Abstract
The Corona Virus was first started in the Wuhan city, China in December of 2019. It belongs to the Coronaviridae family, which can infect both animals and humans. The diagnosis of coronavirus disease-2019 (COVID-19) is typically detected by Serology, Genetic Real-Time reverse transcription–Polymerase Chain Reaction (RT-PCR), and Antigen testing. These testing methods have limitations like limited sensitivity, high cost, and long turn-around time. It is necessary to develop an automatic detection system for COVID-19 prediction. Chest X-ray is a lower-cost process in comparison to chest Computed tomography (CT). Deep learning is the best fruitful technique of machine learning, which provides useful investigation for learning and screening a large amount of chest X-ray images with COVID-19 and normal. There are many deep learning methods for prediction, but these methods have a few limitations like overfitting, misclassification, and false predictions for poor-quality chest X-rays. In order to overcome these limitations, the novel hybrid model called “Inception V3 with VGG16 (Visual Geometry Group)” is proposed for the prediction of COVID-19 using chest X-rays. It is a combination of two deep learning models, Inception V3 and VGG16 (IV3-VGG). To build the hybrid model, collected 243 images from the COVID-19 Radiography Database. Out of 243 X-rays, 121 are COVID-19 positive and 122 are normal images. The hybrid model is divided into two modules namely pre-processing and the IV3-VGG. In the dataset, some of the images with different sizes and different color intensities are identified and pre-processed. The second module i.e., IV3-VGG consists of four blocks. The first block is considered for VGG-16 and blocks 2 and 3 are considered for Inception V3 networks and final block 4 consists of four layers namely Avg pooling, dropout, fully connected, and Softmax layers. The experimental results show that the IV3-VGG model achieves the highest accuracy of 98% compared to the existing five prominent deep learning models such as Inception V3, VGG16, ResNet50, DenseNet121, and MobileNet.
Keywords: Corona virus, COVID-19, Inception V3, VGG16, IV3-VGG, RT-PCR, ResNet50, DenseNet121, MobileNet, Chest X-ray
Introduction
The Coronavirus disease-19, Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) was initially discovered in Wuhan, China in December of 2019 and quickly spread over nations like the United States of America (USA), India, Brazil, France, Turkey, Russia, and others. It has officially announced as a pandemic by the World Health Organization (WHO) [20] in March 2020. Subsequently, forcing billions of people to stay at home and most of the nations announced a lockdown. About 525,080,438 COVID-19-positive cases were confirmed in more than 219 countries until May 19th, 2022, among which 484,920,117 cases were recovered, and 6,294.856 were reported as deaths [20].
Types of corona virus
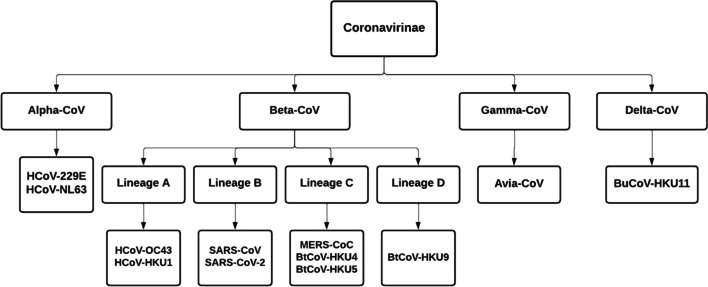
The Coronavirus belongs to the Coronaviridae family and the subgroups of this virus are alpha Coronavirus (α -CoV), beta coronavirus (β-CoV), gamma coronavirus (γ-CoV), and delta coronavirus (δ-CoV) [32]. Various subgroups of the Coronaviridae family are described in Fig. 1. The alpha coronavirus (also called Human Coronavirus) is a positive-sense, single-stranded RNA virus that infects both humans and mammals. Alpha coronavirus is associated with lower and upper respiratory tract diseases and frequently affects young children [22].
Fig. 1.
Types of coronavirus
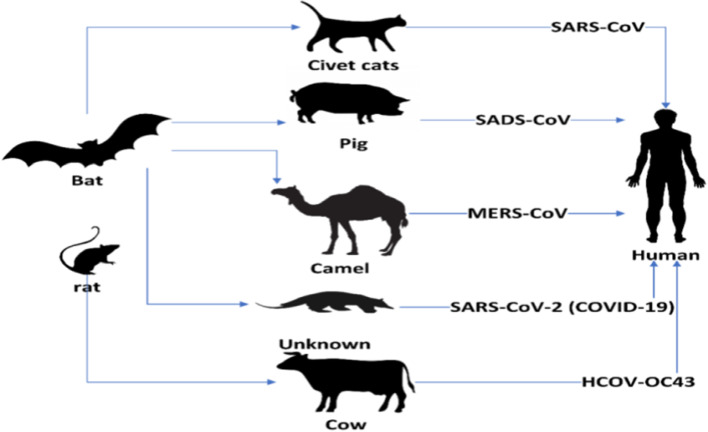
Beta coronavirus causes respiratory and gastrointestinal diseases in humans and most mammals [7]. Rodents and bats are the natural reservoirs of beta coronavirus. SARS-CoV-2 is new to mankind and contaminates from bats to humans as shown in Fig. 2 [13]. It spreads through beads of salivation or sneezing [31]. While 95% of the infected patients are surviving, 5% rest are in critical condition [14]. In more serious cases, the infection can cause shortness of breath, multi-organ failure, chest pressure, and finally death.
Fig. 2.
Transmission of coronavirus
The BtCoV-HKU4, BtCoV-HKU5, and BtCoV-HKU9 are called Bat coronavirus discovered in Hong Kong [10]. The gamma coronavirus is derived from the avian that infects birds [28] and the delta coronavirus is derived from pig gene pools that infect birds and some mammals [29].
