Abstract
Clonal hematopoiesis (CH), in which hematopoietic stem and progenitor cell (HSPC) clones and their progeny expand in the circulating blood cell population, occurs following the acquisition of somatic driver mutations. Individuals diagnosed with clonal hematopoiesis of indeterminate potential (CHIP) carry somatic mutations in hematological malignancy-associated driver genes, historically at or above a variant allele frequency of 2%, but do not exhibit abnormal blood cell counts or any other symptoms of hematologic disease. However, CHIP is associated with moderately increased risk of hematological cancer and a greater likelihood of cardiovascular and pulmonary disease. Recent advances in the resolution of high-throughput sequencing experiments suggest CHIP is much more prevalent in the population than once thought, particularly among those aged 60 and over. Although CHIP does elevate the risk of eventual hematological malignancy, only one in 10 individuals with CHIP will receive such a diagnosis; the problem lies in the continued difficulty in accurately separating the 10% of CHIP patients who are most likely to be in a premalignant state from those who are not, given the heterogeneity of this condition and the etiology of the associated hematological cancers. Concerns over the risk of eventual malignancies must be balanced with growing recognition of CH as a common age-dependent occurrence, and efforts to better characterize and differentiate oncogenic clonal expansion from that which is much more benign. In this review, we discuss evolutionary dynamics of CH and CHIP, the relationship of CH to aging and inflammation, and the role of the epigenome in promoting potentially pathogenic or benign cellular trajectories. We outline molecular mechanisms that may contribute to heterogeneity in the etiology of CHIP and the incidence of malignant disease among individuals. Finally, we discuss epigenetic markers and modifications for CHIP detection and monitoring with the potential for translational applications and clinical utility in the near future.
Keywords: acute myeloid leukemia, hematological neoplasm
INTRODUCTION
Although clonal hematopoiesis of indeterminant potential (CHIP) has been identified as a risk factor for hematological cancers, cardiovascular and pulmonary disease, clonal hematopoiesis (CH) has come to be recognized as a universal mechanism of aging that produces an age-dependent increase in cell mosaicism (Acuna-Hidalgo et al. 2017). The largest risk factors for developing CHIP are chronological age and previous exposure to cytotoxic (e.g., chemotherapeutic) agents (Miller and Steensma 2020). Clonal hematopoiesis confers some putatively elevated risk but is more likely to be benign in cases in which clones are not large in size, do not carry multiple mutations, and do not have mutations in particularly high-risk drivers (e.g., TP53, IDH1, IDH2, JAK2) (Miller and Steensma 2020). CHIP occurs when hematopoietic stem and progenitor cells (HSPCs) acquire driver mutations that promote their clonal proliferation, resulting in certain clonal cell lineages making up a disproportionate fraction of circulating blood cells (Watson et al. 2020). Individuals with CHIP do not exhibit abnormal blood cell counts or other symptoms of hematologic disease (Jaiswal and Ebert 2019; Valent et al. 2019). In contrast, CH refers to the more canonical phenomenon of clonal expansion among hematopoietic stem cells. Most clones will carry evolutionarily neutral mutations, which are typically associated with smaller clone size and are more likely to be of benign effect (Miller and Steensma 2020). Only one in 10 individuals with CHIP will eventually receive an associated cancer diagnosis, but the consequences of a prognosis that either overlooks premalignant disease or results in unnecessary treatment are significant, emphasizing the need for continued study of this condition.
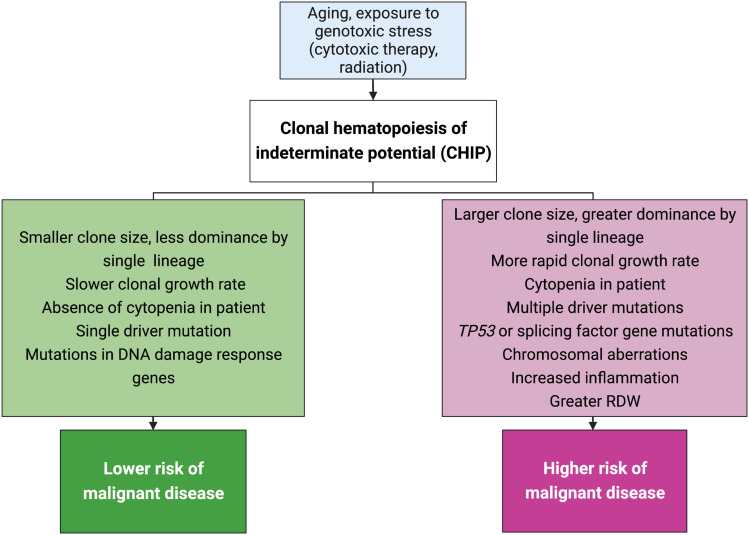
A challenge inherent to understanding any aspect of human health influenced by aging is the marked heterogeneity in how biological change manifests over time in different individuals. This naturally includes age-related change in the human hematopoietic system, in addition to variation in the risk conferred by such change depending on the individual. Reliably distinguishing high-risk CHIP diagnoses (i.e., in those who are likely to develop leukemia or are predisposed to cardiovascular disease [CVD] and/or pulmonary disease) from low-risk cases remains an important step in effective preventative efforts and management of CHIP as a condition. Although no single definitive biomarker exists to differentiate the trajectory of benign, canonically age-related clonal hematopoiesis (ARCH) from that which will eventually progress to hematological malignancy, a number of risk factors have been associated with the likelihood of malignancy and can be used as indicators for closer or more frequent monitoring, or the possibility of therapeutic intervention, such as clone size; the identity, type, and count of driver mutations; and mutations in splicing factor genes, among other factors (Fig. 1; Miller and Steensma 2020; Robertson et al. 2022).
Figure 1.
Risk factors for clonal hematopoiesis of indeterminate potential (CHIP) and for malignant conversion. Risk factors for CHIP are shown in the light blue box at the top of the flowchart. A CHIP diagnosis in a patient with the characteristics shown in the green box (left side of chart) are those associated with lower risk of subsequent conversion to malignancy, whereas those shown in the pink box are generally associated with higher risk. (RDW) Red cell distribution width, measured from complete blood cell counts. Figure created with BioRender.com.
In this short review, we discuss the evolutionary processes and genetic and epigenetic mechanisms at play in the progression (or lack thereof) of CHIP to malignant disease, as well as its implications in the context of aging and inflammation. We conclude by briefly detailing methods that hold potential in the biomarker research space, many of which are grounded in the idea that the transition to hematological malignancy is accompanied by predictable patterns of genetic and epigenetic change that may enable the eventual identification of high-risk cellular states and early detection of malignancy in peripheral blood.
