Abstract
Myosin X forms an antiparallel dimer and moves processively on actin bundles. How the antiparallel dimer affects the stepping mechanism of myosin X remains elusive. Here, we generated several chimeras using domains of myosin V and X and performed single-molecule motility assays. We found that the chimera containing the motor domain from myosin V and the lever arm and antiparallel coiled-coil domain from myosin X has multiple forward step sizes and moves processively, similar to full-length myosin X. The chimera containing the motor domain and lever arm from myosin X and the parallel coiled-coil from myosin V takes steps of ∼40 nm at lower ATP concentrations but was nonprocessive at higher ATP concentrations. Furthermore, mutant myosin X with four mutations in the antiparallel coiled-coil domain failed to dimerize and was nonprocessive. These results imply that the antiparallel coiled-coil domain is necessary for multiple forward step sizes of myosin X.
Motor proteins including myosin, kinesin, and dynein are powered by ATP hydrolysis and exert mechanical force to perform many biological functions including intracellular transport of organelles, cytokinesis, muscle contraction, cell movement, and mechanotransduction.1,2 Myosin moves on actin filaments, whereas kinesin and dynein move on microtubules.1 The myosin superfamily consists of at least 79 classes of motor proteins.3 Generally, myosin is composed of the motor domain, lever arm, coiled-coil domain, and targeting (or cargo-binding) domain.2,4 Most unconventional myosins including myosin V and VI form parallel dimers and move processively on actin filaments.5−10 The parallel dimerization of these unconventional myosins allows two motor domains of the dimerized motor to be juxtaposed parallelly on a single actin filament, which enables these myosins to move on the actin filament processively.11 Recently, the native coiled-coil domain of myosin X was found to form an antiparallel dimer using hydrophobic and charge–charge interaction.11,12 However, how the antiparallel coiled-coil domain affects the stepping mechanisms of myosin X is still poorly understood.
Myosin X, as an unconventional myosin, moves processively on fascin-bundled actin (actin bundles)12−15 and transports specific cargoes including β-integrin and Mena/VASP to filopodial tips effectively.16,17 Thus, myosin X is necessary for the formation and elongation of filopodia which are finger-like plasma membrane protrusions containing actin bundles in cells.17−19 Myosin X plays important roles in cancer metastasis20,21 and neuritogenesis.22 Furthermore, myosin X is also involved in cytoskeleton reorganization23 and cell migration.24
In addition to the antiparallel coiled-coil domain, myosin X contains an N-terminal motor domain, three calmodulin (CaM)-binding sites (IQ motifs), a stable single alpha helical (SAH) domain, and a C-terminal cargo-binding domain (Figure 1A).12,15,25 Three IQ motifs and the SAH domain constitute a flexible lever arm of myosin X,1,26,27 which amplifies changes in the motor domain of myosin X to generate a power stroke. Like other unconventional myosins including myosin V7,28 and myosin VI,5,29 myosin X was reported to walk via a hand-over-hand model.12,14,15,30,31 Interestingly, recent single-molecule motility assays revealed that myosin X has unique stepping behaviors including multiple forward step sizes and frequent backward stepping.12,15,25,32 What domain causes the unique multiple step sizes of myosin X is an important question but remains elusive.
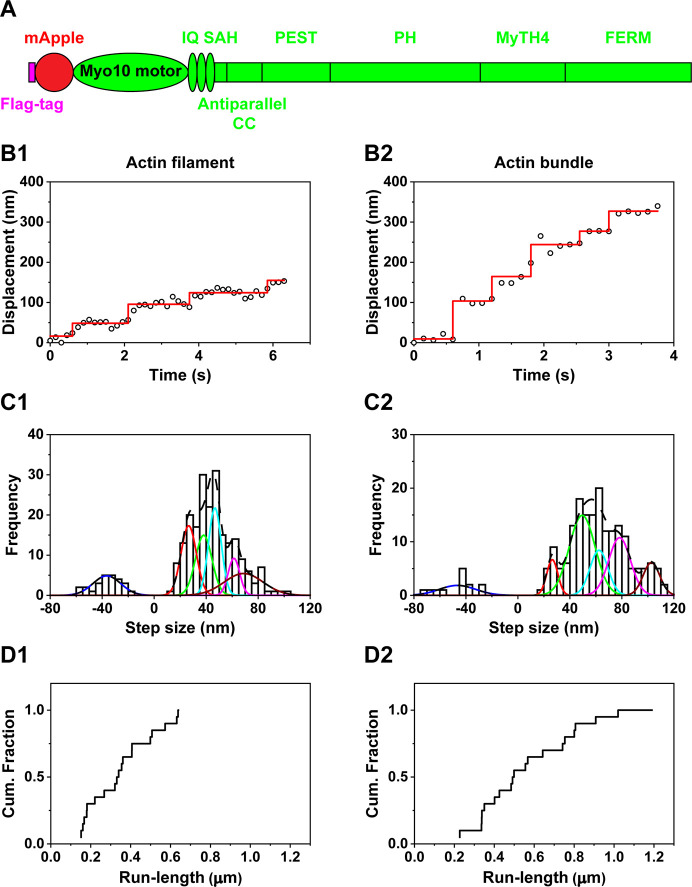
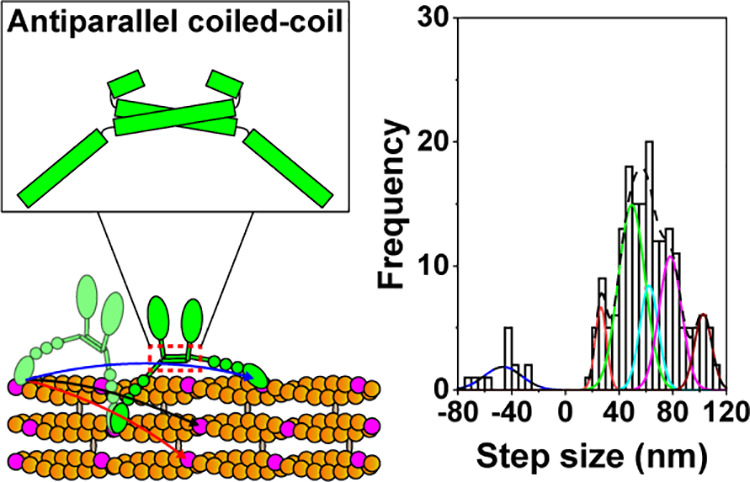
Figure 1.
