ABSTRACT
Macroautophagy/autophagy, an evolutionarily conserved degradative process essential for cell homeostasis and development in eukaryotes, involves autophagosome formation and fusion with a lysosome/vacuole. The soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) proteins play important roles in regulating autophagy in mammals and yeast, but relatively little is known about SNARE function in plant autophagy. Here we identified and characterized two Arabidopsis SNAREs, AT4G15780/VAMP724 and AT1G04760/VAMP726, involved in plant autophagy. Phenotypic analysis showed that mutants of VAMP724 and VAMP726 are sensitive to nutrient-starved conditions. Live-cell imaging on mutants of VAMP724 and VAMP726 expressing YFP-ATG8e showed the formation of abnormal autophagic structures outside the vacuoles and compromised autophagic flux. Further immunogold transmission electron microscopy and electron tomography (ET) analysis demonstrated a direct connection between the tubular autophagic structures and the endoplasmic reticulum (ER) in vamp724-1 vamp726-1 double mutants. Further transient co-expression, co-immunoprecipitation and double immunogold TEM analysis showed that ATG9 (autophagy related 9) interacts and colocalizes with VAMP724 and VAMP726 in ATG9-positive vesicles during autophagosome formation. Taken together, VAMP724 and VAMP726 regulate autophagosome formation likely working together with ATG9 in Arabidopsis.
Abbreviations: ATG, autophagy related; BTH, benzo-(1,2,3)-thiadiazole-7-carbothioic acid S-methyl ester; Conc A, concanamycin A; EM, electron microscopy; ER, endoplasmic reticulum; FRET, Förster/fluorescence resonance energy transfer; MS, Murashige and Skoog; MVB, multivesicular body; PAS, phagophore assembly site; PM, plasma membrane; PVC, prevacuolar compartment; SNARE, soluble N-ethylmaleimide-sensitive factor attachment protein receptor; TEM, transmission electron microscopy; TGN, trans-Golgi network; WT, wild-type.
KEYWORDS: Arabidopsis, ATG9, autophagy, SNARE, VAMP724, VAMP726
Introduction
Macroautophagy (hereafter referred to as autophagy) is a well-defined and highly conserved catabolic process among all eukaryotes, which plays essential roles in degradation and recycling of intracellular materials such as protein and organelles, as well as protecting cells from pathogen attacks and unfavorable conditions [1-3].
Soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) proteins are a large protein family known to control fusion events in membrane trafficking. In yeast and mammalian cells, there are more than 60 evolutionarily conserved members. In the case of Arabidopsis, there are 60 SNAREs [4]. SNAREs have been reported to play important roles in the autophagy pathway, including regulations of autophagosome formation and autophagosome-vacuole/lysosome fusion. In yeast, several SNAREs (Sec9, Sso2, Tlg2, Sec22, and Ykt6) are required for the homotypic fusion of Atg9-containing vesicles, the autophagic precursors [5], whereas Vam3, Vam7, Vti1, and Ykt6 form a SNARE complex to mediate autophagosome-vacuole fusion [6-8]. In mammalian cells, the homotypic fusion of phagophore precursors is regulated by VAMP7, STX7, STX8, and VTI1B [9], whereas the autophagosome-lysosome fusion is driven by STX17 by forming a SNARE complex with SNAP29 and VAMP8 [10]. Independently of STX17, YKT6 also forms a complex with SNAP29 and STX7 to mediate autophagosome-lysosome fusion [11].
Despite the findings on the roles of SNAREs in regulating the autophagy pathways in both yeast and animal systems, relatively little is known about the function of SNAREs in plant autophagy. Only one Arabidopsis SNARE, VTI12, has been shown to play a role in autophagy, possibly in mediating fusion between autophagosomes and the vacuole [12]. Silencing of VPS45, a regulator of VTI12, also leads to early senescence phenotype [13]. In this study, we hypothesized that the Arabidopsis genome contains other SNAREs that function in mediating the autophagy pathway in plants. We therefore used a screening system of transient expression of protoplasts to identify SNAREs functioning in the autophagy pathway for further functional characterization in plants.
YFP fusions with the Arabidopsis SNAREs were transiently co-expressed individually with the autophagosome marker mCherry-ATG8e in Arabidopsis protoplasts. Using this approach, we identified two Arabidopsis SNAREs VAMP724 and VAMP726 that show colocalization with mCherry-ATG8e upon their transient co-expression in Arabidopsis protoplasts. Using Arabidopsis mutants, we further showed that depletion of VAMP724 and VAMP726 leads to defects in the autophagy pathway. Mutants of VAMP724 and VAMP726 displayed sensitivity towards nutrient-starved conditions, whereas YFP-ATG8e showed the formation of abnormal autophagic structures outside the vacuoles and compromised autophagic flux in the mutants. By using live-cell imaging, immunogold TEM and ET analysis on the mutant cells, we further showed that the abnormal autophagic structures have direct connection with the endoplasmic reticulum (ER). Interestingly, VAMP724 and VAMP726 showed partial colocalization with ATG9 (autophagy related 9) in ATG8e-positive autophagosomes upon their co-expression in Arabidopsis protoplasts. Further co-immunoprecipitation (IP), Förster/fluorescence resonance energy transfer (FRET) and immunogold TEM analysis demonstrated that VAMP724 and VAMP726 are closely associated with ATG9 and ATG9-containing vesicles, likely working together in regulating autophagosome formation in Arabidopsis.
Results
VAMP724 and VAMP726 knockout mutants are sensitive to nutrient-limited conditions
Previous studies showed that autophagic SNAREs STX17 and YKT6 colocalize with the autophagsosome marker MAP1LC3/LC3 in mammalian cells under starvation conditions [10,11]. To identify other autophagy-related SNAREs in Arabidopsis, we first used our well-established transient expression system of Arabidopsis suspension culture protoplasts [14,15] to screen and identify autophagy-related SNARE candidates. YFP fusions with various plant SNAREs were co-expressed individually with the autophagosome marker mCherry-ATG8e in Arabidopsis protoplasts for subsequent analysis of colocalization by confocal laser scanning microscopy (CLSM). Here we used the screening of VAMP72 family SNAREs as examples (Figure S1). Most of the YFP-tagged SNAREs showed a separation from the autophagosome marker mCherry-ATG8e, but VAMP72-family SNARE proteins, VAMP724 and VAMP726, showed a partial colocalization with mCherry-ATG8e as puncta (indicated by arrows as examples in Figure S1), indicating that these two SNARE proteins are likely to function in the autophagic pathway in plants, they are thus selected for further characterization.
Both VAMP724 and VAMP726 are R-SNAREs that belong to the VAMP72 SNARE family, which consists of 7 members [16]. To study the possible roles of VAMP724 and VAMP726 in plant autophagy, we used a loss-of-function approach. T-DNA insertional mutants of VAMP724 (vamp724-1, SAIL_569_E12 and vamp724-2, SALK_032219) and VAMP726 (vamp726-1, SALK_082690 and vamp726-2, SALK_147419) were identified (Figure S2A). Real-time PCR analysis using gene-specific primers revealed an extremely low accumulation of VAMP724 and VAMP726 transcripts in vamp724-1 and vamp726-1 mutants, respectively (Figure S2B). However, VAMP724 and VAMP726 transcripts were detectable at a relatively high level in vamp724-2 and vamp726-2 mutants, respectively. In addition, a detached leaf assay was performed on the mutants [17,18]. Accelerated senescence of the detached leaves was observed in vamp724-1 and vamp726-1 mutants (Figure S2C). Therefore, vamp724-1 and vamp726-1 were chosen for further studies. Considering that VAMP724 and VAMP726 were homologous proteins that belonged to the VAMP72 clade and shared 70.8% identity in amino acid sequence, they might be functionally redundant in Arabidopsis. Therefore, a double mutant vamp724-1 vamp726-1 was also generated by crossing of the two mutants, followed by genotyping selection of the F2 generation. Phenotypic analysis was performed on vamp724-1, vamp726-1, and vamp724-1 vamp726-1 under normal growth conditions. All three mutant plants showed similar phenotypes in comparison to the wild-type (WT) plants at various developmental stages, including a 6-day-old seedling stage on plate, 3-week-old and 6-week-old plant stage on soil (Figure S2D,E).
