Abstract
ProTherm 2.0 is the second release of the Thermodynamic Database for Proteins and Mutants that includes numerical data for several thermodynamic parameters, structural information, experimental methods and conditions, functional and literature information. The present release contains >5500 entries, an ~67% increase over the previous version. In addition, we have included information about reversibility of data, details about buffer and ion concentrations and the surrounding residues in space for all mutants. A WWW interface enables users to search data based on various conditions with different sorting options for outputs. Further, ProTherm has links with other structural and literature databases, and the mutation sites and surrounding residues are automatically mapped on the structures and can be directly viewed through 3DinSight developed in our laboratory. The ProTherm database is freely available through the WWW at http://www.rtc.riken.go.jp/protherm.html
INTRODUCTION
Thermodynamic data for proteins are important to understand the mechanism of protein folding and stability. In recent years, the accumulation of thermodynamic data has been steadily increasing. Pfeil (1) collected a set of data for several thermodynamic parameters from experimental studies that had been published up to 1996. Kawabata et al. (2) constructed a Protein Mutant Database (PMD) for literature information, which covers natural and artificial mutants of proteins. Recently, we have developed an electronically accessible database, ProTherm (3), which includes several aspects of thermodynamic data (unfolding Gibbs free energy change, enthalpy change, heat capacity change, transition temperature, activity etc.), structural information (secondary structure, accessibility etc.), measuring methods, experimental conditions and literature information. The current release 2.0 of ProTherm contains over 5500 entries that cover the up-to-date experimental data. We have developed a WWW interface to facilitate searching the database and sorting outputs.
MAIN DEVELOPMENTS IN VERSION 2.0
• Release 2.0 contains 5542 entries, 67% more than release 1.0 (Table 1).
Table 1. Increase of entries in ProTherm for different datasets.
| Dataset | Release 1.0 | Release 2.0 | % Increase |
|---|---|---|---|
| Total | 3317 | 5542 | 67.1 |
| Solvent accessibilitya | |||
| Buried | 1317 | 1854 | 40.8 |
| Partially buried | 918 | 1105 | 20.4 |
| Exposed | 826 | 1098 | 32.9 |
| Mutation type | |||
| Wild type | 429 | 1599 | 272.7 |
| Single | 2434 | 3312 | 36.1 |
| Double | 353 | 480 | 36.0 |
| Multiple | 101 | 151 | 49.5 |
| Secondary structure | |||
| Helix | 1241 | 1594 | 28.4 |
| Strand | 789 | 1071 | 35.7 |
| Turn | 242 | 402 | 66.1 |
| Coil | 846 | 1078 | 27.4 |
| Measurement | |||
| Circular dichroism | 1709 | 2450 | 43.4 |
| Fluorescence | 1052 | 1455 | 38.3 |
| Calorimetry (DSC) | 489 | 1397 | 185.7 |
| Method | |||
| Thermal | 1628 | 2869 | 76.2 |
| GdnHCl | 1094 | 1697 | 55.1 |
| Urea | 573 | 942 | 64.4 |
| Literature | |||
| Number of proteins | 61 | 194 | 218.0 |
| Number of articles | 245 | 599 | 144.5 |
aBuried: ASA <20%; partially buried: 20% < ASA < 50%; exposed: ASA >50%.
• Includes additional information on reversibility for all data.
• Details about buffers and ions and their concentrations for each entry.
• Information about the surrounding residues around each mutant in space for a specific radius (e.g. 0–4 and 4–8 Å).
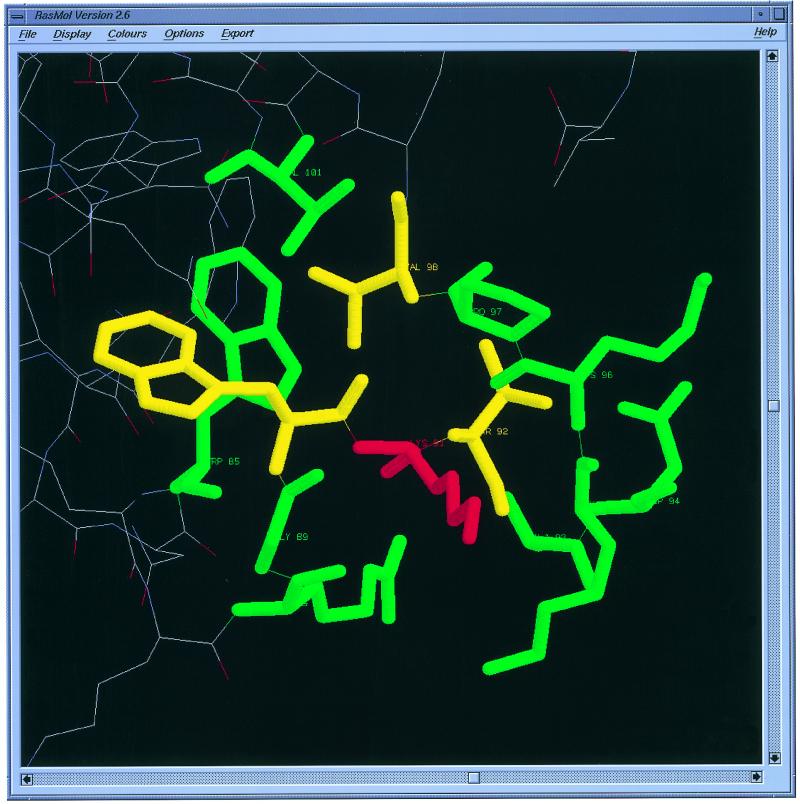
• Display option to view all mutants and surrounding residues through RasMol (4). An example is shown in Figure 1.
