Abstract.
Purpose
Non-alcoholic fatty liver disease (NAFLD) is an increasing global health concern, with a prevalence of 25% worldwide. The rising incidence of NAFLD, an asymptomatic condition, reinforces the need for systematic screening strategies in primary care. We present the use of non-expert acquired point-of-care ultrasound (POCUS) B-mode images for the development of an automated steatosis classification algorithm.
Approach
We obtained a Health Insurance Portability and Accountability Act compliant dataset consisting of 478 patients [body mass index , age ], imaged with POCUS by non-expert health care personnel. A U-Net deep learning (DL) model was used for liver segmentation in the POCUS B-mode images, followed by patch extraction of liver parenchyma. Several DL models including VGG-16, ResNet-50, Inception V3, and DenseNet-121 were trained for binary classification of steatosis. All layers of each tested model were unfrozen, and the final layer was replaced with a custom classifier. Majority voting was applied for patient-level results.
Results
On a hold-out test set of 81 patients, the final DenseNet-121 model yielded an area under the receiver operator characteristic curve of 90.1%, sensitivity of 95.0%, and specificity of 85.2% for the detection of liver steatosis. Average cross-validation performance in models using patches of liver parenchyma as input outperformed methods using complete B-mode frames.
Conclusions
Despite minimal POCUS acquisition training, and low-quality B-mode images, it is possible to detect steatosis using DL algorithms. Implementation of this algorithm in POCUS software may offer an accessible, low-cost steatosis screening technology, for use by non-expert health care personnel.
Keywords: point-of-care ultrasound, liver, steatosis, non-alcoholic fatty liver disease, deep learning
1. Introduction
Non-alcoholic fatty liver disease (NAFLD) is a global health concern, with a prevalence rate of 25.2% worldwide.1 NAFLD is the most common chronic liver disease in Canada, affecting 20% of the Canadian population.2 The rising incidence of NAFLD, an asymptomatic condition has occurred in parallel with the rise in diabetes and obesity.1 Steatosis is the first stage of liver disease, characterized by the storage of excess macrovesicular fat in the liver. Steatosis presents no clinical symptoms, but it causes the liver to become vulnerable to further injury, including liver inflammation and scarring. Over decades, clinically significant non-alcoholic steatohepatitis (NASH) can silently progress to liver cirrhosis, associated with mortality and requiring consideration of liver transplantation.3 Thus, early diagnosis is crucial to implementing therapeutic strategies that prevent further disease progression.
Current diagnostic pathways rely on incidental findings, and specialty level care where transient elastography is applied, or diagnostic ultrasound (US) acquired by a trained radiologist. Existing diagnostic technologies include Fibroscan© controlled attenuation parameter (CAP), shear wave elastography, biopsy, magnetic resonance elastography, diagnostic B-mode US, and magnetic resonance imaging-proton density fat fraction (MRI-PDFF). These diagnostic techniques are only available at specialty level care, and global consensus standards do not exist for the screening of NAFLD.4
There is an increased interest in non-invasive NAFLD diagnostic methods, and research in the liver US and deep learning (DL) field is rapidly emerging. Existing research successfully automates steatosis detection in B-mode US images using machine learning (ML) methods.5–13 To our best knowledge, all previous works used high-quality diagnostic US machines, and highly curated datasets, acquired by expert radiologists or sonographers,6,8–12,14–16 or researchers formally trained in liver US.5 US machines included the GE VivisE9 US System (GE Healthcare INC, Horten, Norway),14,15 Siemens Acuson S1000 (Siemens, Issaquah, Washington, United States),10 Siemens Acuson S2000 (Siemens, Issaquah, Washington, United States)5,8 etc. Furthermore, many previous studies report that US scan views and angles were acquired with specific anatomical features in view such as the renal cortex or specific vasculature.6–8,15 These scan views require expert US skills to capture correctly and contain clear visualizable markers for steatosis diagnosis, such as the hepatorenal index or vascular blunting.17,18 Implementation of previous work in the clinical setting may help reduce inter- and intra-user variability in the analysis of diagnostic US. It may also serve as a diagnostic aid to the radiologists reviewing US images. However, previous work has been limited by the need for expensive, traditional diagnostic US machines, and expert radiographer or sonographer image acquisition.
The increasing prevalence of NAFLD warrants the need for screening tools in primary care settings.4,19 Point-of-care US (POCUS) devices are low-cost, portable, and accessible to primary care physicians (PCPs), rendering these ideal for primary care-based detection of steatosis. However, POCUS systems have reduced image quality, which makes even qualitative B-mode assessments of liver fat challenging. In this paper, we describe a method for automated steatosis detection using highly accessible POCUS and DL methods. We demonstrate that using images acquired from portable and inexpensive POCUS by non-expert health care professionals (HCP) with minimal training, we can yield comparable classification accuracy of NAFLD to that of US images acquired using traditional, expensive diagnostic US hardware. Our fully automated pipeline for steatosis detection, including liver segmentation, automated patch extraction, and DL transfer learning models for classification, has the potential to enhance patient care at the primary care level.
2. Materials and Methods
The study was conducted in accordance with the ethical guidelines of the Declaration of Helsinki of the World Medical Association. The use of this data has been approved by the Institutional Review Board (IRB) and Research Ethics Board of all clinical partners of Oncoustics and approved for secondary use by the research ethics board of the University of Guelph. Data from the Beijing You’an Hospital in Beijing, China and the National Hepatology and Tropical Medicine Research Institute in Cairo, Egypt were used in this research.