Defining Clonal Hematopoiesis and Risk of Hematologic Malignancy
Although early evidence of hematopoietic clonality dates back to studies from the early 1960s examining X-inactivation in females (Beutler et al. 1962; Lyon 1962), next-generation sequencing (NGS) technologies have enabled deeper examination and characterization of the specific loci involved in CH and CHIP over the last decade (Jaiswal et al. 2014; Steensma et al. 2015; for review, see Köhnke and Majeti 2021). In 2014, three large, independent cohort studies discovered an unexpectedly high prevalence of somatic mutations in genes associated with hematological neoplasia but in the absence of cytopenia (abnormal blood cell counts) or other clinical evidence of malignant or nonmalignant hematological disease (Genovese et al. 2014; Jaiswal et al. 2014; Xie et al. 2014), thus identifying the phenomenon of ARCH (Jaiswal et al. 2014) and setting the stage for the definition of CHIP 1 year later (Steensma et al. 2015). Although the risk of progression to malignancy is 0.5%–1% per year (Steensma et al. 2015), the risk of cancer is estimated to be 10 times greater in individuals with CHIP compared to the general population (Genovese et al. 2014; Jaiswal et al. 2014; for review, see Hoermann et al. 2020).
Historically, CHIP has been diagnosed by the presence of a hematological driver mutation at a variant allele frequency (VAF) of at least 2% (Steensma and Ebert 2020; Köhnke and Majeti 2021). Research into CHIP-associated malignant disease has mostly used deep, high-throughput sequencing of bulk samples to determine whether VAFs for driver mutations meet the aforementioned 2% threshold. However, with the advancements in the breadth and depth of high-throughput sequencing experiments in recent years, clonal variants are being identified at lower and lower VAFs (Acuna-Hidalgo et al. 2017; Uddin et al. 2022); whereas the increased resolution is generally desirable, the risk conferred by these mutations at very low VAFs are often hard to assess because of the heterogeneous nature of the disease and an unknown magnitude of effect (Robertson et al. 2022).
Acute myeloid leukemia (AML) is the most common malignancy in CHIP patients who go on to develop cancer. AML is diagnosed when HSPC clonal lineages overproliferate and eventually HSPCs themselves fail to normally differentiate, leading to the accumulation of immature leukocytes (myeloblasts) in bone marrow (Stone et al. 2004; Köhnke and Majeti 2021). Preleukemic clonal abnormalities can precede overt leukemia by several decades, suggesting that early-life driver mutations do not appreciably alter homeostasis of the hematopoietic system earlier in life. However, cooperating mutations acquired later on can operate synergistically in cells containing the early mutations to catalyze clonal expansion and/or oncogenic activity (Genovese et al. 2014; Gao et al. 2021). Indeed, founding mutations acquired during embryogenesis typically do not trigger carcinogenesis until subsequent mutations arise, in accordance with Knudson's two-hit hypothesis (Knudson 1971; Tucker and Friedman 2002).
A challenge associated with CHIP is the variability of its presentation and potential pathogenicity, which has made it difficult to identify reliable prognostic indicators to appropriately risk stratify patients. However, over the last decade, studies have cataloged CH and CHIP mutations and begun to decipher how the life history of clonally expanded cells influences each individual's risk of malignant disease. An ultimate goal after identifying who is at greatest risk of a diagnosis such as AML is to determine the most effective means of extending the health and functionality of the hematopoietic and immune systems so as to avoid such conditions.
Evolutionary Dynamics and the Mechanisms of Clonal Expansion
Hematopoietic stem cells are produced in the bone marrow compartment, but they are also found in the bloodstream and circulating throughout all organs and tissues. Hematopoietic stem cells give rise to multipotent progenitor cells that differentiate along two blood cell lineages, myeloid and lymphoid (Niroula et al. 2021). Myeloid progenitor cells can become red blood cells, platelets, or innate immune cells such as macrophages/monocytes, neutrophils, basophils, and eosinophils (Kondo 2010; de Haan and Lazare 2018). Lymphoid progenitors become natural killer (NK) cells, T or B lymphocytes (de Haan and Lazare 2018). Over time, HSPCs randomly acquire somatic mutations that act as substrates for natural selection, leading to clonal expansion and sometimes CHIP (Kar et al. 2022). Importantly, CH-associated mutations in myeloid and lymphoid cells tend to be different from one another, displaying distinct patterns of distribution throughout the genome. Each is highly predictive of its respective lineage-specific malignancy (Niroula et al. 2021). More than 70 genes have been found to be recurrently mutated in myeloid malignancies (e.g., AML, myelodysplastic syndrome [MDS], myeloproliferative neoplasms [MPN]), but the most common mutations are overwhelmingly found in DNMT3A and TET2 (Buscarlet et al. 2017; Niroula et al. 2021; Robertson et al. 2022). In contrast, the 235 most frequently mutated genes in lymphoid malignancies (e.g., chronic lymphocytic leukemia [CLL], small lymphocytic leukemia [SLL], Hodgkin lymphoma, diffuse large B-cell lymphoma [DLBCL], follicular lymphoma) are comparatively more evenly dispersed throughout the genome (Niroula et al. 2021).
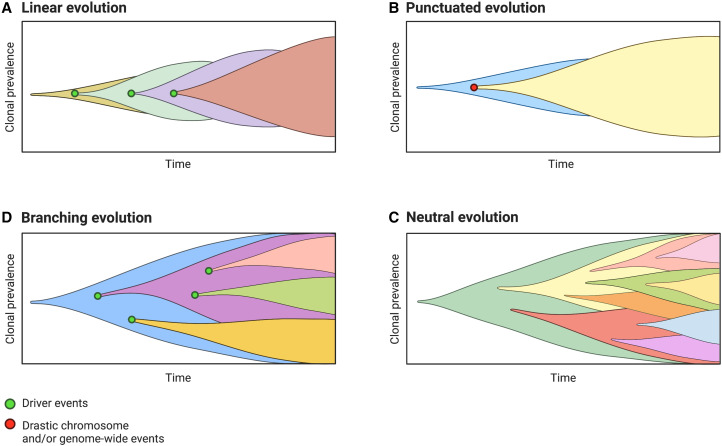
The study of human cancer as an evolutionary process has illuminated our understanding of oncogenesis at the molecular level and across deeper spans of somatic “evolutionary time” (at the cellular scale) (Fidler et al. 1978). Evolutionary models such as those depicted in Figure 2 provide a framework for reconstructions of cancer type–specific molecular time lines and temporal trends in mutational patterns using data from a single time point (e.g., Alexandrov et al. 2020; PCAWG Evolution & Heterogeneity Working Group et al. 2020; Landi et al. 2021). Such models are useful as typically only one biopsy or blood sample is available from a patient (Black and McGranahan 2021). Although the majority of somatic mutations are neutral (“passenger mutations”) (Fig. 2C), some provide a fitness advantage by enhancing survival or proliferative capacity. Under the tenets of Darwinian evolution, rapidly proliferating clonal cells outcompete their neighbors within their microenvironmental niche and can therefore achieve clonal dominance in the bloodstream (Jaiswal and Ebert 2019). However, it should be recognized that all models meant to approximate aspects of human biology are abstractions of reality used to understand complex systems and identify general patterns through some degree of simplification (Davis et al. 2017; Enderling and Wolkenhauer 2021). In reality, the processes underlying cancer development may be best fit by different evolutionary models depending on the type, stage, and molecular landscape of the developing tumor, and thus the most accurate model may change depending on the time and local context. Undergoing cytotoxic therapies (e.g., treatment for previous cancer) can alter the adaptive landscape and selective pressures in the microenvironmental niche of the bone marrow compartment, and such changes in clonal evolutionary dynamics should then be taken into account when considering treatment regimens to ensure the greatest rate of success. Unsurprisingly, there is active and ongoing debate surrounding which model of clonal evolution reflects the most accurate depiction of the underlying processes.
