Abstract
Lysosomes are membrane-bound organelles for biomolecule degradation and recycling. They also serve as a nutrient sensing and signaling center to maintain cell and tissue homeostasis. Lysosomal properties alter in response to developmental or environmental cues, but these changes are hard to track in vivo. Employing C. elegans as a model system, we have developed assays to examine and quantify lysosome properties in vivo, including lysosome maturation, acidification and cleavage activity. These assays can be used to reveal alterations of lysosomal activity during C. elegans development and in stress conditions.
Keywords: Lysosomes, C. elegans, Maturation, Acidification, Cleavage activity
INTRODUCTION
Lysosomes are membrane-bound organelles for degradation and recycling of biomolecules. Cargos derived from multiple trafficking routes are sent to lysosomes for digestion (Cullen and Steinberg 2018). The resulting catabolites are reused for cell growth (Gonzalez et al. 2020). Lysosomes also serve as a signaling center for nutrient sensing and thus the maintenance of cell homeostasis (Lawrence and Zoncu 2019). mTOR and AMPK are activated on the lysosome surface under different conditions to regulate downstream signaling pathways (Gonzalez et al. 2020).
Lysosome was discovered by the biochemist Dr. de Duve in the 1950s, starting from his group’s study of a hexose phosphatase. They noted that an acid phosphotase, mainly as a control for enzyme activity measurement, was less active when using a mild homogenization process compared to a drastic homogenization. Then, they found out that it worked under pH 5 and its enzymatic activities were associated with a specific fraction of membrane surrounded particles. Later, four other acidic hydrolases were found to be enclosed within the same subcellular compartment, i.e. lysosome (de Duve 2005; Sabatini and Adesnik 2013). As the degradative organelle in cells, lysosomes contain over 60 hydrolases, which work in an acidic environment (pH 4.5–5.5). Lysosomes integrate and receive substrates from autophagic, endocytic and phagocytic pathways. The substrates of lysosomal hydrolases consist of various biomolecules including proteins, lipids, glycogen, and nucleic acids, which are degraded in the lysosomal lumen.
After degradation, the resulting catabolites are transported out of lysosomes by various membrane transporters and reused as building blocks for cell growth. Some of the catabolites can function as signaling molecules to regulate activity of the mTORC complex (Lawrence and Zoncu 2019). Lysosomal transporters include solute transporters that mediate ions flux across lysosome membranes, and transporters specific for amino acids, lipids and nucleotides (Huizing and Gahl 2020; Xiong and Zhu 2016). For example, V-ATPase is a proton pump responsible for maintaining the acidic lumen of lysosomes. We found that lysosomal activity is greatly promoted at the C. elegans molting stage by upregulation of the V-ATPase gene expression (Miao et al. 2020). Moreover, amino acid transporters are important for maintaining lysosome function and homeostasis (Gan et al. 2019; Liu et al. 2012). Defects in lysosomal acidity, hydrolases or membrane transporters cause substrate accumulation in the lysosomal lumen, which may lead to lysosome storage diseases (Liu et al. 2012, 2018; Nixon 2016; Parkinson-Lawrence et al. 2010).
Abnormal lysosome activity can be detected by biochemical approaches such as measurement of substrate amounts and/or enzyme digestion activity in purified lysosome fraction. To better appreciate the aberrations of lysosomal function in vivo, instead, we sought to investigate lysosome function and regulation in the model organism C. elegans (Li et al. 2016; Liu et al. 2012, 2018; Miao et al. 2020; Sun et al. 2020). Importantly, lysosomes exhibit variable morphological patterns in different tissues, at different developmental stages and during aging process in C. elegans (Li et al. 2016; Miao et al. 2020; Sun et al. 2020). Here, we present protocols for assaying lysosome activity in C. elegans. Particularly, we develop fluorescent reporters to trace lysosome maturation and acidification and we utilize western blot analysis to determine substrate cleavage, which indicates lysosomal degradation activity.