Motility of full-length myosin X. (A) Schematic of full-length myosin X structure. Full-length myosin X contains mApple-fused motor, 3 IQs for calmodulin-binding, single alpha helical (SAH), proline, glutamate, serine and threonine enriched domain (PEST), pleckstrin homology (PH), myosin tail homology 4 (MyTH4), Band 4.1 protein, Ezrin, Radixin, Moesin (FERM) domains. (B) Representative traces of full-length myosin X moving on actin filaments (B1) and actin bundles (B2) at 2 μM ATP. (C) Step size histograms of full-length myosin X moving on actin filaments (C1) and actin bundles (C2) at 2 μM ATP. The step size histogram of full-length myosin X moving on actin filaments was fitted with the sum of 6 Gaussian components centered at −36.4 ± 10.7 (± SD), 26.6 ± 5.9, 38.1 ± 6.3, 46.7 ± 4.9, 61.4 ± 4.4, and 68.8 ± 15.0 nm (n = 239 steps). The histogram of full-length myosin X moving on actin bundles was fitted with the sum of 6 Gaussian components entered at −46.5 ± 14.0, 26.5 ± 4.2, 49.5 ± 10.0, 62.4 ± 6.9, 78.2 ± 8.4, and 102.7 ± 6.4 (n = 196 steps). These fittings indicate that full-length myosin X has multiple forward step sizes. (D) Cumulative run-length of full-length myosin X on actin filaments (D1) and actin bundle (D2) at 2 mM ATP. Average run-length of myosin X on actin filaments was 0.37 ± 0.17 μm (N = 20 myosin X molecules). Average run-length of myosin X on actin bundles was 0.59 ± 0.26 μm (N = 20).
To address this question, we first performed in vitro single-molecule motility assays of full-length myosin X, which contains the mApple (a red fluorescence protein)-fused motor domain, 3 IQ motifs, the SAH domain, and the cargo-binding domain (Figure 1A). We immobilized actin filaments and bundles to coverslips using NEM myosin and added full-length myosin X containing mApple-fused motor domain without ATP to allow myosin X to dimerize on actin filaments and bundles, using the previously published dimerization method for full-length myosin VI.6 We observed the motility of myosin X after the addition of ATP. Using a method called fluorescence imaging with one-nanometer accuracy (FIONA),7,33−35 we located the centroid of mApple-fused motor domains of full-length myosin X and tracked their motion. Full-length myosin X was highly processive on actin filaments and actin bundles at 2 μM ATP (Figure 1B). Using the widely used MATLAB program (see the Supporting Information for methods), we detected the steps of myosin X on actin filaments and actin bundles. Full-length myosin X had a broad distribution of step sizes on actin filaments and bundles, similar to the previous results of myosin X.12,15,25 The broad step size of full-length myosin X on actin filaments was better fitted with the sum of 6 Gaussian components centered at −36.4 ± 10.7 (± standard deviation (SD)), 26.6 ± 5.9, 38.1 ± 6.3, 46.7 ± 4.9, 61.4 ± 4.4, and 68.8 ± 15.0 nm (Figure 1C1; Table 1), indicating that myosin X has multiple forward step sizes on actin filaments. The broad step size of myosin X on actin bundles was better fitted with the sum of 6 Gaussian components centered at −46.5 ± 14.0, 26.5 ± 4.2, 49.5 ± 10.0, 62.4 ± 6.9, 78.2 ± 8.4, and 102.7 ± 6.4 nm (Figure 1C2; Table 1), indicating that full-length myosin X has multiple forward step sizes on actin bundles.
Table 1. Step Size, Run-Length, and Frequency of Backward Steps of Full-Length Myosin X, Myosin V, and Chimeras on Actin Filaments and Actin Bundles.
| Step size
(nm) (2 μM ATP) |
Run-length (μm) (2 mM ATP) |
||||||
|---|---|---|---|---|---|---|---|
| Construct | Mean | SD | n | Mean | SD | N | % backward steps |
| Full-length myosin X on actin filaments | –36.4 | 10.7 | 239 | 0.37 | 0.17 | 20 | 11% |
| 26.6 | 5.9 | ||||||
| 38.1 | 6.3 | ||||||
| 46.7 | 4.9 | ||||||
| 61.4 | 4.4 | ||||||
| 68.8 | 15.0 | ||||||
| Full-length myosin X on actin bundles | –46.5 | 14.0 | 196 | 0.59 | 0.26 | 20 | 7% |
| 26.5 | 4.2 | ||||||
| 49.5 | 10.0 | ||||||
| 62.4 | 6.9 | ||||||
| 78.2 | 8.4 | ||||||
| 102.7 | 6.4 | ||||||
| Myosin V on actin filaments | –35.3 | 13.7 | 176 | 0.83 | 0.40 | 20 | 5% |
| 36.3 | 11.1 | ||||||
| 69.3 | 6.8 | ||||||
| Myosin V on actin bundles | –26.2 | 12.1 | 169 | 0.64 | 0.26 | 20 | 3% |
| 36.9 | 10.3 | ||||||
| 66.9 | 5.4 | ||||||
| Chimera with the motor domain from myosin V and the lever arm and antiparallel coiled-coil domain from myosin X on actin filaments | –40.1 | 11.9 | 130 | 1.10 | 0.35 | 31 | 5% |
| 26.1 | 0.5 | ||||||
| 42.9 | 9.8 | ||||||
| 59.4 | 2.1 | ||||||
| 71.7 | 4.8 | ||||||
| 90.7 | 5.4 | ||||||
| 115.8 | 4.3 | ||||||
| Chimera with the motor domain from myosin V and the lever arm and antiparallel coiled-coil domain from myosin X on actin bundles | –46.9 | 9.2 | 138 | 1.12 | 0.43 | 28 | 9% |
| 33.7 | 1.2 | ||||||
| 45.0 | 10.5 | ||||||
| 56.8 | 1.7 | ||||||
| 73.6 | 9.3 | ||||||
| 94.3 | 2.2 | ||||||
| 115.9 | 11.4 | ||||||
| Chimera with the motor and lever arm domain from myosin X and the parallel coiled-coil domain from myosin V on actin filaments | –30.9 | 15.5 | 208 | No processive motility | 9% | ||
| 40.3 | 17.5 | ||||||
| Chimera with the motor and lever arm domain from myosin X and the parallel coiled-coil domain from myosin V on actin bundles | –28.0 | 18.8 | 202 | No processive motility | 4% | ||
| 40.6 | 11.6 | ||||||
| 72.7 | 5.9 | ||||||
Among multiple step sizes, some step sizes were larger than the previously reported step sizes.12 The combined analyses of step sizes and fluorescence intensities showed that myosin X took the larger step size after the photobleaching of one mApple in one motor domain, similar to the previous report of myosin X containing Alexa Fluor 488 molecules in each monomer.31 Furthermore, some larger step sizes may be related to not detecting fast steps during our exposure time of the EMCCD camera. Recent real-time two-color tracking of two motor domains labeled with two spectrally distinct fluorescent molecules showed frequent fast steps of myosin X, which were not resolved with an exposure time of 0.1 s.15
Moreover, full-length myosin X showed high processivity on actin filaments and actin bundles at physiological ATP concentration (2 mM ATP) (Figure 1D; Table 1). Run-length of full-length myosin X on actin bundles was larger than those of full-length myosin X on actin filaments (Table 1, p = 0.0021, Student’s t test). The larger run length of myosin X suggests the selectivity of myosin X for actin bundles.