Figure 2.
(From top left, clockwise, A–D). Models proposed to explain somatic evolution and clonal lineage dynamics over time. Colors represent clonal lineages—that is, cells sharing the same genetic variants and common cellular ancestor. Green dots indicate molecular driver events that prompt the expansion of new clonal lineages through natural selection for clones with a fitness advantage. A red dot shows major chromosome-level or genome-wide event. (A) Model of linear evolution; monoclonal model of tumorigenesis. Successive driver mutations result in the establishment of successive clonal lineages that expand one after another via selective sweeps. (B) Model of punctuated evolution; major chromosomal events (e.g., DNA copy-number aberrations, whole-genome duplication) occur in a rapid, early burst, followed by a steady clonal expansion. (C) Model of neutral evolution; clones arise stochastically and natural selection does not shape intratumoral cell population dynamics; a form of branching evolution in which lineages are lost via genetic drift and intratumoral heterogeneity is high. (D) Model of branching evolution; clones arise from the same common ancestor but diverge into multiple clonal lineages and continue to evolve simultaneously. Figure created with BioRender.com.
Other evolutionary forces and factors beyond the influence of natural selection affect cancer cell clonal growth and size. Clonal growth dynamics are affected not only by mutation, but by genetic drift, population size, and the genetic background in which a mutant allele arises (Watson et al. 2020; Ní Leathlobhair and Lenski 2022). Thus, it is also important to remember not all clonal and subclonal mutational lineages that occur at a high prevalence are the product of positive selection. Depending on the age and size of clonally expanded lineages, and the particular point in time at which they are sampled, it is possible for deleterious alleles to become prevalent, or, alternatively, to “drop out” at random via genetic drift, although it is notable that drift is probably not the dominant force shaping cancer genomes (Watson et al. 2020; Mitchell et al. 2022).
Although specific mutations involved in CHIP have been established, the link between genotype, associated cellular phenotype, and the nature and consequences of the fitness advantages conferred is complex. Here, longitudinal analyses prove highly advantageous in resolving ambiguity regarding the ground truth of the evolutionary dynamics of clonal growth, a sacrifice that is inherent to estimating molecular time lines rather than directly profiling them (Robertson et al. 2022). Results from recent work suggest the relationship between fitness and clonal growth is nuanced, not always linear, and is often difficult to capture in the absence of longitudinal sampling to reconstruct clonal evolution. High fitness variants that occur at low frequencies can play a role in disease predisposition and severity, despite going undetected using the traditional threshold of at least 2% VAF (Robertson et al. 2022), although clone size and growth rate are nonetheless generally significant predictors of malignant conversion (Miller and Steensma 2020). Much still remains to be learned about the evolutionary dynamics and sources of interindividual variation in CHIP and its growth trajectories, but it appears that prevalent variants in genes associated with hematologic malignancies are only one part of a larger, more intricate biological narrative. An analysis of nearly 100,000 peripheral blood samples found that both noncoding mutations in the germline and coding mutations acquired somatically in TET2 interact to moderate how CHIP manifests in a given individual, highlighting how inherited and acquired genetic variation can affect risk of oncogenesis (Bick et al. 2020). Additionally, the same authors found differences in the inflammatory phenotypes under CHIP depending on the identity of the mutated driver gene, with TET2 CHIP mutations associating with higher expression of IL-1β, whereas those in JAK2 and SF3B1 were associated with production of IL-18 (Bick et al. 2020).
CHIP-associated driver mutations display unique trends of clonal expansion over time. Fabre et al. (2022) found that DNMT3A-mutant clones are more likely to expand earlier in life, but that HSPC lineages with mutations in DNMT3A and TP53 exhibited the slowest rate of annual growth (5%), with clonal lineages carrying TET2, ASXL1, PPM1D, and SF3B1 mutations expanding about twice as rapidly, and those with mutations in splicing factors SRSF2, PTPN11, and U2AF1 growing at an average rate of 15%–20% annually (Fabre et al. 2022); the rapid growth rate of the latter set may contribute to worse prognoses and disease severity observed for CH-associated mutations in splicing factor genes (Hou et al. 2016). One particular somatic variant, SRSF2P95H, was found to facilitate much more rapid expansion than other mutations in the same gene, whereas most somatic variants that occurred within the same gene were associated with similar rates of clonal growth (Fabre et al. 2022). Truncating and missense mutations in the same gene had similar effects on rates of clonal expansion in most cases, with the exception of clones carrying TP53 missense mutations, which grew 10% more rapidly than those with truncating mutations in the same gene; the reverse was found for missense mutations in CBL, where clones with truncating mutations expanded 11% more rapidly (Fabre et al. 2022). Notably, the growth rate of nearly all clonal lineages slowed at later stages of life, highlighting the nonlinear dynamics and resulting challenge of discerning trajectories of molecular and cellular changes that occur over the long course of a human life span (Black and McGranahan 2021; Fabre et al. 2022).
Interactions between Aging and Clonal Hematopoiesis
Somatic mutations that lead to clonal expansion are a universal feature of biological aging, not just in blood but in nearly all other tissues (Kar et al. 2022). Numerous studies have shown clone size is positively associated with risk of carcinogenesis and other adverse health events (Jaiswal et al. 2014, 2017; Abelson et al. 2018; Desai et al. 2018; Bolton et al. 2019). Even in young adulthood, establishment of clonal lineages in the bloodstream is already beginning to occur (Acuna-Hidalgo et al. 2017), although notably, such mutations are not examples of CHIP (because of their occurrence at too low a VAF) but of CH more generally.
Age is the predominant risk factor for cancer and other chronic, noncommunicable diseases, which are sometimes referred to as “diseases of aging” (Wick et al. 2000). Although the majority of patients who meet the criteria for CHIP will not develop malignant hematological disease (Valent et al. 2017), CHIP has also been causally linked to elevated risk of atherosclerotic cardiovascular disease (for review, see Hoermann et al. 2020) and increased incidence of type 2 diabetes (Fuster et al. 2020) and chronic obstructive pulmonary disease (Miller et al. 2022), all of which also become more common with increasing age. Although causal relationships between CH, aging, and malignant and other diseases have been challenging to disentangle, distinctions in clonal lineage dynamics are emerging that may help to explicitly delineate the different manifestations and trajectories of CH and their downstream implications.
Between the ages of 60 and 70, there is a massive decline in hematopoietic clonal diversity (Mitchell et al. 2022), and the proportion of circulating blood cells derived from single hematopoietic lineages (i.e., cells all derived from the same HSPC ancestor) can reach as high as 60% (Young et al. 2016; Zink et al. 2017). Initial estimates put the prevalence of CH among those over age 70 at 10%–20%, but subsequent work suggests that it may in fact be a ubiquitous feature of human aging after the sixth or seventh decade of life (Young et al. 2016; Zink et al. 2017; Uddin et al. 2022). A study by Mitchell et al. (2022) used deep sequencing of clonal colonies grown from single HSPCs from donors aged 0 to 81 yr and determined that clonal lineages with VAFs >1% were universally present in those >70 yr of age.
