Abstract
Objective:
This study aimed to investigate the role and mechanism of circular RNA PVT1 (circPVT1) in patients with acute myeloid leukemia (AML).
Materials and Methods:
The expression of circPVT1 in 23 patients with de novo AML (not acute promyelocytic leukemia, not APL) and cell lines were detected by RT-qPCR. Loss of function assays were carried out to explore the influence of silenced circPVT1 on the proliferation, migration, and apoptosis in the THP-1 cell line. CCK-8 assays, trans-well assays, and annexin V/PI staining assays were performed to assess proliferation, migration, and apoptosis, respectively.
Results:
CircPVT1 was highly expressed in AML patients and myeloid cell lines compared to healthy controls. Higher expression of circPVT1 was related to shorter overall survival (OS) and relapse-free survival (RFS) in AML patients. Cell viability and migration were inhibited and apoptosis was increased when circPVT1 was knocked down in THP-1 cells. Knockdown of circPVT1 resulted in marked suppression of c-Myc protein with no significant change in mRNA levels. We also found that circPVT1 knockdown markedly increased the phosphorylation of c-Myc Thr-58, which was responsible for c-Myc degradation. Silencing of c-Myc caused a significant decrease in CXCR4 mRNA and protein expression, whereas the overexpression of c-Myc caused the opposite result, suggesting that CXCR4 is a target molecule of c-Myc. Finally, we found that overexpression of c-Myc could partially reverse circPVT1 knockdown-induced anti-tumor effects on THP-1 cells in vitro.
Conclusion:
Our findings showed that circPVT1 was highly expressed in AML patients and was related to shorter OS and RFS. CircPVT1 may exert an oncogenic effect in THP-1 cells by stabilizing c-Myc protein expression and downstream target CXCR4 expression. These data indicate that circPVT1 may be a promising therapeutic target for AML.
Keywords: Acute myeloid leukemia, CircPVT1, c-Myc, CXCR4
Abstract
Amaç:
Bu çalışma, akut myeloid lösemili (AML) hastalarda dairesel RNA PVT1’in (circPVT1) rolünü ve mekanizmasını araştırmayı amaçladı.
Gereç ve Yöntemler:
De novo AML’li (akut promyelositik lösemi olmayan, APL olmayan) 23 hastada circPVT1 ifadesi ve hücre dizileri RT-qPCR ile saptandı. THP-1 hücre hattında susturulmuş circPVT1’in hücre çoğalması, göçü ve ölümü üzerindeki etkisini araştırmak için fonksiyon kaybı analizleri yapıldı. Sırasıyla hücre çoğalması, göçü ve yok edilmesini değerlendirmek için CCK-8 deneyleri, trans-kuyu deneyleri ve annexin V/PI boyama deneyleri yapıldı.
Bulgular:
CircPVT1, sağlıklı kontrollerle karşılaştırıldığında AML hastalarında ve miyeloid hücre hatlarında yüksek oranda ifade edildi. CircPVT1’in daha yüksek ifadesi, AML hastalarında daha kısa genel sağkalım (GS) ve hastalıksız sağkalım (HsS) ile ilişkiliydi. THP-1 hücrelerinde CircPVT1 devre dışı bırakıldığında hücre canlılığı ve göçü inhibe edildi ve ölümü arttı. CircPVT1’in yıkılması, mRNA seviyelerinde önemli bir değişiklik olmadan c-Myc protein ifadesinin belirgin bir şekilde bastırılmasına neden oldu. Ayrıca CircPVT1 yıkımının, c-Myc bozulmasından sorumlu olan c-Myc Thr-58’in fosforilasyonunu belirgin şekilde artırdığını da bulduk. c-Myc’nin susturulması, CXCR4 mRNA’sında ve protein ifadesinde önemli bir azalmaya neden olurken, c-Myc’nin aşırı ifadesi ise CXCR4’ün c-Myc’nin bir hedef molekülü olarak belirgin ifadesi ile tam zıt etkiye neden oldu. Son olarak, c-Myc’nin aşırı ifadesinin, in vitro THP-1 hücreleri üzerindeki CircPVT1 ifadesinin azaltılması ile uyarılan anti-tümör etkilerini kısmen tersine çevirebileceğini bulduk.
Sonuç:
Bulgularımız, CircPVT1’in AML hastalarında yüksek oranda ifade edildiğini ve daha kısa GS ve HsS ile ilişkili olduğunu gösterdi. CircPVT1, c-Myc protein ifadesinin stabilizasyonu ve hedef CXCR4 ifadesinin baskılanması ile THP-1 hücrelerinde onkojenik etkide artış sağlayabilir. Bu veriler, CircPVT1’in AML için umut verici bir terapötik hedef olabileceğini göstermektedir.
Introduction
Acute myeloid leukemia (AML) is a highly aggressive heterogeneous hematological malignancy of myeloid blasts characterized by clonal expansion of leukemic stem cells. The median age at diagnosis is 67 years [1], with half of patients diagnosed at 70 years or older [2]. Over the past decades, the outcomes of transplantation and some targeted therapeutics have progressed significantly beyond those of conventional chemotherapy [3,4]. However, the 5-year overall survival (OS) rate remains at 30% and at only 10% for older patients [5,6]. This highlights the need for novel therapeutic options for patients for AML.
Increasing reports have indicated that the dysregulation of epigenetic mechanisms plays critical roles in leukemogenesis [7,8,9]. Circular RNAs (circRNAs), an emerging type of cellular long non-coding RNAs (lncRNAs), have attracted much attention recently. They are covalently closed loop-like structure with their 5′ and 3′ ends joined together [10]. This circular structure enhances the stability of circRNAs because of resistance to cellular linear RNA decay machineries [11]. Evidence has suggested that circRNAs play critical roles in cell proliferation, apoptosis, cell cycle regulation, and metastasis [11,12]. The plasmacytoma variant translocation 1 (PVT1) gene is located on chromosome band 8q24.21 and encodes for both linear lncRNA and circRNAs isoforms [13]. Recently, Chen and Chen [14] reported that circular RNA PVT1 (circPVT1) expression was upregulated in de novo AML patients in correlation with shorter survival. However, the mechanisms of circPVT1 in AML remain unclear.