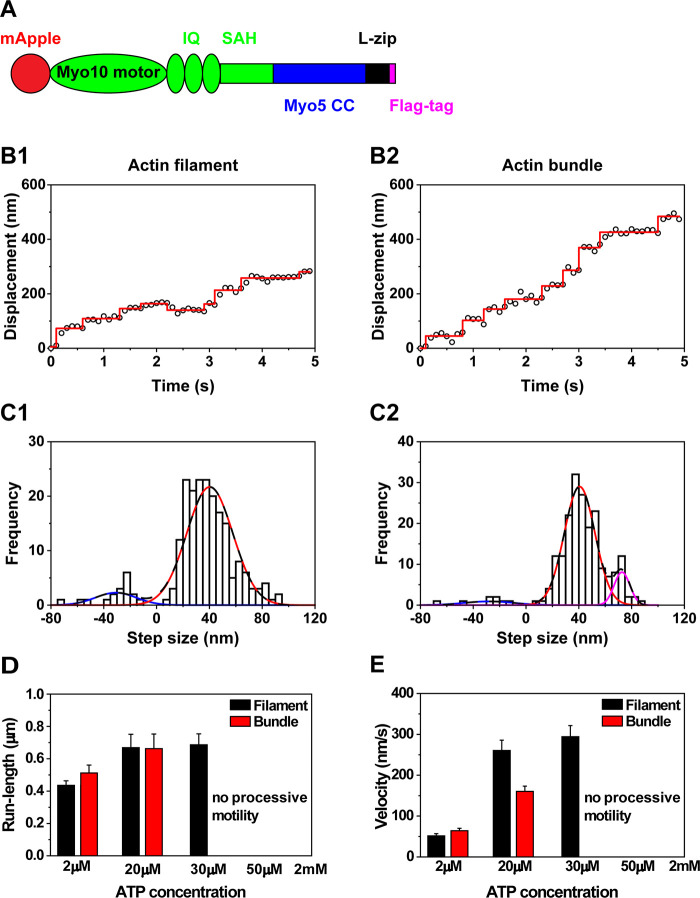
To study the effect of the antiparallel coiled-coil domain on the stepping mechanism of myosin X, we generated chimeras which either replaced the motor domain of myosin X with that of myosin V or replaced the antiparallel coiled-coil of myosin X with the parallel coiled-coil of myosin V. First, we constructed the mApple-fused chimera which replaced the motor domain and the first IQ motif of myosin X with those of myosin V (Figure 2A). This chimera retains the flexible lever arm and antiparallel coiled-coil domain of myosin X. In vitro single-molecule motility assay of the chimera with antiparallel coiled-coil domain and analyses using FIONA showed processive motion on actin filaments (Figure 2B1) and actin bundles (Figure 2B2) at 2 μM ATP. The chimera with the antiparallel coiled-coil domain had a broad distribution of step sizes on actin filaments and bundles (Figure 2C). The broad step size of the chimera with the antiparallel coiled-coil domain on actin filaments was better fitted with the sum of 7 Gaussian components centered at −40.1 ± 11.9, 26.1 ± 0.5, 42.9 ± 9.8, 59.4 ± 2.1, 71.7 ± 4.8, 90.7 ± 5.4, and 115.8 ± 4.3 nm on actin filaments (Figure 2C1; Table 1), which implies that the chimera with the antiparallel coiled-coil domain has multiple forward step sizes on actin filaments, similar to full-length myosin X. The step size histogram of the chimera with the antiparallel coiled-coil domain on actin bundles was better fitted with the sum of 7 Gaussian components centered at −46.9 ± 9.2, 33.7 ± 1.2, 45.0 ± 10.5, 56.8 ± 1.7, 73.6 ± 9.3, 94.3 ± 2.2, and 115.9 ± 11.4 nm on actin bundles (Figure 2C2; Table 1), implying that the chimera with the antiparallel coiled-coil domain has multiple forward step sizes on actin bundles similar to full-length myosin X. The similar multiple forward step sizes between the chimera with the antiparallel coiled-coil domain and full-length myosin X suggest that the antiparallel coiled-coil domain plays an important role in the step size of myosin. Furthermore, the chimera with the antiparallel coiled-coil domain showed high processivity on actin filaments with average run-length of 1.10 ± 0.35 (N = 31) (Figure 2D1; Table 1) and on actin bundles with average run-length of 1.12 ± 0.43 (N = 28) (Figure 2D2; Table 1) at a physiological ATP concentration. The similar run-length of the chimera with the antiparallel coiled-coil domain in contrast to full-length myosin X can be explained by the previously published structures of the motor domain of myosin X12 and myosin V36 revealing that myosin X has a lever arm swing that will favor stepping off on the actin filament (thereby encountering another filament in a bundle), while the myosin V motor tends to have a lever arm swing that favors stepping on the same actin filament.