Additionally, ancestral mutations of clonally expanded lineages appear to have originated before participants reached age 40, in accordance with the two-hit model (Knudson 1971; Mitchell et al. 2022). This observation naturally begs the question of how clonal expansion is held in check in most individuals despite the presence of known hematologic driver mutations, and by what mechanisms this apparent homeostatic state is able to be maintained. In the same study by Mitchell et al. clonal lineages were found to undergo rapid expansion after age 60, and clonal diversity collapsed as the influence of natural selection faded (i.e., with advancing age). The authors posit that, given HSPC driver mutations conferring fitness advantages occur at a constant rate over an individual's lifetime, other age-related physiological and/or microenvironmental changes must occur as a result of accumulated molecular damage that enable these HSPC lineages to clonally expand.
Finally, although ARCH and CHIP are sometimes used synonymously in the literature, several recent publications by leading authors in the field maintain the distinction (Jaiswal and Ebert 2019; Jaiswal 2020; Köhnke and Majeti 2021), likely because the difference, although nuanced and not completely understood, remains biologically meaningful. As such, Jaiswal (2020) proposes that while the current diagnostic criteria for CHIP may be redefined in the future, for now they help to define a “sensible working definition” to assist in the design of experimental research. There have also been efforts to clarify and promote more consistent use of CHIP-related terminology; this includes calls for a more granular distinction of CHIP from CHOP (clonal hematopoiesis of oncogenic potential), the latter of which distinguishes itself by the specific types and number of mutations, both of which are related to an increase in malignancy risk (Valent et al. 2019).
The Role of Inflammation in Clonal Hematopoiesis
Inflammation naturally increases with age, a condition referred to as “inflamm-aging” (Xia et al. 2016; Franceschi et al. 2018). Although we know that clonal expansion is not exclusive to blood but occurs in many other tissues (e.g., skin, Martincorena et al. 2015; bronchus, Yoshida et al. 2020; endometrium, Suda et al. 2018, Moore et al. 2020; and esophagus, Martincorena et al. 2018), it has been best studied in the context of the hematopoietic system, in part because of the accessible nature of peripheral blood. Blood cells are not constrained by the structural and geographical limitations of solid tissues, and immune cells produced by HSPCs reside in most tissues of the body, enabling them to interact with and potentially influence the inflammatory condition of different tissue-specific microenvironments (Jaiswal and Ebert 2019).
A two-step process has been proposed to explain the role of inflammation in the etiology of preleukemic and/or myelodysplastic disease: First, signaling patterns are altered via disruption to supramolecular organizing centers. This is followed by proinflammatory changes within the bone marrow microenvironment that promote expansion of clonal HSPC populations (Trowbridge and Starczynowski 2021). From an evolutionary perspective, one way that clonal HSPC populations have been shown to gain a competitive advantage over normal HSPCs is through noncanonical activation of the transcription factor NF-κB, which inhibits canonical functions related to innate immunity, inflammation, and communicating signals of pathogen invasion to mobilize cellular resistance (Salminen and Kaarniranta 2010). Inflammation may thus act as a selective pressure that favors the persistence of HSPCs with somatic driver mutations.
The most common CHIP mutations occur in genes with epigenetic regulatory function (Buscarlet et al. 2017): DNMT3A is a methyltransferase involved in de novo cytosine methylation, and TET2 is a methylcytosine dioxygenase that facilitates demethylation by initiating the conversion of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC) preferentially in a CpG context (Zhang et al. 2016; Buscarlet et al. 2017; Zink et al. 2017; Tulstrup et al. 2021). Cell lineages carrying mutations in DNMT3A or TET2 display atypical responses to inflammation: DNMT3A mutant cells show resilience to inflammatory conditions that are expected to cause normal HSPCs to differentiate and lose their self-renewal capacity (SanMiguel et al. 2022), whereas Tet2 mutant cells have shown increased differentiation in the presence proinflammatory cytokines and more rapid myeloproliferation in response to microbial-driven inflammation in mice (Meisel et al. 2018; Köhnke and Majeti 2021; McClatchy et al. 2022).
Why some CHIP clonal variants are adapted to inflammatory environmental conditions remains incompletely understood, although recent work has begun to illuminate more of the evolutionary dynamics that govern the etiology of CHIP. A study by SanMiguel et al. (2022) used a knockout mouse model to identify a CHIP mutation (Dnmt3aR878H/+) whose fitness advantage is mediated by TNFR1, a proinflammatory cytokine receptor for tumor necrosis factor-alpha (TNF-α) (SanMiguel et al. 2022). In humans, TET2 mutations engender dominance of clonal HSPC lineages by facilitating cell-intrinsic resistance to TNF-α while simultaneously perpetuating the inflammatory microenvironmental conditions that select for such mutations (Trowbridge and Starczynowski 2021). Collectively, these lines of evidence support a role for the proinflammatory extrinsic milieu as an evolutionary pressure that selects for mutant HSPCs and facilitates clonal expansion. Inflammation related to CHIP is also expected to be a key contributor to higher risk of cardiovascular and pulmonary disease in this patient population (Jaiswal et al. 2017; Miller and Steensma 2020).
CURRENT APPROACHES TO THE STUDY AND DIAGNOSIS OF CHIP
Although many of the mechanisms contributing to increased cancer risk have been identified and are known to be overlapping with those that underlie age-related decline, what triggers these mechanisms and how they interact to produce variation in health and disease outcomes is not well-understood. Hematologic driver mutations are typically acquired many years before AML or other malignancies may be diagnosed (Jaiswal and Ebert 2019).Whole-exome sequencing (WES) or targeted gene panels have traditionally been used to diagnose CHIP, using the aforementioned criterion of the presence of a hematologic driver mutation with a VAF of at least 2% in apparently healthy individuals (Steensma et al. 2015; Valent et al. 2019).
Technologies like targeted error-corrected sequencing are now available and enable easy detection of clonal mutations that exist at VAFs of 0.1%, and may be sensitive enough to detect mutant VAFs as low as 0.00001% (Young et al. 2016; Hoermann et al. 2020). Although such mutations are not considered to be indicative of CHIP and are common in the aging population, there is interest in elucidating the relationship between prevalent mutations and the molecular mechanisms that help promote or prevent disease (Valent et al. 2019). To fully differentiate premalignant and benign forms of CH and CHIP more specifically, a comprehensive understanding of HSPC genetics and epigenetic dynamics in individuals who never develop malignant or other severe disease would shed additional light on the steps that make up the trajectory that leads (or does not lead) to adverse health outcomes.
