Abstract
Alterations in epigenetic regulators are increasingly recognized as early events in tumorigenesis; thus, patients with acquired or inherited variants in epigenetic regulators may be at increased risk for developing multiple types of cancer. DNMT3A overgrowth syndrome (DOS), caused by germline pathogenic variants in the DNA methyltransferase gene DNMT3A, has been associated with a predisposition toward development of hematopoietic and neuronal malignancies. DNMT3A deficiency has been described to promote keratinocyte proliferation in mice. Although altered DNA methylation patterns are well-recognized in melanoma, the role of DNA methyltransferases in melanoma pathogenesis is not clear. We report the case of an adult DOS patient with a germline DNMT3A loss-of-function mutation, who developed an early-onset melanoma with regional lymph node metastatic disease. Exome sequencing of the primary tumor identified an additional acquired, missense DNMT3A mutation in the dominant tumor clone, suggesting that the loss of DNMT3A function was relevant for the development of this tumor.
Keywords: cutaneous melanoma, overgrowth
INTRODUCTION
DNA methylation is an epigenetic process that is established by the de novo DNA methyltransferases DNMT3A and DNMT3B, and maintained by DNMT1 (Li et al. 1992; Okano et al. 1998). DNMT3A overgrowth syndrome (MIM #615879) consists of a constellation of clinical manifestations with three principal features, including overgrowth (tall stature, increased head circumference, elevated body mass index), impaired intellectual development, and characteristic facial features, and is associated with de novo germline heterozygous mutations in the DNA methyltransferase gene DNMT3A (Tatton-Brown et al. 2014). Since its first description, the number of patients with documented DNMT3A overgrowth syndrome (DOS) has grown to more than 300 at present, whereas the repertoire of clinical manifestations has expanded to include obesity, cardiac defects, umbilical hernia, hypotonia, joint hypermobility, seizures, behavioral disorders, and other phenotypes (Hollink et al. 2017; Kosaki et al. 2017; Shen et al. 2017; Xin et al. 2017; Tatton-Brown et al. 2018; Jeffries et al. 2019; Balci et al. 2020; Ferris et al. 2021; Smith et al. 2021; Cecchi et al. 2022).
In addition to its syndromic features, DOS patients are at higher risk for certain malignancies, including hematologic malignancies (Hollink et al. 2017; Smith et al. 2021). Clonal hematopoiesis is caused most frequently by somatic mutations in DNMT3A, which are also a common feature of normal karyotype acute myeloid leukemia (AML) (Ley et al. 2010, 2013; Jaiswal et al. 2014; Xie et al. 2014; Genovese et al. 2015). Protein altering mutations at arginine 882 typically confer dominant-negative DNA methyltransferase activity, resulting in focal, canonical DNA hypomethylation patterns in DOS patient blood cells and in AML cells (Ley et al. 2010; Kim et al. 2013; Russler-Germain et al. 2014; Smith et al. 2021). DNMT3A haploinsufficiency has been shown to lead to a milder DNA hypomethylation phenotype, and corresponding preclinical models are predisposed to develop spontaneous and oncogene-induced leukemia (Meyer et al. 2016; Cole et al. 2017). Although other tumor types have been described in DOS patients, skin cancer has not yet been reported. We present the first known case of a DOS patient who developed early-onset melanoma, associated with a second acquired DNMT3A mutation, suggesting a role for loss-of-function DNMT3A mutations in melanoma initiation.