Figure 2.
Motility of chimera with the motor domain from myosin V and the lever arm and antiparallel coiled-coil domain from myosin X. (A) Schematic of the chimera which contains the motor and the first IQ domain from myosin V (blue) and the second and third IQs and antiparallel coiled-coil domain from myosin X (green). (B) Representative traces of the chimera with the antiparallel coiled-coil domain moving on actin filaments (B1) and actin bundles (B2) at 2 μM ATP. (C) Step size histograms of the chimera with the antiparallel coiled-coil domain moving on actin filaments (C1) and actin bundles (C2) at 2 μM ATP. The step size histogram of the chimera on actin filaments was fitted with the sum of 7 Gaussian components centered at −40.1 ± 11.9, 26.1 ± 0.5, 42.9 ± 9.8, 59.4 ± 2.1, 71.7 ± 4.8, 90.7 ± 5.4, and 115.8 ± 4.3 nm (n = 130 steps). The step size histogram of the chimera on actin bundles was fitted with the sum of 7 Gaussian components centered at −46.9 ± 9.2, 33.7 ± 1.2, 45.0 ± 10.5, 56.8 ± 1.7, 73.6 ± 9.3, 94.3 ± 2.2, and 115.9 ± 11.4 nm (n = 138 steps). These fittings indicate that the chimera with the antiparallel coiled-coil domain has multiple step sizes. (D) Run-length of chimera with the antiparallel coiled-coil domain moving on actin filaments (D1) and actin bundles (D2) at 2 mM ATP. The average run-length on actin filaments was 1.10 ± 0.35 μm (N = 31). The average run-length on actin bundle was 1.12 ± 0.43 μm (N = 28).
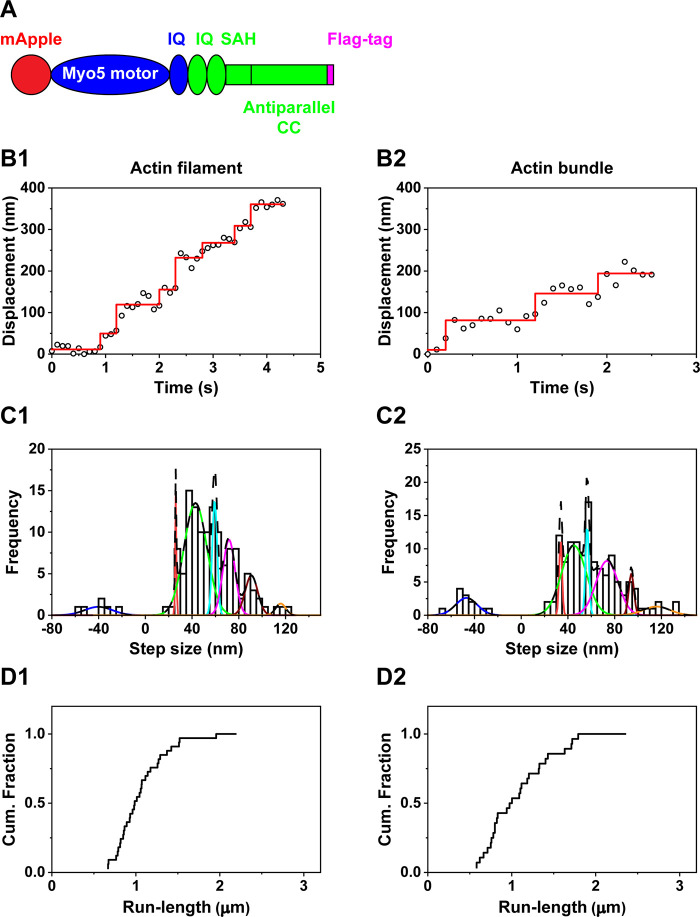
To further examine the effects of the coiled-coil domain on the step sizes, we constructed another chimera which contains the motor domain and the flexible lever arm from myosin X and the parallel coiled-coil domain from myosin V, followed by a leucine zipper (Figure 3A). The chimera with the parallel coiled-coil domain showed processive motion on actin filaments (Figure 3B1) and actin bundles at 2 μM ATP (Figure 3B2). The step size histogram of the chimera with the parallel coiled-coil domain on actin filaments was fitted with the sum of 2 Gaussian components, which showed −30.9 ± 15.5 nm in the backward step and 40.3 ± 17.5 nm in the forward step (Figure 3C1). The single forward step size of the chimera with the parallel coiled-coil domain on actin filaments is in contrast to multiple forward step sizes of full-length myosin X and the previous chimera with the antiparallel coiled-coil domain on actin filaments. The step size histogram of the chimera with the parallel coiled-coil domain on actin bundles was fitted with the sum of 3 Gaussian components, showing −28.0 ± 18.8 nm in the backward step and 40.6 ± 11.6 nm and 72.7 ± 5.9 nm in the forward step (Figure 3C2). The second step size in the forward step was roughly double the first forward step size, implying that the second large step size is related to the motion of one motor domain of the chimera after mApple in the other motor domain photobleached, similar to the observation of our full-length myosin X and other myosin.31 Interestingly, the forward step sizes of the chimera with the parallel coiled-coil domain on actin filaments (40.3 nm) and actin bundles (40.6 nm) were larger than the distance between the helical repeats of actin filaments (36 nm). To test whether the observed single forward step size of the chimera with the parallel coiled-coil domain is closely related to the parallel coiled-coil domain of myosin V, we performed in vitro single-molecule motility assays of mApple-fused myosin V, which contains the rigid lever arm and parallel coiled-coil domain (Supporting Figure 1). Myosin V was processive on actin filaments (Supporting Figure 1A1) and actin bundles (Supporting Figure 1A2). The step size histogram of myosin V on actin filaments was fitted with the sum of 3 Gaussian components, which show −35.3 ± 13.7 nm in the backward step and 36.3 ± 11.1 and 69.3 ± 6.8 nm in the forward steps (Supporting Figure 1B1). The step size histogram of myosin V on actin bundles was also fitted with the sum of 3 Gaussian components, showing −26.2 ± 12.1 nm in the backward step and 36.9 ± 10.3 nm and 66.9 ± 5.4 nm in the forward step (Supporting Figure 1B2). The second forward step sizes of myosin V (69.3 and 66.9 nm) were almost double the first forward step size (∼36 nm), suggesting that the second forward step sizes might be caused by the motion of one motor domain of myosin V after mApple in the other motor domain photobleached, similar to full-length myosin X and other chimeras. The single step size of myosin V and the chimera with the parallel coiled-coil domain suggests that the parallel coiled-coil domain may cause the single step size on actin filaments and bundles. Although the chimera with the parallel coiled-coil domain showed processive motion at 2 μM ATP, this camera was nonprocessive at a physiological ATP concentration. To find the concentration of ATP at which the chimera with the parallel coiled-coil domain does not take processive motion, we performed in vitro single-molecule motility assay at varying ATP concentrations. We found that the chimera with the parallel coiled-coil domain was processive on actin filaments only up to 30 μM ATP and on actin bundles only up to 20 μM ATP (Figure 3D; Table 2). The velocity of this chimera increased up to 30 μM ATP on actin filaments and up to 20 μM ATP on actin bundles (Figure 3E; Table 2).