MYC is a common oncogene that encodes the transcription factor c-Myc protein and is involved in many important biological functions, such as migration, adhesion, proliferation, and apoptosis [15,16,17]. The c-Myc protein has been reported to be overexpressed in many cancers, including AML [16,18,19]. MYC is located only 53 kb upstream of the PVT1 gene [20]. High expression of PVT1 is capable of stabilizing c-Myc expression by preventing phosphorylation [21]. Some data have suggested that circPVT1 is also able to stabilize c-Myc protein expression [12,13]. However, the relationship and mechanism between circPVT1 and c-Myc in cases of AML remain unclear.
In the present study, we demonstrate that circPVT1 expression is significantly increased in AML patients. CircPVT1 is shown to promote CXCR4 protein expression by stabilizing c-Myc protein expression, thus regulating the proliferation, migration, and apoptosis of THP-1 cells.
Materials and Methods
Patients and Treatments
A total of 23 patients with de novo AML (not acute promyelocytic leukemia, not APL) and 20 healthy bone marrow donors admitted to The First Affiliated Hospital of Zhejiang Chinese Medical University from January 2019 to December 2021 were enrolled in this study. AML was diagnosed according to the AML classification of the World Health Organization. The bone marrow mononuclear cells (BMMCs) of samples were isolated using the Ficoll-Hypaque gradient centrifugation method and stored at -80 °C for further analysis. In addition, demographic and clinical data were collected, including age, gender, white blood cell count, French-American-British classification, cytogenetic characteristics, molecular abnormalities, risk stratification, and outcomes. Written informed consent was provided by all participants. This study was approved by the Research Ethics Committee of the relevant hospital.
All patients received treatment according to the 2021 Chinese guidelines for the diagnosis and treatment of adult AML (not APL) [22]. Younger patients (<60 years old) with AML received induction therapy in the form of standard 3+7 regimens, such as IA (idarubicin and cytarabine), DA (daunorubicin and cytarabine), or HAA (homoharringtonine, aclarubicin, and cytarabine) regimens. Patients in complete remission (CR) were offered consolidation treatments of intermediate to high doses of cytarabine or allogeneic hematopoietic stem cell transplantation (allo-HSCT) according to their risk classifications. Patients without CR received reinduction therapy followed by consolidation chemotherapy or allo-HSCT. Based on their age, performance status, and comorbidities, elderly patients (≥60 years old) received intensive therapies (such as 3+7 regimens with intermediate-dose cytarabine) or non-intensive therapies (low-intensity chemotherapy, hypomethylating agents, venetoclax, or other new drugs).
Follow-Up and Survival Assessment
To assess the predictive effect of circPVT1 expression levels on prognosis, we collected clinical outcome information for all patients. All patients were followed until death or the end of this study in August 2022. OS was defined as the time from diagnosis to the time of death by any cause or last follow-up. Relapse-free survival (RFS) was defined as the time from diagnosis to the time of relapse or death from any cause or last-follow up.
Cell Cultures and Transfection
The AML THP-1 cell line was a gift from the Institute of Hematology at the China Academy of Chinese Medical Sciences (Beijing, China). THP-1 cells were cultured in PRMI-1640 (Gibco Invitrogen, Grand Island, NY, USA) containing 10% fetal bovine serum (FBS; Gibco, USA) at 37 °C with 5% CO2.
The small interfering RNA targeting circPVT1 (si-circPVT1) or c-Myc (si-Myc) and a negative control (si-NC) were designed and constructed by GenePharma Co., Ltd. (Shanghai, China). The si-circPVT1, si-Myc, and si-NC were then inserted into pGPU6 plasmid vectors. The c-Myc overexpression plasmid (oe-Myc) and control overexpression plasmid (oe-NC) were constructed using the pcDNA3.1 plasmid vector (Invitrogen, Waltham, MA, USA). The plasmids were transfected into THP-1 cells with Lipofectamine 3000 (Thermo Fisher Scientific, Waltham, MA, USA). After transfection, the expression of circ-PVT1 or c-Myc in THP-1 cells was determined by RT-qPCR at 24 h. After 48 h, cells were collected for the analysis described below.
Reverse Transcription Quantitative PCR (RT-qPCR)
Total RNA was isolated using the TRIzol reagent (Sangon, Shanghai, China) from either patient samples or cultured cells. Linear RNA was removed from the total RNA with RNase R (Sigma, St. Louis, MO, USA). Reverse transcription was then performed using the HiFiScript cDNA Synthesis Kit (CWBIO, Jiangsu, China). RT-qPCR was carried out with the Bio-Rad CFX Connect PCR thermal cycler (Bio-Rad, Hercules, CA, USA) using SYBR Premix Ex Taq II (Takara, Shiga, Japan) according to the manufacturer’s guidelines. The relative levels of RNAs were measured with GAPDH as an internal control using the 2-△△t method. The primers of circPVT1 (chr8: 129108763-129113499, 411 bp) were described in a previous study [23]. The primers used here for RT-qPCR are listed in Table 1.
Table 1. Primer sequences.

Western Blot
Cells were lysed using RIPA lysis buffer (Beyotime Institute of Biotechnology, Shanghai, China) supplemented with protease and phosphatase inhibitors for 30 min on ice and then centrifuged at 12,000x g for 30 min at 4 °C. Protein was quantified using the BCA Protein Assay Kit (Solarbio, Beijing, China), and then 30 µg of protein was loaded and separated via 10% SDS PAGE and transferred to PVDF membranes (Millipore, Darmstadt, Germany). Membranes were blocked with 5% non-fat milk at room temperature for 90 min. The membranes were then incubated with primary antibodies against c-Myc (1:500; Affinity Biosciences, Cincinnati, OH, USA), CXCR4 (1:250; Santa Cruz Biotechnology, Santa Cruz, CA, USA), and GAPDH (1:3000, Affinity Biosciences, USA) at 4 °C overnight. Following three rinses with TBST for 10 min, the membranes were incubated with HRP-conjugated goat anti-rabbit IgG secondary antibody (1:5000, Affinity Biosciences, USA) at room temperature for 1 h. Protein bands were visualized with Western Lightning Plus enhanced chemiluminescence reagent (PerkinElmer, Waltham, MA, USA). Results were analyzed with a Tanon 5500 chemiluminescence device (Tanon Co. Ltd., Shanghai, China).