RESULTS
Clinical Presentation and Family History
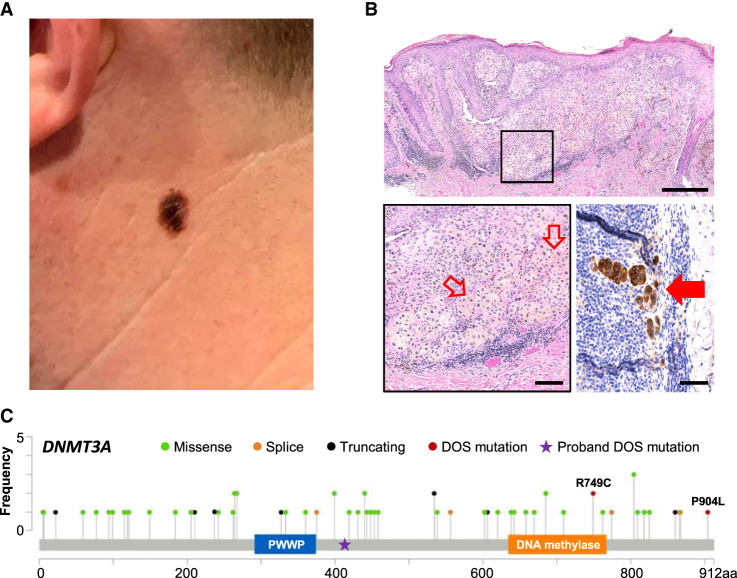
The proband is a male diagnosed with DNMT3A overgrowth syndrome, with a de novo heterozygous insertion in the DNMT3A gene (c.1238dupG) resulting in DNMT3AF414fs*; peripheral blood cells from this patient had been analyzed in a prior report demonstrating a haploinsufficiency phenotype for DNA methylation (Smith et al. 2021). He is the second child of four, and his three siblings are unaffected. He has mild developmental delay and attention deficit hyperactivity disorder, as well as aortic root dilatation. At 20 yr of age, he developed a dermatofibrosarcoma protuberans on the lower back that was surgically resected. At 34 yr of age, he presented with a growing, pigmented skin lesion on his left neck (Fig. 1A). A punch biopsy through one part of the lesion revealed a compound melanocytic proliferation composed of severely atypical epithelioid cells arranged as single cells and nests at all levels of the epidermis and as dermal nests that lack maturation with tumor depth. Ulceration is not present, and mitoses number up to 2/mm2. These findings are compatible with malignant melanoma, invasive to a thickness of 1.3 mm (Fig. 1B, top and bottom left panels). Additional histologic levels reveal an associated nevus. Wide local excision with sentinel lymph node biopsy revealed metastatic melanoma in two out of three lymph nodes sampled in the left parotid tail without extracapsular extension (Fig. 1B, bottom right panel), whereas positron emission tomography-computed tomography (PET-CT) and brain magnetic resonance imaging (MRI) were negative for overt metastatic disease, consistent with stage IIIA (pT2aN2aM0) disease. He then received adjuvant immune checkpoint blockade with pembrolizumab and has more than 8 mo of follow-up without evidence of recurrent or metastatic disease.
Figure 1.
Invasive melanoma in a DNMT3A overgrowth syndrome (DOS) patient. (A) Clinical appearance of a thin plaque with irregular borders and heterogeneous pigmentation on the left lateral neck. (B) Hematoxylin and eosin (H&E)-stained (top) punch biopsy specimen consisting of atypical melanocytes with at the dermal–epidermal junction, as well as pagetoid spread and an invasive dermal component, measuring 1.3 mm in thickness (scale = 250 µm) with higher magnification of the invasive component with a mitotic cell highlighted by hollow red arrows (scale = 100 µm; boxed inset and lower left panel) and MART1 stain highlighting metastatic melanoma (solid red arrow) in the sentinel lymph node (scale = 50 µm; bottom right). Sequencing was performed on the other half of this bisected specimen. (C) DNMT3A variants in melanoma curated from cBioPortal (Cerami et al. 2012; Gao et al. 2013). Proband F414fs* germline mutation is notated with a star, whereas the acquired DNMT3AR749C melanoma mutation is additionally a mutation previously described in DOS patients.
Genomic Analyses
We performed whole-exome sequencing using genomic DNA derived from the biopsy sample of the primary melanoma, which redemonstrated the patient's constitutional DNMT3Ac.1238dupG mutation with a variant allele frequency (VAF) of 0.5. Tumor-specific, somatically acquired variants were defined by comparison to a previously obtained whole-exome data set from the peripheral blood of the same patient several years before (Smith et al. 2021). This analysis identified an acquired BRAFc.1799_1800delinsAC mutation corresponding to the BRAFV600D, which is rarely found in melanoma (0.4%; Greaves et al. 2013) compared to the canonical melanoma oncogene BRAFV600E, found in >50% of cutaneous melanomas (Cancer Genome Atlas Network et al. 2015). Importantly, we also detected an acquired missense variant in DNMT3Ac.2245C > T causing an arginine to cysteine change at amino acid 749 (R749C), which is known to cause methyltransferase loss-of-function (Christian et al. 2020) and which is associated with DOS (Tatton-Brown et al. 2018). The BRAFV600D and DNMT3AR749C mutations both had a VAF of 0.12, suggesting that 24% of the cells in the sample contained these two mutations; this frequency is consistent with the tumor burden in this sample (∼25% of total cells, see Fig. 1B), suggesting that both mutations were in the dominant melanoma clone in the skin lesion. NF1 and NRAS did not harbor somatic mutations. Interestingly, we also detected a somatic GNAQR183Q mutation (previously described in uveal melanomas) in the dominant clone (Table 1; Robertson et al. 2017).