Figure 3.
Motility of chimera with the motor and lever arm domain from myosin X and the parallel coiled-coil domain from myosin V on actin filaments and actin bundles. (A) Schematic of chimera which contains the motor and lever arm domain from myosin X (green) and the parallel coiled-coil domain from myosin V (blue). (B) Representative traces of the chimera with the parallel coiled-coil domain moving on actin filaments (B1) and actin bundles (B2) at 2 μM ATP. (C) Step size histogram of the chimera with the parallel coiled-coil domain moving on actin filaments (C1) and actin bundles (C2) at 2 μM ATP. The step size histogram with the parallel coiled-coil domain on actin filaments was fitted the sum of 2 Gaussian components centered at −30.9 ± 15.5 and 40.3 ± 17.5 nm (n = 208). The step size histogram on actin bundles was fitted with the sum of 3 Gaussian components centered at −28.0 ± 18.8, 40.6 ± 11.6, and 72.7 ± 5.9 nm (n = 202). (D) Run-length of the chimera with the parallel coiled-coil domain at different ATP concentrations. Chimera with the parallel coiled-coil domain was nonprocessive at high ATP concentrations. (E) Velocity of the chimera at different ATP concentrations.
Table 2. Run-Length and Velocity of Chimera with the Motor and Lever Arm Domain from Myosin X and the Parallel Coiled-Coil Domain from Myosin V on Actin Filaments and Actin Bundles at Different ATP Concentration.
| Run-length
(μm) |
Velocity (nm/s) |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Actin track | ATP concentration | Mean | SD | SE | N | Mean | SD | SE | N |
| Actin filaments | 2 μM | 0.43 | 0.13 | 0.03 | 23 | 57.2 | 24.9 | 5.2 | 23 |
| 20 μM | 0.67 | 0.37 | 0.08 | 20 | 288.7 | 138.1 | 30.9 | 20 | |
| 30 μM | 0.71 | 0.31 | 0.07 | 21 | 307.4 | 133.3 | 29.1 | 21 | |
| 2 mM | No processive motility | No processive motility | |||||||
| Actin bundles | 2 μM | 0.51 | 0.24 | 0.05 | 24 | 63.9 | 23.8 | 4.9 | 24 |
| 20 μM | 0.67 | 0.42 | 0.09 | 22 | 174.2 | 68.0 | 14.5 | 22 | |
| 30 μM | No processive motility | No processive motility | |||||||
| 2 mM | No processive motility | No processive motility | |||||||
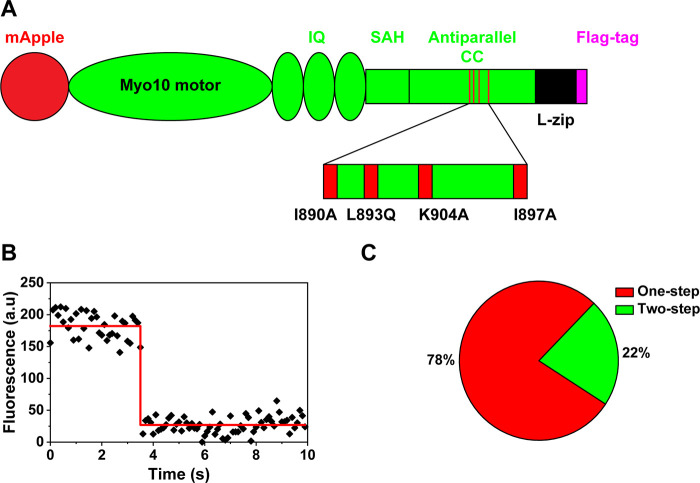
To confirm the importance of the antiparallel coiled-coil in the stepping of myosin X, we constructed mutant myosin X, which disrupts interactions between the antiparallel coiled-coil domain of each monomeric myosin X. Recent structural analyses showed that the conserved residues, including I890, L893, I897, L900, and K904, induce strong hydrophobic and charge–charge interaction between the antiparallel coiled-coil domain in each monomeric myosin X.11 We replaced these residues with alanine or glutamine (I890A, L893Q, I897A, and K904A) to disrupt the hydrophobic interaction in the antiparallel coiled-coil domain of myosin X (Figure 4A). In vitro single-molecule motility assay showed that mutant myosin X was not processive on actin filaments and actin bundles. To find whether the nonprocessive motion is caused by the failure of dimerization, we analyzed the photobleaching steps of mutant myosin X using strong laser power.37 Around 80% of mutant myosin X showed one-step photobleaching (Figure 4B,C), which implies that the disruption of the antiparallel coiled-coil domain of myosin X prevents the formation of dimers and destroys processive motion on actin filaments and actin bundles.
Figure 4.
Mutations in the antiparallel coiled-coil domain disrupt the dimerization of myosin X. (A) Schematic of mutant myosin X which contains four mutations in the antiparallel coiled-coil domain. (B) Representative fluorescent intensity trace of mutant myosin X showing one-step photobleaching. (C) Prevalence of one-step and two-step photobleaching of mutant myosin X (N = 40 molecules).