Types of COVID-19 diagnosis
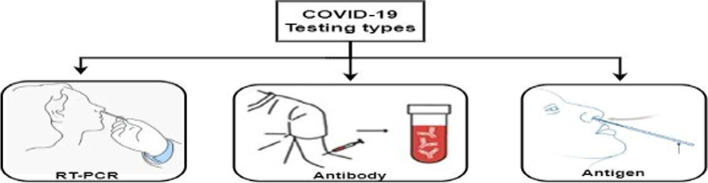
The diagnosis of COVID-19 is typically detected by Serology (Antibody), Genetic Real-Time reverse transcription–Polymerase ChainReaction(RT-PCR), and Antigen testing. Various types of diagnosis of COVID-19 are shown in Fig. 3.
Fig. 3.
Types of COVID-19 testing
Genetic testing (RT-PCR)
The Reverse Transcription - Polymerase Chain Reaction (RT-PCR) is a gold standard for detecting the presence of coronavirus [9]. It can reliably detect a virus in the early days of infection [27]. This test will report whether the person is currently infected by the virus or not. This testing method has limitations like incorrect sampling, availability, specificity, cost, and long turnaround time [1]. However, many countries are not able to provide sufficient RT-PCR testing kits.
Serology testing (Antibody)
Antibody testing for COVID-19 diagnosis provides qualitative detection of IgG and/or IgM from blood, human serum, or plasma samples [18]. This test can detect only whether the person is previously infected or not, and also how the immune system has responded to the infection. This method cannot show the current presence of the infection (like RT-PCR). After symptoms occur, it takes 1-3 weeks to develop antibodies [23]. This testing method is expensive and time taking.
Antigen testing
This testing method is similar to the RT-PCR, which collects samples from nasal swabs to detect the presence or absence of coronavirus infection in the body. It takes less time and is relatively cheap when compared to the other tests [19]. This testing method might miss an active coronavirus infection in comparison to RT-PCR tests.
There are many limitations in the above testing techniques like incorrect predictions, taking more time for results, and high cost. So implementing an automatic detection method is needed at this time. Therefore, silicosis diagnosis (X-rays and CT scans (Computed Tomography)) are introduced for screening and identifying Coronavirus cases [16]. The features of coronavirus in X-ray are Ground Glass Opacities, Consolidation, Bilateral Distribution, and Peripheral Distribution [8]. Therefore, X-rays are a well-known, quick, effective, broadly accessible analytic imaging procedure and compact in nature.
This paper is coordinated as follows. Section 2, presents the related work on different deep-learning techniques used to detect COVID-19. In Section 3, the chest X-ray dataset is shown. Section 4 describes the hybrid CNN model formation. Section 5 provides a step-by-step process of the proposed IV3-VGG model. Section 6 describes the results and performance of the IV3-VGG method and Section 7 presents the comparative analysis of a proposed method with some existing CNN models. Finally, the conclusion of the work is presented.
Related work
Mohamed Loey et al. [13] proposed a model called “Generative Adversarial Network (GAN) with deep transfer learning for coronavirus detection”. The authors collected COVID-19 chest X-ray images from Dr. Joseph Cohen in a github repository. The Pneumonia, COVID-19, and Normal images are collected from the identifying medical diagnoses, Kaggle. Pre-processing is done with GAN and then three deep transfer models such as Restnet18, Alexnet, and Googlenet are applied for training. In the first scenario, Restnet (normal, COVID-19, pneumonia, bacterial) model achieved 80.6% accuracy. In the second scenario, the Alexnet (normal, COVID-19, bacterial) model achieved 85.2% accuracy while in the third scenario, Googlenet (normal and COVID-19) achieved 100% in testing accuracy. The Googlenet achieved the highest accuracy, but an over-fitting problem is occurring.
Lamia Nabil Mahdy et al. [14] proposed multilevel thresholding followed by Support Vector Machine (SVM). This framework begins with a patient’s X-ray visualizing and enhancing the contrast of the input image using a median filter. The Otsu objective function-based multi-level image segmentation threshold is applied for splitting the grey images into several distinct areas and SVM has been implemented for the classification of infected lungs from non-infected X-ray images. This framework gets an accuracy of 97.48%. The input dataset consists of 15 normal images from the Montgomery County X-ray Set and 25 COVID-19 images from Covid-chest-Xray-dataset-master in a Github repository. This model was done with only 40 images.
Arpan Mangal et al. [15] proposed CovidAID (COVID-19 Artificial Intelligence Detector) to detect COVID-19 patients using Chest X-Ray dataset. The model consists of pre-trained CheXNet with a 121-layer DenseNet followed by a fully connected layer. Optimizer Adam is used in the training stage. Then, the network weights are initialized, and the whole network is trained using the hyper parameters. This model classifies four different parameters like normal, bacterial, viral pneumonia, and COVID-19 with 90.5% accuracy.
Mesut Togacara et a [26] proposed a new model with Fuzzy Color and Stacking techniques. The Fuzzy Color technique is used for removing noise. The Stacking technique is used to create a new dataset for better data quality images. The MobileNetV2 and Squeeze Net models are used to train the dataset and Support Vector Machine (SVM) is used for the classification. The framework gives a 99.27% accuracy to classify the dataset. The dataset is collected from Covid-chest-Xray-dataset in a Github repository. It failed to focus on distinguishing patients, showing COVID-19 rather than pneumonia symptoms.
The DarkCovidNet model was proposed by TulinOzturket et al. [17] and features 17 convolutional layers, each of which has a convolutional with a Leaky Rectified Linear Unit (LeakyReLU) and batch normalization. All pooling layers employ the Max-pool procedure. Training is carried out over 100 iterations, and performance is assessed using 5-fold cross-validation. In the binary class, accuracy rates were 98.08% and 87.02%, respectively. 125 COVID-19 images, 500 normal photos, and 500 images of pneumonia should be collected from the ChestX-ray8 database. For X-rays of inadequate quality, this model produces predictions that are inaccurate.
For the purpose of COVID-19 prediction, Thejeshwar et al. [25] introduce the KE Sieve Neural Network model. Pre-processing, feature extraction, and the KE Sieve algorithm are the three parts of this procedure. Joseph Paul Cohen, Chest-Xray-Pneumonia, and Kaggle compiled the dataset. The dataset is unbalanced; however, this model can distinguish between coronavirus, normal, and viral with 98.07% accuracy.