Trans-Well Assays
Trans-well assays were performed for the detection of the migration ability of THP-1 cells using 8-µm-pore polycarbonate membrane inserts (Sigma-Aldrich, St. Louis, MO, USA). Briefly, 5x105 cells suspended in 100 µL of medium were loaded into the upper chamber while 600 µL of medium containing 100 ng/mL SDF-1α or not (baseline control) was added to the lower chamber. Four hours later, cells that had migrated into the lower chamber were collected and counted by flow cytometry (BD Pharmingen, San Jose, CA, USA) at high flow for 20 s. Results were reported as migration index values, representing the ratio of the number of cells that migrated in the presence of the chemoattractant to the number of cells that migrated without the chemoattractant.
Cell Proliferation Assay
Cell proliferation was assessed using the Cell Counting Kit-8 (CCK-8) assay (MedChemExpress, Monmouth Junction, NJ, USA). In brief, 100 µL of the suspension (1x104 cells per well) was seeded into 96-well plates and cultured in a CO2 incubator at 37 °C for 24, 48, or 72 h. Subsequently, 10 µL of CCK-8 solution was added to each well. After incubation for 4 h, the absorbance was quantified with a microplate reader (Molecular Devices, San Jose, CA, USA) at 450 nm.
Apoptosis Assay
The Annexin V-FITC Apoptosis Detection Kit (BD, San Jose, CA, USA) was used for apoptosis assays. According to the manufacturer’s protocol, THP-1 cells were harvested, resuspended in 100 µL of binding buffer, and incubated with 5 µL of annexin V-FITC and 10 µL of propidium iodide (PI) for 15 min in the dark at room temperature. Subsequently, 400 µL of binding buffer was added and stained cells were evaluated by flow cytometry after 1 h.
Statistical Analysis
Statistical analysis was performed using IBM SPSS Statistics 22.0 (IBM Corp., Armonk, NY, USA). OS and RFS were calculated using the Kaplan-Meier method and the groups were compared using the log-rank test. All other data were recorded as values of mean ± standard deviation from at least three individual experiments. Comparisons were performed by one-way analysis of variance (ANOVA) or two-tailed, unpaired Student t-tests. Values of p<0.05 were considered to be significant.
Results
Expression of CircPVT1 in AML Patients and Its Clinical Significance
We first evaluated the expression levels of circPVT1 in BMMCs from 23 patients with de novo AML (clinical data are provided in Table 2) and 20 healthy control volunteers by RT-qPCR. We found that circPVT1 was significantly upregulated in AML patients compared to the healthy control group (p<0.01; Figure 1a). At the same time, we also measured the expression levels of circPVT1 in AML cell lines and normal control CD34+ cells. As shown in Figure 1b, circPVT1 was highly expressed in most AML cell lines (KG-1a, HL-60, and THP-1) compared to normal control cells. Since the expression of circPVT1 was highest in patients with the M5 subgroup of AML and in THP-1 cells, the THP-1 cell line was selected for the following experiments.
Table 2. Clinical characteristics of patients with acute myeloid leukemia. Molecular abnormalities testing panel included 72 common gene mutations in myeloid tumors and 42 common acute leukemia fusion genes. Patients were stratified into high (>1.326) and low (<1.326) expression groups according to relative circPVT1 expression.
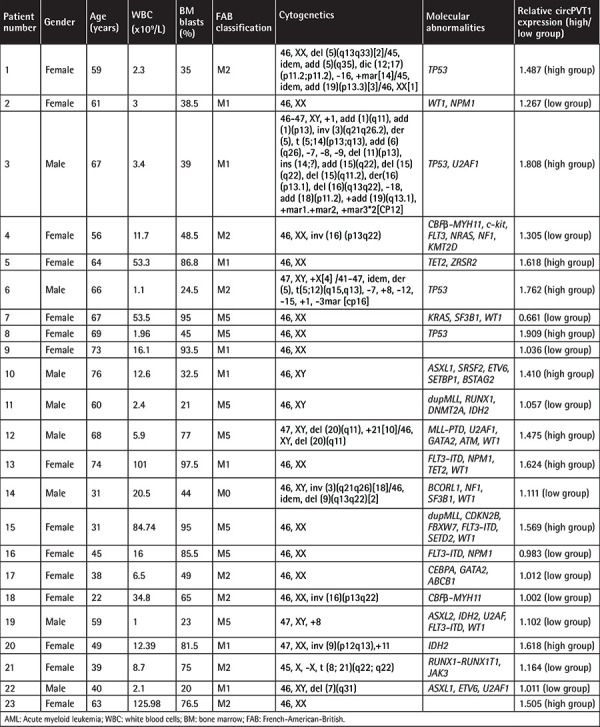
Figure 1.
CircPVT1 was overexpressed in acute myeloid leukemia (AML) in correlation with poor outcomes. a) Comparison of circPVT1 expression in bone marrow mononuclear cells (BMMCs) from AML patients (n=23) and healthy donors (n=20). b) Comparison of circPVT1 in AML cell lines and normal control CD34+ cells from healthy donors. c) Comparison of estimated 3-year overall survival between patients with relatively high circPVT1 expression and relatively low circPVT1 expression (18.2±14.8% vs. 62.5±15.5%, p=0.027). d) Comparison of estimated 3-year relapse-free survival between patients with relatively high circPVT1 expression and relatively low circPVT1 expression (18.2±11.6% vs. 29.2±16.2%, p=0.047). *: p<0.05, **: p<0.01.