MATERIALS AND EQUIPMENT
Strains
Strains of C. elegans are cultured and maintained using standard protocols (Brenner 1974). The N2 Bristol strain is used as the wild-type strain. Standard microinjection methods were used to generate transgenic animals carrying extrachromosomal arrays (qxEx). Genome-integrated arrays (qxIs) were acquired by γ-irradiation to achieve stable expression from arrays with low copy numbers. The following strains were used: N2, daf-2(e1370), cup-5(bp510), qxIs257 (Pced-1NUC-1::CHERRY), qxIs612 (PhsNUC-1::sfGFP::CHERRY), and qxIs750 (PhsNUC-1::pHTomato).
Buffers
• M9 (1 L): KH2PO4 3 g, Na2HPO4 6 g, NaCl 5 g, add ddH2O to 1 L, then add 1 mL 1 mol/L MgSO4 after autoclave;
• SDS-PAGE sample buffer (Sangon).
Equipment
• Confocal microscope: LSM 880 Meta plus Zeiss Axiovert zoom (Carl Zeiss) with 488 (emission filter BP 503–530) and 543 (emission filter BP 560–615) lasers;
• Chemi-luminescence detection system (CLiNX Science instruments).
Software
• LSM Image Browser and ZEN software (Carl Zeiss Inc.);
• Volocity (PerkinElmer);
• Image J;
• GraphPad Prism (GraphPad Software).
OVERVIEW OF THE EXPERIMENTAL DESIGN
This protocol includes three sections. The first section provides a detailed procedure to track lysosomal maturation and acidification. The second section presents the steps for detecting lysosomal acidity, and the last section describes the method to measure lysosomal degradation activity.
In the first part of the protocol, we express NUC-1::sfGFP::CHERRY transiently using the heat-shock (hs) promoter in worms and we follow the delivery of the tandem fusion protein to lysosomes in the hypodermis at different time points (Fig. 1A, B). The GFP fluorescence is visible in endosomes but is quenched in lysosomes due to the acidic environment, whereas CHERRY fluorescence is visible in both compartments (Fig. 1A). Lysosomal maturation is indicated by gradual disappearance of GFP in the CHERRY-positive structures. We quantify colocalization of GFP and CHERRY using Volocity software in wild type and lysosome-defective cup-5 mutants.