Figure 1.
Display of protein mutants and surrounding residues by RasMol. As an example, we show the surrounding residues of Lys 91 in Ribonuclease H (2RN2). The central residue, surrounding residues within 4 Å and surrounding residues between 4 and 8 Å are shown in red, yellow and green, respectively.
FEATURES AVAILABLE AT THE ProTherm SITE
All details about search options, tutorials and database statistics may be accessed by clicking the text links of the home page.
Each entry in the database contains: (i) structural information, (ii) thermodynamic data obtained from thermal and denaturant denaturation experiments, (iii) experimental methods and conditions, (iv) functional and (v) literature information.
The solvent accessibilities of all residues were computed using the program ASC (5,6) as described in our earlier article (7).
DATABASE STATISTICS
Details about the increase of data in release 2.0 for secondary structures, mutation types, various regions of solvent accessibility (ASA), different experimental measurements and methods are presented in Table 1. We observed a substantial increase of data in most of the classified groups.
ACCESS TO ProTherm USING THE WWW
The ProTherm database can be directly accessed online using the WWW server at http://www.rtc.riken.go.jp/protherm.html . At present, cross references to Enzyme Code, EC (http://www.expasy.ch/sprot/enzyme.html ); Protein Mutant Database, PMD (ftp://ftp.nig.ac.jp/pub/db/mutant/ ) (2); Protein Data Bank, PDB (http://www.tcsb.org/pdb/ ) (8); 3DinSight, integrated database for structure, function and property of biomolecules (http://www.rtc.riken.go.jp/3DinSight.html ) (9) and MEDLINE PUBMED (http://www.ncbi.nlm.nih.gov/Entrez/medline.html ) can be directly accessed through the WWW server.
CITATION OF ProTherm
Users of ProTherm are asked to cite this article in their publication, including the URL, http://www.rtc.riken.go.jp/protherm.html . Suggestions and other materials for inclusion in the database are welcome and should be sent to the corresponding author.
Acknowledgments
ACKNOWLEDGEMENTS
MMG wishes to thank JISTEC for providing financial support in the form of an STA fellowship and RIKEN for local hospitality to develop and maintain the database.
REFERENCES
- 1.Pfeil W. (1998) Protein Stability and Folding: A Collection of Thermodynamic Data. Springer, NY.
- 2.Kawabata T., Ota,M. and Nishikawa,K. (1999) Nucleic Acids Res., 27, 355–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gromiha M.M., An,J., Kono,H., Oobatake,M., Uedaira,H. and Sarai,A. (1999) Nucleic Acids Res., 27, 286–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sayle R.A. and Milner-White,E.J. (1995) Trends Biochem. Sci., 20, 374–376. [DOI] [PubMed] [Google Scholar]
- 5.Eisenhaber F. and Argos,P. (1993) J. Comp. Chem., 14, 1272–1280. [Google Scholar]
- 6.Eisenhaber F., Lijnzaad,P., Argos,P., Sander,C. and Scharf,M. (1995) J. Comp. Chem., 16, 273–284. [Google Scholar]
- 7.Gromiha M.M., Oobatake,M., Kono,H., Uedaira,H. and Sarai,A. (1999) Protein Eng., 12, 549–555. [DOI] [PubMed] [Google Scholar]
- 8.Bernstein F.C., Koetzle,T.F., Williams,G.J., Meyer,E.E.,Jr, Brice,M.D., Rodgers,J.R., Kennard,O., Shimanouchi,T. and Tasumi,M. (1977) J. Mol. Biol., 112, 535–542. [DOI] [PubMed] [Google Scholar]
- 9.An J., Nakama,T., Kubota,Y. and Sarai,A. (1998) Bioinformatics, 14, 188–195. [DOI] [PubMed] [Google Scholar]