The traditional stepping mechanism of unconventional myosins on actin filaments was based on the studies of Myosin V, which contains the rigid 6IQ and stable parallel coiled-coil domain.38 The parallel dimerization of myosin V enables two heads of the dimerized myosin V to be juxtaposed to a single actin filament with at least one head attached to the actin filament to prevent myosin V from detaching actin filaments and terminating the processive run. Thus, the parallel dimerization of myosin V is crucial for its processive motion on actin filaments. This traditional stepping model applies to other unconventional myosins containing parallel coiled-coil domains. In this study, we showed that the unique antiparallel coiled-coil domain of myosin X allows the multiple forward step sizes and the processive motion on actin filaments and bundles.
Multiple forward step sizes by the antiparallel coiled-coil domain of myosin X allow myosin X to move on several actin filaments in an actin bundle, which can reduce the possibility of being obstructed by proteins or organelles in the crowded cellular environment during transport. Moreover, multiple forward step sizes of myosin X can shorten the searching time of the motor domain using many nearby available binding sites on actin bundles. Owing to these advantages, myosin X transports unique membrane-localized cargoes including β-integrin on actin bundles effectively to filopodia tips,39 which is required for the formation and elongation of filopodia19 and is crucial for fast growth of neurites in neurons and filopodia in cancer cells. Thus, the antiparallel coiled-coil domain optimizes functions of myosin X in cells.
Interestingly, the chimera with the motor domain and flexible lever arm from myosin X and the parallel coiled-coil domain and leucine zipper from myosin V was not processive at a physiological ATP concentration. A major difference between the parallel and antiparallel coiled-coil domain is that the former positions two heads above the actin filament whereas the latter places two heads in close proximity to the actin surface. When the antiparallel coiled-coil domain is coupled with the flexible lever arm of myosin X, it allows the head to explore a large volume near the filament surface, which is particularly advantageous for movement on an actin bundle. However, when this explored volume is moved away from the actin filament surface by a parallel coiled-coil domain, the time needed to find the preferred binding site is simply too long to support processive movement. This nonprocessive motion of this chimera with the parallel coiled-coil domain at higher ATP concentrations may be exacerbated by off-axis stepping of myosin X causing the forward step size of this chimera to be larger than the distance of helical repeat of actin filaments (36 nm). That is, the discrepancy between the single forward step size of this chimera and the helical repeat of actin filaments may provide less time to reposition the motor domain to the preferred binding sites on actin filaments before the other motor domain binds ATP and detaches from actin filaments at higher ATP concentrations. The previous forced antiparallel dimer of myosin X was not processive on actin filaments at high ATP concentration, although this antiparallel dimer of myosin X showed processive motion with two step sizes of 20 and 40 nm at a lower ATP level.32 Furthermore, myosin V mutant with only 2IQ motifs, whose step size was smaller than the helical repeat, was weakly processive at 10 μM ATP but not processive at 2 mM ATP.40 Ultimately, if the coiled-coil domain positions the unbound head so that an actin-binding site takes longer to find than the dissociation of the bound head by ATP, processive motion is destroyed. We suggest that the antiparallel coiled-coil domain allows the multiple step sizes of myosin X by positioning both heads close to the surface of an actin filament (or filaments in a bundle), which allows the flexible lever arm to explore a number of binding sites. In contrast, substituting a parallel coiled-coil domain constrains the number of potential actin-binding sites available by moving the unbound head away from the filament surface.
Understanding about the relationship between the structures and stepping mechanisms of molecular motors has contributed to engineering nanoscale molecular devices.32,41−44 For example, an engineered bidirectional myosin from the domains of myosin V and VI was shown to change the moving direction depending on the Ca2+ level.45 Thus, our finding that the antiparallel coiled-coil domain of myosin X allows the multiple step sizes will contribute to designing new nanoscale devices for carrying cargoes inside the densely crowded cells.
As a summary, the antiparallel coiled-coil domain of myosin X allows multiple forward step sizes on actin filaments and bundles. Substitution of a parallel coiled-coil domain greatly constrains the possible step sizes and destroys processive motion at higher ATP concentrations. The disruption of this antiparallel coiled-coil domain prevents the formation of myosin X dimer and destroys processive motion. Our findings about the effects of the antiparallel coiled-coil domain on the stepping behaviors of myosin X provide new insights into the relationship between the structures and the stepping mechanisms of molecular motors, which will contribute to designing new nanoscale machines for transporting cargoes.
Acknowledgments
We thank Jing Li for sharing data. We also thank members of Park lab for helpful discussion and comments. We would like to thank Jocyndra Wright and Tianming Lin of the Sweeney lab for their technical contributions. This work was supported by grants from the Research Grants Council of Hong Kong (16101518, N_HKUST613/17 to H.P.), the Innovation and Technology Commission (ITCPD/17-9 to H.P.), and NIH grant (DC009100 to H.L.S.).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jpclett.3c00512.
Additional supporting figures about motility results of single myosin V and PSF; detailed experimental methods for single-molecule motility assays, generation and purification of constructs, polymerization of actin filaments and bundles, total internal reflection microscopy, and data analysis methods (PDF)
Author Present Address
† School of Material Science and Engineering, Zhejiang Sci-Tech University, Hangzhou 310018, China
Author Contributions
# Y.Z. and M.S.C. contributed equally to this work. Q.Q.N., H.L.S., and H.P. designed the experiments. Q.Q.N., Y.Z., M.S.C., X.Q., and H.C.M.C. performed the experiments. Q.Q.N., Y.Z., M.S.C., X.Q., and H.C.M.C. analyzed the data. Q.Q.N., H.L.S., and H.P. interpreted the data. X.L. and H.L.S provided proteins. Q.Q.N., H.L.S., and H.P. wrote the manuscript. H.L.S. and H.P. supervised students.
The authors declare no competing financial interest.