Figure 1.
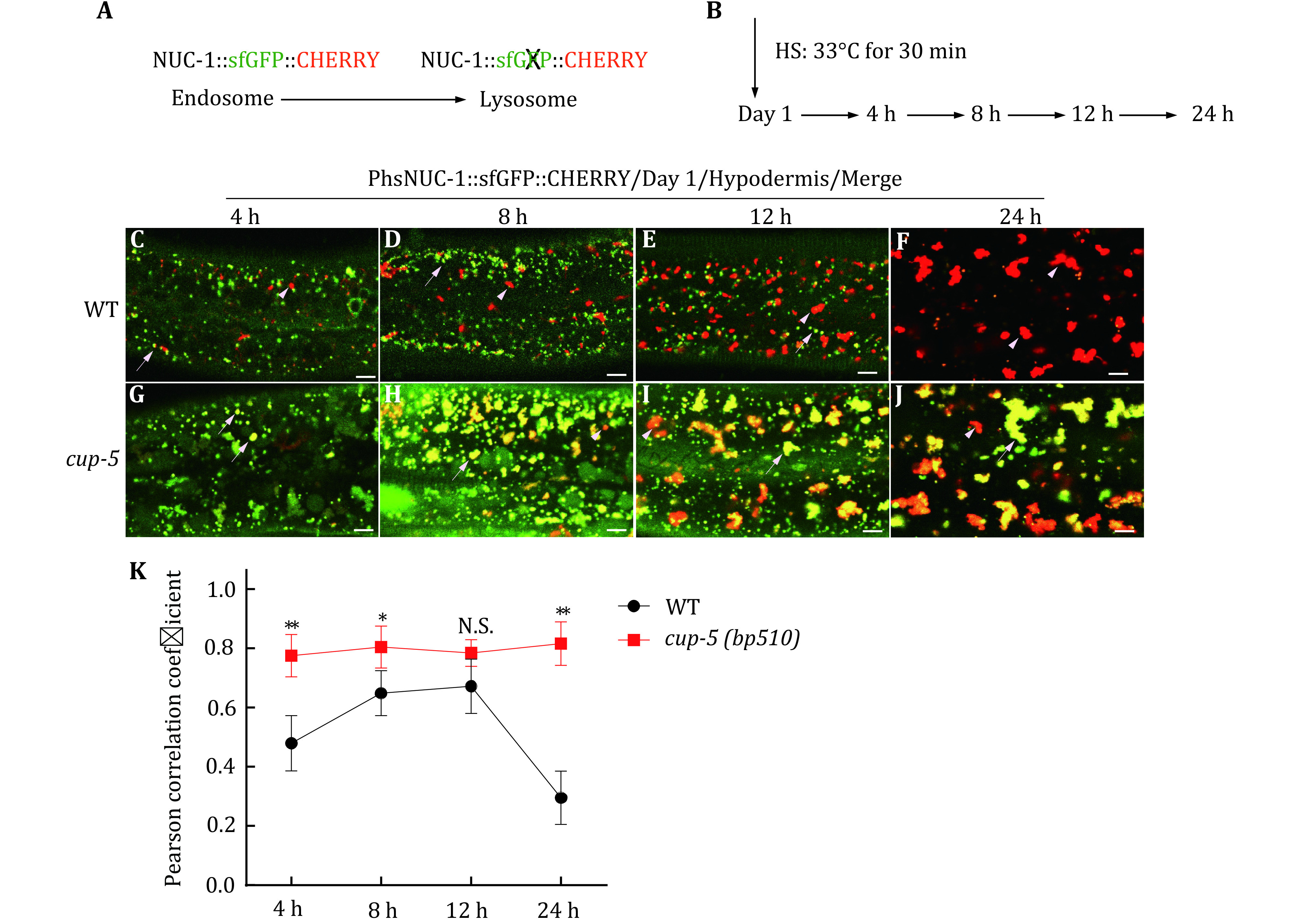
Lysosome maturation assay. A, B Diagram of the assay to examine lysosome maturation in the hypodermis using heat-shock (HS) induction of NUC-1::sfGFP::CHERRY. C–J Merged confocal fluorescence images of the hypodermis in wild type (WT) and cup-5(bp510) expressing NUC-1::sfGFP::CHERRY at 4, 8, 12 and 24 h post HS. White arrowheads indicate structures labeled by CHERRY only and white arrows indicate overlap of GFP and CHERRY. K Overlap of GFP and CHERRY is quantified by Pearson’s correlation coefficient. At least five animals were scored at each time point in each strain. Data are shown as mean ± SD. Two-way ANOVA with Tukey’s multiple comparison test was used to compare mutant datasets with the wild type. *p < 0.05, **p < 0.01; N.S.: no significance. Scale bars: 5 μm
In the second part, we analyze lysosomal acidity. We fused the pH-sensitive fluorescent protein pHTomato with NUC-1, and we transiently express NUC-1::pHTomato using the heat-shock (hs) promoter. pHTomato has a pKa close to 7.8 and thus exhibits increased fluorescence when the pH is increased (Li and Tsien 2012). We measure pHTomato fluorescence intensity in each lysosome using Volocity software. Loss of the lysosomal Ca2+ channel CUP-5 impairs lysosome activity and acidity (Hersh et al. 2002; Miao et al. 2020; Sun et al. 2011; Treusch et al. 2004), whereas lysosomal acidity increases in the long-lived daf-2 mutant worms (Sun et al. 2020). We assay the lysosomal acidity in wild-type, cup-5 and daf-2 worms.
