Abstract
The PRINTS database houses a collection of protein family fingerprints. These are groups of motifs that together are diagnostically more potent than single motifs by virtue of the biological context afforded by matching motif neighbours. Around 1200 fingerprints have now been created and stored in the database. The September 1999 release (version 24.0) encodes ~7200 motifs, covering a range of globular and membrane proteins, modular polypeptides and so on. In addition to its continued steady growth, we report here several major changes to the resource, including the design of an automated strategy for database maintenance, and implementation of an object-relational schema for more efficient data management. The database is accessible for BLAST, fingerprint and text searches at http://www.bioinf.man.ac.uk/dbbrowser/PRINTS/
INTRODUCTION
Pattern databases are well-established tools for sequence analysis. Several distinct databases now exist, reflecting differences in their underlying pattern-recognition techniques. Nevertheless, the methods share a common principle: i.e., in each approach, information in the sequence databanks is distilled into some kind of discriminator that facilitates family diagnosis. Today, the most widely-used pattern databases include: PROSITE, which houses regular expressions and a few profiles (1); the BLOCKS databases, which store aligned, weighted motifs, or blocks (2); Pfam, which offers a range of hidden Markov models (HMMs) (3); and PRINTS, which provides groups of aligned, un-weighted sequence motifs, or fingerprints (4). Diagnostically, each database has different strengths and weaknesses, and hence different areas for optimum application. The resources also tend to differ in terms of family coverage. Thus, for best results, search strategies should ideally combine them all.
The fingerprinting method arose from the need for a reliable technique for detecting members of large, highly divergent protein super-families (5,6). The idea was to exploit the most conserved regions within sequence alignments to build diagnostic signatures of family membership. In a database search, there would then be a greater chance of identifying a distant relative, whether or not all parts of a signature were matched (providing the motifs were found in the correct order and the distances between them were consistent with those expected of true neighbouring motifs). The ability to tolerate mismatches, both at the level of residues within individual motifs, and at the level of motifs within entire fingerprints, rendered fingerprinting a powerful diagnostic approach.
Since 1993, to complement other pattern resources, we have made a range of protein fingerprints available in the PRINTS database (4). Here, we report substantial changes to the resource in terms of its underlying data source and its management strategy, yielding a new streamlined system termed PRINTS-S.
SOURCE DATABASE AND METHODS
The data source for PRINTS was OWL (7), but PRINTS-S exploits a SWISS-PROT/TrEMBL (8) composite, in order to bring the resource in line with its companion pattern databases, all of which are based on SWISS-PROT, or SWISS-PROT and TrEMBL. The current release was built from SWISS-PROT37 and TrEMBL9, with updates to February 22, 1999 (fragments excluded); incremental updates were based on SWISS-PROT37 and TrEMBL10, with updates to June 25, 1999.
Fingerprinting is an iterative process that commences with manual sequence alignment and excision of conserved motifs [e.g., using SOMAP (9) or CINEMA (10)]. The motifs are used to trawl the source database independently using routines first developed in the ADSP suite (5,6). The scanning algorithm interprets the motifs essentially as a series of frequency matrices—i.e., identity searches are made, with no mutation or other similarity data to weight the results. Diagnostic performance is enhanced by iterative database scanning. The motifs therefore mature with each database pass, as more sequences are matched and assimilated into the process. Full potency is gained from the mutual context provided by motif neighbours, allowing sequence identification even when parts of the signature are absent. Nevertheless, only sequences that match all motifs are allowed to contribute to a final fingerprint.
Database format
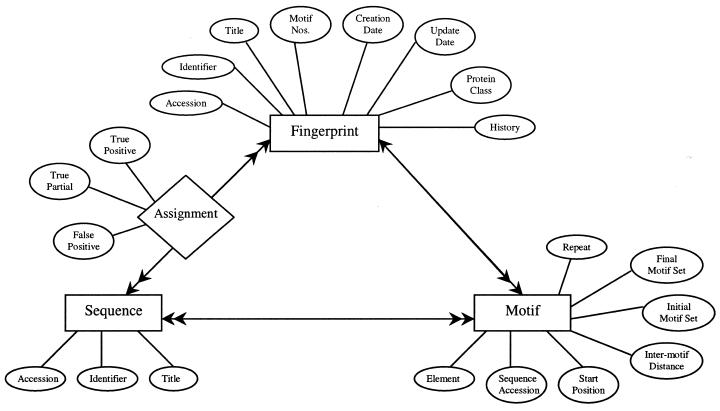
PRINTS was formerly built as a single ASCII (text) file. With the continued growth of the database, however, maintenance was becoming inefficient and error-prone. We have therefore designed an object-relational schema, which places existing database fields (e.g., relating to motifs, sequence data, true and false assignments, etc.) into separate but related tables. The underlying model, which constitutes the heart of PRINTS-S, is illustrated in Figure 1. Adopting such a management system reduces redundancy, maintains consistency and facilitates routine maintenance. It also permits more complex queries, and allows us to support both new display and flat-file formats; at the same time, we can continue to support the original flat-file format, should this be necessary for existing dependent applications.