Hypersensitive phenotypes towards nutrient-limited conditions were observed in well-characterized autophagy-defective mutants like atg5-1, atg7-2, as well as the mutant of SNARE VTI12 [12,19,20]. We thus wanted to find out if the depletion of VAMP724 or VAMP726 would lead to similar phenotypes. Firstly, we performed a nitrogen starvation assay as described previously [21-24]. Plants (WT and various mutants) were grown on nitrogen-rich (N+) Murashige and Skoog (MS) plates for 6 days and then transferred to either fresh nitrogen-rich (N+) medium or nitrogen-deficient (N-) medium for an additional 14 days before observation. WT (control), vamp724-1, vamp726-1, vamp724-1 vamp726-1 and atg7-2 were tested. Indeed, similar to the atg7-2 mutant, both vamp724-1 and vamp726-1 mutants showed enhanced sensitivity to nitrogen starvation as compared to WT plants, while the double mutant vamp724-1 vamp726-1 showed a more enhanced effect than the single mutants, as indicated by more-yellowish appearance and lower chlorophyll levels (Figure 1A,C).
Figure 1.
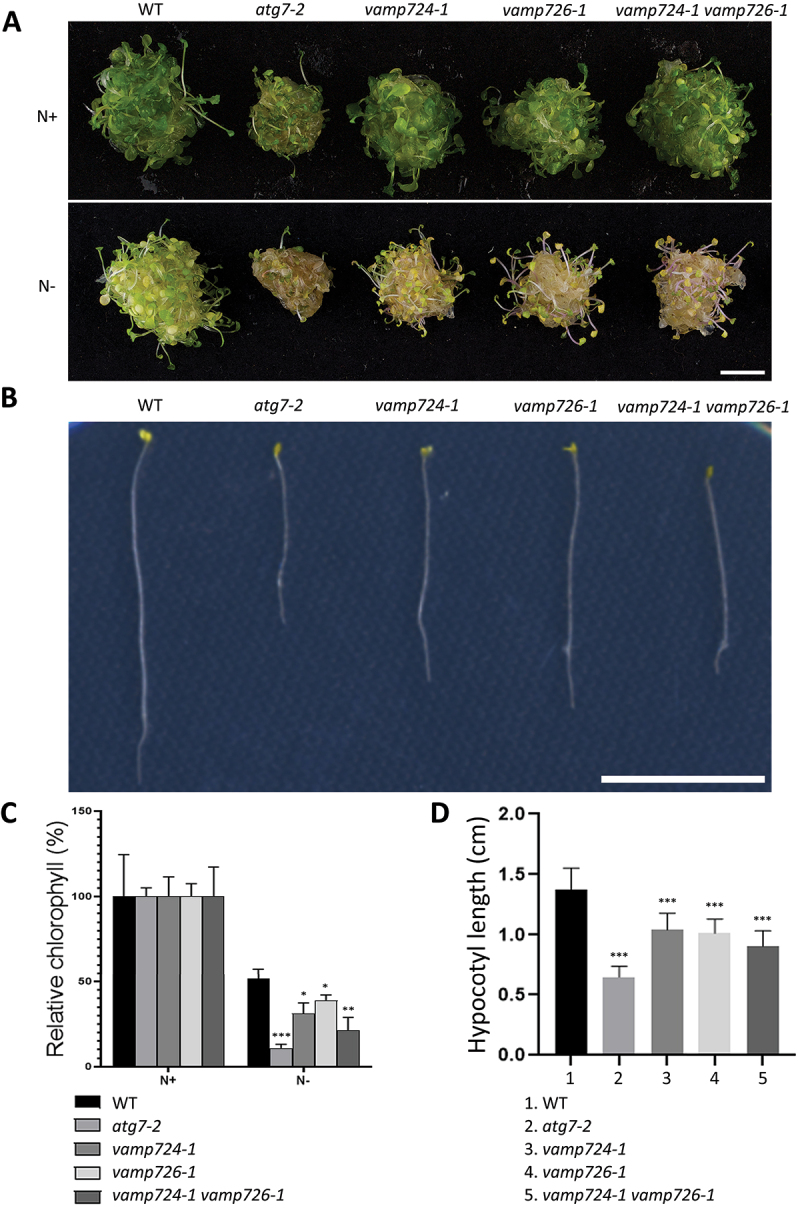
The VAMP724 and VAMP726 knock-out (KO) mutants display sensitive phenotypes under nutrient-deficient conditions. (A) vamp724-1 or vamp726-1 mutants showed enhanced sensitivity to nitrogen starvation. 6-day-old seedlings of WT and various indicated mutants from MS plate (+N) were transferred to fresh MS medium (+N) or N-deficient (–N) medium for 2 weeks before photography. Bar: 1 cm. (B) Phenotype of suppressed hypocotyl elongation. Seedlings of WT and various indicated mutants were grown in continuous darkness on MS solid medium without sucrose (-C) for 1 week before photography. Representative photographs of the etiolated seedlings are shown. Bar: 1 cm. (C) Relative chlorophyll contents of seedlings from WT and various mutants shown in (A). Each seedling bundle contained 30 seedlings and the results were obtained from three independent experiments (error bars ± SD). *P < 0.05, **P < 0.01, ***P < 0.001, two-sided Student’s t-test. (D) Quantification of hypocotyl length shown in (B). Results were obtained from three independent experiments of 10 seedlings each (error bars ± SD). *P < 0.05, **P < 0.01, ***P < 0.001, two-sided Student’s t-test.
Previous studies showed that carbon-starved etiolated autophagy-defected mutant seedlings displayed shorter hypocotyls in comparison to WT plants, and the suppressed elongation was presumably due to impaired mobilization of nutrient reserves when autophagy was blocked [25,26]. Thus, an etiolated hypocotyl elongation test was also performed. WT (control), vamp724-1, vamp726-1, vamp724-1 vamp726-1 and atg7-2 were kept in darkness for 7 days from the onset of germination on MS plates without sucrose (C-) prior to observation. Similar to the atg7-2 mutant, the lengths of etiolated hypocotyls of both vamp724-1 and vamp726-1 mutants were shorter than those of the WT, whereas the double mutant vamp724-1 vamp726-1 appeared even shorter (Figure 1B). Further statistical analysis confirmed these observations (Figure 1D). Taken together, mutation of VAMP724 or VAMP726 resulted in enhanced sensitivity to nutritional stress.
Depletion of VAMP724 and VAMP726 affected autophagy pathway in Arabidopsis, forming abnormal autophagic structures
To further investigate the roles of VAMP724 and VAMP726 in the autophagy pathway at subcellular level, we next transformed the autophagosome marker YFP-ATG8e into the three mutants of vamp724-1, vamp726-1, and vamp724-1 vamp726-1. Upon autophagic induction by benzo-(1,2,3)-thiadiazole-7-carbothioic acid S-methyl ester (BTH, a salicylic acid agonist that triggers the autophagic pathway in Arabidopsis), the YFP-ATG8e-labeled puncta increased in all three mutants (vamp724-1, vamp726-1, and vamp724-1 vamp726-1) (Figure 2A, middle and Figure 2B), which was similar to that in WT plants. However, extended tubular structures decorated by YFP-ATG8e were also observed in these three mutants (indicated by arrows as examples in Figure 2A, middle), which were rarely seen in WT plants (Figure 2C).