In the third part, the lysosomal degradation activity is analyzed by examining cleavage of the NUC-1::CHERRY fusion protein and CPL-1 processing. CHERRY is cleaved from the NUC-1::CHERRY fusion protein by proteases in lysosomes, and the amount of CHERRY protein is visualized by western blot and quantified to indicate the degradation activity of lysosomes (Miao et al. 2020; Sun et al. 2020). Cathepsin L (CPL-1) is synthesized as an inactive pro-enzyme, which is converted to the active mature form in lysosomes through proteolytic removal of the pro-domain (Stoka et al. 2016). The processing of endogenous CPL-1 can be examined by western blot and quantified to indicate the degradation activity of lysosomes. These assays are performed in wild-type, cup-5 and daf-2 worms.
STEP-BY-STEP PROCEDURE
Lysosome acidification and maturation assay
(1) Worms are synchronized to the L4 stage and transferred to fresh NGM plates containing OP50. Worms are further cultured for 24 h to reach day 1 of adulthood.
(2) Worms at day 1 of adulthood are heat-shocked at 33 °C for 30 min to induce transient expression of NUC-1::sfGFP::CHERRY.
(3) The worms are then incubated at 20 °C for 4, 8, 12 and 24 h (Fig. 1B). At each time-point, worms are picked, and the hypodermis is imaged by confocal fluorescence microscopy (Fig. 1C–J).
(4) The colocalization of CHERRY and GFP is presented as the Pearson correlation coefficient score, which is calculated from the images by Volocity software (PerkinElmer). At least five worms are analyzed in each strain at each time point.
We found that the fluorescence of GFP, but not CHERRY, reduced gradually and disappeared completely at 24-h post heat-shock (HS) treatment. In wild type, the Pearson correlation coefficient increased at 8-h post HS and reduced after 12-h post HS (Fig. 1K). This indicates a gradual maturation of lysosomes in the C. elegans hypodermis. In cup-5, lysosomal acidification was impaired and the GFP signal was visible even at 24-h post HS (Fig. 1G–K). In our previous study, we used this assay to examine lysosome maturation during C. elegans larval development (Miao et al. 2020). We found that GFP disappeared faster at molt compared with the intermolt stage, which suggests that the speed of lysosome maturation increases during molting (Miao et al. 2020).
Lysosome acidity assay
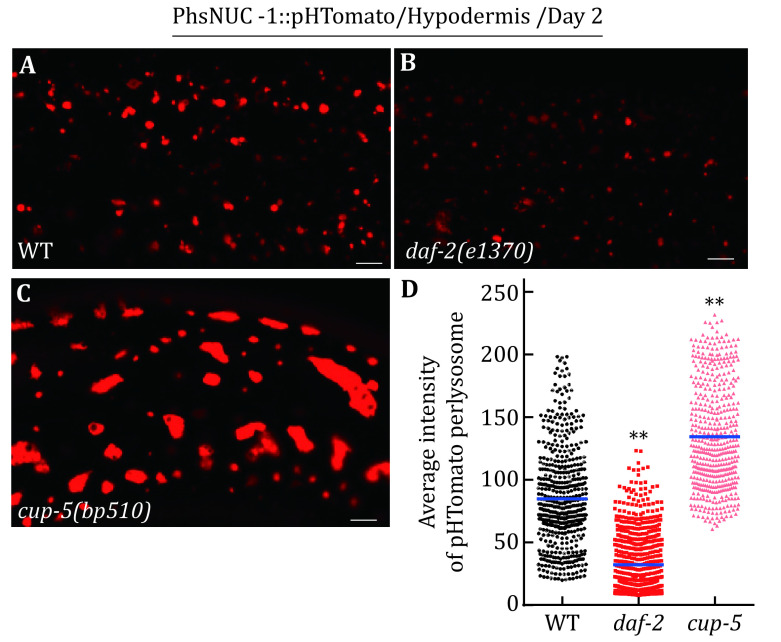
(1) Gravid adult worms are bleached, and the released embryos are placed in M9 buffer for 24 h to reach the larvae 1 (L1) stage. The synchronized L1 worms are transferred to NGM plates containing OP50 and grown to adults.