2.1. Study Subjects and Data Acquisition
Data were acquired by Oncoustics (Toronto, Ontario, Canada), a Canadian start-up focused on accessible, non-invasive tests using POCUS and ML. All data were obtained through IRB-approved protocols at local data acquisition sites; all patients were consented before data acquisition. The dataset used for training and testing consists of 478 patients [body mass index (BMI) , age ]. Non-identifying demographic, anthropometric and POCUS data were collected with Health Insurance Portability and Accountability Act compliance. Patients attending a standard-of-care Fibroscan exam were approached by research staff before or after their appointment. The research study was explained to patients individually, in a private setting, and patients had the opportunity to ask questions. If patients agreed to participate, they signed a consent form and underwent the POCUS scan either on the same date of their Fibroscan appointment or were scheduled to have the POCUS exam on a later date within month. Patients between the ages of 18 and 75, with suspected or confirmed liver disease from various etiologies, are included in this research. Patients taking any medications or participating in a clinical trial that may alter the state of liver tissue between the clinical standard assessment and the POCUS scan were excluded. Patients with concomitant liver diseases, such as fibrosis, cirrhosis, hepatocellular carcinoma (HCC), cysts, or nodules, were excluded. In addition, cases with missing or incomplete demographic, anthropometric, or diagnostic information were excluded from the study.
Clinical standard assessment of steatosis was based on the CAP score, available on the FibroScan® elastography system (Echosens, France). The presence of hepatic steatosis was confirmed through Fibroscan evaluations, taking place within weeks of the POCUS scan. Echosens provides interpretation guides for CAP measurements and cut-off values that vary with etiologies.20 There is a large range of CAP values between 222 and 331 in which the steatosis stage can be S0, S1, S2, or S3 dependent on the etiology and NAFLD/NASH status of the patient, which is unknown in this patient population.20 Thus, only patients with S0 and S3 level steatosis were included in the experiments described in this paper. As a standard, the meta-analysis (muti-etiology) cut-off values were used to label patients as having steatosis. The high-end cut-off used to label patients without steatosis (S0) was 238 and the low-end cut-off for patients presenting steatosis was 290 (S3). For the 341 of the 478 patients in the dataset who had hepatitis B virus (HBV), the relevant, adjusted cut-offs were used. The adjusted cut-offs were 222 and 274, with a higher concordance to biopsy results, as validated by Chen et al.21 Likewise, for the 14 hepatitis C virus (HCV) patients, the adjusted cut-offs were 222 and 290.22
Of the 478 patients included in this study, 221 had no evidence of hepatic steatosis, and 257 had S3 level steatosis according published Fibroscan interpretations as described above. The inclusion and exclusion criteria are summarized in Table 1 below.
Table 1.
Inclusion and exclusion criteria used for patient recruitment and study inclusion.
| Inclusion criteria | 1. Patients between the ages of 18 and 75 with suspected or confirmed liver disease |
| 2. Patients who have received a Fibroscan assessment as part of their standard of care within <1 month of the oncoustics scan | |
| 3. Patients with the ability to understand and willingness to sign a written informed consent document | |
| Exclusion criteria | Recruitment exclusion: |
| 1. Patients <18 or >75 years of age | |
| 2. Patients unable to consent | |
| 3. Patients participating in a clinical trial that may alter the state of liver tissue in the time between the Fibroscan assessment and POCUS scan | |
| Study exclusion: | |
| 1. Patients with concomitant liver diseases, such as fibrosis, cirrhosis, HCC, cholangiocarcinoma, metastatic cancer, cysts, or nodules | |
| 2. Patients who have received a liver biopsy <1 month prior to the POCUS scan | |
| 3. Patients with missing or incomplete demographic, anthropometric or clinical information | |
| 4. Patients with S1 or S2 level steatosis according to CAP interpretation guides |
There were 2370 patients recruited, for a broad clinical research study by Oncoustics. In total, 1892 patients were excluded to meet the exclusion criteria for steatosis detection research discussed in this paper. None of the patients included in this study had concomitant liver diseases, and thus they represent a population with early-stage, reversible liver disease. Figure 1 below presents the flow chart of patients after applying inclusion and exclusion criteria.
Fig. 1.