Figure 2.
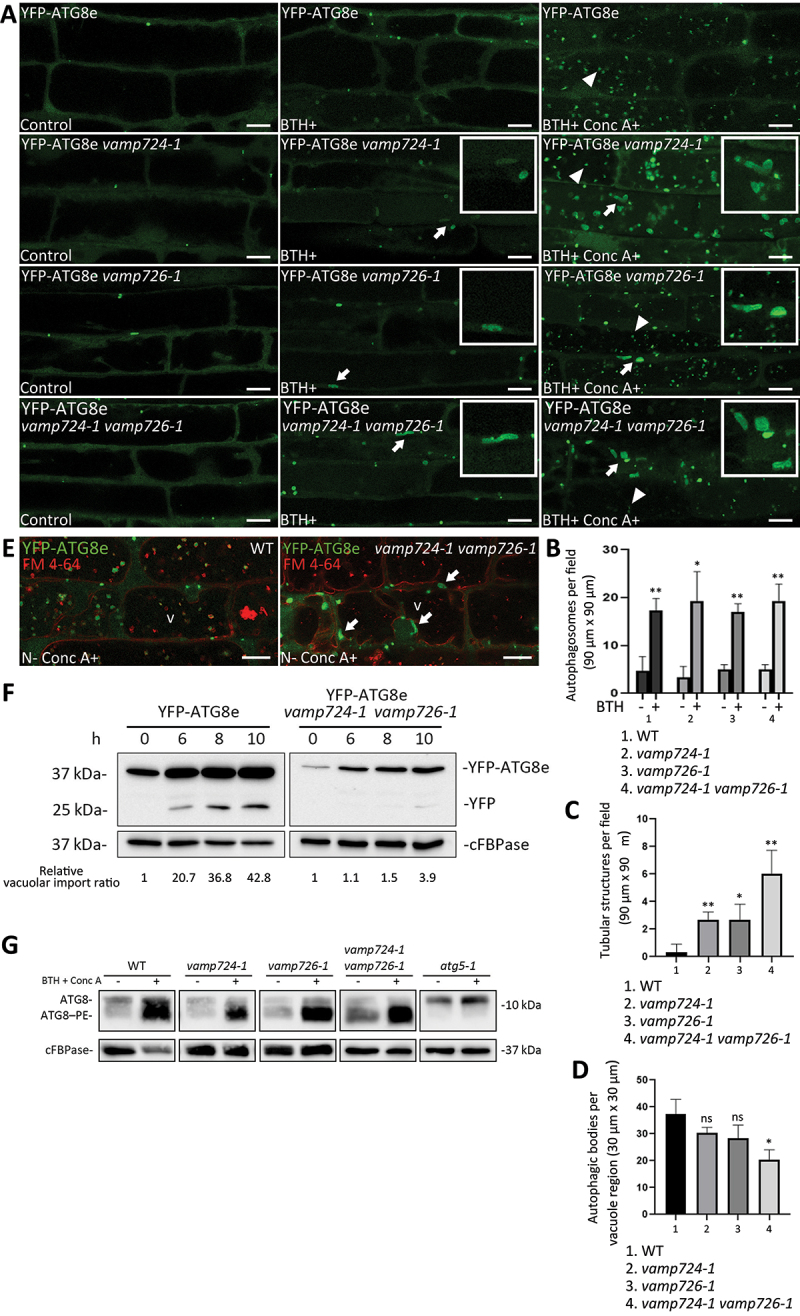
Depletion of VAMP724 and VAMP726 affects the autophagy pathway in Arabidopsis. (A) Detection of YFP-ATG8e-positive autophagic tubules in vamp724-1 and vamp726-1 mutants upon BTH-induced autophagy. 5-day-old seedlings of WT, as well as vamp724-1 and vamp726-1 mutants expressing YFP-ATG8e were incubated in medium without BTH (Left panel, Control), with BTH (Middle panel) or with BTH+Conc A (Right panel) for 6 h, followed by confocal imaging. Arrows indicated examples of the ATG8e-positive tubular structures with enlargement in the insets, while arrowheads indicated examples of autophagic bodies inside the vacuoles. Bar: 10 μm. (B) Quantification of the numbers of YFP-ATG8e-positive autophagosomes in BTH-treated or untreated WT, vamp724-1, vamp726-1, and vamp724-1 vamp726-1 shown in (A). Results were obtained from three independent experiments (error bars ± SD). *P < 0.05, **P < 0.01, two-sided Student’s t-test. (C) Quantification of the numbers of YFP-ATG8e-positive tubular structures in WT, vamp724-1, vamp726-1, and vamp724-1 vamp726-1 upon BTH treatments shown in (A). Results were obtained from three independent experiments (error bars ± SD). *P < 0.05, **P < 0.01, two-sided Student’s t-test. (D) Quantification of the numbers of autophagic bodies inside the vacuoles of WT, vamp724-1, vamp726-1, and vamp724-1 vamp726-1 upon BTH+Conc A treatments shown in (A). Results were obtained from three independent experiments (error bars ± SD). *P < 0.05, ns: not significant (P > 0.05), two-sided Student’s t-test. (E) 5-day-old seedlings of YFP-ATG8e and YFP-ATG8e vamp724-1 vamp726-1 were incubated in MS medium without nitrogen (N-) for 24 h in dark, followed by FM 4-64 uptake for 1 h and subsequent Conc A treatment for 6 h prior to confocal imaging. v, vacuole. Bar: 10 μm. (F) Immunoblot detection of vacuolar turnover of YFP-ATG8e WT and YFP-ATG8e vamp724-1 vamp726-1 upon BTH-induced autophagy. Five-day-old seedlings of YFP-ATG8e and YFP-ATG8e vamp724-1 vamp726-1 were subjected to BTH treatments, followed by total protein extraction at indicated time points and subsequent immunoblot analysis with GFP antibodies whereas cFBPase antibodies were used as a loading control. For quantification analysis, protein band intensities were quantified using ImageJ software and all values were normalized according to the protein loading control (cFBPase) before calculation of vacuolar import ratio (YFP:total). Relative vacuolar import ratio of different time points was normalized to time 0 (which is set to 1). (G) Immunoblot detection of the ATG8 lipidation levels in WT, vamp724-1, vamp726-1, vamp724-1 vamp726-1, and atg5-1. 5-day-old seedlings of WT and various mutants were incubated in medium without (control) or with BTH and Conc A for 6 h, followed by extraction of membrane proteins fractions and subsequent immunoblot analysis with ATG8 antibodies, whereas cFBPase antibodies were used as a loading control.