Table 1.
Selected melanoma tumor somatic invariants identified by exome sequencing
| Gene | Chromosome | VAF | HGVS DNA reference | HGVS protein reference | Variant type | Predicted effect |
|---|---|---|---|---|---|---|
| BRAF | 7 | 0.124 | ENST00000646891.1:c.1799_1800delinsAC | ENSP00000493543.1:p.Val600Asp | Deletion-insertion | Missense |
| DNMT3A | 2 | 0.121 | ENST00000264709.7:c.2245C>T | ENSP00000264709.3:p.Arg749Cys | Substitution | Missense |
| GNAQ | 9 | 0.163 | ENST00000286548.8:c.548G>A | ENSP00000286548.4:p.Arg183Gln | Substitution | Missense |
(VAF) Variant allele frequency, (HGVS) Human Genome Variation Society.
DISCUSSION
Multiple cancers exhibit altered DNA methylation patterns, typified by global DNA hypomethylation and focal hypermethylation (Jones and Baylin 2007; Ehrlich 2009). In melanoma, DNA methylation is thought to play a complex role in regulating the expression of canonical tumor suppressor genes, as well as genes that modulate homeostatic processes including cellular proliferation, metabolism, and immune response (Micevic et al. 2017). However, the contributions of individual DNA methyltransferases to skin cancer pathogenesis are incompletely understood. In preclinical studies, Dnmt3b deficiency was shown to prevent melanoma development in Braf- and Pten-deficient murine melanocytes through modulation of the mTOR pathway (Micevic et al. 2016). Studies in the B16 murine melanoma line, on the other hand, suggested a requirement for functional Dnmt3a in melanoma survival and proliferative capacity (Deng et al. 2009; Kim et al. 2018a).
In human melanomas, somatic DNMT3A variants have been detected in 3.5% (65/1849) of the sequenced melanoma samples in cBioportal (Berger et al. 2012; Cerami et al. 2012; Hodis et al. 2012; Krauthammer et al. 2012; Gao et al. 2013; Snyder et al. 2014; Van Allen et al. 2014, 2015; Cancer Genome Atlas Network et al. 2015; Hugo et al. 2016; Catalanotti et al. 2017; Liu et al. 2019; Shoushtari et al. 2021), which are distributed throughout the gene; they include the R749C and P904L variants described in DOS patients (Fig. 1C; Tatton-Brown et al. 2018), as well as multiple nonsense mutations predicted to cause early termination prior to the methyltransferase domain. In the present case, the proband has a germline, truncated, loss-of-function mutation in DNMT3A. Methylation analysis of his peripheral blood cells revealed a hypomethylation phenotype that was consistent with that of DNMT3A haploinsufficiency (Smith et al. 2021). The acquired missense DNMT3A mutation in the dominant clone (which is also known to cause loss of function) suggests a strong selective pressure for near-total DNMT3A loss of function that probably represented the initiating event for this tumor. Interestingly, there was a notable absence of the canonical melanoma BRAF/NRAS/NF1 driver mutations; instead, we detected a noncanonical BRAFV600D mutation that, although rare in melanoma (0.4% of cases), retains sensitivity to dabrafenib kinase inhibition in cell lines, implicating its role as an oncogenic driver (Gentilcore et al. 2013; Greaves et al. 2013). In the present case, we also identified a GNAQR138Q variant that has been previously described in uveal melanoma and that exhibits activation of the MAPK pathway in vitro, although not as strongly as the more common GNAQR209 variants (Shirley et al. 2013). The G protein α-subunit GNAQ is mutated in 46% of uveal melanomas and 84% of blue nevi samples (van Raamsdonk et al. 2009) and is less frequently (2.4%) altered in cutaneous melanoma, suggesting that DNMT3A loss of function may be more impactful for melanoma development in certain contexts.