Supplementary Material
References
- Sweeney H. L.; Holzbaur E. L. F. Motor Proteins. Cold Spring Harb. Perspect. Biol. 2018, 10 (5), a021931. 10.1101/cshperspect.a021931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney H. L.; Houdusse A. Structural and functional insights into the Myosin motor mechanism. Annu. Rev. Biophys. 2010, 39, 539–557. 10.1146/annurev.biophys.050708.133751. [DOI] [PubMed] [Google Scholar]
- Kollmar M.; Muhlhausen S. Myosin repertoire expansion coincides with eukaryotic diversification in the Mesoproterozoic era. BMC Evol. Biol. 2017, 17 (1), 211. 10.1186/s12862-017-1056-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syamaladevi D. P.; Spudich J. A.; Sowdhamini R. Structural and functional insights on the Myosin superfamily. Bioinform. Biol. Insights. 2012, 6, BBI.S8451. 10.4137/BBI.S8451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz A.; Park H.; Safer D.; Yang Z.; Chen L. Q.; Selvin P. R.; Sweeney H. L. Myosin VI steps via a hand-over-hand mechanism with its lever arm undergoing fluctuations when attached to actin. J. Biol. Chem. 2004, 279 (36), 37223–37226. 10.1074/jbc.C400252200. [DOI] [PubMed] [Google Scholar]
- Park H.; Ramamurthy B.; Travaglia M.; Safer D.; Chen L. Q.; Franzini-Armstrong C.; Selvin P. R.; Sweeney H. L. Full-length myosin VI dimerizes and moves processively along actin filaments upon monomer clustering. Mol. Cell 2006, 21 (3), 331–336. 10.1016/j.molcel.2005.12.015. [DOI] [PubMed] [Google Scholar]
- Yildiz A.; Forkey J. N.; McKinney S. A.; Ha T.; Goldman Y. E.; Selvin P. R. Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science 2003, 300 (5628), 2061–2065. 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- Rock R. S.; Rice S. E.; Wells A. L.; Purcell T. J.; Spudich J. A.; Sweeney H. L. Myosin VI is a processive motor with a large step size. Proc. Natl. Acad. Sci. U.S.A. 2001, 98 (24), 13655–13659. 10.1073/pnas.191512398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale R. D. Myosin V motor proteins: marching stepwise towards a mechanism. J. Cell Biol. 2003, 163 (3), 445–450. 10.1083/jcb.200308093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale R. D.; Milligan R. A. The way things move: looking under the hood of molecular motor proteins. Science 2000, 288 (5463), 88–95. 10.1126/science.288.5463.88. [DOI] [PubMed] [Google Scholar]
- Lu Q.; Ye F.; Wei Z.; Wen Z.; Zhang M. Antiparallel coiled-coil-mediated dimerization of myosin X. Proc. Natl. Acad. Sci. U.S.A. 2012, 109 (43), 17388–17393. 10.1073/pnas.1208642109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ropars V.; Yang Z.; Isabet T.; Blanc F.; Zhou K.; Lin T.; Liu X.; Hissier P.; Samazan F.; Amigues B.; Yang E. D.; Park H.; Pylypenko O.; Cecchini M.; Sindelar C. V.; Sweeney H. L.; Houdusse A. The myosin X motor is optimized for movement on actin bundles. Nat. Commun. 2016, 7, 12456. 10.1038/ncomms12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy S.; Ricca B. L.; Norstrom M. F.; Courson D. S.; Brawley C. M.; Smithback P. A.; Rock R. S. A myosin motor that selects bundled actin for motility. Proc. Natl. Acad. Sci. U.S.A. 2008, 105 (28), 9616–9620. 10.1073/pnas.0802592105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y.; Sato O.; Ruhnow F.; Arsenault M. E.; Ikebe M.; Goldman Y. E. Single-molecule stepping and structural dynamics of myosin X. Nat. Struct. Mol. Biol. 2010, 17 (4), 485–491. 10.1038/nsmb.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X.; Yoo H.; Man Cheng H. C.; Nguyen Q. Q.; Li J.; Liu X.; Prunetti L.; Chen X.; Liu T.; Sweeney H. L.; Park H. Simultaneous tracking of two motor domains reveals near simultaneous steps and stutter steps of myosin 10 on actin filament bundles. Biochem. Biophys. Res. Commun. 2020, 525, 94–99. 10.1016/j.bbrc.2020.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuo H.; Ikebe M. Myosin X transports Mena/VASP to the tip of filopodia. Biochem. Biophys. Res. Commun. 2004, 319 (1), 214–220. 10.1016/j.bbrc.2004.04.167. [DOI] [PubMed] [Google Scholar]
- Berg J. S.; Cheney R. E. Myosin-X is an unconventional myosin that undergoes intrafilopodial motility. Nat. Cell Biol. 2002, 4 (3), 246–250. 10.1038/ncb762. [DOI] [PubMed] [Google Scholar]
- Bohil A. B.; Robertson B. W.; Cheney R. E. Myosin-X is a molecular motor that functions in filopodia formation. Proc. Natl. Acad. Sci. U.S.A. 2006, 103 (33), 12411–12416. 10.1073/pnas.0602443103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T. M.; Tokuo H.; Gonda K.; Higuchi H.; Ikebe M. Myosin-X induces filopodia by multiple elongation mechanism. J. Biol. Chem. 2010, 285 (25), 19605–19614. 10.1074/jbc.M109.093864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arjonen A.; Kaukonen R.; Ivaska J. Filopodia and adhesion in cancer cell motility. Cell Adh Migr. 2011, 5 (5), 421–430. 10.4161/cam.5.5.17723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R.; Chen J.; Zhang X.; Zhai Y.; Qing X.; Xing W.; Zhang L.; Malik Y. S.; Yu H.; Zhu X. Elevated expression of myosin X in tumours contributes to breast cancer aggressiveness and metastasis. Br. J. Cancer. 2014, 111 (3), 539–550. 10.1038/bjc.2014.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent E. W.; Kwiatkowski A. V.; Mebane L. M.; Philippar U.; Barzik M.; Rubinson D. A.; Gupton S.; Van Veen J. E.; Furman C.; Zhang J.; Alberts A. S.; Mori S.; Gertler F. B. Filopodia are required for cortical neurite initiation. Nat. Cell Biol. 2007, 9 (12), 1347–13459. 10.1038/ncb1654. [DOI] [PubMed] [Google Scholar]