To further examine the effect of VAMP724 or VAMP726 depletion on the autophagic flux, we next treated the plants with BTH and concanamycin A (Conc A), a V-ATPase inhibitor, which leads to vacuole deacidification, and thus prevention of vacuolar degradation of autophagic bodies [22,27-29]. After BTH+Conc A treatment for 6 h, numerous autophagic bodies (indicated by arrowheads) accumulated within the vacuole of the WT plant (Figure 2A, right). Similar results were also observed in these three mutants (Figure 2A, right), albeit the numbers of the autophagic bodies were fewer in the vamp724-1 vamp726-1 double mutant (Figure 2D). ATG8e-positive tubular structures could also be observed in these three mutants after BTH+Conc A treatment, especially in vamp724-1 vamp726-1 double mutant (Figure 2A, right, indicated by arrows). To confirm whether these abnormal structures can also be formed in other autophagy conditions like nitrogen starvation (N-), we performed N- treatment on vamp724-1 vamp726-1 mutant, followed by confocal imaging analysis and quantification. Indeed, similar YFP-ATG8e-positive autophagic tubules were also observed in vamp724-1 vamp726-1 double mutant under nitrogen starvation (Figure S3A), which is similar to BTH induction (Figure S3B). To distinguish whether the tubular structures reside within or outside the vacuole, we performed an FM 4-64 uptake study in vamp724-1 vamp726-1 seedlings to label the tonoplast under nitrogen starvation condition, followed by Conc A treatment and confocal imaging analysis. As shown in Figure 2E, abnormal autophagosomes (indicated by arrows) were clearly detected outside the tonoplast, with less autophagic bodies accumulated inside the vacuole compared to that of WT cells, implying that the delivery of the abnormal autophagosomes to the vacuole is compromised in vamp724-1 vamp726-1 mutant. It is very likely that the abnormal autophagosomes also reside outside the vacuoles in single mutants of vamp724-1 and vamp726-1.
To further confirm these results, we performed a YFP-ATG8e processing assay, which reflects the delivery of autophagic membrane to the vacuole [20,30]. Protein fractions isolated from YFP-ATG8e and YFP-ATG8e vamp724-1 vamp726-1, with or without BTH treatment at different time points, were subjected to immunoblot analysis with GFP antibodies. Consistent with the confocal observations, much less YFP core was detected in YFP-ATG8e vamp724-1 vamp726-1 as compared with that of the YFP-ATG8e WT, which was also supported by quantification analysis (Figure 2F). These results demonstrated that the transport of autophagosomes to the vacuole was partially defective in the vamp724-1 vamp726-1 double mutant.
Moreover, we conducted a lipidation assay to examine the phosphatidylethanolamine-conjugated ATG8 (ATG8–PE) levels in the WT plants vs. the mutants. The ATG8–PE is a faster migrating species upon SDS-PAGE of membrane preparations in the presence of urea [19,26,30]. Therefore, membrane fractions of WT, vamp724-1, vamp726-1, and vamp724-1 vamp726-1, as well as the autophagy deficiency mutant atg5-1, in which ATG8–PE could not be detected [19], were extracted and subjected to immunoblot analysis with ATG8 antibodies. As shown in Figure 2G, vamp724-1, vamp726-1, and vamp724-1 vamp726-1 showed similar ATG8–PE levels as compared to the WT, suggesting that VAMP724 and VAMP726 are not necessary for the lipidation of ATG8.
Autophagosome-related tubules in vamp724-1 vamp726-1 mutants are in association with the ER membrane
Interestingly, the morphology of the ATG8-positive tubular structures in VAMP724- or VAMP726-deficient mutants were similar to those observed in autophagy defected mutants such as atg9-3, atg11-1 and atg13ab [26,30]. Previous studies have demonstrated a close relationship between the ER and phagophore membrane [28,30,31]. We thus hypothesized that these YFP-ATG8e-labeled structures might also be related to the ER. We thus tried to visualize the two organelles and analyze their correlation under WT or mutant backgrounds. Since the results obtained so far showed that more severe autophagy-defective phenotypes and more abnormal tubular structures were observed in the vamp724-1 vamp726-1 double mutants, possibly due to an additional effect, we next generated YFP-ATG8e CNX-mRFP WT and YFP-ATG8e CNX-mRFP vamp724-1 vamp726-1 transgenic plants and subjected to BTH treatment to induce autophagy, followed by confocal imaging analysis (Figure 3). In vamp724-1 vamp726-1 mutant, the YFP-ATG8e-labeled ring-like signals were detected in close proximity to the CNX-mRFP signal (Figure 3B), similar to previous studies [28,30,31], indicating that the tubular structures are related to the ER. The result is further confirmed by transient expression experiment using the leaf protoplast system (Figure S3D). We co-expressed the autophagosome marker YFP-ATG8e and the ER marker CNX-mRFP in leaf protoplasts isolated from WT or vamp724-1 vamp726-1 mutant. In vamp724-1 vamp726-1 mutant cells, the YFP-ATG8e-labeled ring-like signals were detected in close proximity to the CNX-mRFP signal (Figure S3D), consistent with the former results (Figure 3). Notably, we found that the YFP-ATG8e-labeled tubules were surrounded by CNX-mRFP signals (Figure 3B and S3D), suggesting that these tubular structures may be the consequence of defective autophagosome formation.
Figure 3.
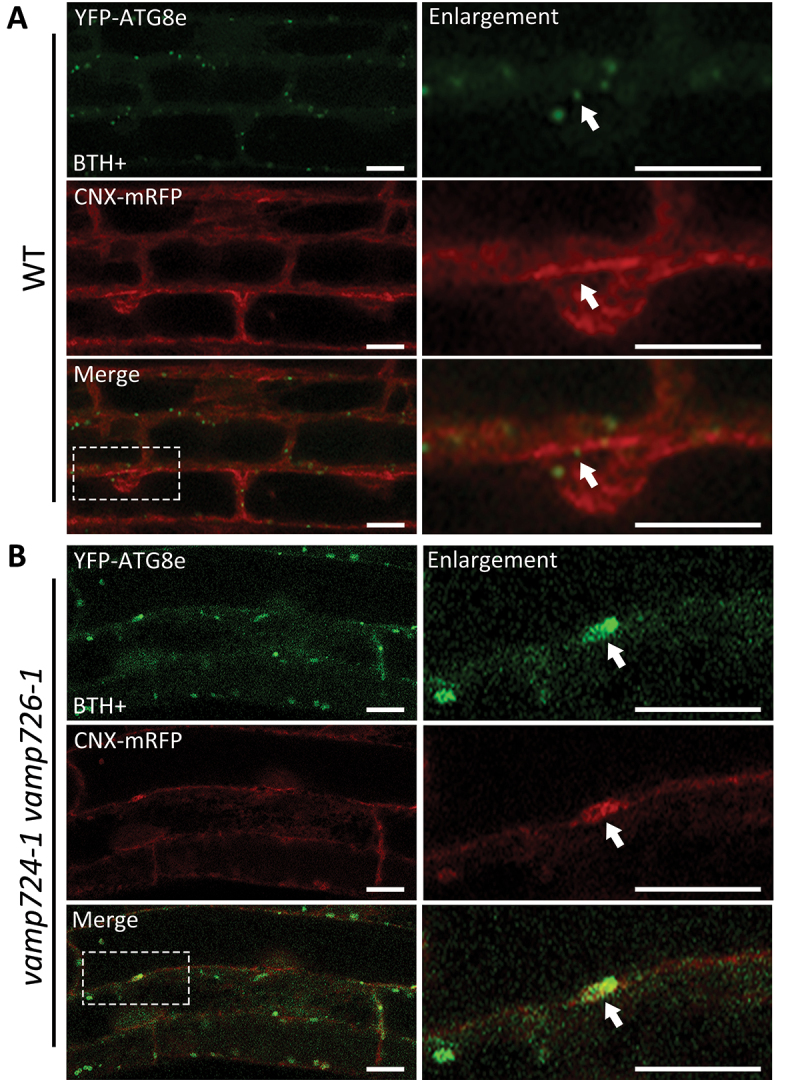
Autophagosome-related tubular structures are associated with the ER membranes in vamp724-1 vamp726-1 double mutant. (A) YFP-ATG8e-positive structures are formed in a close proximity to the ER membrane in WT. 5-day-old YFP-ATG8e CNX-mRFP seedlings were incubated in MS medium with BTH for 6 h before confocal imaging analysis. The dashed square regions are enlarged and shown respectively in the right panels. Arrows indicated examples of association between autophagosome and the ER marker. (B) YFP-ATG8e-positive tubules are accompanied with the ER membrane in vamp724-1 vamp726-1 double mutant. 5-day-old YFP-ATG8e CNX-mRFP vamp724-1 vamp726-1 seedlings were incubated in MS medium with BTH for 6 h before confocal imaging analysis. The dashed square regions are enlarged and shown respectively in the right panels. Arrows indicated examples of association between the YFP-ATG8e-positive tubules and the ER. Bar: 10 μm.