Niroula et al. (2021) examined mosaic chromosomal aberrations (mCAs) associated with myeloid and lymphoid driver mutations. Using lineage-specific genetic aberrations in conjunction with complete blood count data, the authors were able to predict risk of developing myeloid malignancies, CLL and SLL. This may be a viable means of identifying individuals with CHIP who are at high risk of malignant conversion in a clinical setting. Utility of such an assay would be limited to known CHIP-associated mCAs and driver mutations that occur above a certain frequency, and would fail to identify individuals carrying rare, high-fitness CHIP variants. Uddin et al. (2022) recently published a cost-effective assay for CHIP detection (single-molecule molecular inversion probe sequencing, or smMIPS) targeting the 11 most commonly affected CHIP genes in addition to four hotspots for recurrent mutations. Results from Robertson et al. (2022) expand on the findings of Uddin et al. (2022), providing finer-grained molecular details on CHIP clonal dynamics. They have proposed a novel algorithmic approach, LiFT, for CHIP detection that is better able to detect relevant variants compared to VAF thresholding at ≥2%. Using LiFT, Robertson et al. (2022) were able to identify high-fitness CHIP mutations existing at very low variant allele frequencies.
Abelson et al. (2018) proposed two models for predicting transition to AML, one based on the somatic mutations in pre-AML and benign ARCH cases, and the other on clinical data commonly stored in electronic health records (e.g., complete blood count data). The timing, number, and type of mutations in CHIP were all relevant factors to disease prognosis, and cases of benign and malignant CHIP have different mutational profiles (for review, see Hoermann et al. 2020). Abelson et al. (2018) identified 95 individuals who went on to develop AML but were sampled (on average 6.3 yr) prior to diagnosis, out of more than 500,000 samples that underwent targeted sequencing from a large European cohort (Sellar et al. 2018). They found that mutations in most hematologic driver genes increase AML risk by approximately twofold for every 5% increase in clone prevalence, and that mutations in TP53 and spliceosome genes such as U2AF1 were associated with especially augmented risk of AML (Abelson et al. 2018). Additionally, mutations in putative driver genes as well as an increased number of mutations were associated with heightened AML risk (Abelson et al. 2018). They further found that red cell distribution width (RDW) was positively associated with elevated risk of AML, and developed a model using blood count data to predict the development of AML between 6 and 12 mo prior to disease diagnosis (Abelson et al. 2018). Although the sensitivity of the blood count–based model was too limited (26%) to be clinically useful in isolation, the study is promising and offers a conceptual foundation for the development of predictive models using novel data types (Sellar et al. 2018).
Epigenetic Mechanisms with Biomarker Potential for Investigating Clonal Hematopoiesis
The recent advancements and declining cost of next-generation sequencing technologies have underscored the potential for integrating new types of NGS data to better understand and monitor the dynamics of CHIP over time. It is important to note that although the costs of NGS are declining, WGS or related genome-wide assays have not been widely available to all patients, and such data may be more useful for research purposes than it is directly to patients at this time, because of the uncertainty that comes with such a clinical diagnosis.
The advent of single-cell, multiomics technologies (such as scNMT-seq [Clark et al. 2018], which generates single-cell nucleosome occupancy, DNA methylation, and gene expression data simultaneously) have further expanded our capacity to more precisely decipher the information content of the epigenome. This offers a novel opportunity to marry mutational signature and timing data (e.g., PCAWG Evolution & Heterogeneity Working Group et al. 2020) with methods that produce data on gene expression (RNA-seq), DNA methylation (whole-genome bisulfite sequencing [WGBS]), chromatin accessibility (e.g., ATAC-seq, MNase-seq), histone modifications and/or transcription factor binding (ChIP-seq, CUT&RUN, CUT&TAG), and distal chromatin looping involving three dimensional changes in chromatin structure and organization (e.g., HI-C, ChIA-PET) (Chawla et al. 2021) to interrogate the molecular effects of different mutational signatures. This will also expand our understanding of how these mutations mechanistically enhance leukemia risk. For example, the dynamic nature of DNA methylation can be used to model patterns of molecular change, the rate at which such changes are occurring, assuming canonical patterns of DNA methylation status and change over time can be established at informative CpG sites (Gabbutt et al. 2022). Lower-density clusters of CpG sites exhibiting intermediate methylation may begin to show a “W”-shaped methylation fraction distribution indicative of increasingly heterogenous methylation among clonal cells (Gabbutt et al. 2022). Monitoring epigenetic activity, such as the degree and pace of methylation change, under CHIP may reveal novel variation indicative of a potentially pathogenic change. Buscarlet et al. (2017) found that the ratio of 5mC to 5hmC is indicative of TET2 zygosity, consistent with the role of TET2 in 5mC demethylation; deviations from the expected ratio could be used as an indicator of potential loss of function mutations in this gene. Finally, allele-specific methylation and allele switching of methylation are both more common in tumor tissue, pointing to other ways that epigenetic modifications may be used to identify pertinent deviations from expectations indicative of increased disease risk (Do et al. 2020).
FUTURE DIRECTIONS
The cost of whole-genome sequencing (WGS) has historically been prohibitive to its use in clinical practice, with some exceptions. However, as the price of WGS continues its rapid descent and the “<$100 genome” moves closer to reality, WGS may soon be a feasible option for clinical screening, monitoring, and prevention for a much wider array of individuals.
However, although WGS and NGS technologies in general have rapidly declined in cost over the past several years, these may not be the optimal choice for CHIP screening, because of the unknown risk conferred by many low-frequency variants that may be detectable but not particularly useful to clinicians or their patients, as well as the inability to separate the premalignant cases from those that are benign or increase risk of nonmalignant disease with high certainty. Such knowledge is, again, essential to avoiding over- and undertreatment and achieving the best cost/benefit ratio for patients to help them maintain their quality of life (Strom 2016).
Nonetheless, substantial advances in the understanding of hematological malignancies have been made in recent years. Bick et al. (2020) recently published on characteristics of three specific subgroups of mutations that tend to malfunction under CHIP in different ways (e.g., genes that acquire mutations due to loss of function in genes that maintain genomic integrity vs. those implicated in up-regulated self-renewal capacity). Further characterization of the nuances of this CHIP and its potential molecular subtypes should pave the way for enhanced understanding of shared and unique features of CHIP etiology that may lead to more informed and efficacious treatment and monitoring protocols.
Epigenetic analyses aimed at detecting or monitoring CHIP and risk of transition to malignancy will benefit substantially from using single-cell sequencing whenever possible, given the confounding influence of cell type composition in bulk NGS analyses of epigenetic data (Black and McGranahan 2021). Use of mutational signatures associated with specific types of cancer from genomic studies in conjunction with transcriptomic and epigenomic modalities at relevant loci may elucidate how different mutations can mechanistically affect dynamic and regulatory activity involved in cancer development (e.g., Ocsenas and Reimand 2022). The association between CHIP and nonmalignant, noncardiovascular disease also warrants further investigation, as intriguing relationships have been identified between CHIP mutations and insulin resistance (Fuster et al. 2020), as well as a surprising, potentially protective effect of CHIP against Alzheimer's disease (Bouzid et al. 2021). Future analyses should aim to further identify and connect cellular phenotypes that promote clonal expansion to underlying genetic and epigenetic variation to build a comprehensive portrait of CHIP dynamics, aging, and cancer risk.