(2) C. elegans adults (1-day post L4) expressing PhsNUC-1::pHTomato are collected and heat shocked at 33 °C for 30 min.
(3) Worms are incubated at 20 °C for 24 h before examination in order to let the fusion protein enter into lysosomes.
(4) Worms are examined by fluorescence microscopy. The average intensity of pHTomato per lysosome in the hypodermis is measured by Volocity software. At least five worms and over 200 lysosomes in each worm are analyzed in each strain.
(5) Raw data is loaded and analyzed in Graphpad. The paired t-test is performed to compare different datasets.
We performed this assay in wild-type, cup-5 and daf-2 worms (Fig. 2). We found that pHTomato intensity increased significantly in cup-5 lysosomes but decreased in daf-2 lysosomes (Fig. 2D). This indicates that lysosomal acidity is increased in daf-2 worms but reduced in cup-5 mutants.
Figure 2.
Lysosome acidity assay. A–C Confocal fluorescence images of the hypodermis at adult day 2 in wild type (WT), daf-2(e1370) and cup-5(bp510) expressing NUC-1::pHTomato controlled by the heat-shock (hs) promoter. Scale bars: 5 μm. D The average intensity of pHTomato per lysosome is quantified. At least 20 animals were scored in each strain. Data are shown as mean ± SD. The paired t-test was performed to compare the mutant datasets with the wild type. **p < 0.001
[CRITICAL STEP] It is important that same laser power and exposure time are used to capture images of lysosomes in different strains. It is recommended to image different groups of samples at the same time. The average fluorescence intensity in multiple individual lysosomes (>200) is quantified to compare the different strains.
Lysosome degradation activity assay
Quantification of NUC-1::CHERRY cleavage
(1) Worms are synchronized and cultured at 20 °C as described in the section of Lysosome acidity assay.
(2) Over 50 adults (1-day post L4) are collected and washed three times with M9 buffer to remove OP50 as much as possible.
(3) The worms are lysed in sample buffer by 2–3 rounds of freezing and boiling treatment. The resulting worm lysate is centrifuged at 14,000 r/min for 10 min. The supernatant is resolved by SDS-PAGE. NUC-1::CHERRY and CHERRY are detected by anti-CHERRY antibodies (SUNGENE BIOTECH, China, 1:1000). α-tubulin antibody (Sigma) is used at 1:5000 as an internal control.
(4) The amount of NUC-1::CHERRY and CHERRY is quantified by determining the total intensity of NUC-1::CHERRY and CHERRY bands using Image J software. CHERRY cleavage is calculated by dividing the amount of CHERRY by the total amount of NUC-1::CHERRY and CHERRY. At least three independent experiments are performed and analyzed.
Examination and quantification of CPL-1 processing
(1) Worm synchronization and sample preparation are done as described in the above section. The resulting worm lysate is analyzed by Western blot and CPL-1 is detected by anti-CPL antibodies (Antibody core, NIBS, 1:1000). α-tubulin antibody (Sigma) is used at 1:5000 as an internal control.
(2) The band intensity of the mature (lower band) and pro- (higher band) forms of CPL-1 is quantified by Image J software. CPL-1 processing is calculated by dividing the amount of mature CPL-1 by the amount of total CPL-1 (both pro- and mature forms). At least three independent experiments are performed and analyzed.
We used the assays detailed above to examine lysosome activity in wild-type, cup-5(bp510) and daf-2(e1370) worms. Notably, CHERRY cleavage reduced significantly in cup-5 mutants but increased in daf-2 worms compared with wild-type animals (Fig 3A, B). Similarly, CPL-1 processing was reduced significantly in cup-5 mutants but increased in daf-2 worms (Fig. 3C, D). These results suggest that lysosomal degradation activity is increased in daf-2 but impaired in cup-5 mutants.