The Inception V3 deep learning model with a transfer learning technique was proposed by SohaibAsif et al. [5]. The Inception V3 model is used for training while the chest X-rays are used as input. The classification accuracy for the normal, COVID-19, viral, and pneumonia classes is greater than 96%. The images are compiled in a Github repository from the Covid-chest-Xray-dataset, the Italian Society of Medical and Interventional Radiology (SIRM), and the Radiography Database. For low-resolution photos, it sometimes produces inaccurate findings.
A combination of CNN and LSTM models was suggested by Alzubiet et al. [3] for the production and analysis of deep image captions. In this paradigm, there are two steps. The Inception model is used in the first stage, while the LSTM model is used in the second. With the aid of a dense layer, these two models were combined. Using the Inception model and LSTM to extract the key insights from the provided caption, this model encodes the input image. The proposed method for caption creation was also contrasted with other approaches like GRU and bi-dimensional LSTM models by the authors.
In order to address the issues surrounding the Coronal virus, Alzubiet et al. [2] designed the COBERT system. This model searches and accesses the 59K COVID-19 works of literature using the Coronavirus Open Research Dataset Challenge (CORD-19). The reader employs the Bidirectional Encoder Representation from Transformers (BERT) to polish the sentences from the filtered papers after the COBERT gets 500 articles based on the TF-IDF score for the input query. The ranker then receives the sentences and compares the values of the logit function to produce a short response to the inquiry.
Using a boosted neural network, Jafar A. Alzubier et al. [4] created a classification model for diagnosing lung cancer illness. This network was split into two modules by the authors. Utilize the combined Newton-Rapsons Maximum Likelihood and Minimum Redundancy (MLMR) preprocessing method in the first module to cut down on classification time. You will use the Boosted Weighted Optimised Neural Network to categorize the Lung Cancer Disease in the second module.
By using the Reddit social network, Bonifazi, Gianluca, et al. [28] suggested a brand-new method for gathering insightful data about COVID-19. The authors used a three-step process to implement this strategy. Apply the dynamic and semi-automatic categorization in the first stage to extract the data for COVID-19 from Reddit postings. The second phase is creating fake subreddits utilizing uniform themes. The last step is to locate the COVID-19 user communities online.
A CNN model called "CorDet" was suggested by Hussain, Emtiaz, et al. [11] to identify COVID-19 in chest X-ray and CT scan pictures. For two categories, three classifications, and four classifications, such as COVID, Normal, non-COVID viral pneumonia, and non-COVID bacterial pneumonia, this model was created to offer a precise diagnosis. The authors of the model explained its results, and a doctor agreed with them.
To determine whether COVID-19 was present in chest X-ray pictures, Ismael, Aras M., and AbdulkadirEngür [12] built three deep CNN models, including deep feature extraction, fine-tuning, and an end-to-end trained CNN model. The three CNN models were applied to 180 COVID and 200 regular chest X-ray pictures by the authors. The ResNet50 model's deep features were retrieved, and an SVM classifier using a linear kernel function yielded a score of 94.7% accuracy.
Dataset
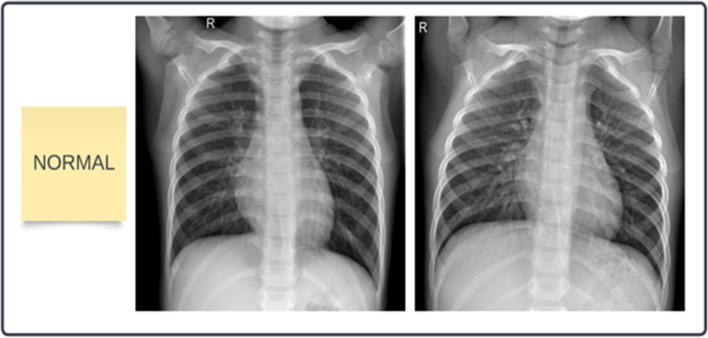
In this framework, the dataset of 243 images is collected from the COVID-19 Radiography Database [21] which is created by a team of researchers from the University of Dhaka, Qatar, and Doha University, and Bangladesh, along with Malaysia and Pakistan medical doctors. This dataset has coronavirus-positive and normal images. Out of 243 X-rays, 121 are COVID-19 positive and 122 are normal images. The dataset is divided into two parts i.e. training set and testing set. The training set contains 200 images and the testing set contains 43 images. The samples of COVID-19 and normal chest X-ray images are presented in Figs. 4 and 5.
Fig. 4.
Samples of COVID-19 Chest X-ray images
Fig. 5.
Samples of normal Chest X-ray images.
Model formations
Inception V3
A CNN-based network for categorization is Inception V3 [24]. It employs inception modules, which are 42 layers deep and consist of a concatenated layer with 1 × 1, 3 × 3, and 5 × 5 convolutions. Reduce the number of parameters while accelerating the training rate. The GoogLeNet model is another name for Inception 3. The benefits of Inception V3 include the following. Factorization into Smaller Convolutions
Auxiliary Classifiers- used to combat the vanishing gradient problem in very deep networks.
Reduction of Grid Size
VGG16
The ImageNet project, a sizable visual database project used in the development of visual object identification software, uses the VGG16 (Visual Geometry Group) architecture, a straightforward and widely used convolutional neural network architecture. Very Deep Convolutional Networks for Large-Scale Image Recognition is an idea put out by K. Simonyan and A. Zisserman from the University of Oxford. There are 16 convolutional layers in it. VGG16 is frequently utilised out of the box for a variety of applications because it is freely accessible online. Medical imaging techniques like X-rays and MRIs can be utilised to diagnose diseases using VGG16. It may also be used to reading street signs while driving.
There are many deep learning models for prediction, but these models have a few limitations like overfitting, misclassification, and false predictions for poor-quality chest X-rays. In this regard, the novel hybrid model Inception V3 with VGG16 is proposed for COVID-19 prediction using chest X-rays.