Most cases of DOS-associated malignancies have been described in the pediatric and young adult populations (Hollink et al. 2017; Tatton-Brown et al. 2018; Ferris et al. 2021). Melanoma is typically a disease diagnosed in later life, with a median age of 65 yr at diagnosis (SEER21). Melanoma in males in their fourth decade accounted for 2.8% of all cases of invasive melanomas diagnosed between 2001 and 2015; in this limited group, only a small fraction (6.2%) of patients exhibited metastatic disease to regional lymph nodes (Paulson et al. 2020). It is intriguing that the proband had previously developed a rare skin cancer at age 25—dermatofibrosarcoma protuberans—a cutaneous sarcoma with an estimated incidence of 4.2 cases per million in the United States, and a peak incidence in the fourth and fifth decades of life (Criscione and Weinstock 2007). Although the development of two uncommon cancers is not direct evidence that DNMT3A haploinsufficiency generates a premalignant state for skin tumors, preclinical models have suggested that Dnmt3a deficiency causes proliferative priming as a potential mechanism (Rinaldi et al. 2017; Chen et al. 2021). These findings suggest that dermatologic surveillance and preventative measures (counseling UV protection), may be an important part of comprehensive care for patients with DOS.
Finally, an important consideration in DOS and other constitutive disorders is that the molecular alterations that promote tumorigenesis in these patients may also have systemic implications that have a bearing on therapeutic response. There are two adjuvant therapies currently in use for resected stage III melanoma—immunotherapy versus targeted therapy for BRAFV600E-mutated melanoma (Long et al. 2017; Eggermont et al. 2018). Preclinical studies demonstrate that Dnmt3a-dependent de novo DNA methylation reduces the efficacy of PD1 checkpoint blockade by promoting CD8 T-cell exhaustion, whereas Dnmt3a inhibition can aid in T-cell rejuvenation (Ghoneim et al. 2017). Although the role of checkpoint blockade in tumors occurring in DOS patients is currently is unknown, DNMT3A haploinsufficiency could potentially affect its efficacy.
METHODS
Genome Sequencing and Data Analysis
A tissue core was obtained from paraffin-embedded tissue from the primary melanoma tumor biopsy specimen, using a 1-mm punch biopsy tool. Genomic DNA was extracted, and sequencing libraries were generated with IDT xGen exome kit version 1, and then sequenced on the Illumina Novaseq 6000 platform using an S4 flow cell. Sequence data was aligned against reference sequence hg38 using BWA-MEM (Li 2013). The aligned reads were sorted, deduplicated, and run through base quality score recalibration (BQSR). Sequence variants were called against reference blood from this patient, which was sequenced in a prior report (UPN 228211; Smith et al. 2021). Structural variants (SVs) and large indels were detected using Manta (Chen et al. 2016). Single-nucleotide variants (SNVs) and small indels were detected using VarScan2 (Koboldt et al. 2012), Strelka2 (Kim et al. 2018b), and MuTect2 (Cibulskis et al. 2013). Variants with a population frequency in gnomAD of >0.1% were removed, as were those in regions of low-quality mapping and low coverage (<20×). Variant annotation was performed with the Variant Effect Predictor, version 95 (McLaren et al. 2016). The entire somatic pipeline is available as a CWL workflow at https://github.com/genome/analysis-workflows (CommitID: 3e653e78fea91cf9c487534ceca1db328b6b68e0; commitURL: https://github.com/genome/analysis-workflows/commit/3e653e78fea91cf9c487534ceca1db328b6b68e0).
ADDITIONAL INFORMATION
Data Deposition and Access
Sequencing data for this patient was deposited to dbGaP (phs000159). Tha variants were submitted to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) and can be found under accession numbers SCV003914810–SCV003914813 for NM_022552.5:c.1238dupG, NM_004333.6:c.1799_1800delinsAC, NM_022552.5:c.2245C>T, and NM_002072.4:c.548G>A, respectively.
Ethics Statement
Patient data, samples, and photos were collected and analyzed with written patient consent under the IRB approved protocol #201011766, which explicitly allows for potentially identifying genomic studies, including genome sequencing, and approved by the Human Research Protection Office at Washington University School of Medicine.
Acknowledgments
We thank the patient and his family for contributing to this study and his physician Michael E. Gifford, MD for procuring the tumor tissue sample in this study.
Author Contributions
T.J.L. conceptualized the project, D.Y.C. and T.J.L. wrote the primary draft, L.A.S., S.M.R., C.A.M., and D.Y.C. performed the formal analysis, E.J.D., S.E.H., and L.A.C. provided clinical support, and D.Y.C., L.A.S., S.M.R., E.J.D., C.A.M., and T.J.L. edited the article.