Figure 3.
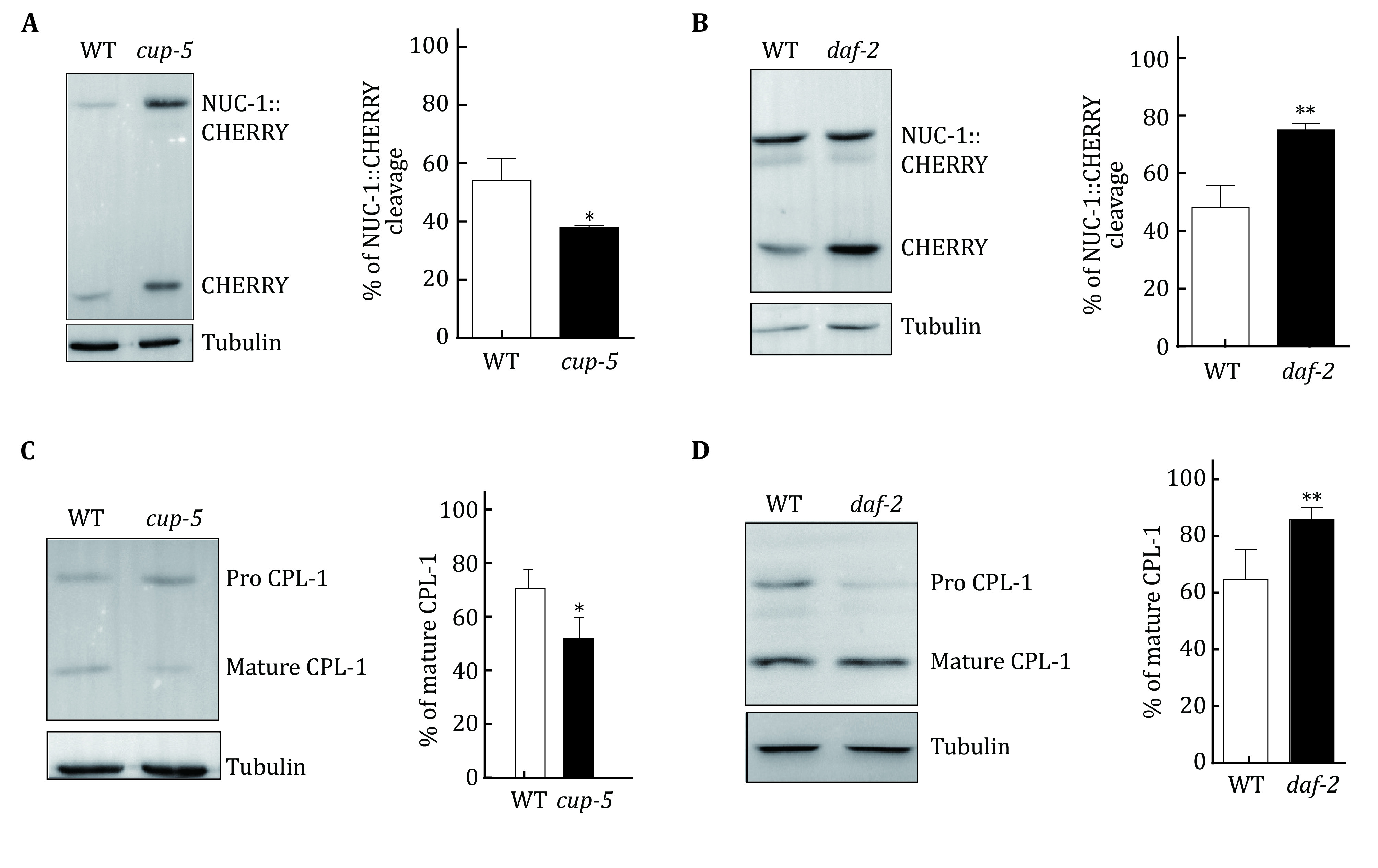
Lysosome degradation activity assay. A, B Western blot analysis of NUC-1::CHERRY cleavage in wild type (WT), cup-5(bp510) and daf-2(e1370) at day 1 of adulthood. The percentage of cleaved CHERRY was quantified and is shown in the right panels. C, D Western blot analysis of CPL-1 processing in wild type (WT), cup-5(bp510) and daf-2(e1370) at day 1 of adulthood. The percentage of mature CPL-1 was quantified and is shown in the right panels. Three independent experiments were performed. Data are shown as mean ± SD. The paired t-test was performed to compare the mutant datasets with the wild type. *p < 0.05, **p < 0.001
[CRITICAL STEP] The bands resolved by Western blot should not be overexposed. For different antibodies or different protein abundance, the exposure time should be tested and optimized. In each experiment, equal amounts of total proteins are loaded, which are normalized by the levels of tubulin.