To assess the clinical significance of circPVT1, AML patients were stratified into a high-expression group (>1.326) and a low-expression group (<1.326) based on circPVT1 expression before treatment, separated by the median level. As shown in Figures 1c and 1d, higher expression of circPVT1 was associated with shorter 3-year OS (18.2±14.8% vs. 62.5±15.5%, p=0.027) and 3-year RFS (18.2±11.6% vs. 29.2±16.2%, p=0.047) by Kaplan-Meier analysis. Our results indicated that circPVT1 expression levels were upregulated in AML patients in association with poor prognosis.
CircPVT1 Knockdown Inhibits Cell Viability and Migration and Induces Cell Apoptosis
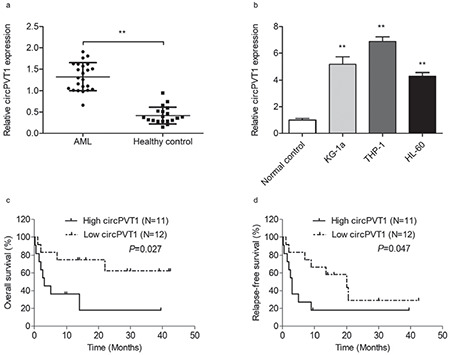
To evaluate the biological roles of circPVT1 in AML, we performed loss-of-function experiments with the THP-1 cell line by transfection with special siRNA. We designed three siRNA sequences, respectively referred to as si-circPVT1 #1, #2, and #3, but only si-circPVT1 #2 showed satisfactory knockdown efficiency. Therefore, si circPVT1 #2 was used for the following transfections and referred to as si-circPVT1. The expression of circPVT1 was significantly downregulated in the si-circPVT1 group compared to the control groups, indicating successful knockdown (p<0.01; Figure 2a). We then performed experiments to monitor the biological activities of THP-1 cells, including proliferation, migration, and apoptosis. CCK-8 assays showed that cell viability in the si-circPVT1 group was obviously weakened compared to that in the si-NC or control group (p<0.01; Figure 2b). Trans-well assays indicated that the silencing of circPVT1 markedly suppressed cell migration (p<0.01; Figure 2c). Meanwhile, as detected by annexin V/PI staining, the knockdown of circPVT1 induced apoptosis in THP-1 cells (p<0.01, Figure 2d).
Figure 2.
CircPVT1 knockdown suppressed cell proliferation and migration while inducing cell apoptosis. a) The efficiency of circPVT1 knockdown in THP-1 cells was verified by RT-qPCR. b) CCK-8 analysis revealed that circPVT1 knockdown suppressed proliferation of THP-1 cells. c) Trans-well assays showed that circPVT1 knockdown inhibited cell migration. d) Flow cytometry was performed to analyze cell apoptosis at 48 h in THP-1 cells. Data showed that circPVT1 knockdown induced cell apoptosis. *: p<0.05, **: p<0.01.
CircPVT1 Stabilizes c-Myc Protein Expression
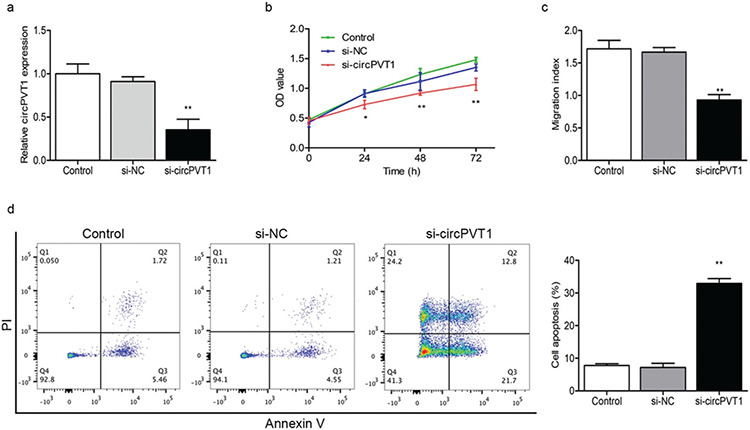
It is known that the human PVT1 and MYC genes are both located at the 8q24 locus [20], and many reports have provided strong evidence that PVT1 promotes c-Myc protein expression by inhibiting the degradation and phosphorylation of the Thr-58 site on c-Myc [24,25,26]. Thus, we investigated the relationship between circPVT1 and c-Myc. When circPVT1 was knocked down by siRNA, decreased c-Myc protein expression (Figure 3a) was found without significant changes in mRNA levels (Figure 3b). At the same time, our results showed increased phosphorylated c-Myc Thr-58 expression after the knockdown of circPVT1 (p<0.01; Figure 3a). These results indicated that circPVT1 may stabilize c-Myc protein expression by decreasing the c-Myc phosphorylation level at Thr-58 to prevent its degradation.
Figure 3.
CircPVT1 knockdown affected the stabilization of c-Myc. a) The knockdown of circPVT1 reduced c-Myc protein and increased phosphorylated c-Myc Thr-58 protein expression in THP-1 cells. b) The knockdown of circPVT1 did not affect the mRNA level of c-Myc.
CircPVT1 Regulates Cell Proliferation, Migration, and Apoptosis by Modulating the Expression of CXCR4
PVT1 was reported to participate in the downstream signaling pathway of c-Myc by regulating key molecules [20]. Therefore, we investigated the possibility of a similar mechanism for cirPVT1. Our previous study revealed that the CXCR4 protein mediates biological functions such as leukemia cell migration, proliferation, and apoptosis [27,28]. It was also reported that blocking c-Myc expression significantly downregulated CXCR4 expression [29]. This suggests that CXCR4 is one of the potential targets of c-Myc. We further explored whether circPVT1 promotes leukemogenesis by stabilizing c-Myc expression and regulating downstream CXCR4. First, we verified the efficiency of c-Myc knockdown and overexpression by using RT-qPCR and western blotting (Figures 4a and 4c). Our results showed that overexpression of c-Myc markedly reduced CXCR4 mRNA and protein expression, while downregulation of c-Myc caused the opposite results (Figure 4b and 4c). Meanwhile, we found that the knockdown of circPVT1 inhibited CXCR4 expression (Figures 4d and 4e), while simultaneous overexpression of c-Myc could partially reverse CXCR4 expression (Figures 4d and 4e). In functional experiments, we found that the knockdown of circPVT1 suppressed the proliferation and migration and induced the apoptosis of THP-1 cells, which was partially reversed by c-Myc overexpression (Figures 5a-5d). These results indicate that circPVT1 regulated THP-1 cell proliferation, migration, and apoptosis by stabilizing c-Myc protein expression and its downstream target molecule CXCR4.