Funding
This study was supported by grants from the National Institutes of Health CA237727 (to D.Y.C.), CA211782 (to C.A.M.), and CA101937 and CA197561 (to T.J.L.) and from the Barnes Jewish Foundation (to T.J.L.).
Competing Interest Statement
The authors have declared no competing interest.
REFERENCES
- Balci TB, Strong A, Kalish JM, Zackai E, Maris JM, Reilly A, Surrey LF, Wertheim GB, Marcadier JL, Graham GE, et al. 2020. Tatton-Brown–Rahman syndrome: six individuals with novel features. Am J Med Genet A 182: 673–680. 10.1002/ajmg.a.61475 [DOI] [PubMed] [Google Scholar]
- Berger MF, Hodis E, Heffernan TP, Deribe YL, Lawrence MS, Protopopov A, Ivanova E, Watson IR, Nickerson E, Ghosh P, et al. 2012. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature 485: 502–506. 10.1038/nature11071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network; Akbani R, Akdemir KC, Aksoy AB, Albert M, Ally A, Amin SB, Arachchi H, Arora A, Auman TJ, et al. 2015. Genomic classification of cutaneous melanoma. Cell 161: 1681–1696. 10.1016/j.cell.2015.05.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalanotti F, Cheng DT, Shoushtari AN, Johnson DB, Panageas KS, Momtaz P, Higham C, Won HH, Harding JJ, Merghoub T, et al. 2017. PTEN loss-of-function alterations are associated with intrinsic resistance to BRAF inhibitors in metastatic melanoma. JCO Precis Oncol 1: PO.16.00054. 10.1200/PO.16.00054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchi AC, Haidar A, Marin I, Kwartler CS, Prakash SK, Milewicz DM. 2022. Aortic root dilatation and dilated cardiomyopathy in an adult with Tatton-Brown–Rahman syndrome. Am J Med Genet A 188: 628–634. 10.1002/ajmg.a.62541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. 2012. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2: 401–404. 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Schulz-Trieglaff O, Shaw R, Barnes B, Schlesinger F, Källberg M, Cox AJ, Kruglyak S, Saunders CT. 2016. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics 32: 1220–1222. 10.1093/bioinformatics/btv710 [DOI] [PubMed] [Google Scholar]
- Chen DY, Ferguson IM, Braun KA, Sutton LA, Helton NM, Ramakrishnan SM, Smith AM, Miller CA, Ley TJ. 2021. Dnmt3a deficiency in the skin causes focal, canonical DNA hypomethylation and a cellular proliferation phenotype. Proc Natl Acad Sci 118: e2022760118. 10.1073/pnas.2022760118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian DL, Wu DY, Martin JR, Moore JR, Liu YR, Clemens AW, Nettles SA, Kirkland NM, Papouin T, Hill CA, et al. 2020. DNMT3A haploinsufficiency results in behavioral deficits and global epigenomic dysregulation shared across neurodevelopmental disorders. Cell Rep 33: 108416. 10.1016/j.celrep.2020.108416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, Getz G. 2013. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 31: 213–219. 10.1038/nbt.2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole CB, Russler-Germain DA, Ketkar S, Verdoni AM, Smith AM, Bangert CV, Helton NM, Guo M, Klco JM, O'Laughlin S, et al. 2017. Haploinsufficiency for DNA methyltransferase 3A predisposes hematopoietic cells to myeloid malignancies. J Clin Invest 127: 3657–3674. 10.1172/JCI93041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criscione VD, Weinstock MA. 2007. Descriptive epidemiology of dermatofibrosarcoma protuberans in the United States, 1973 to 2002. J Am Acad Dermatol 56: 968–973. 10.1016/j.jaad.2006.09.006 [DOI] [PubMed] [Google Scholar]
- Deng T, Kuang Y, Wang L, Li J, Wang Z, Fei J. 2009. An essential role for DNA methyltransferase 3a in melanoma tumorigenesis. Biochem Biophys Res Commun 387: 611–616. 10.1016/j.bbrc.2009.07.093 [DOI] [PubMed] [Google Scholar]
- Eggermont A, Blank CU, Mandala M, Long GV, Atkinson V, Dalle S, Haydon A, Lichinitser M, Khattak A, Carlino MS, et al. 2018. Adjuvant pembrolizumab versus placebo in resected stage III melanoma. N Engl J Med 378: 1789–1801. 10.1056/NEJMoa1802357 [DOI] [PubMed] [Google Scholar]