Proposed method
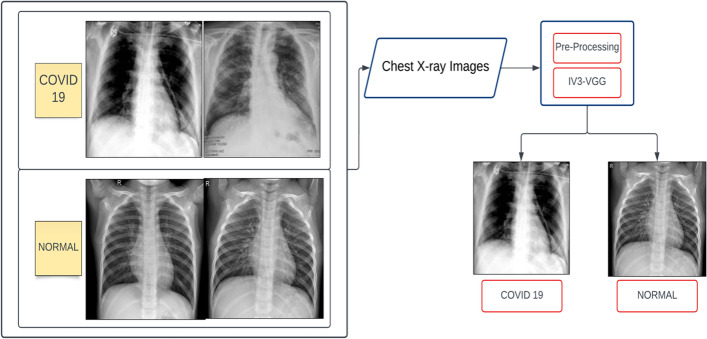
The proposed method is the combination of two Convolutional Neural Networks namely Inception V3 and VGG16 models. This model overcomes the limitations such as misclassification, incorrect predictions for poor-quality X-ray images, and the lowest accuracy in the existing models. This model is divided into two modules namely pre-processing and the IV3-VGG. The chest X-rays are considered as an input for the pre-processing stage. The block diagram of the proposed IV3-VGG is shown in Fig. 6.
Fig. 6.
Block diagram of proposed IV3-VGG method
Pre-processing is an important phase in image classification and prediction. Some of the images have different sizes and different colour intensities. So have to convert all the images to the same size of 320 × 320 pixels to increase processing time. The pre-processed images are forwarded to IV3-VGG hybrid model.
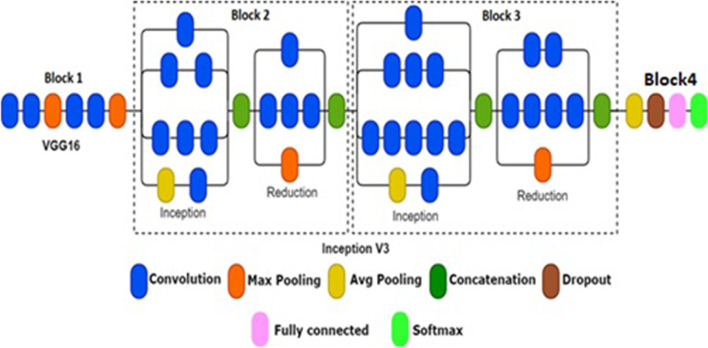
The second module i.e. IV3-VGG consists of four blocks. The architecture of IV3-VGG is shown in Fig. 7. The first block is considered for VGG-16 and blocks 2 and 3 are considered for Inception V3 networks and block 4 consists of four layers namely Avg pooling, dropout, fully connected, and Softmax layers. In general, the VGG-16 is the Convolutional Neural Network model with 16 layers. These layers have a combination of convolution and pooling operations. But in block 1 only the first 6 layers of VGG16 are considered.
Fig. 7.
Proposed IV3-VGG architecture
Avg-pooling and Max-pooling techniques are used in the proposed architecture for dimensionality reduction and feature extraction purposes. Avg-pooling extracts the features smoothly by selecting the average pixel of the batch, and max-pooling extracts the main features of an image by selecting the maximum pixel of a batch. Concatenation is used to concatenate several input units into a single unit of output. The general structure of the proposed IV3-VGG method is as described in Table 1.
Table 1.
The general structure of IV3-VGG
In block 1, consider only six layers of VGG16. Out of six layers, four layers are 3 x 3 convolutional layers and two are 2 x 2 max-pooling layers. The architecture of block 1 i.e. VGG is shown in Fig. 8. The outcome of block 1 is the input for block 2.
Fig. 8.
Architecture of block 1
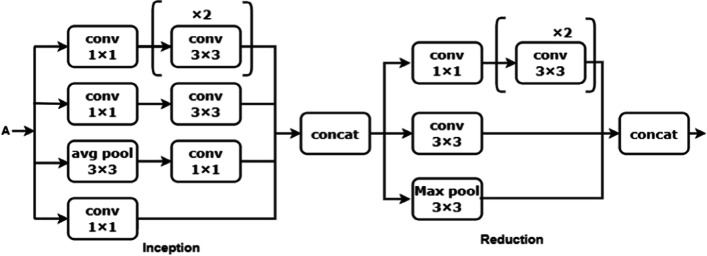
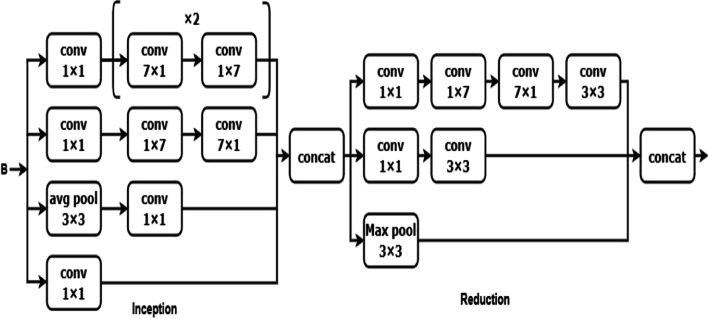
Inception V3 model is implemented in blocks 2 and 3. Block 2 consists of one inception and one reduction block with convolution, pooling, and concatenate operations. The 1 × 1 convolution is lower than the 3 × 3 convolution, can reduce the number of input channels, and accelerate training speed [24]. The 1 × 1 and 3 × 3 convolution layers are used to extract the image's low-level features like edges, corners, and lines. The inception block has four 1 x 1, three 3 x 3, and avg-pooling of kernel size 3 x 3 which are concatenated and sent to the reduction block. The reduction block is used to avoid a representational bottleneck. This reduction block consists of three 3 x 3, one 1 x 1 convolutions, and max-polling layers. Block 2 makes the network efficient and less expensive. The architecture of block 2 is shown in Fig. 9.
Fig. 9.