To gain further insights into these tubular structures at the ultrastructural level, immunogold-TEM analysis was performed using ultrathin sections prepared from autophagy-induced YFP-ATG8e vamp724-1 vamp726-1 transgenic plants after high pressure freezing/freeze substitution (HPF/FS). Large, extended tubular structures were observed (Figure 4A-B as indicated by open arrows). The autophagic identity of these tubular structures was confirmed by immunolabeling with anti-ATG8 antibodies (indicated by arrowheads) in BTH-treated root cells of YFP-ATG8e vamp724-1 vamp726-1 seedlings (Figure S4A-F). These tubular autophagic structures likely represent the YFP-ATG8e-positive tubular structures in the confocal observations (Figure 2A, middle). We also carried out double-immunogold labeling using anti-ATG8 and anti-calreticulin (ER marker) [32] to examine the possible correlation between the autophagosome-related tubular structures and ER membranes (Figure 4C-E and S4G-J). Consistent with the confocal observations, both antibodies (10-nm gold particles for anti-ATG8 and 6-nm gold particles for anti-calreticulin) labeled the same autophagosome-like structures with rough ER nearby. The sequestration of apparent cargoes by these tubular autophagosomes can also be observed (Figure S4E, S4F, and S4I).
Figure 4.
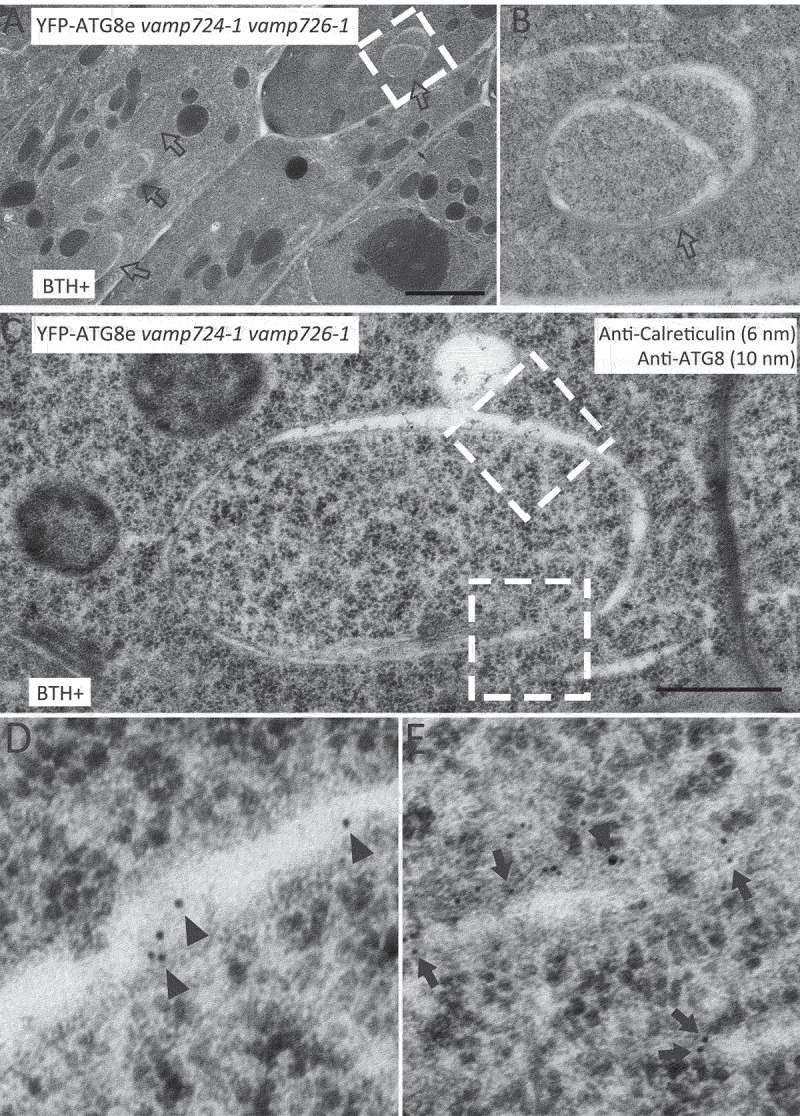
Immunogold-TEM analysis of tubular autophagic structures in root cells of YFP-ATG8e vamp724-1 vamp726-1 upon autophagy induction. Ultrathin sections were prepared from high-pressure frozen/freeze-substituted root cells of YFP-ATG8e vamp724-1 vamp726-1 seedlings upon 6 h BTH treatment, followed by immunogold labeling with indicated antibodies. (A) An overview of BTH-treated root cells of transgenic YFP-ATG8e vamp724-1 vamp726-1 seedlings. Open arrows indicate examples of the tubular autophagic structures. Bar: 2 μm. (B) Enlargement of the dashed box shown in (A). (C) Double immunogold labeling with anti-ATG8 and anti-calreticulin antibodies on autophagosome-related tubular structures in BTH-treated root cells of transgenic YFP-ATG8e vamp724-1 vamp726-1 seedlings. Bar: 500 nm. (D and E) Enlargement of the dashed boxes shown in (C). Arrowheads and arrows indicate gold particles for anti-ATG8 (10 nm) and anti-calreticulin (6 nm), respectively.
A direct connection between developing autophagic membranes and the ER in vamp724-1 vamp726-1 mutant is displayed by three-dimensional (3D) electron tomographic reconstruction
The results above suggested a close proximity between the autophagic tubule and the ER membrane in vamp724-1 vamp726-1 mutants. Previous studies indicated that there are only one or few connections between the ER and phagophore or autophagosome [30,33]. It is challenging to observe the direct connection between the ER and autophagosome using conventional 2D TEM analysis. We thus performed 3D electron tomography (ET) analysis. A representative example of the tubule-like autophagosome structure in autophagy-induced vamp724-1 vamp726-1 mutant is shown in Figure 5A. A 3D tomographic model was generated using the 3dmod program of the IMOD software package (Figure 5B,C). The multi-layer autophagosome contains tubules likely representing the ER fragments, as well as another cup-shaped phagophore. A connection between the autophagosome and ER was observed (Figure 5D-G, indicated by arrowhead). Interestingly, connection between multivesicular body (MVB) and autophagosome was also observed (Figure 5H-K, indicated by arrow). To investigate the dynamics between autophagosomes and MVBs, autophagosome marker YFP-ATG8e and MVB marker mRFP-VSR2 were transiently co-expressed in leaf protoplasts isolated from WT or vamp724-1 vamp726-1 mutants, followed by a time-lapse confocal imaging analysis. Indeed, possible fusion events between autophagosomes and MVBs were observed in vamp724-1 vamp726-1, while the autophagosomes are separated from MVBs in WT (Figure S5). We hypothesize that the connection between the two organelles (Figure 5H-K) might represent a fusion event. A 3D ET analysis was also performed in the BTH-treated WT root cells to investigate the relationship between autophagosome and the ER membrane (Figure S6A-E). A close proximity but no direct connection between the autophagosome and the ER membrane was observed (Figure S6F-G). Taken together, these data demonstrate a direct connection between the autophagic tubular structures and the ER in the vamp724-1 vamp726-1 mutant, implying that VAMP724 and VAMP726 are involved in autophagosome progression from the ER membrane.