FUTURE PERSPECTIVES
Lysosome maturation, acidity and degradation activity are important properties that can be monitored in combination to indicate the overall functionality of lysosomes in various biological processes. In this study, we provide detailed methods to assay lysosomal maturation, acidity and degradation activity in C. elegans. It is worth noting that in our study, lysosomal maturation and acidity assays are mainly performed in the hypodermis, a multi-nuclear syncytium that extends throughout most of the animal. As heat-shock treatment induces transient expression of reporters in multiple C. elegans tissues, these assays can be performed in other tissues if they are accessible to microscopy imaging. Moreover, lysosomal maturation and acidity assays can be performed in both larval and adult stages. However, we have not examined lysosomal acidity and maturation in small-sized cells such as neurons or embryonic cells. More sophisticated methods may be developed to examine lysosome property in small cells.
Conflict of interest
Xin Li, Yanan Sun and Xiaochen Wang declare that they have no conflict of interest.
Acknowledgements
Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40OD010440). This work was supported by the National Natural Science Foundation of China (3163001, 91754203), the Ministry of Science and Technology (2016YFA0500203), and the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB19000000) to X. Wang. X. Li is supported by the National Natural Science Foundation of China (31671401).
Compliance with Ethical Standards
Human and animal rights and informed consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
- Brenner S The genetics of Caenorhabditis elegans. Genetics. 1974;77(1):71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen PJ, Steinberg F To degrade or not to degrade: mechanisms and significance of endocytic recycling. Nat Rev Mol Cell Biol. 2018;19(11):679–696. doi: 10.1038/s41580-018-0053-7. [DOI] [PubMed] [Google Scholar]
- de Duve C The lysosome turns fifty. Nat Cell Biol. 2005;7(9):847–849. doi: 10.1038/ncb0905-847. [DOI] [PubMed] [Google Scholar]
- Gan Q, Wang X, Zhang Q, Yin Q, Jian Y, Liu Y, Xuan N, Li J, Zhou J, Liu K, Jing Y, Wang X, Yang C The amino acid transporter SLC-36.1 cooperates with PtdIns3P 5-kinase to control phagocytic lysosome reformation. J Cell Biol. 2019;218(8):2619–2637. doi: 10.1083/jcb.201901074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez A, Hall MN, Lin SC, Hardie DG AMPK and TOR: the Yin and Yang of cellular nutrient sensing and growth control. Cell Metab. 2020;31(3):472–492. doi: 10.1016/j.cmet.2020.01.015. [DOI] [PubMed] [Google Scholar]
- Hersh BM, Hartwieg E, Horvitz HR The Caenorhabditis elegans mucolipin-like gene cup-5 is essential for viability and regulates lysosomes in multiple cell types. Proc Natl Acad Sci USA. 2002;99(7):4355–4360. doi: 10.1073/pnas.062065399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huizing M, Gahl WA Inherited disorders of lysosomal membrane transporters. Biochim Biophys Acta Biomembr. 2020;1862(12):183336. doi: 10.1016/j.bbamem.2020.183336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence RE, Zoncu R The lysosome as a cellular centre for signalling, metabolism and quality control. Nat Cell Biol. 2019;21(2):133–142. doi: 10.1038/s41556-018-0244-7. [DOI] [PubMed] [Google Scholar]