Figure 4.
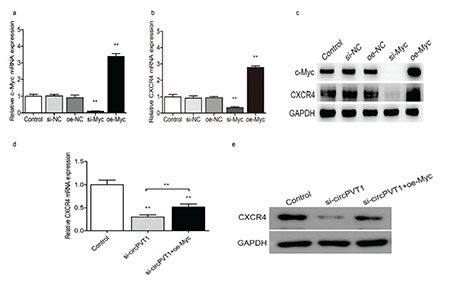
CircPVT1 knockdown decreased the expression of CXCR4, a downstream target of c-Myc. a) The efficiencies of c-Myc knockdown and overexpression in THP-1 cells were verified by RT-qPCR. b) c-Myc knockdown and overexpression affected CXCR4 mRNA levels. c) c-Myc knockdown and overexpression affected CXCR4 protein expression. CircPVT1 knockdown also reduced CXCR4 mRNA d) and protein e) expression, while simultaneous overexpression of c-Myc partially reversed this effect. *: p<0.05, **: p<0.01.
Figure 5.
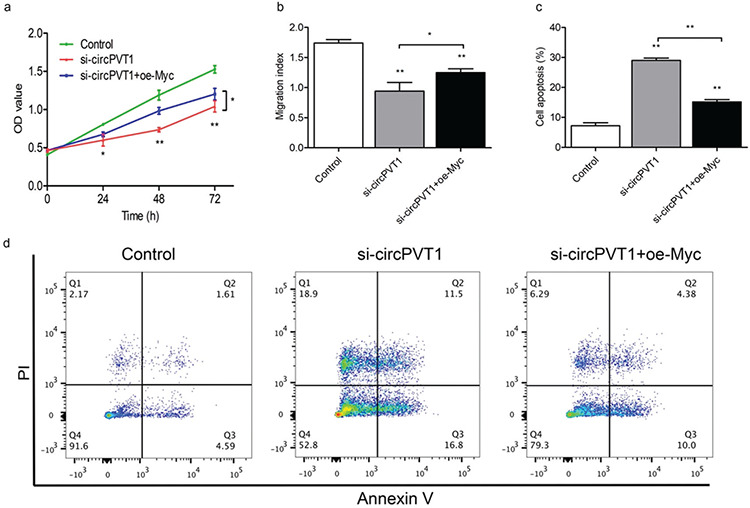
c-Myc overexpression partially reversed the effects of circPVT1 knockdown on cell proliferation, migration, and apoptosis. THP-1 cells were transfected with small interfering RNA targeting circPVT1 (si-circPVT1) or si-circPVT1 + c-Myc overexpression plasmid (oe-Myc). CCK-8 assays a), trans-well assays b), and annexin V/PI staining assays (c, d) were then performed to evaluate cell proliferation, migration, and apoptosis, respectively. *: p<0.05, **: p<0.01.
Discussion
AML is a deadly myeloid malignancy associated with poor outcomes. However, improved understanding of the molecular mechanisms in leukemogenesis has yielded some novel therapeutic targets [3]. In recent years, lncRNAs have been reported to play important roles in the progression of malignancies [30]. Some lncRNAs have already been proposed as diagnostic or prognostic markers, and some even serve as therapeutic targets [30,31,32]. CircRNAs have become a popular topic of research in the field of tumors [33,34], but their roles in hematological tumors have rarely been reported, and especially in cases of AML. In the present study, we focused on the role and mechanism of circPVT1 in AML.
The human PVT1 gene is located on chromosome band 8q24.21, which was identified as a locus of risk for many cancers including leukemia [20,35]. The PVT1 gene was reported to encode varieties of linear and circular lncRNAs, as well as 6 microRNAs [13]. LncRNA PVT1 is highly expressed in APL and promotes progression by modulating proliferation, differentiation, and cell cycle arrest [36,37]. In other AML subtypes, controversial results have been reported regarding PVT1 expression [13,38,39]. Only two studies to date have reported the expression of circPVT1 in AML patients and these results were inconsistent [12,14]. Hu et al. [12] reported that circPVT1 expression was significantly upregulated in ALL (48 patients) but not AML (20 patients) compared to normal controls (40 healthy volunteers). However, as reported by Chen and Chen [14], circPVT1 was upregulated in 68 AML patients. This discrepancy may be due to the fact that different studies examined different circPVT1 isoforms. In our study, we found that circPVT1 was highly expressed in AML (not APL) patients. Higher expression levels of circPVT1 were correlated with shorter OS and RFS in AML patients. Our data indicate that circPVT1 may potentially serve as a new indicator of prognosis in AML.
CircPVT1 can act as an miRNA sponge to regulate gene expression and promote tumorigenesis in various types of cancer [12,40,41]. For example, circPVT1 promotes cell proliferation by regulating microRNA (miR)-125 in gastric cancer [40], and it also regulates cell proliferation and apoptosis by sponging up miR-149 in epithelial ovarian cancer [41]. Another cancer-promoting mechanism of PVT1 involves interaction with its neighbor gene MYC. MYC, a proto-oncogene localized about 53 kb upstream of PVT1 [42], has a pivotal function in genomic instability, gene amplification, cellular proliferation, and the repression of apoptosis [43]. Previous studies have confirmed that the protein c-Myc, a key transcription factor encoded by the MYC gene, showed increased expression in various tumors, including AML [15,16,17]. It is currently believed that both linear and circular PVT1 can protect c-Myc from degradation [13,21]. Hu et al. [12] reported that circPVT1 may regulate proliferation and apoptosis through promotion of neighbor gene c-Myc and Bcl-2 protein expression in ALL cell lines. To date, however, no study has been reported on the mechanism of circPVT1 in AML.