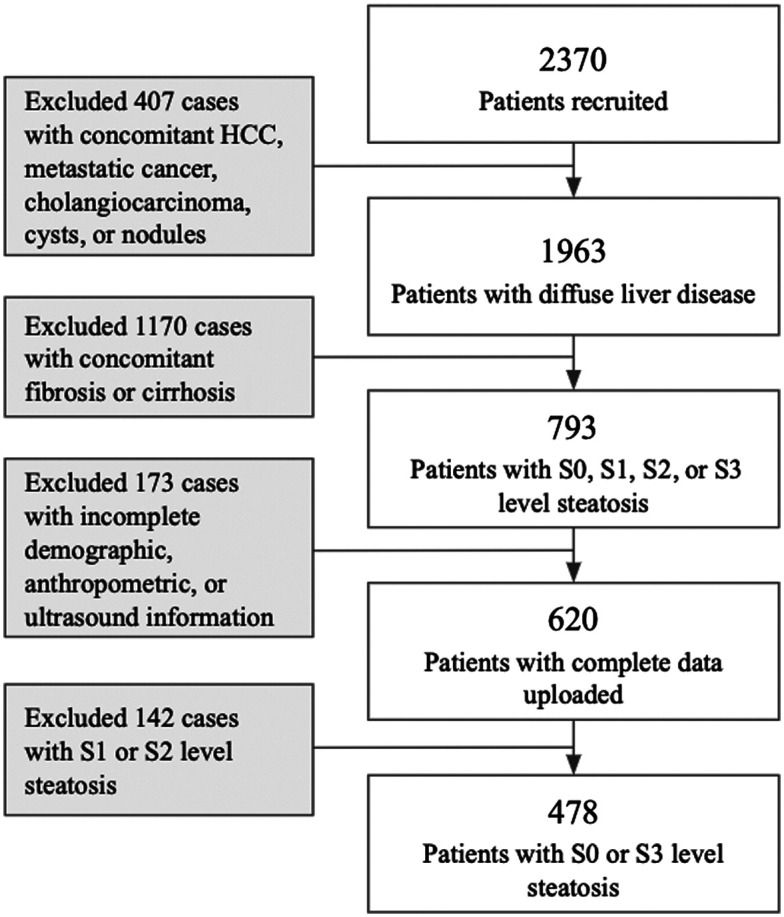
Patient exclusion flow chart.
Disease history, and potential etiologies as well as age, sex, and BMI of the 478 included patients, are presented below in Table 2. The majority of the study population (71.34%) had a medical history of HBV.
Table 2.
Study participant characteristics: demographic, anthropometric, and clinical information.
| Information | Details | Values (All) | Values (S0) | Values (S3) |
|---|---|---|---|---|
| Number of patients | 478 | 221 | 257 | |
| Sex, number of patients | Male/female | 271/207 | 89/132 | 182/75 |
| Age, years | Mean ± standard deviation | 40.97 ± 10.61 | 40.70 ± 10.32 | 41.22 ± 10.86 |
| (Range) | (18 to 70) | (18 to 70) | (18 to 70) | |
| Median | 40 | 40 | 40 | |
| BMI, | Mean ± standard deviation | 23.60 ± 3.55 | 21.40 ± 2.51 | 25.49 ±3.21 |
| (Range) | (17 to 40) | (17 to 29) | (18 to 40) | |
| Median | 23 | 21 | 26 | |
| Disease history, number of patients | HBV | 341 | 164 | 177 |
| Elevated ALT/ASTa | 101 | 39 | 62 | |
| HCV | 14 | 6 | 8 | |
| Other | 12 | 3 | 9 | |
| Healthy volunteers | 10 | 9 | 1 | |
| Steatosis grade, number of patients (based on Fibroscan CAP ranges) | S0 | 221 | 221 | 0 |
| S3 | 257 | 0 | 257 | |
| Concurrent fibrosis/cirrhosis (based on Fibroscan kPa ranges) | 0 | 0 | 0 |
ALT, alanine transaminase; AST, aspartate aminotransferase.
All HCPs performing data collection were non-experts in sonography and did not have formal US training. Personnel involved in data acquisition included nurses, hepatologists, general physicians, research students, and other HCPs inexperienced in sonography. The Clarius C3 Multi-purpose Scanner (Vancouver, British Columbia, Canada) has a convex transducer, with 192 piezoelectric crystals which emit signals at a 3 MHz frequency. The scanner was equipped with dedicated Oncoustics liver pre-sets, such that the acquiring HCP would not have control over US settings. Signal depth was set to 15 cm, and frequency, brightness, and time-gain compensation could not be adjusted. The frame rate was set to 5 Hz, and POCUS B-mode video recordings were timed to be 3 to 5 s in length. HCPs were trained by Oncoustics to collect standardized and easy-to-acquire scan views including subcostal transverse (SCT), intercostal (IC1, IC2, etc.,), and subcostal mid-axillary line (SCS-MAL). A fanning motion was applied whilst recording the 3 to 5 s B-mode videos. There were neither imaging requirements regarding detailed liver anatomy, vasculature nor surrounding organs, such as gallbladder or kidney. 10 B-mode frames were automatically selected for each patient using the U-Net segmentation algorithm described further in Sec. 2.2.1. The 10 frames with largest segmentations by number of pixels classified as liver were passed on to the patch extraction and classification algorithms. Liver images captured from the subcostal (SCT and SCS-MAL) scan angles often had a higher image quality and increased liver visibility. In contrast to the IC scans, the subcostal scan views are not obstructed by the ribcage, and therefore had reduced shadowing artifacts. Thus, the subcostal (SCT and SCS-MAL) scan angles were isolated and used as input for frame-level and patch-level DL algorithms.