- Ehrlich M. 2009. DNA hypomethylation in cancer cells. Epigenomics 1: 239–259. 10.2217/epi.09.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris MA, Smith AM, Heath SE, Duncavage EJ, Oberley MJ, Freyer D, Wynn R, Douzgou S, Maris JM, Reilly AF, et al. 2021. DNMT3A overgrowth syndrome is associated with the development of hematopoietic malignancies in children and young adults. Blood 139: 461–464. 10.1182/blood.2021014052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. 2013. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6: pl1. 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovese G, Jaiswal S, Ebert BL, McCarroll SA. 2015. Clonal hematopoiesis and blood-cancer risk. N Engl J Med 372: 1071–1072. 10.1056/NEJMc1500684 [DOI] [PubMed] [Google Scholar]
- Gentilcore G. et al. 2013. Effect of dabrafenib on melanoma cell lines harbouring the BRAFV600D/R mutations. BMC Cancer 13: 17. 10.1186/1471-2407-13-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoneim HE, Fan Y, Moustaki A, Abdelsamed HA, Dash P, Dogra P, Carter R, Awad W, Neale G, Thomas PG, et al. 2017. De novo epigenetic programs inhibit PD-1 blockade-mediated T cell rejuvenation. Cell 170: 142–157.e19. 10.1016/j.cell.2017.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves WO, Verma S, Patel KP, Davies MA, Barkoh BA, Galbincea JM, Yao H, Lazar AJ, Aldape KD, Medeiros LJ, et al. 2013. Frequency and spectrum of BRAF mutations in a retrospective, single-institution study of 1112 cases of melanoma. J Mol Diagn 15: 220–226. 10.1016/j.jmoldx.2012.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat J-P, Nickerson E, Auclair D, Li L, Place C, et al. 2012. A landscape of driver mutations in melanoma. Cell 150: 251–263. 10.1016/j.cell.2012.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollink I, van den Ouweland AM, Beverloo BH, Arentsen-Peters S, Zwaan MC, Wagner A. 2017. Acute myeloid leukaemia in a case with Tatton-Brown–Rahman syndrome: the peculiar DNMT3A R882 mutation. J Med Genet 54: 805. 10.1136/jmedgenet-2017-104574 [DOI] [PubMed] [Google Scholar]
- Hugo W, Zaretsky JM, Sun L, Song C, Moreno B, Hu-Lieskovan S, Berent-Maoz B, Pang J, Chmielowski B, Cherry G, et al. 2016. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell 165: 35–44. 10.1016/j.cell.2016.02.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, Lindsley CR, Mermel CH, Burtt N, Chavez A, et al. 2014. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med 371: 2488–2498. 10.1056/NEJMoa1408617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffries AR, Maroofian R, Salter CG, Chioza BA, Cross HE, Patton MA, Dempster E, Temple IK, Mackay DJG, Rezwan FI, et al. 2019. Growth disrupting mutations in epigenetic regulatory molecules are associated with abnormalities of epigenetic aging. Genome Res 29: 1057–1066. 10.1101/gr.243584.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. 2007. The epigenomics of cancer. Cell 128: 683–692. 10.1016/j.cell.2007.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SJ, Zhao H, Hardikar S, Singh AK, Goodell MA, Chen T. 2013. A DNMT3A mutation common in AML exhibits dominant-negative effects in murine ES cells. Blood 122: 4086–4089. 10.1182/blood-2013-02-483487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim G, Kim J, Lim S, Lee KY, Kim O, Choi HS. 2018a. SUV39H1/DNMT3A-dependent methylation of the RB1 promoter stimulates PIN1 expression and melanoma development. FASEB J 32: 5647–5660. 10.1096/fj.201700645RRRRR [DOI] [PubMed] [Google Scholar]
- Kim S, Scheffler K, Halpern AL, Bekritsky MA, Noh E, Källberg M, Chen X, Kim Y, Beyter D, Krusche P, et al. 2018b. Strelka2: fast and accurate calling of germline and somatic variants. Nat Methods 15: 591–594. 10.1038/s41592-018-0051-x [DOI] [PubMed] [Google Scholar]
- Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, Wilson RK. 2012. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 22: 568–576. 10.1101/gr.129684.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaki R, Terashima H, Kubota M, Kosaki K. 2017. Acute myeloid leukemia-associated DNMT3A p.Arg882His mutation in a patient with Tatton-Brown–Rahman overgrowth syndrome as a constitutional mutation. Am J Med Genet A 173: 250–253. 10.1002/ajmg.a.37995 [DOI] [PubMed] [Google Scholar]