An outstanding challenge in this field is the uncertainty surrounding what a diagnosis of CHIP ultimately means for an otherwise healthy patient. It is unknown whether CH plays a consequential or even causal role in age-related functional and systemic decline in humans after age 70. Both clinicians and their patients must weigh the costs and benefits of having such knowledge and the option for intervention with the degree of uncertainty inherent to a CHIP diagnosis and the generally low risk of transition to malignancy. This is why personalized approaches that consider a person's age, medical history, and any other relevant health factors are the best path forward. As research in this field continues to progress, we expect to see additional longitudinal studies that simultaneously track genetic and epigenetic changes in individuals over time, which will reveal more about the complex and heterogenous dynamics of this phenomenon.
ADDITIONAL INFORMATION
Ethics Statement
No patients or patient data were involved, accessed, or analyzed for the purpose of composing this review article.
Acknowledgments
We thank the reviewers for their thoughtful and constructive feedback.
Competing Interest Statement
P.T.S. consults for Natera, Twinstrand, Laboratory Corporation of America, Foresight Diagnostics, and Foundation Medicine.
Referees
Robert Bowman
Rafael Bejar
REFERENCES
- Abelson S, Collord G, Ng SWK, Weissbrod O, Mendelson Cohen N, Niemeyer E, Barda N, Zuzarte PC, Heisler L, Sundaravadanam Y, et al. 2018. Prediction of acute myeloid leukaemia risk in healthy individuals. Nature 559: 400–404. 10.1038/s41586-018-0317-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acuna-Hidalgo R, Sengul H, Steehouwer M, van de Vorst M, Vermeulen SH, Kiemeney LALM, Veltman JA, Gilissen C, Hoischen A. 2017. Ultra-sensitive sequencing identifies high prevalence of clonal hematopoiesis-associated mutations throughout adult life. Am J Hum Genet 101: 50–64. 10.1016/j.ajhg.2017.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov LB, Kim J, Haradhvala NJ, Huang MN, Tian Ng AW, Wu Y, Boot A, Covington KR, Gordenin DA, Bergstrom EN, et al. 2020. The repertoire of mutational signatures in human cancer. Nature 578: 94–101. 10.1038/s41586-020-1943-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutler E, Yeh M, Fairbanks VF. 1962. The normal human female as a mosaic of X-chromosome activity: studies using the gene for C-6-PD-deficiency as a marker. Proc Natl Acad Sci 48: 9–16. 10.1073/pnas.48.1.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bick AG, Weinstock JS, Nandakumar SK, Fulco CP, Bao EL, Zekavat SM, Szeto MD, Liao X, Leventhal MJ, Nasser J, et al. 2020. Inherited causes of clonal haematopoiesis in 97,691 whole genomes. Nature 586: 763–768. 10.1038/s41586-020-2819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black JRM, McGranahan N. 2021. Genetic and non-genetic clonal diversity in cancer evolution. Nat Rev Cancer 21: 379–392. 10.1038/s41568-021-00336-2 [DOI] [PubMed] [Google Scholar]
- Bolton KL, Ptashkin RN, Gao T, Braunstein L, Devlin SM, Kelly D, Patel M, Berthon A, Syed A, Yabe M, et al. 2019. Oncologic therapy shapes the fitness landscape of clonal hematopoiesis. Nat Genet 52: 1219–1226. 10.1038/s41588-020-00710-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouzid H, Belk JA, Jan M, Qi Y, Sarnowski C, Wirth S, Ma L, Chrostek M, Ahmad H, Nachun D, et al. 2021. Clonal hematopoiesis is associated with protection from Alzheimer's disease. Hematology 10.1101/2021.12.10.21267552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buscarlet M, Provost S, Zada YF, Barhdadi A, Bourgoin V, Lépine G, Mollica L, Szuber N, Dubé M-P, Busque L. 2017. DNMT3A and TET2 dominate clonal hematopoiesis and demonstrate benign phenotypes and different genetic predispositions. Blood 130: 753–762. 10.1182/blood-2017-04-777029 [DOI] [PubMed] [Google Scholar]
- Chawla A, Nagy C, Turecki G. 2021. Chromatin profiling techniques: exploring the chromatin environment and its contributions to complex traits. Int J Mol Sci 22: 7612. 10.3390/ijms22147612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SJ, Argelaguet R, Kapourani C-A, Stubbs TM, Lee HJ, Alda-Catalinas C, Krueger F, Sanguinetti G, Kelsey G, Marioni JC, et al. 2018. scNMT-seq enables joint profiling of chromatin accessibility DNA methylation and transcription in single cells. Nat Commun 9: 781. 10.1038/s41467-018-03149-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis A, Gao R, Navin N. 2017. Tumor evolution: linear, branching, neutral or punctuated? Biochim Biophys Acta 1867: 151–161. 10.1016/j.bbcan.2017.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan G, Lazare SS. 2018. Aging of hematopoietic stem cells. Blood 131: 479–487. 10.1182/blood-2017-06-746412 [DOI] [PubMed] [Google Scholar]
- Desai P, Mencia-Trinchant N, Savenkov O, Simon MS, Cheang G, Lee S, Samuel M, Ritchie EK, Guzman ML, Ballman KV, et al. 2018. Somatic mutations precede acute myeloid leukemia years before diagnosis. Nat Med 24: 1015–1023. 10.1038/s41591-018-0081-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do C, Dumont ELP, Salas M, Castano A, Mujahed H, Maldonado L, Singh A, DaSilva-Arnold SC, Bhagat G, Lehman S, et al. 2020. Allele-specific DNA methylation is increased in cancers and its dense mapping in normal plus neoplastic cells increases the yield of disease-associated regulatory SNPs. Genome Biol 21: 153. 10.1186/s13059-020-02059-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enderling H, Wolkenhauer O. 2021. Are all models wrong? Comp Sys Onco 1: e1008. 10.1002/cso2.1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre MA, de Almeida JG, Fiorillo E, Mitchell E, Damaskou A, Rak J, Orrù V, Marongiu M, Chapman MS, Vijayabaskar MS, et al. 2022. The longitudinal dynamics and natural history of clonal haematopoiesis. Nature 606: 335–342. 10.1038/s41586-022-04785-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidler IJ, Gersten DM, Hart IR. 1978. The biology of cancer invasion and metastasis. Adv Cancer Res 28: 149–250. 10.1016/s0065-230x(08)60648-x [DOI] [PubMed] [Google Scholar]
- Franceschi C, Garagnani P, Parini P, Giuliani C, Santoro A. 2018. Inflammaging: a new immune–metabolic viewpoint for age-related diseases. Nat Rev Endocrinol 14: 576–590. 10.1038/s41574-018-0059-4 [DOI] [PubMed] [Google Scholar]
