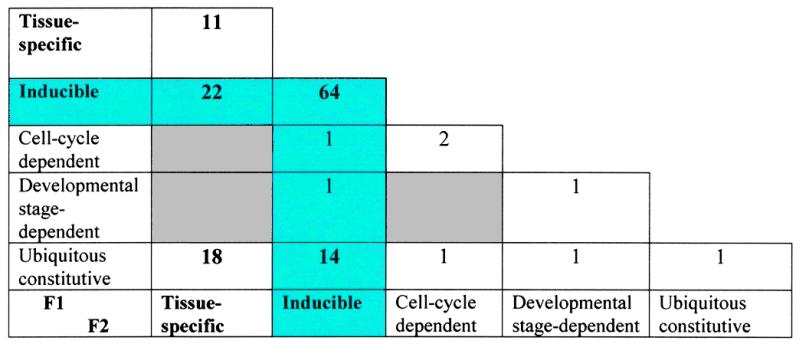
Functional properties of factors F1 and F2 are given in columns and rows. The figures represent the number of CEs of each type. CEs containing at least one binding site for an inducible factor are highlighted with a blue background. CEs of certain functions have not yet been described (grey cells).
DATABASE STRUCTURE AND CONTENT
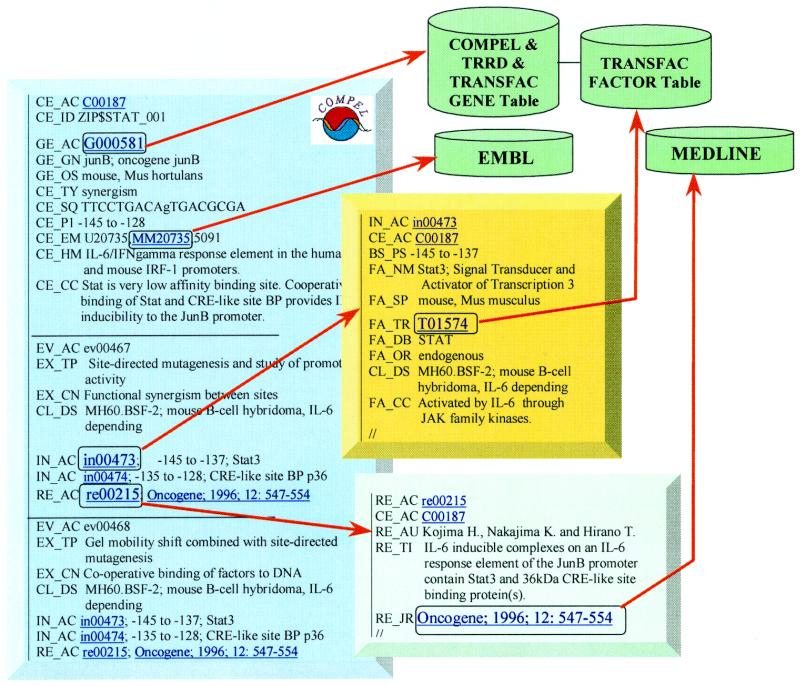
The relational model of COMPEL has been previously described (2,11). It now comprises 16 different tables. COMPEL has been made publicly available and distributed in three ASCII flat-files: (i) Composite Elements, (ii) Interactions and (iii) References. Examples of COMPEL entries and links between the three tables are illustrated in Figure 1. A detailed description of the fields is given in the database documentation available at http://compel.bionet.nsc.ru/compel/description.html . COMPEL is closely linked to other databases on transcriptional regulation, TRANSFAC and TRRD. The file Composite Elements is connected with the GENE tables in the TRANSFAC and TRRD databases (8,9). TRRD contains the field ‘CE’ which in turn refers to COMPEL by its accession number (4). The file Interactions is connected with the TRANSFAC FACTOR table. Most of the CEs are linked to the EMBL databank (12), and all references to the original papers are linked to MEDLINE (Fig. 1). The content of the current release is shown in Table 3.
Figure 1.
Example entries of the COMPEL database. An entry of the table Composite Elements is shown in the blue rectangle, an Interactions entry in the yellow box and a References record in the green box. Green cylinders symbolize external databases.
Table 3. Content of the COMPEL release 3.0.
| Tables | Number of entries | |
|---|---|---|
| Composite Elements | ||
| Composite elements | 178 | |
| Genes | 112 | |
| Transcription factors | 178 | |
| Evidences | 481 | |
| Links to EMBL | 146 | |
| Interactions | 488 | |
| References | 171 | |
WWW INTERFACE
WWW search and browse options are now available for COMPEL release 3.0. The browsing is provided by the type of DNA binding domains of transcription factors involved. For example, by clicking the ZIP tag one will get a list of all CEs in COMPEL comprising at least one binding site for a transcription factor of leucine zipper family (AP-1, CREB and others). From this list one could retrieve any individual COMPEL entry supplied by all necessary hyperlinks to interaction entries, references, as well as to the foreign databases TRRD, TRANSFAC, EMBL and MEDLINE (see Fig. 1). The search machine enables the user to retrieve COMPEL entries by gene name, species, name of transcription factor, DNA-binding domain as well as to make a full-text query. Simple Boolean operations on two terms are available.