2.2. DL Segmentation and Classification of POCUS B-Mode Images
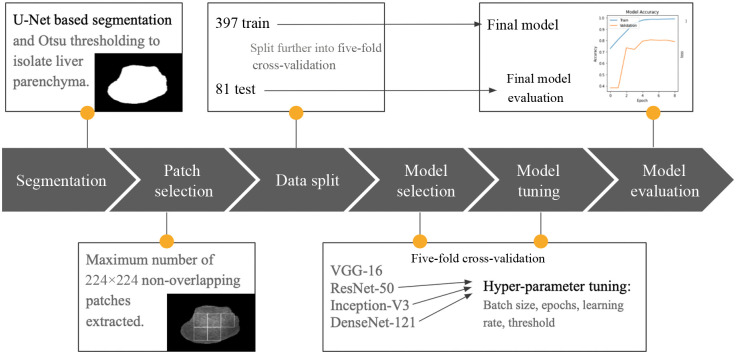
A U-Net segmentation algorithm was used to segment liver tissue in each B-mode frame; this was followed by a patch extraction algorithm that selects patches within the segmentation mask. Various pre-trained DL architectures including visual geometry group (VGG)-16, residual network (ResNet)-50, Inception V3, and DenseNet-121 were initialized with ImageNet weights, and re-trained on the POCUS training dataset. All layers of the described DL architectures were unfrozen when training. A model was selected and tuned using five-fold cross validation. After tuning the model, the five cross-validation folds were grouped into a single training set used to train the model, and the 17% set-aside test set was used for final evaluation. The pipeline of our method is displayed in Fig. 2 and details are further described below.
Fig. 2.
Overview of model development methodology.
2.2.1. Adapted U-net segmentation algorithm and frame selection
A separate, independent dataset of 140 patients and to 40 annotated frames per patient were used to train and evaluate the segmentation network. These 140 annotated patients are entirely separate from the 478 patients used in the classification study. The dataset was also acquired by Oncoustics, using the same Clarius C3 transducer and Oncoustics pre-set imaging parameters. Images were annotated primarily by a consulting abdominal sonographer, as well as trained Oncoustics employees. Expert sonographers and Oncoustics employees performing manual segmentation were trained to identify liver tissue by looking for the liver boundary, the homogenous texture of the parenchyma, and vasculature throughout the tissue. Frames that were substantially affected by artifacts, in which liver tissue was not clearly visible, were not manually segmented. These frames were still used to train the segmentation model and served as training examples that included no clearly identifiable liver. Images were down-sampled to a size of , prior to U-Net model training and prediction.
The U-Net architecture is highly validated for medical imaging segmentation, producing results comparable to expert radiologist segmentation.23 The predicted output of a U-Net algorithm is a probability map, where the value of each pixel represents the probability that the corresponding pixel in the input image belongs to the object. We used a U-Net with five down-sampling convolutional layers, five up-sampling layers, and five corresponding skip connections between layers, similar to the original U-Net proposed by Ronneberger et al.24 Weights were randomly initialized. The Adam optimizer was used with a learning rate of 1e-5. Stratified five-fold cross-validation was implemented to train and evaluate the network. The resultant U-Net successfully segmented liver tissue, with Dice scores ranging between 0.78 and 0.89 in the cross-validation folds. Resultant masks were up-sampled to the original B-mode size using bicubic interpolation.
This basic U-Net segmentation algorithm (previously trained on a separate dataset of 140 patients from a similar POCUS dataset) was used to segment liver tissue in the dataset described in Sec. 2.1, for which there are no available ground-truth segmentations. For purposes of this research, the resultant segmented frames were evaluated visually, and the first author verified that liver parenchyma was correctly segmented.
We used Otsu thresholding25 to convert the output image of varying intensities into a binary mask to be used for segmentation. Through Otsu thresholding, a unique threshold intensity is calculated for each frame. Otsu thresholding25 selects an optimal threshold intensity that minimizes intra-class variance and maximizes inter-class variance according to
| (1) |
where is the inter-class variance, is the fraction of pixels in the background, is the fraction of pixels in the foreground, is the mean intensity of pixels in the background, and is the mean intensity of pixels in the foreground. The top 10 frames were selected per patient, based on the largest liver segmentations by number of pixels, yielding 4780 frames.
2.2.2. Liver tissue patch extraction within B-mode frames
After segmentation of all 4780 B-mode frames in the dataset using the U-Net based method, a patch extraction algorithm was applied. Patch-based strategies were successful in studies by Han et al.,5 Cao et al.13, Chen et al.,21 Reddy et al.,10 and Sanabria et al.12 As described in Sec. 1, the aforementioned studies had expert radiologists and sonographers manually select regions of interest (ROIs) of homogenous liver tissue, or manually annotate the liver region and randomly select patches within its border.12 We developed a simple patch extraction algorithm, which divided the full US frame into non-overlapping patches of size . The patches slide vertically and horizontally in increments of 50 pixels, and the position in which the maximum number of non-overlapping patches fit within the segmentation boundary was selected. Two to seven patches were segmented from each of the 4780 training, validation, and testing frames, yielding frames per patient and a total of 21,503 patches.
2.2.3. Steatosis classification
Two classification approaches were employed, including patch-based classification and frame-based classification. We considered 83% (397) of patients for training and validation and 17% (81) for test data and performed a patient-wise stratified split with all samples from an individual patient assigned to either train or test. The test data were selected such that it would be representative of the global prevalence of NAFLD, with of patients having confirmed steatosis. The training data was further divided into five stratified cross-validation folds. All samples from an individual patient were assigned to a single fold, such that frames from patients were never separated between training, validation, and test sets. 10 frames per patient were used in all experiments; however, the number of patches varied per frame and per patient.