Figure 1.
PRINTS-S entity relationship diagram. In order to transpose the current PRINTS databank to a relational database, various models were developed on the basis of existing data fields and their properties. This diagram depicts the PRINTS data space modelled using three entities: fingerprint, motif and sequence (rectangular boxes). Each entity has a relationship with another, as represented by a connecting arrow. A single arrow-head denotes a ‘single relationship’ and a double arrow-head denotes a ‘many relationship’: e.g., one fingerprint has many motifs, and several motifs belong to one fingerprint. The many-to-many relationship between entities ‘fingerprint’ and ‘sequence’ is special, as highlighted by means of a diamond. The diamond represents a function (i.e., a relationship with a property), in this case ‘assignment’: sequences are given assignments dependent on their relationship with the fingerprint (i.e., they may be true positive, true partial or false positive). Ellipses denote specific entity attributes.
Content, update and growth
Release 24.0 (September 1999) contains 1210 entries, encoding ~7200 individual motifs. A complete content list is available from the distribution sites and from the Web site.
PRINTS has been released in major and minor versions: the former denote database expansions (i.e., the addition of new material to the resource); the latter reflect updates of existing entries to bring results in line with the current version of the underlying data source. To date, there have been 24 major and five minor releases. A major or minor version is made available quarterly—in the last year, we have achieved four major and one minor release.
The principal obstacle to the frequency of expansions, and particularly of updates, is the time-consuming nature of the approach. Deriving a fingerprint is laborious, involving both swift computational and slow manual aspects—the latter are necessary to validate the results and to provide useful family annotations. The value of manually-input annotations has tended to justify the sacrifice of speed, setting the database apart from the growing number of automatically-derived family resources [e.g., ProDom (11) and DOMO (12)], for which there are no annotations and no result validation. However, although we have achieved regular major releases, the full database had not been updated for 3 years. To address this issue, we implemented a semi-automatic protocol, which has allowed us to update the entire database. The process was not fully automated because of the complexity of the task, and because we wished to minimise false assignments that might compromise fingerprint quality.
Access and distribution
PRINTS-S is accessible for interactive use via the Web. The interface allows strict keyword searching of database code, accession number, text, sequence, etc.; more powerful queries can be built using a combination of regular expressions and logical operators. Such queries are made possible by calls to the underlying query language, SQL, the syntax of which is conveniently hidden from the user beneath the Web interface.
For local installation, original- and new-format (InterPro-compatible) flat-files may be retrieved from the anonymous-ftp servers at Manchester (ftp://ftp.bioinf.man.ac.uk/pub/prints ), HGMP-RC (ftp://ftp.hgmp.mrc.ac.uk/pub/database/prints ), EBI (ftp://ftp.ebi.ac.uk/pub/databases/prints ), EMBL (ftp://ftp.embl-heidelberg.de/ftp/pub/databases/prints ) and NCBI (ftp://ncbi.nlm.nih.gov/repository/PRINTS ).
Search software
Two main tools are provided for searching the database: (i) a BLAST server allows similarity searches against sequences matched in the current version of the database (13); and (ii) the fingerPRINTScan suite allows sequence searches against fingerprints contained in the current release—probability- and expect-values are calculated to assign a measure of confidence to both complete and partial matches (14). FingerPRINTScan, which is now used within the EDITtoTrEMBL suite as part of the EBI’s automatic protocol to annotate TrEMBL (15), is a powerful diagnostic tool, affording greater specificity than the BLAST implementation (13). The diagnostic performance of these approaches is contrasted in the supplementary material given at http://www.bioinf.man.ac.uk/dbbrowser/nar/printss.html
Derivative databases
A major strength of PRINTS is that its motifs are stored in the form of un-gapped, local sequence alignments. This allows different implementations to be established with alternative scoring methods. Thus, a BLOCKS-format version of the resource that exploits BLOCKS scoring methods is available at the Fred Hutchinson Cancer Research Center (2). In addition, the protein function identification resource (IDENTIFY) at Stanford overlays a permissive regular expression approach over PRINTS’ multiply-aligned motifs, offering different levels of stringency from which to infer the significance of matches (16). Derivative databases are useful as they provide different perspectives on the same data: they afford the opportunity to validate results where there are equivalent matches in more than one resource; and they offer the chance to make diagnoses that may have been missed by the original implementation.