CONNECTED PROGRAMS
CEs collected in the COMPEL database can be effectively used for creating new computer programs for searching potential CEs in DNA sequences. In COMPEL, the most numerous CEs are NFAT elements that belong to the ZIP$REL type (Table 1). These CEs, consisting of binding sites for the transcription factors NFATp/c and AP-1, were found in promoters and enhancers of cytokine genes that are induced during immune response in activated T-, B- and mast cells (for review, see 15). Structural features of experimentally confirmed NFAT CEs were taken into consideration for constructing a computer routine for potential CEs prediction (16). The program is available through the WWW at http://transfac.gbf.de/dbsearch/funsitep/s_comp.html and http://compel.bionet.nsc.ru/FunSite/CompelScan.html . One should launch the sequence under study and adjust the cut-off values for two individual sites (NFATp/c and AP-1) as well as for the CS (default cut-off values are provided). The program lists the positions of found CEs and of the individual sites constituting the CEs along with the corresponding score values. Additional information about clusters of potential CEs (if any) is given in the report. This program is a reliable tool to generate experimentally testable hypotheses about potential CEs within regulatory regions of genes which are up-regulated during immune response. The test revealed a high specificity of the method for the regulatory regions of these genes in comparison with promoters of genes irrelevant for immune response (such as muscle-specific genes) (16). Presently, the program is being adapted for the prediction of other types of CEs.
Acknowledgments
ACKNOWLEDGEMENTS
Different parts of this work were funded by the German Bundesministerium für Bildung, Wissenschaft, Forschung und Technologie (FANGREB project and project no. X224.6), by the Russian Ministry of Sciences and the Siberian Branch of Russian Academy of Sciences, by the North Atlantic Treaty Organisation (grant no. 951149) as well as by BIOBASE Ltd (Braunschweig, Germany).
REFERENCES
- 1.Perier R.C., Junier,Th., Bonnard,C. and Bucher,P. (1999) Nucleic Acids Res., 27, 307–309. Updated article in this issue: Nucleic Acids Res. (2000), 28, 302–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heinemeyer T., Chen,X., Karas,H., Kel,A.E., Kel,O.V., Liebich,I., Meinhardt,T., Reuter,I., Schacherer,F. and Wingender,E. (1999) Nucleic Acids Res., 27, 318–322. Updated article in this issue: Nucleic Acids Res. (2000), 28, 316–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kel A.E., Kolchanov,N.A., Kel,O.V., Romashencko,A.G., Ananko,E.A., Ignatieva,E.V., Merkulova,T.I., Podkolodnaya,O.A., Stepanenko,I.L., Kochetov,A.V., Kolpakov,F.A., Podkolodnyi,N.L. and Naumochkin,A.N. (1997) Mol. Biol. (Mosk)., 31, 626–636. [PubMed] [Google Scholar]
- 4.Kolchanov N.A., Ananko,E.A., Podkolodnaya,O.A., Ignatieva,E.V., Stepanenko,I.L., Kel-Margoulis,O.V., Kel,A.E., Merkulova,T.I., Goryachkovskaya,T.N., Busigina,T.N., Kolpakov,F.A., Podkolodny,N.L., Naumochkin,A.N. and Romashchenko,A.G. (1999) Nucleic Acids Res., 27, 303–306. Updated article in this issue: Nucleic Acids Res. (2000), 28, 298–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kel O.V., Romaschenko,A.G., Kel,A.E., Naumochkin,A.N. and Kolchanov,N.A. (1995) Proceedings of the 28th Annual Hawaii International Conference on System Scienses [HICSS]. Biotechnology Computing, IEE Computer Society Press, Los Alamitos, CA, Vol. 5, pp. 42–51.
- 6.Kel O.V., Romaschenko,A.G., Kel,A.E., Wingender,E. and Kolchanov,N.A. (1995) Nucleic Acids Res., 23, 4097–4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kel O.V., Romaschenko,A.G., Kel,A.E., Wingender,E. and Kolchanov,N.A. (1997) Mol. Biol. (Mosk)., 31, 498–512. [Google Scholar]
- 8.Wingender E., Kel,A.E., Kel,O.V., Karas,H., Heinemeyer,T., Dietze,P., Knüppel,R., Romaschenko,A.G. and Kolchanov,N.A. (1997) Nucleic Acids Res., 25, 265–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karas H., Kel,A.E., Kel,O.V., Kolchanov,N.A. and Wingender,E. (1997) Mol. Biol. (Mosk)., 31, 531–539. [PubMed] [Google Scholar]
- 10.Heinemeyer T., Wingender,E., Reuter,I., Hermjakob,H., Kel,A.E., Kel,O.V., Ignatieva,E.V., Ananko,E.A., Podkolodnaya,O.A., Kolpakov,F.A., Podkolodny,N.L. and Kolchanov,N.A. (1998) Nucleic Acids Res., 26, 362–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kel-Margoulis O.V., Kel,A.E., Frisch,M., Romaschenko,A.G., Kolchanov,N.A., and Wingender,E. (1998) Proceedings of the First International Conference on Bioinformatics of Genome Regulation and Structure, (BGRS ’98), ICG, Novosibirsk, Russia, Vol. 1, pp. 54–57.
- 12.Stoesser G., Tuli,M.A., Lopez,R. and Sterk P. (1999) Nucleic Acids Res., 27, 18–24. Updated article in this issue: Nucleic Acids Res. (2000), 28, 19–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diamond M.I., Miner,J.N., Yoshinaga,S.K. and Yamamoto K.R. (1990) Science, 249, 1266–1272. [DOI] [PubMed] [Google Scholar]
- 14.Wingender E. (1997) Mol. Biol. (Mosk)., 31, 483–497. [PubMed] [Google Scholar]
- 15.Rao A., Luo,C. and Hogan,P.G. (1997) Annu. Rev. Immunol., 15, 707–747. [DOI] [PubMed] [Google Scholar]
- 16.Kel A., Kel-Margoulis,O., Babenko,V. and Wingender,E. (1999) J. Mol. Biol., 288, 353–376. [DOI] [PubMed] [Google Scholar]