- Krauthammer M, Kong Y, Ha B, Evans P, Bacchiocchi A, McCusker JP, Cheng E, Davis MJ, Goh G, Choi M, et al. 2012. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet 44: 1006–1014. 10.1038/ng.2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, Kandoth C, Payton JE, Baty J, Welch J, et al. 2010. DNMT3A mutations in acute myeloid leukemia. N Engl J Med 363: 2424–2433. 10.1056/NEJMoa1005143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley T, Miller C, Ding L, Raphael BJ, Mungall AJ, Robertson A, Hoadley K, Triche TJ, Laird PW, et al. 2013. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 368: 2059–2074. 10.1056/NEJMoa1301689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Arxiv 10.48550/arXiv.1303.3997 [DOI] [Google Scholar]
- Li E, Bestor TH, Jaenisch R. 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69: 915–926. 10.1016/0092-8674(92)90611-F [DOI] [PubMed] [Google Scholar]
- Liu D, Schilling B, Liu D, Sucker A, Livingstone E, Jerby-Arnon L, Zimmer L, Gutzmer R, Satzger I, Loquai C, et al. 2019. Integrative molecular and clinical modeling of clinical outcomes to PD1 blockade in patients with metastatic melanoma. Nat Med 25: 1916–1927. 10.1038/s41591-019-0654-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long GV, Hauschild A, Santinami M, Atkinson V, Mandalà M, Chiarion-Sileni V, Larkin J, Nyakas M, Dutriaux C, Haydon A, et al. 2017. Adjuvant dabrafenib plus trametinib in stage III BRAF-mutated melanoma. N Engl J Med 377: 1813–1823. 10.1056/NEJMoa1708539 [DOI] [PubMed] [Google Scholar]
- McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GRS, Thormann A, Flicek P, Cunningham F. 2016. The ensembl variant effect predictor. Genome Biol 17: 122. 10.1186/s13059-016-0974-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer SE, Qin T, Muench DE, Masuda K, Venkatasubramanian M, Orr E, Suarez L, Gore SD, Delwel R, Paietta E, et al. 2016. DNMT3A haploinsufficiency transforms FLT3ITD myeloproliferative disease into a rapid, spontaneous, and fully penetrant acute myeloid leukemia. Cancer Discov 6: 501–515. 10.1158/2159-8290.CD-16-0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micevic G, Muthusamy V, Damsky W, Theodosakis N, Liu X, Meeth K, Wingrove E, Santhanakrishnan M, Bosenberg M. 2016. DNMT3b modulates melanoma growth by controlling levels of mTORC2 component RICTOR. Cell Rep 14: 2180–2192. 10.1016/j.celrep.2016.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micevic G, Theodosakis N, Bosenberg M. 2017. Aberrant DNA methylation in melanoma: biomarker and therapeutic opportunities. Clin Epigenet 9: 34. 10.1186/s13148-017-0332-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M, Xie S, Li E. 1998. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet 19: 219–220. 10.1038/890 [DOI] [PubMed] [Google Scholar]
- Paulson KG, Gupta D, Kim TS, Veatch JR, Byrd DR, Bhatia S, Wojcik K, Chapuis AG, Thompson JA, Madeleine MM, et al. 2020. Age-specific incidence of melanoma in the United States. JAMA Dermatol 156: 57–64. 10.1001/jamadermatol.2019.3353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldi L, Avgustinova A, Martín M, Datta D, Solanas G, Prats N, Benitah S. 2017. Loss of Dnmt3a and Dnmt3b does not affect epidermal homeostasis but promotes squamous transformation through PPAR-γ. Elife 6: e21697. 10.7554/eLife.21697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson GA, Shih J, Yau C, Gibb EA, Oba J, Mungall KL, Hess JM, Uzunangelov V, Walter V, Danilova L, et al. 2017. Integrative analysis identifies four molecular and clinical subsets in uveal melanoma. Cancer Cell 32: 204–220.e15. 10.1016/j.ccell.2017.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russler-Germain DA, Spencer DH, Young MA, Lamprecht TL, Miller CA, Fulton R, Meyer MR, Erdmann-Gilmore P, Townsend RR, Wilson RK, et al. 2014. The R882H DNMT3A mutation associated with AML dominantly inhibits wild-type DNMT3A by blocking its ability to form active tetramers. Cancer Cell 25: 442–454. 10.1016/j.ccr.2014.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W, Heeley JM, Carlston CM, Acuna-Hidalgo R, Nillesen WM, Dent KM, Douglas GV, Levine KL, Bayrak-Toydemir P, Marcelis CL, et al. 2017. The spectrum of DNMT3A variants in Tatton-Brown–Rahman syndrome overlaps with that in hematologic malignancies. Am J Med Genet A 173: 3022–3028. 10.1002/ajmg.a.38485 [DOI] [PubMed] [Google Scholar]