Various pre-trained DL architectures, including VGG-16, ResNet-50, Inception V3, and DenseNet-121, were initialized with ImageNet weights and re-trained on the POCUS dataset. Pre-trained DL architectures and weights were used as a starting point for the model, as they have been trained on large datasets, and are known to be highly generalizable to new image classification tasks. Transfer learning-based architectures expect three-channel RGB images as input, thus the grayscale B-mode images were concatenated such that there would be three identical channels per sample. The final classification layer of each model was replaced with new layers including global average pooling, flattening, and batch normalization. Adam optimizer was used with a fixed learning rate and a binary cross-entropy loss function was implemented at the final node. Dropout and batch normalization layers, as well as early stopping, were implemented to reduce model complexity, improve stability, and promote generalization of the model. After obtaining classification results, majority voting was applied to the patches for patient-level results. The same transfer learning architectures were tested with full B-mode images as input for comparison to the patch-based method, and majority voting was applied to the frames for patient-level results.
We fine-tuned by unfreezing the entire model, thereby allowing all weights and biases to update during training. Hyperparameters were tuned using random search. Learning rates were adjusted for all models starting at 0.1 and incrementally reduced by a factor of 10 until stable learning was observed at a rate of 1e-5. Models were tested with batch sizes of 16, 32, and 64. Batch size 32 consistently yielded superior results in early experimentation and was held constant while comparing architectures. The threshold was adjusted such that the average cross-validation sensitivity was higher than specificity. Optimal thresholds were found to be 0.46, 0.53, and 0.52 for Res-Net 50, DenseNet-121, and Inception V3, respectively.
The 397 patients used for the S0 versus S3 model five-fold cross-validation were then combined into a single training set, which was used to train the final model. No further hyperparameter tuning or changes to the model architecture were performed at this stage. The finalized model was then evaluated on an independent test set of 81 patients.
2.2.4. Statistical analysis and reporting metrics
We computed the receiver operator characteristic (ROC) curves to assess the model discrimination of healthy liver tissue (S0) from fatty liver tissue (S3). A ROC presents true positive and false positive rates for a range of thresholds. The area under the ROC curve (AUROC) is a widely used metric for assessing the accuracy of a binary classification model. We selected a threshold on the ROC curve at the upper left corner, where the true positive rate is high and the false positive rate is low. We calculated sensitivity and specificity to further evaluate the models. Sensitivity represents the proportion of true positives while specificity represents the proportion of true negatives. We calculated the AUROC values for all models and compared performance between models trained with full B-mode images against models trained with patches of liver tissue using the maximum. We performed a one-way analysis of variance (ANOVA) test to compare AUROC, sensitivity, and specificity between three DL architectures. The final model was selected based on the maximum sensitivity value at the optimal ROC threshold. All statistical analysis was done in Python using the Scikit-learn module.
3. Results
First, average cross-validation AUROCs using the patch-based algorithm are compared to the initial full B-mode image models for steatosis detection. The comparisons are summarized in Table 3 below.
Table 3.
Comparison of average fivefold cross-validation AUROCs in transfer learning models trained with full B-mode images and patch-based images of liver parenchyma using data from 397 training/validation patients. The higher result between full-image and patch inputs for each model is bolded.
| Model | Input B-mode | AUROC |
|---|---|---|
| VGG-16 | Full image | 0.773 ± 0.073 |
| Patches | 0.808 ± 0.061 | |
| ResNet-50 | Full image | 0.762 ± 0.108 |
| Patches | 0.830 ± 0.035 | |
| Inception V3 | Full image | 0.757 ± 0.130 |
| Patches | 0.818 ± 0.048 | |
| DenseNet-121 | Full image | 0.770 ± 0.116 |
| Patches | 0.824 ± 0.032 |
Our results indicate that the patch-based method outperforms the method based on full B-mode images, in all four models. Standard deviation values were also lower in the patch-based method.
Five-fold cross-validation results after fine-tuning ResNet-50, DenseNet-121, and Inception V3 are presented in Table 4 below. Patient-level results are clinically relevant and are obtained using majority voting. DenseNet-121 and Inception V3 present nearly identical average cross-validation results. A one-way ANOVA test was performed to compare patient level AUROC, sensitivity, and specificity between the three models and resulted in -values of 0.033, 0.033, and 0.087 and a -values of 0.968, 0.967, and 0.918, respectively. Therefore, the performance difference between models was not statistically significant. DenseNet-121 was selected due to its high sensitivity results at both the patch level and patient level.
Table 4.
ResNet-50, DenseNet-121, and Inception V3 B-Mode patches average cross-validation patient-level results after hyperparameter tuning using data from 397 training/validation patients. The highest AUROC, sensitivity, and specificity are bolded.
| Model | Results reported | AUROC | Sensitivity | Specificity |
|---|---|---|---|---|
| ResNet-50 | Patch level | 0.726 ± 0.027 | 0.783 ± 0.070 | 0.720 ± 0.044 |
| Patient level | 0.829 ± 0.036 | 0.816 ± 0.071 | 0.842 ± 0.054 | |
| Inception V3 | Patch level | 0.722 ± 0.017 | 0.697 ± 0.097 | 0.747 ± 0.099 |
| Patient level | 0.825 ± 0.030 | 0.826 ± 0.089 | 0.824 ± 0.105 | |
| DenseNet-121 | Patch level | 0.727 ± 0.021 | 0.710 ± 0.079 | 0.743 ± 0.071 |
| Patient level | 0.824 ± 0.032 | 0.827 ± 0.061 | 0.822 ± 0.084 |
All 397 (S0 and S3) training examples were grouped and used to train the final DenseNet-121 model, which was evaluated once on the independent test set of 81 patients. The 81 patients used for testing are a set-aside subset of the participants described in Table 2. The test set included 20 patients with S3-level steatosis and 61 patients with no steatosis (S0). Details regarding their demographic, anthropometric and clinical information are described in Table 5.