Figure 5.
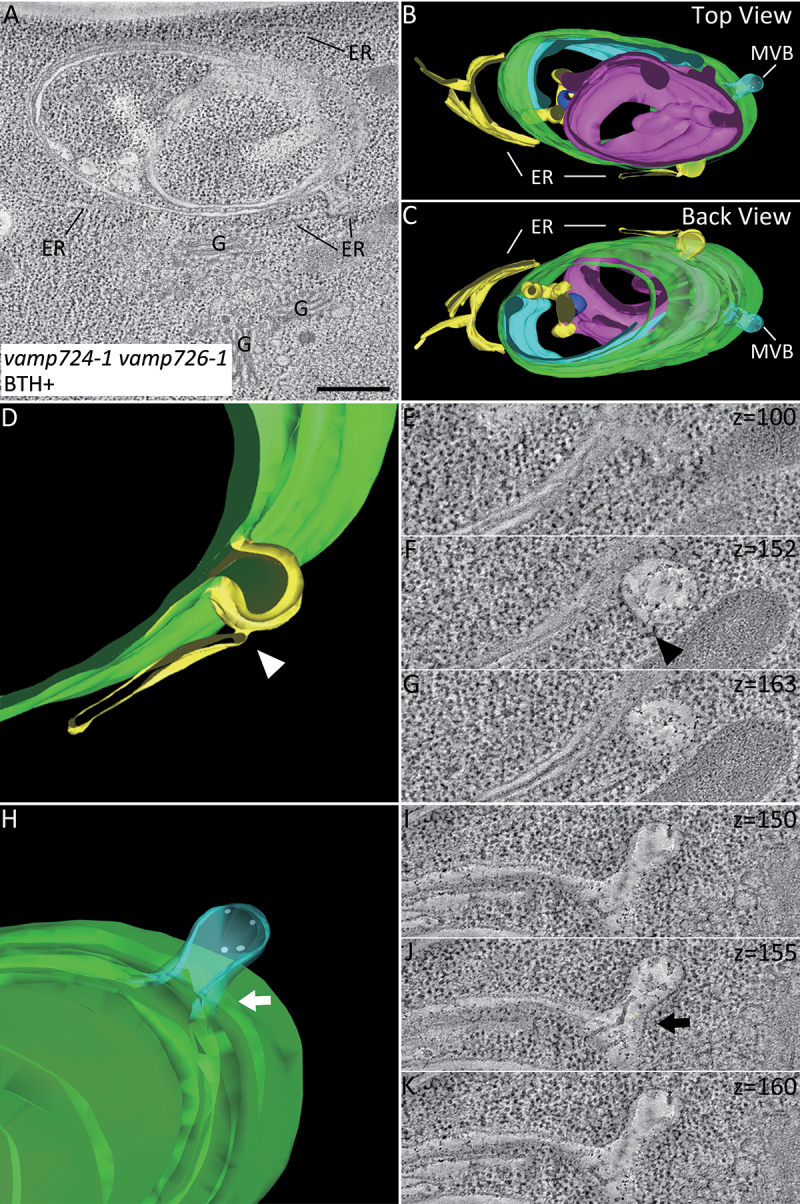
A 3D Electron tomography (ET) analysis reveals direct connection between the autophagic tubule and the ER membranes in root cells of vamp724-1 vamp726-1 mutant upon autophagy induction. A 3D tomographic analysis was performed on the high-pressure frozen root tip cells of vamp724-1 vamp726-1 mutant after 6-h BTH treatment. (A) Tomographic slice showing a representative example of the abnormal autophagic structure in BTH-treated root cells of vamp724-1 vamp726-1 mutant. (B and C) The 3D tomographic models of the autophagic structure shown in (A). (D to G) The 3D model and tomographic slices showing direct connection between the autophagic tubule and the ER as indicated by the arrowhead. (H to K) The 3D model and tomographic slices showing connection between MVB and autophagosome as indicated by the arrow. ER, endoplasmic reticulum; G, Golgi apparatus. MVB, multivesicular body. Bar: 500 nm.
VAMP724 and VAMP726 partially colocalized with an autophagosome marker upon autophagy induction in transgenic plants
VAMP724 and VAMP726 partially colocalized with ATG8e in Arabidopsis protoplasts (Figure S1). We also generated transgenic Arabidopsis plants expressing YFP-VAMP724 and YFP-VAMP726, driven by a constitutive ubiquitin (UBQ) promoter, followed by crossing into the autophagosome marker line mCherry-ATG8e to generate double transgenic Arabidopsis plants YFP-VAMP724 mCherry-ATG8e and YFP-VAMP726 mCherry-ATG8e. Under normal condition, both VAMP724 and VAMP726 displayed plasma membrane (PM) plus puncta patterns, showing rare colocalization with the limiting mCherry-ATG8e puncta (Figure 6A). However, upon autophagic induction by BTH treatment, both VAMP724 and VAMP726 showed partial colocalization with the mCherry-ATG8e-labeled puncta (Figure 6B) and the colocalization ratio increased significantly (Figure 6C). To examine the possible effects of VAMP724 and VAMP726 overexpression on autophagosome formation and autophagic flux, transgenic seedlings of mCherry-ATG8e, YFP-VAMP724 mCherry-ATG8e, and YFP-VAMP726 mCherry-ATG8e were subjected to autophagic induction by BTH treatment, followed by confocal imaging and quantification analysis. The results showed that, upon BTH induction, the numbers of autophagosomes formed in YFP-VAMP724 mCherry-ATG8e, and YFP-VAMP726 mCherry-ATG8e were similar to that of the control mCherry-ATG8e (Figure S7F). No abnormal autophagic structures were observed in BTH-treated root cells of YFP-VAMP724 mCherry-ATG8e and YFP-VAMP726 mCherry-ATG8e (Figure 6B), indicating that overexpression of VAMP724 and VAMP726 did not affect autophagosome formation. Quantification of the numbers of autophagic bodies showed that, upon BTH + Conc A treatments, the numbers of autophagic bodies inside the vacuoles in YFP-VAMP724 mCherry-ATG8e and YFP-VAMP726 mCherry-ATG8e were similar to that of the control mCherry-ATG8e (Figure S7E and S7G), which was further confirmed by immunoblot detection of vacuolar turnover of mCherry-ATG8e in these transgenic plants (Figure S7H). In addition, upon BTH treatment, the ratio of vacuolar import of mCherry-ATG8e in YFP-VAMP724 mCherry-ATG8e and YFP-VAMP726 mCherry-ATG8e showed no significant difference from the control mCherry-ATG8e (Figure S7I). Taken together, the autophagic flux was not affected by over-expression of VAMP724 and VAMP726.
Figure 6.