- Shirley MD, Tang H, Gallione CJ, Baugher JD, Frelin LP, Cohen B, North PE, Marchuk DA, Comi AM, Pevsner J. 2013. Sturge–Weber syndrome and port-wine stains caused by somatic mutation in GNAQ. N Engl J Med 368: 1971–1979. 10.1056/NEJMoa1213507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoushtari AN, Chatila WK, Arora A, Sanchez-Vega F, Kantheti HS, Zamalloa JAR, Krieger P, Callahan MK, Warner AB, Postow MA, et al. 2021. Therapeutic implications of detecting MAPK-activating alterations in cutaneous and unknown primary melanomas. Clin Cancer Res 27: 2226–2235. 10.1158/1078-0432.CCR-20-4189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AM, LaValle TA, Shinawi M, Ramakrishnan SM, Abel HJ, Hill CA, Kirkland NM, Rettig MP, Helton NM, Heath SE, et al. 2021. Functional and epigenetic phenotypes of humans and mice with DNMT3A overgrowth syndrome. Nat Commun 12: 4549. 10.1038/s41467-021-24800-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, Walsh LA, Postow MA, Wong P, Ho TS, et al. 2014. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med 371: 2189–2199. 10.1056/NEJMoa1406498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatton-Brown K, Seal S, Ruark E, Harmer J, Ramsay E, Del Vecchio Duarte S, Zachariou A, Hanks S, O'Brien E, Aksglaede L, et al. 2014. Mutations in the DNA methyltransferase gene DNMT3A cause an overgrowth syndrome with intellectual disability. Nat Genet 46: 385–388. 10.1038/ng.2917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatton-Brown K, Zachariou A, Loveday C, Renwick A, Mahamdallie S, Aksglaede L, Baralle D, Barge-Schaapveld D, Blyth M, Bouma M, et al. 2018. The Tatton-Brown–Rahman syndrome: a clinical study of 55 individuals with de novo constitutive DNMT3A variants. Wellcome Open Res 3: 46. 10.12688/wellcomeopenres.14430.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Allen EMV, Wagle N, Sucker A, Treacy DJ, Johannessen CM, Goetz EM, Place CS, Taylor-Weiner A, Whittaker S, Kryukov GV, et al. 2014. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov 4: 94–109. 10.1158/2159-8290.CD-13-0617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Allen EMV, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, Sucker A, Hillen U, Foppen MHG, Goldinger SM, et al. 2015. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 350: 207–211. 10.1126/science.aad0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Raamsdonk CD, Bezrookove V, Green G, Bauer J, Gaugler L, O'Brien JM, Simpson EM, Barsh GS, Bastian BC. 2009. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature 457: 599–602. 10.1038/nature07586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, Lu C, Wang J, McLellan MD, Johnson KJ, Wendl MC, McMichael JF, Schmidt HK, Yellapantula V, Miller CA, et al. 2014. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med 20: 1472–1478. 10.1038/nm.3733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin B, Marino TC, Szekely J, Leblanc J, Cechner K, Sency V, Wensel C, Barabas M, Therriault V, Wang H. 2017. Novel DNMT3A germline mutations are associated with inherited Tatton-Brown–Rahman syndrome. Clin Genet 91: 623–628. 10.1111/cge.12878 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Sequencing data for this patient was deposited to dbGaP (phs000159). Tha variants were submitted to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) and can be found under accession numbers SCV003914810–SCV003914813 for NM_022552.5:c.1238dupG, NM_004333.6:c.1799_1800delinsAC, NM_022552.5:c.2245C>T, and NM_002072.4:c.548G>A, respectively.