Architecture of block 2
The output of block 2 is input for the block 3. Block 3 is also the combination of inception and reduction blocks. The 7 × 7 kernel size convolution is used to extract the high-level features like objects and events. A 7 × 7 convolution is replaced with a 7 × 1 and 1× 7 convolution. The inception block is having three sets of [7 x 1 and 1 x 7], four 1 x 1, and 3 × 3 avg-polling layers. This factorization makes the model cheaper than the single 7 x 7 convolution. Then all these layers are concatenated and sent to the reduction block as illustrated in Fig. 10.
Fig. 10.
Architecture of block 3
The output of block 3 is the input for block 4 which consist of four layers. The average pooling layer of block 4 is to calculate the average of all the features of the image. Then the fully connected layer is used to transform multi-scale feature vectors into one-dimensional vectors. However, a 0.3 dropout fraction rate of input units is taken to overcome the issue of overfitting. Finally, the Softmax layer is used as a classifier to represent class output with probability, and the highest probability was chosen as the predicted class.
Results
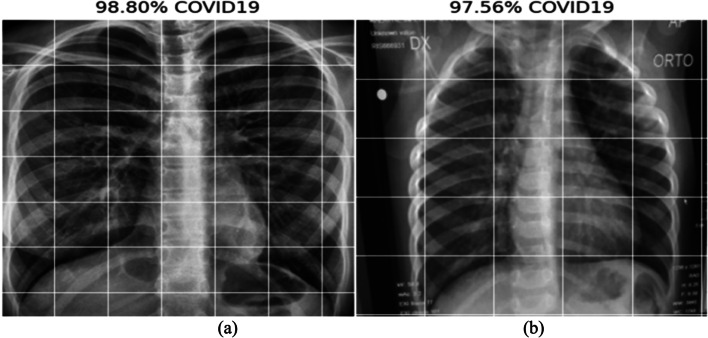
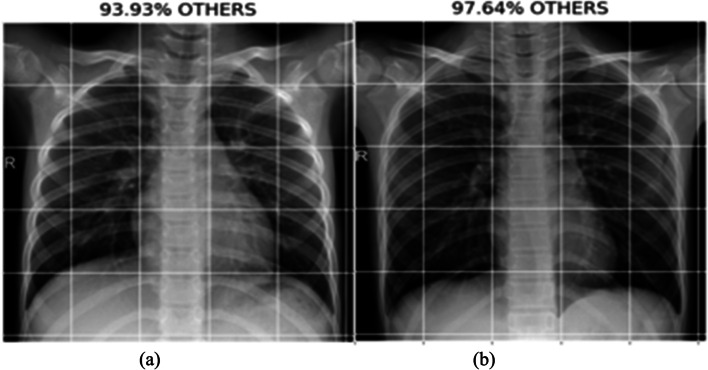
This section provides the results and analysis of the proposed IV3-VGG method. Implementation of IV3-VGG is done using Google Colab, which is a free service that requires no setup to use hosted by Jupyter Notebook. This model is trained with 10 epochs and each epoch has 13 steps. Adam optimizer is used for faster convergence with a 0.0001 learning rate. During the training phase, the best prediction performance is saved in each step based on the lowest loss and highest accuracy. Testing is done with a total of 43 images. Out of 43 images, 21 are COVID-19 images and 22 are normal images. The following Fig. 11 depicts the two sample COVID-19 images predicted as COVID with a probability of 98.80% and 97.56%. In 22 normal tested images, the two sample normal images predicted as Normal/others with a probability of 93.93% and 97.64% are shown Fig. 12.
Fig. 11.
Predicted results of COVID-19 with a probability of (a) 98.80%, and (b) 97.56%
Fig. 12.
Predicted results of normal with a probability of (a) 93.93%, and (b) 97.64%
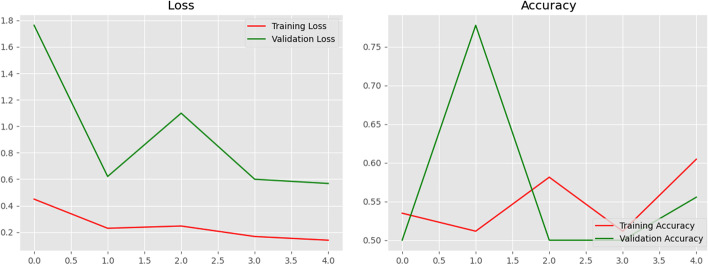
Along with projected normal and COVID 19 case accuracy, the models' performance is also evaluated by contrasting loss and validation accuracy when training and validating the provided dataset. Figure 13 shows a visualisation of the proposed model's training and validation accuracy as well as its training and validation loss. It is observed that validation accuracy is found to be more compared to training accuracy. These loss and accuracy plots can help to develop the model further with greater accuracy and less loss.
Fig. 13.
Graphs of the training, validation loss, and accuracy
Comparative analysis
In a comparative analysis, metrics such as Precision, Recall, and Accuracy are used to evaluate the proposed method. The results are compared with the metrics of the existing methods such as InceptionV3, VGG16, ResNet50, DenseNet121, and MobileNet. The IV3-VGG model achieves promising and high-performing results with an accuracy of 98% compared to these models. DenseNet121 is a popular CNN model, where each layer is connected deeply with another layer. But, this model gets the lowest accuracy of 88%. MobileNet is having depth-wise separable convolutions and achieved 91% accuracy. Another popular CNN model is ResNet50, which consists of 5 stages with convolution and pooling layers, obtaining 93% accuracy. Inception V3 is a widely used CNN model with 48 layers. This model consists of symmetric and asymmetric building blocks, with pooling, convolutional and auxiliary classifiers. The VGG16 is a CNN model which is mainly used in image detection with 16 layers of convolution and max-pooling. Table 2 presents the comparative analysis of IV3-VGG with prominent five Convolutional Neural Network (CNN) models.
Table 2.