Since THP-1 cells showed the highest circPVT1 levels, this cell line was selected for the following experiments to explore the underlying molecular mechanism. We first found that knockdown of circPVT1 inhibited the proliferation and migration capacity of THP-1 cells while inducing apoptosis. Furthermore, the knockdown of circPVT1 caused a decrease in c-Myc protein expression while the mRNA of c-Myc was unaffected. Our results also showed that the knockdown of circPVT1 upregulated the expression of phosphorylated c-Myc Thr-58, which is often confirmed as an important pathway for c-Myc degradation in cases of many solid tumors. Subsequent experiments could use protein proteasome inhibitors to further clarify the underlying mechanism. Our results indicate that circPVT1 can stabilize c-Myc expression by preventing the phosphorylation of c-Myc Thr-58.
As it was previously reported that PVT1 participates in the downstream signaling pathways of c-Myc by regulating key molecules [20], we aimed to determine whether circPVT1 has a similar mechanism in AML. The inhibition of c-Myc expression was previously shown to cause a decrease in CXCR4 protein expression in AML cells, thus effectively eliminating leukemic cells [29], and our previous study found that CXCR4 expression was increased in AML patients and cell lines and that the inhibition of CXCR4 reduced cell proliferation and migration and induced cell apoptosis [28]. In the present study, we found that knockdown or overexpression of c-Myc caused a respective increase or decrease of CXCR4 expression, confirming that CXCR4 is a target molecule of c-Myc. We next investigated whether circPVT1 regulates CXCR4 expression by stabilizing c-Myc, thereby affecting cell proliferation, migration, and apoptosis. Our results confirmed that hypothesis and we found that the knockdown of circPVT1 significantly downregulated CXCR4 expression, while simultaneous overexpression of c-Myc could partially rescue CXCR4 expression. Finally, we found that overexpression of c-Myc could partially reverse the cellular functional changes caused by circPVT1 knockdown; in other words, cell proliferation and migration were partially restored and apoptosis was partially decreased. Thus, circPVT1 may stabilize c-Myc and promote its downstream target molecule of CXCR4, thereby affecting the biological functions of cells.
Conclusion
The findings of this study confirmed that circPVT1 was upregulated in patients with AML compared to healthy controls. Higher circPVT1 expression was also associated with shorter OS and RFS in AML patients. CircPVT1 enhances cell proliferation and migration while inhibiting cell apoptosis by stabilizing c-Myc and promoting its downstream CXCR4 expression in AML. Our data suggest that the novel circular RNA circPVT1 functions as an oncogene and may be a potential therapeutic target for AML.
Footnotes
Ethics
Ethics Committee Approval: This study was approved by the Ethics Committee of Zhejiang Chinese Medical University.
Informed Consent: All included patients signed informed consent forms in accordance with the Declaration of Helsinki.
Authorship Contributions
Surgical and Medical Practices: H-F.Z., Q.W.; Concept: Y.Z.; Design: X-F.S., H-F.Z.; Data Collection or Processing: X-F.S., L-L.H.; Analysis or Interpretation: L.F.; Literature Search: K-L.C., J.M., S-Y.S.; Writing: X-F.S.
Conflict of Interest: No conflict of interest was declared by the authors.
Financial Disclosure: The authors declared that this study received no financial support.
References
- 1.Tallman MS, Wang ES, Altman JK, Appelbaum FR, Bhatt VR, Bixby D, Coutre SE, De Lima M, Fathi AT, Fiorella M, Foran JM, Hall AC, Jacoby M, Lancet J, LeBlanc TW, Mannis G, Marcucci G, Martin MG, Mims A, O'Donnell MR, Olin R, Peker D, Perl A, Pollyea DA, Pratz K, Prebet T, Ravandi F, Shami PJ, Stone RM, Strickland SA, Wieduwilt M, Gregory KM, Hammond L, Ogba N. Acute Myeloid Leukemia, Version 3. 2019, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw. 2019;17:721–749. doi: 10.6004/jnccn.2019.0028. [DOI] [PubMed] [Google Scholar]
- 2.Hjort Jakobsen L, Stidsholt Roug A, Kiesbye Ovlisen A, Werenberg Marcher C, Beier Ommen H, Theilgaard-Monch K, Moller P, Schollkopf C, Kristensen D, Maria Henriette Naur T, Bogsted M, Christoffer El-Galaly T, Tang Severinsen M. Temporal changes in survival among adult patients with acute myeloid leukaemia in the period 2000-2016: a Danish population-based study. Br J Haematol. 2021;193:482–487. doi: 10.1111/bjh.17213. [DOI] [PubMed] [Google Scholar]
- 3.Swaminathan M, Wang ES. Novel therapies for AML: a round-up for clinicians. Expert Rev Clin Pharmacol. 2020;13:1389–1400. doi: 10.1080/17512433.2020.1850255. [DOI] [PubMed] [Google Scholar]
- 4.Pei X, Huang X. New approaches in allogenic transplantation in AML. Semin Hematol. 2019;56:147–154. doi: 10.1053/j.seminhematol.2018.08.007. [DOI] [PubMed] [Google Scholar]
- 5.Shallis RM, Wang R, Davidoff A, Ma X, Zeidan AM. Epidemiology of acute myeloid leukemia: recent progress and enduring challenges. Blood Rev. 2019;36:70–87. doi: 10.1016/j.blre.2019.04.005. [DOI] [PubMed] [Google Scholar]