Table 5.
Independent test set: demographic, anthropometric, and clinical information.
| Information | Details | Values (All) | Values (S0) | Values (S3) |
|---|---|---|---|---|
| Number of patients | 81 | 61 | 20 | |
| Sex, number of patients | Male/female | 46/35 | 30/31 | 16/4 |
| Age, years | Mean ± standard deviation | 38.44 ± 9.28 | 38.71 ± 8.91 | 37.65 ± 10.51 |
| (Range) | (21 to 58) | (21 to 58) | (23 to 54) | |
| Median | 38 | 38 | 36 | |
| BMI, | Mean ± standard deviation | 22.79 ± 3.54 | 21.79 ± 2.85 | 37.65 ± 3.72 |
| (Range) | (17 to 36) | (17 to 29) | (20 to 36) | |
| Median | 22 | 21 | 25 | |
| Disease History, number of patients | HBV | 70 | 53 | 17 |
| Elevated ALT/AST | 9 | 7 | 2 | |
| HCV | 1 | 0 | 1 | |
| Other | 1 | 1 | 0 | |
| Healthy volunteers | 0 | 0 | 0 | |
| Steatosis grade, number of patients (based on Fibroscan CAP ranges) | S0 | 61 | 61 | 0 |
| S3 | 20 | 0 | 20 | |
| Concurrent fibrosis/cirrhosis (based on Fibroscan kPa ranges) | 0 | 0 | 0 |
Testing results are described in Table 6 below. The threshold was selected using the ROC curve, at the upper left corner where the true positive rate is high, and the false positive rate is low. There were no significant relationships observed relating BMI, sex, age, or patient history to the model’s performance.
Table 6.
Final results of DenseNet-121 model on test set of 81 patients.
| Model | Results reported | AUROC | Sensitivity | Specificity |
|---|---|---|---|---|
| DenseNet-121 final test set evaluation | Patient level | 0.901 | 0.950 | 0.852 |
With DenseNet-121, we achieved an AUROC of 0.901, with 95.0% sensitivity and 85.2% specificity for steatosis detection on the hold-out test set of 81 patients.
In a clinical setting, it would be required that B-mode US images for a single patient are de-identified and uploaded to a cloud computer for processing. The series of algorithms, including U-Net based segmentation, Otsu thresholding, frame selection, patch extraction, and DL-based steatosis classification, can run automatically and does not require manual intervention. Excluding upload and download time, processing in a cloud computer with 2 NVIDIA® T4 GPUs, 32 CPUs, and 28.8 GB RAM is 22.64 s for 24 B-mode frames from a single patient. Processing time for each algorithm is presented in Table 7 below.
Table 7.
Sample processing time for data from a single patient with 24 B-mode US frames.
| Algorithm | Processing time (s) |
|---|---|
| U-Net segmentation (inclusive of down-sampling and up-sampling time) | 16.08 |
| Otsu thresholding | 0.04 |
| Frame selection | 0.02 |
| Patch extraction | 1.52 |
| DenseNet-121 steatosis classification | 4.97 |
| Total runtime | 22.64 |
4. Discussion
Our work serves as a proof of concept to support the use of POCUS acquired by non-experts and supported by DL to classify liver steatosis in primary care settings. A fully automated pipeline was developed for frame selection, liver segmentation, patch extraction, and steatosis classification. The final steatosis classification model achieved an AUROC of 0.901 in an independent test set of 81 patients. The test set was representative of the 25.2% global prevalence of NAFLD, with 20 steatosis and 61 healthy patients. The results are comparable to, but do not outperform, previous studies that utilize B-mode images in ML algorithms, which report AUROCs between 0.71 and 1.0.5,6,8–12,14–16 However, all previous works used high quality diagnostic US and highly curated datasets acquired by expert radiologists or sonographers6,8–12,14–16 or researchers formally trained in liver US.5 Furthermore, previous research in the field of automated NAFLD detection with US utilized high-quality images obtained from traditional, expensive, cart-based US systems, such as the Philips EPIQ7 (Philips Ultrasound, Inc., Bothell, Washington, United States).12,16 In contrast, the images in this research have a reduced resolution and quality from POCUS with pre-set parameters, compared to traditional diagnostic US hardware. POCUS systems are inexpensive and accessible to PCPs.
The models trained with B-mode patches of liver tissue outperformed models trained with full B-mode images. It was expected that the model’s classification capabilities may be limited with the full B-mode images from this dataset as they lack key anatomical features that are traditionally used for diagnosis by radiologists, such as the renal cortex. The images in this research are acquired by non-experts, who received minimal training, in contrast to expert radiologists in all previously reported studies.6,8,16 Non-expert HCPs who participated in data acquisition were trained to identify the appearance of basic liver tissue. Correspondingly, our results suggest that it is beneficial for the models to focus on tissue microstructures, rather than full liver anatomy for steatosis detection in our dataset. Furthermore, the standard deviation values were lower when using patches in comparison to when using the full B-mode frame throughout all our experiments. This suggests that learning based on patches is more stable than with full B-mode frames, and there is a lesser likelihood for overfitting.