Comparative analysis of IV3-VGG with prominent five CNN models
| Method | Precision (%) | Recall (%) | Accuracy (%) |
|---|---|---|---|
| IV3-VGG | 98 | 98 | 98 |
| InceptionV3 | 96 | 95 | 95 |
| VGG16 | 96 | 95 | 95 |
| DenseNet121 | 89 | 88 | 88 |
| MobileNet | 90 | 91 | 91 |
| ResNet50 | 94 | 93 | 93 |
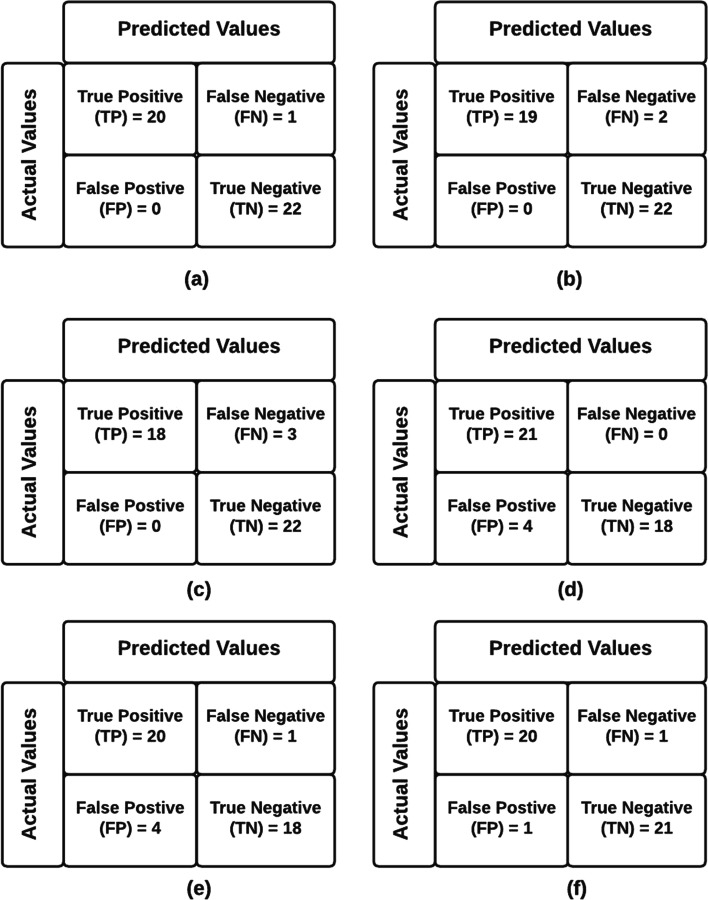
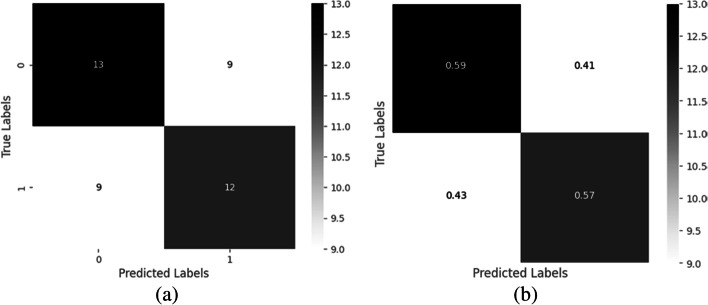
The confusion matrix shows the correct and incorrect label predictions for different classes. It is based on four parameters of True Positive (TP), False Negative (FN), False Positive (FP), and True Negative (TN). Therefore, out of 21 COVID-19 positive X-rays, 20 can predict COVID-19 correctly with a probability of 95%, and out of 22 Normal X-rays, all images are predicted correctly as Normal with a 100% probability. Compare to the remaining prominent models, the proposed IV3-VGG model shows valid results which are shown in Fig. 14. The confusion matrix analysis graphs shown in Fig. 14 are generated for training data consisting of 43 images and the heatmaps maps also are generated by using the same set of data which is depicted in Fig. 15.
Fig. 14.
Confusion matrix analysis of (a) IV3-VGG, (b) VGG16, (c) ResNet50, (d) MobileNet, (e) DenseNet121, and (f) Inception V3
Fig. 15.
Heat map of the analysis confusion matrix a) without and b) with normalization
The performance of a classification model is displayed by the Receiver Operating Characteristics (ROC). It is a 2-dimensional graph. True Positive and False Positive Rates are two metrics that are plotted on this graph. By using the ROC curve to assess a test’s overall diagnostic performance. The optimal cut-off value is also selected to determine if a disease is present or not. The ROC curve is presented in Fig. 16 with a false positive and true positive rate. The true positive rate remained stable following a little increase, demonstrating the effectiveness of the analysis. To compare the results of the current model “IV3-VGG” with the few other existing models; the ROC curves of Resnet50 & InceptionV3 models for COVID-19 prediction [21] are represented in Figs. 17 and 18.
Fig. 16.
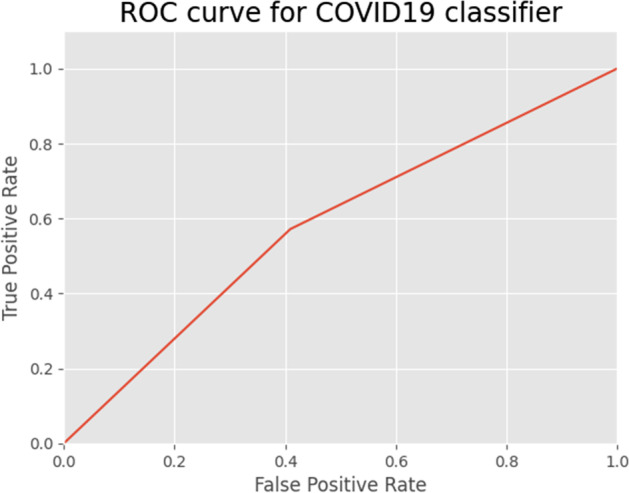
ROC curve for IV3-VGG model
Fig. 17.