Applications
A criticism recently made of pattern databases is that they endeavour to be as general as possible. It was suggested that a classification system capable of diagnosing sub-family relationships within super-families would be useful, but that such a system does not exist (17). In fact, PRINTS departs from other pattern databases precisely because it does provide family- and sub-family-specific fingerprints. Such a hierarchical approach has been used, for example, to resolve G-protein-coupled receptor (GPCR) super-families into their constituent families and receptor sub-types, and to classify a variety of channel proteins, enzymes, etc. FingerPRINTScan was designed to exploit this hierarchical structure, as readily demonstrated by searching the database with a melanocortin type 4 receptor (e.g., MC4R_HUMAN)—the diagnosis returned reveals the sequence to be a member of the rhodopsin-like GPCR super-family and melanocortin family, and it pinpoints the specific receptor sub-type, discriminating it from the closely-related sub-type 5 (see Supplementary Material).
PRINTS now has a central role within the newly-launched InterPro project, an international initiative to unite the efforts of the pattern database providers. InterPro pools the high-level documentation from PRINTS and PROSITE (and minimal annotation in Pfam) into a central compendium of family and domain descriptions, around which satellite the different pattern resources. These maintain their unique analytical flavours, thus offering a range of diagnostic opportunities for a given query. InterPro aims to reduce duplication of effort in the laborious process of annotation, and to facilitate communication between disparate resources, ultimately providing a one-stop shop for the analysis of newly-determined sequences.
CONCLUSION
Creating and annotating family descriptors is time-consuming, so pattern databases have not kept pace with the deluge of sequence data. Nevertheless, as they become more comprehensive, their diagnostic potency ensures that pattern databases like PRINTS will play an increasingly important role as the post-genome quest to assign functional information to raw sequence data gains pace.
SUPPLEMENTARY MATERIAL
See Supplementary Material available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
PRINTS is built and maintained at the University of Manchester with support from the Royal Society (T.K.A. is a Royal Society University Research Fellow). We are grateful for individual support from Astra Charnwood (P.S.), Zeneca Pharmaceuticals/Open Molecule Foundation (J.N.S.), Glaxo Wellcome (W.W.), the BBSRC (J.E.M. and W.W.) and the EC (M.D.R.C.).
REFERENCES
- 1.Hofmann K., Bucher,P., Falquet,L. and Bairoch,A. (1999) Nucleic Acids Res., 27, 215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Henikoff S., Henikoff,J.G. and Pietrokovski,S. (1999) Bioinformatics, 15, 471–479. [DOI] [PubMed] [Google Scholar]
- 3.Bateman A., Birney,E., Durbin,R., Eddy,S.R., Finn,R.D. and Sonhammer,E.L.L. (1999) Nucleic Acids Res., 27, 260–262. Updated article in this issue: Nucleic Acids Res. (2000) 28, 263–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Attwood T.K., Flower,D.R., Lewis,A.P., Mabey,J.E., Morgan,S.R., Scordis,P., Selley,J.N. and Wright,W. (1999) Nucleic Acids Res., 27, 220–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parry-Smith D.J. and Attwood,T.K. (1992) Comp. Appl. Biosci., 8, 451–459. [DOI] [PubMed] [Google Scholar]
- 6.Attwood T.K. and Findlay,J.B.C. (1994) Protein Eng., 7, 195–203. [DOI] [PubMed] [Google Scholar]
- 7.Bleasby A.J., Akrigg,D. and Attwood,T.K. (1994) Nucleic Acids Res., 22, 3574–3577. [PMC free article] [PubMed] [Google Scholar]
- 8.Bairoch A. and Apweiler,R. (1999) Nucleic Acids Res., 27, 49–54. Updated article in this issue: Nucleic Acids Res. (2000) 28, 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parry-Smith D.J. and Attwood,T.K. (1991) Comp. Appl. Biosci., 7, 233–235. [DOI] [PubMed] [Google Scholar]
- 10.Parry-Smith D.J., Payne,A.W.R, Michie,A.D. and Attwood,T.K. (1998) Gene, 11, GC45–GC56. [DOI] [PubMed] [Google Scholar]
- 11.Gouzy J., Corpet,F. and Kahn,D. (1999) Nucleic Acids Res., 27, 263–267. Updated article in this issue: Nucleic Acids Res. (2000) 28, 267–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gracy J. and Argos,P. (1998) Trends Biochem. Sci., 23, 495–497. [DOI] [PubMed] [Google Scholar]
- 13.Wright W., Scordis,P. and Attwood,T.K. (1999) Bioinformatics, 15, 523–524. [DOI] [PubMed] [Google Scholar]
- 14.Scordis P., Flower,D.R. and Attwood,T.K. (1999) Bioinformatics, 15, in press. [Google Scholar]
- 15.Moeller S., Leser,U., Fleischmann,W. and Apweiler,R. (1999) Bioinformatics, 15, 219–227. [DOI] [PubMed] [Google Scholar]
- 16.Nevill-Manning C.G., Wu,T.D. and Brutlag,D.L. (1998) Proc. Natl Acad. Sci. USA, 95, 5865–5871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hofmann K. (1998) In Trends Guide to Bioinformatics, Elsevier Science Ltd, Kidlington, Oxford, UK, pp. 18–21.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.