- 6.Laribi K, Sobh M, Ghez D, Baugier de Materre A. Impact of age, functional status, and comorbidities on quality of life and outcomes in elderly patients with AML: review. Ann Hematol. 2021;100:1359–1376. doi: 10.1007/s00277-020-04375-x. [DOI] [PubMed] [Google Scholar]
- 7.Kogan AA, Lapidus RG, Baer MR, Rassool FV. Exploiting epigenetically mediated changes: acute myeloid leukemia, leukemia stem cells and the bone marrow microenvironment. Adv Cancer Res. 2019;141:213–253. doi: 10.1016/bs.acr.2018.12.005. [DOI] [PubMed] [Google Scholar]
- 8.Crunkhorn S. Epigenetic therapy prevents AML. Nat Rev Drug Discov. 2020;19:168. doi: 10.1038/d41573-020-00020-4. [DOI] [PubMed] [Google Scholar]
- 9.Cai SF, Levine RL. Genetic and epigenetic determinants of AML pathogenesis. Semin Hematol. 2019;56:84–89. doi: 10.1053/j.seminhematol.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patop IL, Wust S, Kadener S. Past, present, and future of circRNAs. EMBO J. 2019;38:e100836. doi: 10.15252/embj.2018100836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71:428–442. doi: 10.1016/j.molcel.2018.06.034. [DOI] [PubMed] [Google Scholar]
- 12.Hu J, Han Q, Gu Y, Ma J, McGrath M, Qiao F, Chen B, Song C, Ge Z. Circular RNA PVT1 expression and its roles in acute lymphoblastic leukemia. Epigenomics. 2018;10:723–732. doi: 10.2217/epi-2017-0142. [DOI] [PubMed] [Google Scholar]
- 13.Ghetti M, Vannini I, Storlazzi CT, Martinelli G, Simonetti G. Linear and circular PVT1 in hematological malignancies and immune response: two faces of the same coin. Mol Cancer. 2020;19:69. doi: 10.1186/s12943-020-01187-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen T, Chen F. The role of circular RNA plasmacytoma variant translocation 1 as a biomarker for prognostication of acute myeloid leukemia. Hematology. 2021;26:1018–1024. doi: 10.1080/16078454.2021.1987649. [DOI] [PubMed] [Google Scholar]
- 15.Khan I, Eklund EE, Gartel AL. Therapeutic vulnerabilities of transcription factors in AML. Mol Cancer Ther. 2021;20:229–237. doi: 10.1158/1535-7163.MCT-20-0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalkat M, De Melo J, Hickman KA, Lourenco C, Redel C, Resetca D, Tamachi A, Tu WB, Penn LZ. MYC deregulation in primary human cancers. Genes (Basel) 2017;8:151. doi: 10.3390/genes8060151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schaub FX, Dhankani V, Berger AC, Trivedi M, Richardson AB, Shaw R, Zhao W, Zhang X, Ventura A, Liu Y, Ayer DE, Hurlin PJ, Cherniack AD, Eisenman RN, Bernard B, Grandori C; Cancer Genome Atlas Network. Pan-cancer alterations of the MYC oncogene and its proximal network across the Cancer Genome Atlas. Cell Syst. 2018;6:282–300. doi: 10.1016/j.cels.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohanian M, Rozovski U, Kanagal-Shamanna R, Abruzzo LV, Loghavi S, Kadia T, Futreal A, Bhalla K, Zuo Z, Huh YO, Post SM, Ruvolo P, Garcia-Manero G, Andreeff M, Kornblau S, Borthakur G, Hu P, Medeiros LJ, Takahashi K, Hornbaker MJ, Zhang J, Nogueras-Gonzalez GM, Huang X, Verstovsek S, Estrov Z, Pierce S, Ravandi F, Kantarjian HM, Bueso-Ramos CE, Cortes JE. MYC protein expression is an important prognostic factor in acute myeloid leukemia. Leuk Lymphoma. 2019;60:37–48. doi: 10.1080/10428194.2018.1464158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bulaeva E, Pellacani D, Nakamichi N, Hammond CA, Beer PA, Lorzadeh A, Moksa M, Carles A, Bilenky M, Lefort S, Shu J, Wilhelm BT, Weng AP, Hirst M, Eaves CJ. MYC-induced human acute myeloid leukemia requires a continuing IL-3/GM-CSF costimulus. Blood. 2020;136:2764–2773. doi: 10.1182/blood.2020006374. [DOI] [PubMed] [Google Scholar]
- 20.Jin K, Wang S, Zhang Y, Xia M, Mo Y, Li X, Li G, Zeng Z, Xiong W, He Y. Long non-coding RNA PVT1 interacts with MYC and its downstream molecules to synergistically promote tumorigenesis. Cell Mol Life Sci. 2019;76:4275–4289. doi: 10.1007/s00018-019-03222-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Umemori M, Kurata M, Yamamoto A, Yamamoto K, Ishibashi S, Ikeda M, Tashiro K, Kimura T, Sato S, Takahashi H, Kitagawa M. The expression of MYC is strongly dependent on the circular PVT1 expression in pure Gleason pattern 4 of prostatic cancer. Med Mol Morphol. 2020;53:156–167. doi: 10.1007/s00795-020-00243-9. [DOI] [PubMed] [Google Scholar]
- 22.Leukemia & Lymphoma Group, Chinese Society of Hematology, Chinese Medical Association. Chinese guidelines for the diagnosis and treatment of adult acute myeloid leukemia (not APL) (2021) Zhonghua Xue Ye Xue Za Zhi. 2021;42:617–623. doi: 10.3760/cma.j.issn.0253-2727.2021.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun X, Luo L, Gao Y. Circular RNA PVT1 enhances cell proliferation but inhibits apoptosis through sponging microRNA-149 in epithelial ovarian cancer. J Obstet Gynaecol Res. 2020;46:625–635. doi: 10.1111/jog.14190. [DOI] [PubMed] [Google Scholar]
- 24.Yao R, Zhang M, Zhou J, Liu L, Zhang Y, Gao J, Xu K. Novel dual-targeting c-Myc inhibitor D347-2761 represses myeloma growth via blocking c-Myc/Max heterodimerization and disturbing its stability. Cell Commun Signal. 2022;20:73. doi: 10.1186/s12964-022-00868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Colombo T, Farina L, Macino G, Paci P. PVT1: a rising star among oncogenic long noncoding RNAs. Biomed Res Int. 2015;2015:304208. doi: 10.1155/2015/304208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu D, Luo P, Wang Q, Ye Y, Wang B. lncRNA PVT1 in cancer: a review and meta-analysis. Clin Chim Acta. 2017;474:1–7. doi: 10.1016/j.cca.2017.08.038. [DOI] [PubMed] [Google Scholar]