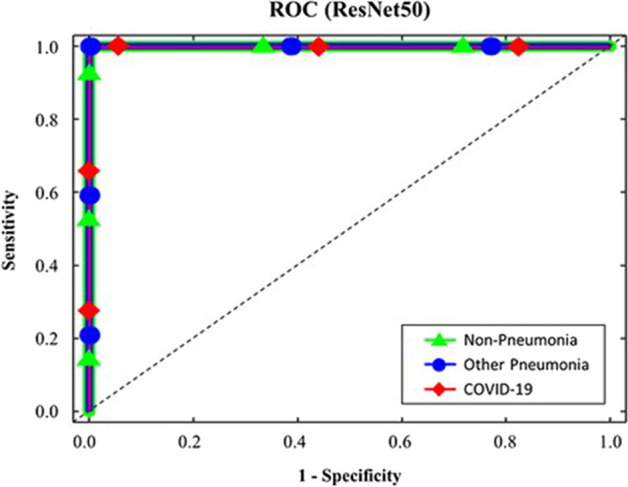
ROC curve for Resnet50 model
Fig. 18.
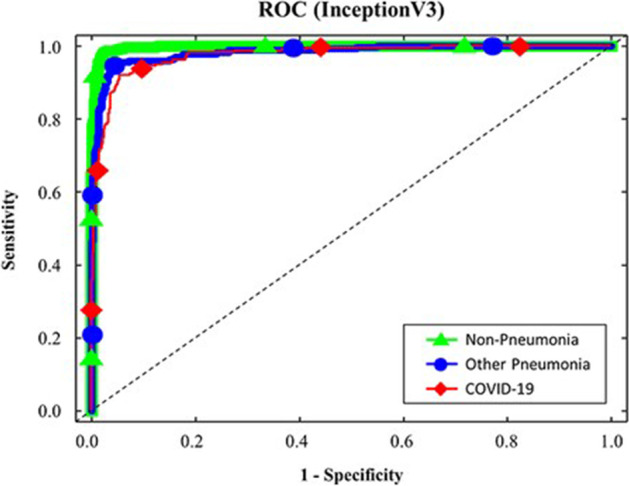
ROC curve for InceptionV3 model
Conclusion
In this framework, a deep learning-based hybrid model called "Inception V3 with VGG16 (IV3-VGG)" is suggested for the X-ray-based prediction of normal health conditions and COVID-19. The suggested approach makes use of pre-processing to prevent inaccurate categorization and predictions for poor chest X-ray image quality. This approach is also trustworthy and effective when it comes to classification and data irregularity. The experimental results demonstrated that the proposed IV3-VGG model outperformed the existing Convolution Neural Network models in terms of precision, recall, and accuracy. In the comparative analysis, the same dataset is used to implement five prominent CNN models namely Inception V3, VGG16, ResNet50, DenseNet121, and MobileNet. The proposed IV3-VGG model achieved the highest precision of 98%, recall of 98%, and accuracy of 98% for discriminating between COVID-19 and Normal images compared to the above five existing CNN models. Out of five CNN models, Inception V3 gives 96% and VGG16 gives 95% accuracy, but Inception V3 and VGG16 models achieve 95% precision and recall. The remaining three models obtain the lowest accuracy of 88% for DenseNet121, ResNet50 at 93%, and MobileNet gets an accuracy of 91%. In future work, the proposed model IV3-VGG is extended for the large datasets of X-rays and also to predict the level or stage of the disease if he/she is suffering from COVID-19. The proposed model can also be extended to predict other types of chronic diseases such as Alzheimer’s disease, Huntington's disease, and Parkinson's disease.
Data availability
Not applicable.
Code availability
Customized code available.
Declarations
Conflicts of interest/Competing interests
• All authors have participated in (a) conception and design, or analysis and interpretation of the data; (b) drafting the article or revising it critically for important intellectual content; and (c) approval of the final version.
• This manuscript has not been submitted to, nor is under review at, another journal or other publishing venue.
• The authors have no affiliation with any organization with a direct or indirect financial interest in the subject matter discussed in the manuscript
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
K. Srinivas, Email: kudipudi72@gmail.com
R. Gagana Sri, Email: gagana.rachamalla@gmail.com
K. Pravallika, Email: kudipudipravallika@gmail.com
K. Nishitha, Email: nishithakommineni0204@gmail.com
Subba Rao Polamuri, Email: psr.subbu546@gmail.com.
References
- 1.Adhikari SP, Meng S, Wu YJ, Mao YP, Ye RX, Wang QZ, ..., Zhou H (2020) Epidemiology, causes, clinical manifestation and diagnosis, prevention and control of coronavirus disease (COVID-19) during the early outbreak period: a scoping review. Infect Dis Poverty 9(1):1–12 [DOI] [PMC free article] [PubMed]
- 2.Alzubi JA, Jain R, Singh A, Parwekar P, Gupta M (2021) COBERT: COVID-19 question answering system using bert. Arab J Sci Eng. 10.1007/s13369-021-05810-5 [DOI] [PMC free article] [PubMed]
- 3.Alzubi JA, Jain R, Nagrath P, Satapathy S, Taneja S, Gupta P (2020) Deep image captioning using an ensemble of CNN and LSTM based deep neural networks J Intell Fuzzy Syst. 10.3233/JIFS-189415
- 4.Alzubi JA, Bharathikannan B, Tanwar S, Manikandan R, Khanna A, Thaventhiran C. Boosted neural network ensemble classification for lung cancer disease diagnosis. Appl Soft Comput. 2019;80:579–591. doi: 10.1016/j.asoc.2019.04.031. [DOI] [Google Scholar]
- 5.Asif S, Wenhui Y (2020) Automatic detection of COVID-19 using X-ray images with deep convolutional neural networks and machine learning. medRxiv
- 6.Bonifazi Gianluca, et al. New Approaches to Extract Information from Posts on COVID-19 Published on Reddit. Int J Inform Technol Decision Making. 2022;2022:1–47. [Google Scholar]
- 7.Dai L, Zheng T, Xu K, Han Y, Xu L, Huang E, Gao GF. A universal design of betacoronavirus vaccines against COVID-19, MERS, and SARS. Cell. 2020;182(3):722–733. doi: 10.1016/j.cell.2020.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ehsan S, and Norbert PJ (2019) Computed tomography approaches, applications, and operations, Springer International Publishing, p 470
- 9.Govindarajan S, Swaminathan R. Differentiation of COVID-19 conditions in planar chest radiographs using optimized convolutional neural networks. Appl Intell. 2021;51(5):2764–2775. doi: 10.1007/s10489-020-01941-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu B, Ge X, Wang LF, Shi Z. Bat origin of human coronaviruses. Virol J. 2015;12(1):1–10. doi: 10.1186/s12985-015-0422-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hussain Emtiaz, et al. CoroDet: A deep learning based classification for COVID-19 detection using chest X-ray images. Chaos Solitons Fractals. 2021;142:110495. doi: 10.1016/j.chaos.2020.110495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ismael AM, Şengür A. Deep learning approaches for COVID-19 detection based on chest X-ray images. Expert Syst Applic. 2021;164:114054. doi: 10.1016/j.eswa.2020.114054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loey Mohamed, Smarandache F, Khalifa M, N. E. Within the lack of chest COVID-19 X-ray dataset: a novel detection model based on GAN and deep transfer learning. Symmetry. 2020;12(4):651. doi: 10.3390/sym12040651. [DOI] [Google Scholar]