- Li Y, Chen B, Zou W, Wang X, Wu Y, Zhao D, Sun Y, Liu Y, Chen L, Miao L, Yang C, Wang X The lysosomal membrane protein SCAV-3 maintains lysosome integrity and adult longevity. J Cell Biol. 2016;215(2):167–185. doi: 10.1083/jcb.201602090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Tsien RW pHTomato, a red, genetically encoded indicator that enables multiplex interrogation of synaptic activity. Nat Neurosci. 2012;15(7):1047–1053. doi: 10.1038/nn.3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Du H, Rutkowski R, Gartner A, Wang X LAAT-1 is the lysosomal lysine/arginine transporter that maintains amino acid homeostasis. Science. 2012;337:351–354. doi: 10.1126/science.1220281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zou W, Yang P, Wang L, Ma Y, Zhang H, Wang X Autophagy-dependent ribosomal RNA degradation is essential for maintaining nucleotide homeostasis during C. elegans. Elife. 2018;7:e36588. doi: 10.7554/eLife.36588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao R, Li M, Zhang Q, Yang C, Wang X An ECM-to-nucleus signaling pathway activates lysosomes for C. elegans larval development. Dev Cell. 2020;52(1):21–37. doi: 10.1016/j.devcel.2019.10.020. [DOI] [PubMed] [Google Scholar]
- Nixon RA New perspectives on lysosomes in ageing and neurodegenerative disease. Ageing Res Rev. 2016;32:1–1. doi: 10.1016/j.arr.2016.11.001. [DOI] [PubMed] [Google Scholar]
- Parkinson-Lawrence EJ, Shandala T, Prodoehl M, Plew R, Borlace GN, Brooks DA Lysosomal storage disease: revealing lysosomal function and physiology. Physiology (Bethesda) 2010;25(2):102–115. doi: 10.1152/physiol.00041.2009. [DOI] [PubMed] [Google Scholar]
- Sabatini DD, Adesnik M Christian de Duve: explorer of the cell who discovered new organelles by using a centrifuge. Proc Natl Acad Sci USA. 2013;110(33):13234–13235. doi: 10.1073/pnas.1312084110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoka V, Turk V, Turk B Lysosomal cathepsins and their regulation in aging and neurodegeneration. Ageing Res Rev. 2016;32:22–37. doi: 10.1016/j.arr.2016.04.010. [DOI] [PubMed] [Google Scholar]
- Sun T, Wang X, Lu Q, Ren H, Zhang H CUP-5, the C. elegans ortholog of the mammalian lysosomal channel protein MLN1/TRPML1, is required for proteolytic degradation in autolysosomes. Autophagy. 2011;7(11):1308–1315. doi: 10.4161/auto.7.11.17759. [DOI] [PubMed] [Google Scholar]
- Sun Y, Li M, Zhao D, Li X, Yang C, Wang X Lysosome activity is modulated by multiple longevity pathways and is important for lifespan extension in C. elegans. Elife. 2020;9:e55745. doi: 10.7554/eLife.55745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treusch S, Knuth S, Slaugenhaupt SA, Goldin E, Grant BD, Fares H Caenorhabditis elegans functional orthologue of human protein h-mucolipin-1 is required for lysosome biogenesis. Proc Natl Acad Sci USA. 2004;101(13):4483–4488. doi: 10.1073/pnas.0400709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J, Zhu MX Regulation of lysosomal ion homeostasis by channels and transporters. Sci China Life Sci. 2016;59(8):777–791. doi: 10.1007/s11427-016-5090-x. [DOI] [PMC free article] [PubMed] [Google Scholar]