The following limitations were identified for this work: first, the labels from previous studies were derived from reliable MRI-PDFF or histology examinations, which are considered the gold-standard for steatosis detection. In contrast, the labels used in this research were derived from the Fibroscan© CAP score, for which the cut-off values for S0, S1, S2, and S3 level steatosis vary significantly and are etiology-dependent.20 Furthermore, the interquartile range/median values, which are a measure of reliability, were absent in this study. Future work should address the labeling limitations from this study by evaluating models with reference to biopsy or MRI-PDFF results. Second, BMIs for patients included in this study were and had limited representation for obese populations. Overall, 60% of diabetic patients and 90% of obese patients have a form of NAFLD.26 Therefore, future studies should include a diverse BMI distribution amongst participants. This study consisted of patient data from China and Egypt. To ensure algorithm generalizability, data from a broad spectrum of ethnicities should be collected for model training and validation. With more accurate labels and a larger dataset, a multi-class classification algorithm for stages S0, S1, S2, and S3 of steatosis may be developed for patients with a wide range of BMIs.
5. Conclusions
Our results demonstrate that a pre-trained DL network can reasonably classify steatosis from normal liver tissues in POCUS B-mode images acquired by non-expert HCPs. A total of 397 patients were used for S0 versus S3 model selection and development with five-fold cross validation. We compared the performance of VGG-16, ResNet-50, Inception V3, and DenseNet-121 pre-trained on the ImageNet dataset and unfrozen for training with B-mode POCUS data. DL algorithms trained with automatically segmented liver tissue and extracted ROI patches had superior performance when compared to algorithms trained with full B-mode images. On an independent test set of 81 patients, the model achieved an AUROC of 0.901, with 95.0% sensitivity and 85.2% specificity for binary steatosis detection.
The successful implementation of this software into POCUS transducers would allow for early diagnosis through affordable and widely accessible screening, by a wide range of HCPs with minimal training. Future work includes incorporating radiofrequency data and multi-class steatosis and fibrosis classification evaluated against liver histology for complete liver tissue characterization.
Acknowledgments
This research was funded by the University of Guelph and Mitacs, a Canadian not-for-profit research organization, which links post-secondary institutions with the private sector. A Toronto start-up, Oncoustics Inc. (Toronto, Ontario, Canada), was the industry partner for the study and provided all data used for training and model evaluation. The study was conducted in accordance with the ethical guidelines of the Declaration of Helsinki of the World Medical Association. The use of this data has been approved by the Institutional Review Board and Research Ethics Board of all clinical partners of Oncoustics and approved for secondary use by the research ethics board of the University of Guelph. All patients were consented before data acquisition.
Biographies
Miriam Naim Ibrahim received her BEng degree from the University of Guelph in 2020 and has since pursued MITACs-funded research focused on NAFLD and ultrasound diagnostics. She has completed her MASc degree from the University of Guelph, co-supervised by the University of Toronto. Her current research interests include sonography, non-invasive tests, and hepatology.
Eranga Ukwatta received his master’s and PhD degrees in electrical and computer engineering and biomedical engineering from Western University, Canada, in 2009 and 2013, respectively. He is currently an associate professor at the School of Engineering, University of Guelph, Canada and an adjunct professor in systems and computer engineering at Carleton University, Canada. He has been an author/coauthor for more than 100 journal articles and conference proceedings. From 2013 to 2015, he was a multicenter postdoctoral fellow at Johns Hopkins University and the University of Toronto. He is also a professional engineer in Canada and a senior member of IEEE. His research interests include artificial intelligence for medical imaging, medical image segmentation and registration, and deep learning for computer-aided diagnosis.
Biographies of the other authors are not available.
Disclosures
This project was funded by Mitacs, and Oncoustics was the industry partner.
Contributor Information
Miriam Naim Ibrahim, Email: mnaimibr@uoguelph.ca.
Raul Blázquez-García, Email: rbgarcia@oncoustics.com.
Adi Lightstone, Email: adi.lightstone@gmail.com.
Fankun Meng, Email: mengfankun@ccmu.edu.cn.
Mamatha Bhat, Email: Mamatha.Bhat@uhn.ca.
Ahmed El Kaffas, Email: aelkaffas@oncoustics.com.
Eranga Ukwatta, Email: erangauk@gmail.com.