- 14.Mahdy LN, Ezzat KA, Elmousalami HH, Ella HA, Hassanien AE. (2020) Automatic x-ray covid-19 lung image classification system based on multi-level thresholding and support vector machine. MedRxiv
- 15.Mangal A, Kalia S, Rajgopal H, Rangarajan K, Namboodiri V, Banerjee S, Arora C. (2020). CovidAID: COVID-19 detection using chest X-ray, arXiv preprint arXiv:2004.09803, 2020
- 16.Minaee S, Kafieh R, Sonka M, Yazdani S, Soufi GJ. Deep-covid: Predicting covid-19 from chest x-ray images using deep transfer learning. Med Image Anal. 2020;65:101794. doi: 10.1016/j.media.2020.101794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ozturk T, Talo M, Yildirim EA, Baloglu UB, Yildirim O, Acharya UR. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput Biol Med. 2020;121:103792. doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pan Y, Li X, Yang G, Fan J, Tang Y, Zhao J, Li Y. Serological immunochromatographic approach in diagnosis with SARS-CoV-2 infected COVID-19 patients. J Infect. 2020;81(1):e28–e32. doi: 10.1016/j.jinf.2020.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Porte L, Legarraga P, Vollrath V, Aguilera X, Munita JM, Araos R, Weitzel T. Evaluation of a novel antigen-based rapid detection test for the diagnosis of SARS-CoV-2 in respiratory samples. Int J Infect Dis. 2020;99:328–333. doi: 10.1016/j.ijid.2020.05.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Punn NS, Agarwal S. Automated diagnosis of COVID-19 with limited posteroanterior chest X-ray images using fine-tuned deep neural networks. Appl Intell. 2021;51(5):2689–2702. doi: 10.1007/s10489-020-01900-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rahman T (2022) COVID-19 Radiography database, https://www.kaggle.com/tawsifurrahman/covid19-radiography-database, accesses on May 7 2022.
- 22.Sastre P, Dijkman R, Camuñas A, Ruiz T, Jebbink MF, Van Der Hoek L, Rueda P. Differentiation between human coronaviruses NL63 and 229E using a novel double-antibody sandwich enzyme-linked immunosorbent assay based on specific monoclonal antibodies. Clin Vacc Immunol. 2021;18(1):113–118. doi: 10.1128/CVI.00355-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spicuzza L, Montineri A, Manuele R, Crimi C, Pistorio MP, Campisi R, Crimi N. Reliability and usefulness of a rapid IgM-IgG antibody test for the diagnosis of SARS-CoV-2 infection: A preliminary report. J Infect. 2020;81(2):e53. doi: 10.1016/j.jinf.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna, Z (2016) Rethinking the inception architecture for computer vision, In 2016 IEEE conference on computer vision and pattern recognition (CVPR), pp. 2818-2826.10.1109/CVPR.2016.308
- 25.Thejeshwar SS, Chokkareddy C, Eswaran K (2020) Precise prediction of COVID-19 in chest X-Ray images using KE sieve algorithm. medRxiv
- 26.Togacar M, Ergen B, Cömert Z. COVID-19 detection using deep learning models to exploit Social Mimic Optimization and structured chest X-ray images using fuzzy color and stacking approaches. Comput Biol Med. 2020;121:103805. doi: 10.1016/j.compbiomed.2020.103805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Kang H, Liu X, Tong Z (2020) Combination of RT‐qPCR testing and clinical features for diagnosis of COVID‐19 facilitates management of SARS‐CoV‐2 outbreak. J Med Virol [DOI] [PMC free article] [PubMed]
- 28.Wang L, Byrum B, Zhang Y. Detection and genetic characterization of deltacoronavirus in pigs, Ohio, USA, 2014. Emerg Infect Dis. 2014;20(7):1227. doi: 10.3201/eid2007.140296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watanabe S, Masangkay JS, Nagata N, Morikawa S, Mizutani T, Fukushi S, Akashi H. Bat coronaviruses and experimental infection of bats, the Philippines. Emerg Infect Dis. 2010;16(8):1217. doi: 10.3201/eid1608.100208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Worldometer (2022), Covid-19 coronavirus pandemic, https://www.worldometers.info/coronavirus/?utm.campaign=homeAdvegas1?, accessed on May 19, 2022
- 31.Yan Y, Shin WI, Pang YX, Meng Y, Lai J, You C, ..., Pang CH (2020) The first 75 days of novel coronavirus (SARS-CoV-2) outbreak: recent advances, prevention, and treatment. Int J Environ Res Public Health 17(7):2323 [DOI] [PMC free article] [PubMed]
- 32.Yasar H, Ceylan M. A new deep learning pipeline to detect Covid-19 on chest X-ray images using local binary pattern, dual tree complex wavelet transform and convolutional neural networks. Appl Intell. 2021;51(5):2740–2763. doi: 10.1007/s10489-020-02019-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.
Customized code available.