- Tokuo H.; Mabuchi K.; Ikebe M. The motor activity of myosin-X promotes actin fiber convergence at the cell periphery to initiate filopodia formation. J. Cell Biol. 2007, 179 (2), 229–238. 10.1083/jcb.200703178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuo H.; Bhawan J.; Coluccio L. M. Myosin X is required for efficient melanoblast migration and melanoma initiation and metastasis. Sci. Rep. 2018, 8 (1), 10449. 10.1038/s41598-018-28717-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato O.; Jung H. S.; Komatsu S.; Tsukasaki Y.; Watanabe T. M.; Homma K.; Ikebe M. Activated full-length myosin-X moves processively on filopodia with large steps toward diverse two-dimensional directions. Sci. Rep. 2017, 7, 44237. 10.1038/srep44237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baboolal T. G.; Sakamoto T.; Forgacs E.; White H. D.; Jackson S. M.; Takagi Y.; Farrow R. E.; Molloy J. E.; Knight P. J.; Sellers J. R.; Peckham M. The SAH domain extends the functional length of the myosin lever. Proc. Natl. Acad. Sci. U.S.A. 2009, 106 (52), 22193–22198. 10.1073/pnas.0909851106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight P. J.; Thirumurugan K.; Xu Y.; Wang F.; Kalverda A. P.; Stafford W. F. 3rd; Sellers J. R.; Peckham M. The predicted coiled-coil domain of myosin 10 forms a novel elongated domain that lengthens the head. J. Biol. Chem. 2005, 280 (41), 34702–34708. 10.1074/jbc.M504887200. [DOI] [PubMed] [Google Scholar]
- Forkey J. N.; Quinlan M. E.; Shaw M. A.; Corrie J. E.; Goldman Y. E. Three-dimensional structural dynamics of myosin V by single-molecule fluorescence polarization. Nature 2003, 422 (6930), 399–404. 10.1038/nature01529. [DOI] [PubMed] [Google Scholar]
- Okten Z.; Churchman L. S.; Rock R. S.; Spudich J. A. Myosin VI walks hand-over-hand along actin. Nat. Struct. Mol. Biol. 2004, 11 (9), 884–887. 10.1038/nsmb815. [DOI] [PubMed] [Google Scholar]
- Ricca B. L.; Rock R. S. The stepping pattern of myosin X is adapted for processive motility on bundled actin. Biophys. J. 2010, 99 (6), 1818–1826. 10.1016/j.bpj.2010.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao J.; Huck D.; Gunther L. K.; Sellers J. R.; Sakamoto T. Actin structure-dependent stepping of myosin 5a and 10 during processive movement. PloS one 2013, 8 (9), e74936 10.1371/journal.pone.0074936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporizzo M. A.; Fishman C. E.; Sato O.; Jamiolkowski R. M.; Ikebe M.; Goldman Y. E. The Antiparallel Dimerization of Myosin X Imparts Bundle Selectivity for Processive Motility. Biophys. J. 2018, 114 (6), 1400–1410. 10.1016/j.bpj.2018.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H.; Toprak E.; Selvin P. R. Single-molecule fluorescence to study molecular motors. Q. Rev. Biophys. 2007, 40 (1), 87–111. 10.1017/S0033583507004611. [DOI] [PubMed] [Google Scholar]
- Selvin P. R.; Lougheed T.; Hoffman M. T.; Park H.; Balci H.; Blehm B. H.; Toprak E.. In vitro and in vivo FIONA and other acronyms for watching molecular motors walk. In Single-Molecule Techniques: A Laboratory Manual; Cold Spring Harbor Press: Cold Spring Harbor, NY, 2008; pp 37–71. [Google Scholar]
- Yildiz A.; Selvin P. R. Fluorescence imaging with one nanometer accuracy: application to molecular motors. Acc. Chem. Res. 2005, 38 (7), 574–582. 10.1021/ar040136s. [DOI] [PubMed] [Google Scholar]
- Coureux P. D.; Sweeney H. L.; Houdusse A. Three myosin V structures delineate essential features of chemo-mechanical transduction. EMBO J. 2004, 23 (23), 4527–4537. 10.1038/sj.emboj.7600458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulbrich M. H.; Isacoff E. Y. Subunit counting in membrane-bound proteins. Nat. Methods 2007, 4 (4), 319–321. 10.1038/nmeth1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell T. J.; Morris C.; Spudich J. A.; Sweeney H. L. Role of the lever arm in the processive stepping of myosin V. Proc. Natl. Acad. Sci. U.S.A. 2002, 99 (22), 14159–14164. 10.1073/pnas.182539599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H.; Berg J. S.; Li Z.; Wang Y.; Lang P.; Sousa A. D.; Bhaskar A.; Cheney R. E.; Stromblad S. Myosin-X provides a motor-based link between integrins and the cytoskeleton. Nat. Cell Biol. 2004, 6 (6), 523–531. 10.1038/ncb1136. [DOI] [PubMed] [Google Scholar]
- Sakamoto T.; Wang F.; Schmitz S.; Xu Y.; Xu Q.; Molloy J. E.; Veigel C.; Sellers J. R. Neck length and processivity of myosin V. J. Biol. Chem. 2003, 278 (31), 29201–29207. 10.1074/jbc.M303662200. [DOI] [PubMed] [Google Scholar]
- Park H.; Li A.; Chen L. Q.; Houdusse A.; Selvin P. R.; Sweeney H. L. The unique insert at the end of the myosin VI motor is the sole determinant of directionality. Proc. Natl. Acad. Sci. U.S.A. 2007, 104 (3), 778–783. 10.1073/pnas.0610066104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsiavaliaris G.; Fujita-Becker S.; Manstein D. J. Molecular engineering of a backwards-moving myosin motor. Nature 2004, 427 (6974), 558–561. 10.1038/nature02303. [DOI] [PubMed] [Google Scholar]
- Liao J. C.; Elting M. W.; Delp S. L.; Spudich J. A.; Bryant Z. Engineered myosin VI motors reveal minimal structural determinants of directionality and processivity. J. Mol. Biol. 2009, 392 (4), 862–867. 10.1016/j.jmb.2009.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saper G.; Hess H. Synthetic Systems Powered by Biological Molecular Motors. Chem. Rev. 2020, 120 (1), 288–309. 10.1021/acs.chemrev.9b00249. [DOI] [PubMed] [Google Scholar]
- Chen L.; Nakamura M.; Schindler T. D.; Parker D.; Bryant Z. Engineering controllable bidirectional molecular motors based on myosin. Nat. Nanotechnol. 2012, 7 (4), 252–256. 10.1038/nnano.2012.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.