References
- 1.Mitra S., De A., Chowdhury A., “Epidemiology of non-alcoholic and alcoholic fatty liver diseases,” Transl. Gastroenterol. Hepatol. 5, 16 (2020). 10.21037/tgh.2019.09.08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Canadian Liver Foundation, “Liver disease in Canada,” 2022, https://www.liver.ca/wp-content/uploads/2017/09/CLF_LiverDiseaseInCanada_Synopsis_E.pdf
- 3.Benedict M., Zhang X., “Non-alcoholic fatty liver disease: an expanded review,” World J Hepatol 9(16), 715–732 (2017). 10.4254/wjh.v9.i16.715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pandyarajan V., et al. , “Screening for nonalcoholic fatty liver disease in the primary care clinic,” Gastroenterol. Hepatol. (N Y) 15(7), 357–365 (2019). [PMC free article] [PubMed] [Google Scholar]
- 5.Han A., et al. , “Noninvasive diagnosis of nonalcoholic fatty liver disease and quantification of liver fat with radiofrequency ultrasound data using one-dimensional convolutional neural networks,” Radiology 295(2), 342–350 (2020). 10.1148/radiol.2020191160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Byra M., et al. , “Transfer learning with deep convolutional neural network for liver steatosis assessment in ultrasound images,” Int. J. Comput. Assist. Radiol. Surg. 13(12), 1895–1903 (2018). 10.1007/s11548-018-1843-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Byra M., et al. , “Liver fat assessment in multiview sonography using transfer learning with convolutional neural networks,” J. Ultrasound Med. 41(1), 175–184 (2022). 10.1002/jum.15693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li B., et al. , “Accurate and generalizable quantitative scoring of liver steatosis from ultrasound images via scalable deep learning,” World J. Gastroenterol. 28(22), 2494–2508 (2022). 10.3748/wjg.v28.i22.2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen J. R., et al. , “Clinical value of information entropy compared with deep learning for ultrasound grading of hepatic steatosis,” Entropy (Basel) 22(9), 1006 (2020). 10.3390/e22091006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reddy D. S., Bharath R., Rajalakshmi P., “Classification of nonalcoholic fatty liver texture using convolution neural networks,” in IEEE 20th Int. Conf. e-Health Networking, Appl. and Serv., Healthcom 2018, Institute of Electrical and Electronics Engineers Inc. (2018). 10.1109/HealthCom.2018.8531193 [DOI] [Google Scholar]
- 11.Biswas M., et al. , “Symtosis: a liver ultrasound tissue characterization and risk stratification in optimized deep learning paradigm,” Comput. Methods Prog. Biomed. 155, 165–177 (2017). 10.1016/j.cmpb.2017.12.016 [DOI] [PubMed] [Google Scholar]
- 12.Sanabria S. J., et al. , “Learning steatosis staging with two-dimensional convolutional neural networks: comparison of accuracy of clinical B-mode with a co-registered spectrogram representation of RF data,” in IEEE Int. Ultrasonics Symp., IUS, IEEE Computer Society; (2020). 10.1109/IUS46767.2020.9251329 [DOI] [Google Scholar]
- 13.Cao W., et al. , “Application of deep learning in quantitative analysis of 2-dimensional ultrasound imaging of nonalcoholic fatty liver disease,” J. Ultrasound Med. 39(1), 51–59 (2020). 10.1002/jum.15070 [DOI] [PubMed] [Google Scholar]
- 14.Byra M., et al. , “Classification of breast lesions using segmented quantitative ultrasound maps of homodyned K distribution parameters,” Med. Phys. 43(10), 5561–5569 (2016). 10.1118/1.4962928 [DOI] [PubMed] [Google Scholar]
- 15.Che H., et al. , “Liver disease classification from ultrasound using multi-scale CNN,” Int. J. Comput. Assist. Radiol. Surg. 16(9), 1537–1548 (2021). 10.1007/s11548-021-02414-0 [DOI] [PubMed] [Google Scholar]
- 16.Gummadi S. M., et al. , “Automated machine learning in the sonographic diagnosis of non-alcoholic fatty liver disease,” Adv. Ultrasound Diagn. Ther. 4(3), 176 (2020). 10.37015/AUDT.2020.200008 [DOI] [Google Scholar]
- 17.Azizaddini S., Mani N., Liver Imaging, StatPearls, Treasure Island, Florida, United States: (2022). [Google Scholar]
- 18.Wu M., Sharma P. G., Grajo J. R., “The echogenic liver: steatosis and beyond,” Ultrasound Q. 37(4), 308–314 (2020). 10.1097/RUQ.0000000000000510 [DOI] [PubMed] [Google Scholar]
- 19.Patel K., Sebastiani G., “Limitations of non-invasive tests for assessment of liver fibrosis,” JHEP Rep. 2(2), 100067 (2020). 10.1016/j.jhepr.2020.100067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Echosens, “Echosens: interpretation guide for fibroscan results,” 2021, https://www.echosens.com/products/my-fibroscan/.
- 21.Chen J., et al. , “Controlled attenuation parameter for the detection of hepatic steatosis in patients with chronic hepatitis B,” Infect. Dis. 48(9), 670–675 (2016). 10.3109/23744235.2016.1165860 [DOI] [PubMed] [Google Scholar]
- 22.Sasso M., et al. , “Novel controlled attenuation parameter for noninvasive assessment of steatosis using Fibroscan®: validation in chronic hepatitis C,” J. Viral Hepat. 19(4), 244–253 (2012). 10.1111/j.1365-2893.2011.01534.x [DOI] [PubMed] [Google Scholar]
- 23.Siddique N., et al. , “U-net and its variants for medical image segmentation: a review of theory and applications,” IEEE Access 9, 82031–82057 (2021). 10.1109/ACCESS.2021.3086020 [DOI] [Google Scholar]
- 24.Ronneberger O., Fischer P., Brox T., “U-net: convolutional networks for biomedical image segmentation,” Lect. Notes Comput. Sci. 9351, 234–241 (2015). 10.1007/978-3-319-24574-4_28 [DOI] [Google Scholar]
- 25.Otsu N., “A threshold selection method from gray-level histograms,” IEEE Trans. Syst. Man Cybern. 9(1), 62–66 (1979). 10.1109/TSMC.1979.4310076 [DOI] [Google Scholar]
- 26.Zhou J. H., et al. , “Noninvasive evaluation of nonalcoholic fatty liver disease: current evidence and practice,” World J. Gastroenterol. 25(11), 1307–1326 (2019). 10.3748/wjg.v25.i11.1307 [DOI] [PMC free article] [PubMed] [Google Scholar]