- Fuster JJ, Zuriaga MA, Zorita V, MacLauchlan S, Polackal MN, Viana-Huete V, Ferrer-Pérez A, Matesanz N, Herrero-Cervera A, Sano S, et al. 2020. TET2-loss-of-function-driven clonal hematopoiesis exacerbates experimental insulin resistance in aging and obesity. Cell Rep 33: 108326. 10.1016/j.celrep.2020.108326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabbutt C, Schenck RO, Weisenberger DJ, Kimberley C, Berner A, Househam J, Lakatos E, Robertson-Tessi M, Martin I, Patel R, et al. 2022. Fluctuating methylation clocks for cell lineage tracing at high temporal resolution in human tissues. Nat Biotechnol 40: 720–730. 10.1038/s41587-021-01109-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao T, Ptashkin R, Bolton KL, Sirenko M, Fong C, Spitzer B, Menghrajani K, Ossa JEA, Zhou Y, Bernard E, et al. 2021. Interplay between chromosomal alterations and gene mutations shapes the evolutionary trajectory of clonal hematopoiesis. Nat Commun 12: 338. 10.1038/s41467-020-20565-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovese G, Kähler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF, Chambert K, Mick E, Neale BM, Fromer M, et al. 2014. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med 371: 2477–2487. 10.1056/NEJMoa1409405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoermann G, Greiner G, Griesmacher A, Valent P. 2020. Clonal hematopoiesis of indeterminate potential: a multidisciplinary challenge in personalized hematology. J Pers Med 10: 94. 10.3390/jpm10030094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou H-A, Liu C-Y, Kuo Y-Y, Chou W-C, Tsai C-H, Lin C-C, Lin L-I, Tseng M-H, Chiang Y-C, Liu M-C, et al. 2016. Splicing factor mutations predict poor prognosis in patients with de novo acute myeloid leukemia. Oncotarget 7: 9084–9101. 10.18632/oncotarget.7000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal S. 2020. Clonal hematopoiesis and non-hematologic disorders. Blood 136: 1606–1614. 10.1182/blood.2019000989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal S, Ebert BL. 2019. Clonal hematopoiesis in human aging and disease. Science 366: eaan4673. 10.1126/science.aan4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, Lindsley RC, Mermel CH, Burtt N, Chavez A, et al. 2014. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med 371: 2488–2498. 10.1056/NEJMoa1408617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal S, Natarajan P, Silver AJ, Gibson CJ, Bick AG, Shvartz E, McConkey M, Gupta N, Gabriel S, Ardissino D, et al. 2017. Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N Engl J Med 377: 111–121. 10.1056/NEJMoa1701719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar SP, Quiros PM, Gu M, Jiang T, Mitchell J, Langdon R, Iyer V, Barcena C, Vijayabaskar MS, Fabre MA, et al. 2022. Genome-wide analyses of 200,453 individuals yield new insights into the causes and consequences of clonal hematopoiesis. Nat Genet 54: 1155–1166. 10.1038/s41588-022-01121-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudson AG. 1971. Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci 68: 820–823. 10.1073/pnas.68.4.820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhnke T, Majeti R. 2021. Clonal hematopoiesis: from mechanisms to clinical intervention. Cancer Discov 11: 2987–2997. 10.1158/2159-8290.CD-21-0901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo M. 2010. Lymphoid and myeloid lineage commitment in multipotent hematopoietic progenitors: roles of bone marrow microenvironment. Immunol Rev 238: 37–46. 10.1111/j.1600-065X.2010.00963.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landi MT, Synnott NC, Rosenbaum J, Zhang T, Zhu B, Shi J, Zhao W, Kebede M, Sang J, Choi J, et al. 2021. Tracing lung cancer risk factors through mutational signatures in never-smokers. Am J Epidemiol 190: 962–976. 10.1093/aje/kwaa234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon MF. 1962. Sex chromatin and gene action in the mammalian X-chromosome. Am J Hum Genet 14: 135–148. [PMC free article] [PubMed] [Google Scholar]
- Martincorena I, Roshan A, Gerstung M, Ellis P, Van Loo P, McLaren S, Wedge DC, Fullam A, Alexandrov LB, Tubio JM, et al. 2015. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 348: 880–886. 10.1126/science.aaa6806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martincorena I, Fowler JC, Wabik A, Lawson ARJ, Abascal F, Hall MWJ, Cagan A, Murai K, Mahbubani K, Stratton MR, et al. 2018. Somatic mutant clones colonize the human esophagus with age. Science 362: 911–917. 10.1126/science.aau3879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClatchy J, Strogantsev R, Wolfe E, Estabrook J, Lin H, Mohammadhosseini M, Davis BA, Eden C, Goldman D, Fleming WH, et al. 2022. The transcriptional and epigenetic reprogramming mediated by chronic IL1β exposure drives self-renewal ability and myeloid priming in TET2 deficient stem and progenitor cells. Cancer Biol 10.1101/2022.09.15.507688 [DOI] [Google Scholar]
- Meisel M, Hinterleitner R, Pacis A, Chen L, Earley ZM, Mayassi T, Pierre JF, Ernest JD, Galipeau HJ, Thuille N, et al. 2018. Microbial signals drive pre-leukaemic myeloproliferation in a Tet2-deficient host. Nature 557: 580–584. 10.1038/s41586-018-0125-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller PG, Steensma DP. 2020. Implications of Clonal Hematopoiesis for Precision Oncology. JCO Precis Oncol 4: 639–646. 10.1200/PO.20.00144 [DOI] [PubMed] [Google Scholar]
- Miller PG, Qiao D, Rojas-Quintero J, Honigberg MC, Sperling AS, Gibson CJ, Bick AG, Niroula A, McConkey ME, Sandoval B, et al. 2022. Association of clonal hematopoiesis with chronic obstructive pulmonary disease. Blood 139: 357–368. 10.1182/blood.2021013531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell E, Spencer Chapman M, Williams N, Dawson KJ, Mende N, Calderbank EF, Jung H, Mitchell T, Coorens THH, Spencer DH, et al. 2022. Clonal dynamics of haematopoiesis across the human lifespan. Nature 606: 343–350. 10.1038/s41586-022-04786-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore L, Leongamornlert D, Coorens THH, Sanders MA, Ellis P, Dentro SC, Dawson KJ, Butler T, Rahbari R, Mitchell TJ, et al. 2020. The mutational landscape of normal human endometrial epithelium. Nature 580: 640–646. 10.1038/s41586-020-2214-z [DOI] [PubMed] [Google Scholar]
- Ní Leathlobhair M, Lenski RE. 2022. Population genetics of clonally transmissible cancers. Nat Ecol Evol 6: 1077–1089. 10.1038/s41559-022-01790-3 [DOI] [PubMed] [Google Scholar]