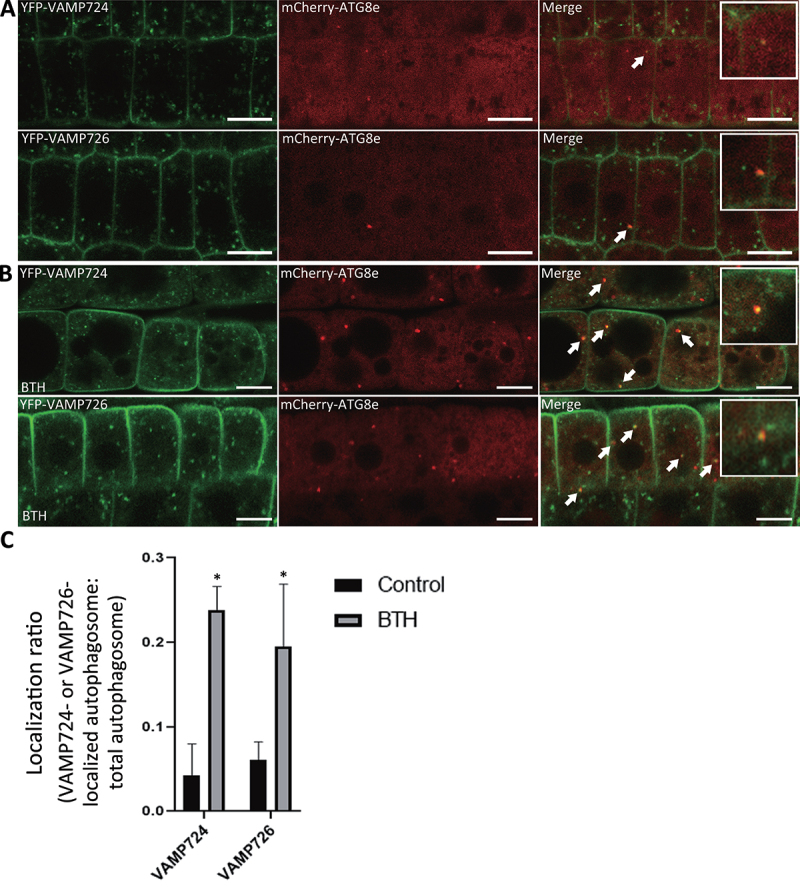
VAMP724 and VAMP726 partially colocalized with the autophagosome marker ATG8e upon autophagy induction in transgenic Arabidopsis plants. Transgenic Arabidopsis plants expressing YFP-VAMP724 or YFP-VAMP726 were crossed into the autophagosome marker line mCherry-ATG8e to generate the double transgenic plants of YFP-VAMP724 mCherry-ATG8e and YFP-VAMP726 mCherry-ATG8e respectively. Root cells of 5-day-old double transgenic seedlings were incubated in medium without (A) or with BTH (B) to induce autophagy, followed by confocal imaging analysis. Arrows indicated examples of colocalization between the two fusion proteins with enlargement in the insets. Bar: 10 μm. (C) Quantification of the colocalization ratio of VAMP724 or VAMP726-colocalized autophagosomes over total autophagosomes shown in (A) and (B). Results were obtained from three independent experiments (error bars ± SD). *P < 0.05, two-sided Student’s t-test.
In addition, under normal growth condition, both YFP-VAMP724 and YFP-VAMP726 showed a dual localization in PM and trans-Golgi network (TGN) (Figure S7A,B), consistent with previous results [34]. Taken together, YFP-VAMP724 and YFP-VAMP726 showed a dual localization in PM and TGN under normal condition, while a population respond to autophagy induction and colocalize with ATG8e.
VAMP724 and VAMP726 associate with ATG9 and are involved in the formation of autophagosome
Similar autophagosome-related tubular structures with direct connection to the ER were previously observed in an ATG9 mutant atg9-3 [30]. ATG9-containing vesicles were also shown to be in close proximity to the TGN and late endosome [30]. In addition, the recruitment of ATG9-containing vesicles to the phagophore assembly site (PAS) is required for the initiation of autophagosome formation and it has been reported that the ATG9-containing vesicles transiently interacted with the ATG8-positive autophagic membrane, moving away after a short period of interaction [30]. Therefore, we hypothesized that there might be a relationship between ATG9 and VAMP724 or VAMP726 in plant autophagy.
To gain an insight into the possible link between ATG9 and VAMP724 or VAMP726 in autophagy, transient expression in Arabidopsis protoplasts was used to analyze the subcellular association among ATG9, ATG8e, and VAMP724 or VAMP726. When ATG9-CFP and mCherry-ATG8e were co-expressed together with YFP-VAMP724 or YFP-VAMP726 respectively, overlapping of the three signals was observed in both cases (Figure 7A). To find out whether the overlapping signals represent the ATG9-containing vesicles or TGN, we next transiently co-expressed ATG9-CFP, YFP-ATG8e, and the TGN marker RFP-SYP61 together in protoplasts for confocal imaging analysis. As shown in Figure S8B, a population of ATG9 puncta was associated with the TGN marker (indicated by arrowheads) but separated from the autophagosome marker ATG8e, whereas the ATG8e-associated ATG9 puncta were separated from the TGN marker SYP61 (indicated by arrows). Thus, the overlapping signals of ATG9, ATG8e, and VAMP724 or VAMP726 were not the TGN but likely representing the transient interaction between the ATG9-containing vesicle and the ATG8e-positive PAS. In addition, the co-immunoprecipitation (Co-IP) assay, using the protein extracts from Arabidopsis cells co-expressing ATG9-5FLAG and VAMP724 or VAMP726, showed that ATG9-5FLAG was immunoprecipitated by both VAMP724 and VAMP726 (Figure 7B), indicating the association of ATG9 and the two SNAREs. Moreover, FRET analysis suggested an in vivo interaction between ATG9 and VAMP724 or VAMP726 as their FRET efficiency was significantly higher than the control (Figure 7C,D).
Figure 7.
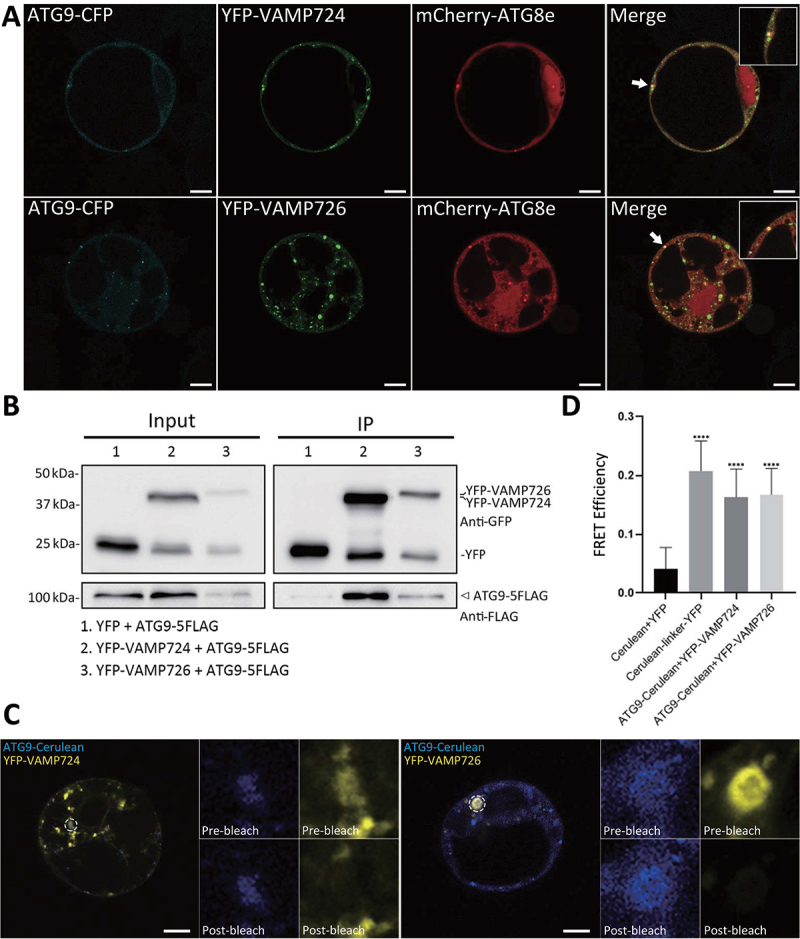
VAMP724 and VAMP726 are associated with ATG9 in Arabidopsis. (A) Subcellular localization analysis among ATG9, ATG8 and VAMP724 or VAMP726. The triple-constructs were transiently expressed together in Arabidopsis protoplasts and incubated for 12 h, followed by confocal imaging analysis. Arrows indicated examples of colocalization among the three fusions proteins with enlargement in the inset. Bar: 10 μm. (B) Immunoprecipitation (IP) assay shows association between ATG9 and VAMP724 or VAMP726. Arabidopsis protoplasts co-expressing ATG9-5FLAG with YFP (lane 1), YFP-VAMP724 (lane 2), or YFP-VAMP726 (lane 3) were subjected to protein extraction and IP with GFP-trap, followed by immunoblot analysis with indicated antibodies. Arrowhead indicates the ATG9 proteins immunoprecipitated by VAMP724 or VAMP726. (C) FRET analysis of the colocalized puncta between VAMP724 and ATG9 (left) or VAMP726 and ATG9 (right). The dashed oval represents the region for photobleaching. Bar: 10 μm. (D) Quantification of FRET efficiency using the acceptor photobleaching approach. 10 individual protoplasts were used for the statistical analysis. Error bars are the S.D. ****P < 0.0001, two-sided Student’s t-test.