- 27.Sheng X, Zhong H, Wan H, Zhong J, Chen F. Granulocyte colony-stimulating factor inhibits CXCR4/SDF-1α signaling and overcomes stromal-mediated drug resistance in the HL-60 cell line. Exp Ther Med. 2016;12:396–404. doi: 10.3892/etm.2016.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheng XF, Hong LL, Li H, Huang FY, Wen Q, Zhuang HF. Long non-coding RNA MALAT1 modulate cell migration, proliferation and apoptosis by sponging microRNA-146a to regulate CXCR4 expression in acute myeloid leukemia. Hematology. 2021;26:43–52. doi: 10.1080/16078454.2020.1867781. [DOI] [PubMed] [Google Scholar]
- 29.Piya S, Mu H, Bhattacharya S, Lorenzi PL, Davis RE, McQueen T, Ruvolo V, Baran N, Wang Z, Qian Y, Crews CM, Konopleva M, Ishizawa J, You MJ, Kantarjian H, Andreeff M, Borthakur G. BETP degradation simultaneously targets acute myelogenous leukemia stem cells and the microenvironment. J Clin Invest. 2019;129:1878–1894. doi: 10.1172/JCI120654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zimta AA, Tomuleasa C, Sahnoune I, Calin GA, Berindan-Neagoe I. Long non-coding RNAs in myeloid malignancies. Front Oncol. 2019;9:1048. doi: 10.3389/fonc.2019.01048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Irimie AI, Braicu C, Sonea L, Zimta AA, Cojocneanu-Petric R, Tonchev K, Mehterov N, Diudea D, Buduru S, Berindan-Neagoe I. A looking-glass of non-coding RNAs in oral cancer. Int J Mol Sci. 2017;18:2620. doi: 10.3390/ijms18122620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lajos R, Braicu C, Jurj A, Chira S, Cojocneanu-Petric R, Pileczki V, Berindan-Neagoe I. A miRNAs profile evolution of triple negative breast cancer cells in the presence of a possible adjuvant therapy and senescence inducer. J BUON. 2018;23:692–705. [PubMed] [Google Scholar]
- 33.Su Y, Zhong G, Jiang N, Huang M, Lin T. Circular RNA, a novel marker for cancer determination (Review) Int J Mol Med. 2018;42:1786–1798. doi: 10.3892/ijmm.2018.3795. [DOI] [PubMed] [Google Scholar]
- 34.Jahani S, Nazeri E, Majidzadeh AK, Jahani M, Esmaeili R. Circular RNA; a new biomarker for breast cancer: a systematic review. J Cell Physiol. 2020;235:5501–5510. doi: 10.1002/jcp.29558. [DOI] [PubMed] [Google Scholar]
- 35.Tong Y, Yu T, Li S, Zhao F, Ying J, Qu Y, Mu D. cumulative evidence for relationships between 8q24 variants and prostate cancer. Front Physiol. 2018;9:915. doi: 10.3389/fphys.2018.00915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zeng C, Yu X, Lai J, Yang L, Chen S, Li Y. Overexpression of the long noncoding RNA PVT1 is correlated with leukemic cell proliferation in acute promyelocytic leukemia. J Hematol Oncol. 2015;8:126. doi: 10.1186/s13045-015-0223-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Izadifard M, Pashaiefar H, Yaghmaie M, Montazeri M, Sadraie M, Momeny M, Jalili M, Ahmadvand M, Ghaffari SH, Mohammadi S, Alimoghaddam K, Ghavamzadeh A. Expression analysis of PVT1, CCDC26, and CCAT1 long noncoding RNAs in acute myeloid leukemia patients. Genet Test Mol Biomarkers. 2018;22:593–598. doi: 10.1089/gtmb.2018.0143. [DOI] [PubMed] [Google Scholar]
- 38.El-Khazragy N, Elayat W, Matbouly S, Seliman S, Sami A, Safwat G, Diab A. The prognostic significance of the long non-coding RNAs “CCAT1, PVT1” in t(8;21) associated acute myeloid leukemia. Gene. 2019;707:172–177. doi: 10.1016/j.gene.2019.03.055. [DOI] [PubMed] [Google Scholar]
- 39.He RQ, Qin MJ, Lin P, Luo YH, Ma J, Yang H, Hu XH, Chen G. Prognostic significance of lncRNA PVT1 and its potential target gene network in human cancers: a comprehensive inquiry based upon 21 cancer types and 9972 cases. Cell Physiol Biochem. 2018;46:591–608. doi: 10.1159/000488627. [DOI] [PubMed] [Google Scholar]
- 40.Chen J, Li Y, Zheng Q, Bao C, He J, Chen B, Lyu D, Zheng B, Xu Y, Long Z, Zhou Y, Zhu H, Wang Y, He X, Shi Y, Huang S. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 2017;388:208–219. doi: 10.1016/j.canlet.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 41.Li M, Chi C, Zhou L, Chen Y, Tang X. Circular PVT1 regulates cell proliferation and invasion via miR-149-5p/FOXM1 axis in ovarian cancer. J Cancer. 2021;12:611–621. doi: 10.7150/jca.52234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parolia A, Cieslik M, Chinnaiyan AM. Competing for enhancers: PVT1 fine-tunes MYC expression. Cell Res. 2018;28:785–786. doi: 10.1038/s41422-018-0064-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abbate LA, Tolomeo D, Cifola I, Severgnini M, Turchiano A, Augello B, Squeo G, P DA, Traversa D, Daniele G, Lonoce A, Pafundi M, Carella M, Palumbo O, Dolnik A, Muehlematter D, Schoumans J, Van Roy N, De Bellis G, Martinelli G, Merla G, Bullinger L, Haferlach C, Storlazzi CT. MYC-containing amplicons in acute myeloid leukemia: genomic structures, evolution, and transcriptional consequences. Leukemia. 2018;32:2152–2166. doi: 10.1038/s41375-018-0033-0. [DOI] [PMC free article] [PubMed] [Google Scholar]