- Niroula A, Sekar A, Murakami MA, Trinder M, Agrawal M, Wong WJ, Bick AG, Uddin MM, Gibson CJ, Griffin GK, et al. 2021. Distinction of lymphoid and myeloid clonal hematopoiesis. Nat Med 27: 1921–1927. 10.1038/s41591-021-01521-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocsenas O, Reimand J. 2022. Chromatin accessibility of primary human cancers ties regional mutational processes and signatures with tissues of origin. PLoS Comput Biol 18: e1010393. 10.1371/journal.pcbi.1010393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- PCAWG Evolution & Heterogeneity Working Group, PCAWG Consortium, Gerstung M, Jolly C, Leshchiner I, Dentro SC, Gonzalez S, Rosebrock D, Mitchell TJ, Rubanova Y, et al. 2020. The evolutionary history of 2,658 cancers. Nature 578: 122–128. 10.1038/s41586-019-1907-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson NA, Latorre-Crespo E, Terradas-Terradas M, Lemos-Portela J, Purcell AC, Livesey BJ, Hillary RF, Murphy L, Fawkes A, MacGillivray L, et al. 2022. Longitudinal dynamics of clonal hematopoiesis identifies gene-specific fitness effects. Nat Med 28: 1439–1446. 10.1038/s41591-022-01883-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salminen A, Kaarniranta K. 2010. Genetics vs. entropy: longevity factors suppress the NF-κB-driven entropic aging process. Ageing Res Rev 9: 298–314. 10.1016/j.arr.2009.11.001 [DOI] [PubMed] [Google Scholar]
- SanMiguel JM, Eudy E, Loberg MA, Young KA, Mistry JJ, Mujica KD, Schwartz LS, Stearns TM, Challen GA, Trowbridge JJ. 2022. Distinct tumor necrosis factor alpha receptors dictate stem cell fitness versus lineage output in Dnmt3a-mutant clonal hematopoiesis. Cancer Discov 12: 2763–2773. 10.1158/2159-8290.CD-22-0086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellar RS, Jaiswal S, Ebert BL. 2018. Predicting progression to AML. Nat Med 24: 904–906. 10.1038/s41591-018-0114-7 [DOI] [PubMed] [Google Scholar]
- Steensma DP, Ebert BL. 2020. Clonal hematopoiesis as a model for premalignant changes during aging. Exp Hematol 83: 48–56. 10.1016/j.exphem.2019.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steensma DP, Bejar R, Jaiswal S, Lindsley RC, Sekeres MA, Hasserjian RP, Ebert BL. 2015. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood 126: 9–16. 10.1182/blood-2015-03-631747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone RM, O'Donnell MR, Sekeres MA. 2004. Acute myeloid leukemia. Hematology 2004: 98–117. 10.1182/asheducation-2004.1.98 [DOI] [PubMed] [Google Scholar]
- Strom SP. 2016. Current practices and guidelines for clinical next-generation sequencing oncology testing. Cancer Biol Med 13: 3–11. 10.20892/j.issn.2095-3941.2016.0004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suda K, Nakaoka H, Yoshihara K, Ishiguro T, Tamura R, Mori Y, Yamawaki K, Adachi S, Takahashi T, Kase H, et al. 2018. Clonal expansion and diversification of cancer-associated mutations in endometriosis and normal endometrium. Cell Rep 24: 1777–1789. 10.1016/j.celrep.2018.07.037 [DOI] [PubMed] [Google Scholar]
- Trowbridge JJ, Starczynowski DT. 2021. Innate immune pathways and inflammation in hematopoietic aging, clonal hematopoiesis, and MDS. J Exp Med 218: e20201544. 10.1084/jem.20201544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker T, Friedman J. 2002. Pathogenesis of hereditary tumors: beyond the “two-hit” hypothesis: pathogenesis of hereditary tumors. Clin Genet 62: 345–357. 10.1034/j.1399-0004.2002.620501.x [DOI] [PubMed] [Google Scholar]
- Tulstrup M, Soerensen M, Hansen JW, Gillberg L, Needhamsen M, Kaastrup K, Helin K, Christensen K, Weischenfeldt J, Grønbæk K. 2021. TET2 mutations are associated with hypermethylation at key regulatory enhancers in normal and malignant hematopoiesis. Nat Commun 12: 6061. 10.1038/s41467-021-26093-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin MM, Nguyen NQ, Yu B, Brody J, Pampana A, Nakao T, Fornage M, Bressler J, Sotoodehnia N, Weinstock J. 2022. Clonal hematopoiesis of indeterminate potential, DNA methylation, and risk for coronary artery disease. Nat Commun 13: 5350. 10.1038/s41467-022-33093-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valent P, Orazi A, Steensma DP, Ebert BL, Haase D, Malcovati L, van de Loosdrecht AA, Haferlach T, Westers TM, Wells DA, et al. 2017. Proposed minimal diagnostic criteria for myelodysplastic syndromes (MDS) and potential pre-MDS conditions. Oncotarget 8: 73483–73500. 10.18632/oncotarget.19008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valent P, Kern W, Hoermann G, Milosevic Feenstra J, Sotlar K, Pfeilstöcker M, Germing U, Sperr W, Reiter A, Wolf D, et al. 2019. Clonal hematopoiesis with oncogenic potential (CHOP): separation from CHIP and roads to AML. Int J Mol Sci 20: 789. 10.3390/ijms20030789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson CJ, Papula AL, Poon GYP, Wong WH, Young AL, Druley TE, Fisher DS, Blundell JR. 2020. The evolutionary dynamics and fitness landscape of clonal hematopoiesis. Science 367: 1449–1454. 10.1126/science.aay9333 [DOI] [PubMed] [Google Scholar]
- Wick G, Jansen-Dürr P, Berger P, Blasko I, Grubeck-Loebenstein B. 2000. Diseases of aging. Vaccine 18: 1567–1583. 10.1016/S0264-410X(99)00489-2 [DOI] [PubMed] [Google Scholar]
- Xia S, Zhang X, Zheng S, Khanabdali R, Kalionis B, Wu J, Wan W, Tai X. 2016. An update on inflamm-aging: mechanisms, prevention, and treatment. J Immunol Res 2016: 1–12. 10.1155/2016/8426874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, Lu C, Wang J, McLellan MD, Johnson KJ, Wendl MC, McMichael JF, Schmidt HK, Yellapantula V, Miller CA, et al. 2014. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med 20: 1472–1478. 10.1038/nm.3733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Gowers KHC, Lee-Six H, Chandrasekharan DP, Coorens T, Maughan EF, Beal K, Menzies A, Millar FR, Anderson E, et al. 2020. Tobacco smoking and somatic mutations in human bronchial epithelium. Nature 578: 266–272. 10.1038/s41586-020-1961-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young AL, Challen GA, Birmann BM, Druley TE. 2016. Clonal haematopoiesis harbouring AML-associated mutations is ubiquitous in healthy adults. Nat Commun 7: 12484. 10.1038/ncomms12484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Su J, Jeong M, Ko M, Huang Y, Park HJ, Guzman A, Lei Y, Huang Y-H, Rao A, et al. 2016. DNMT3A and TET2 compete and cooperate to repress lineage-specific transcription factors in hematopoietic stem cells. Nat Genet 48: 1014. 10.1038/ng.3610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink F, Stacey SN, Norddahl GL, Frigge ML, Magnusson OT, Jonsdottir I, Thorgeirsson TE, Sigurdsson A, Gudjonsson SA, Gudmundsson J, et al. 2017. Clonal hematopoiesis, with and without candidate driver mutations, is common in the elderly. Blood 130: 742–752. 10.1182/blood-2017-02-769869 [DOI] [PMC free article] [PubMed] [Google Scholar]