Next, we also investigated the possible relationship between VAMP726 and ATG9 at the ultrastructural level via immunogold-TEM analysis. We first crossed the ATG9-mCherry line into YFP-VAMP726 to generate double transgenic plants YFP-VAMP726 ATG9-mCherry. We then prepared ultrathin sections from autophagy-induced YFP-VAMP726 ATG9-mCherry after high pressure freezing/freeze substitution (HPF/FS) for double-immunogold labeling using anti-GFP and anti-RFP antibodies to examine the correlation between ATG9 and VAMP726 during autophagy (Figure 8). Indeed, ATG9-positve 10-nm gold particles (indicated by arrowheads) resided on the TGN which was also labeled by VAMP726-positive 6-nm gold particles (indicated by arrows) (Figure 8A,B). In addition, both VAMP726 and ATG9 were detected at the edge of an expanding autophagosome, as labeled by anti-GFP (arrows) and anti-RFP (arrowheads) antibodies, respectively (Figure 8C,D), which is similar to the observation in yeast where Atg9-GFP was confined to the edges of phagophores [35].
Figure 8.
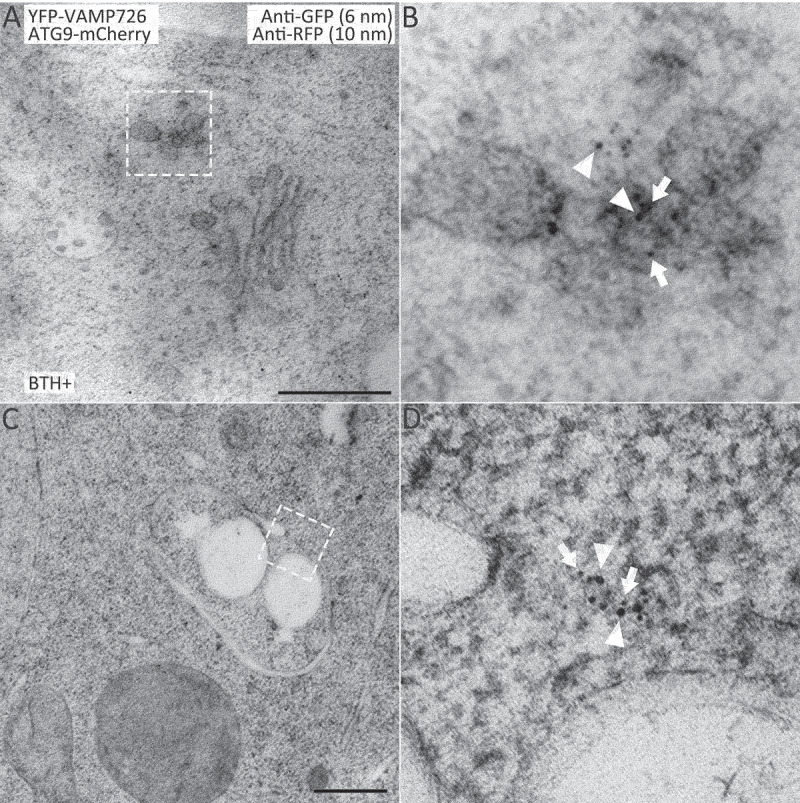
ATG9 resides on TGN and traffics together with VAMP726 to the forming autophagosome upon autophagy-induction. Ultrathin sections were prepared from high-pressure frozen/freeze-substituted root cells of YFP-VAMP726 ATG9-mCherry seedlings upon 6-h BTH treatment, followed by double immunogold labeling with anti-GFP (6-nm gold, indicated by arrows) and anti-RFP antibodies (10-nm gold, indicated by arrowheads). (A) ATG9 resides on the TGN (trans-Golgi network) labeled by VAMP726. (B) Enlargement of the indicated box in (A). (C) A membrane expanding phagophore. (D) Enlargement of the indicated box in (C), both anti-ATG9 and anti-VAMP726 antibodies labeled the edge of an expanding phagophore. Bar: 500 nm.
We speculate two possibilities for the functional relationship between ATG9 and VAMP724 or VAMP726: 1) VAMP724 or VAMP726 may regulate the trafficking of ATG9-containing vesicles, and 2) ATG9-containing vesicles may carry and deliver VAMP724 or VAMP726. We next performed transient expression experiment using the leaf protoplast system (Figure S8C-I). YFP-ATG8e and mCherry-ATG9 were transiently co-expressed in leaf protoplasts isolated from WT or vamp724-1 vamp726-1 mutants, followed by a time-lapse confocal imaging analysis (Figure S8C,D). In WT cells, transient association between ATG9 and ATG8e was observed (Figure S8C), which is consistent with previous observations [30]. However, in vamp724-1 vamp726-1 mutant cells, the ATG9 and ATG8e puncta were relatively separated and the association between ATG9 and ATG8e was decreased (Figure S8D,E). YFP-VAMP724 or YFP-VAMP726 was also transiently co-expressed with the TGN marker RFP-SYP61 in leaf protoplasts isolated from WT or atg9-3 mutant [30]. Both YFP-VAMP724 and YFP-VAMP726 displayed PM and TGN localization in atg9-3 mutant cells, similar to the WT cells (Figure S8F-I). Thus, ATG9 trafficking may be regulated by VAMP724 and VAMP726. Taken together, VAMP724 and VAMP726 are closely associated with ATG9, likely mediating its trafficking for autophagosome formation.
Discussion
In this study, using a combination of genetic, cellular and biochemical approaches, we have demonstrated the role of two SNARE proteins VAMP724 and VAMP726 in the autophagy pathway in Arabidopsis. We have shown that genetic depletion of VAMP724 and VAMP726 results in nutrient-sensitive phenotypes (Figure 1) and reduction of autophagy flux (Figure 2A-F). Confocal and immuno-EM analysis revealed the formation of tubular autophagic structures in the mutants (Figure 2A,E, and 4A). A direct connection between the tubular structure and ER was visualized by 3D-ET analysis (Figure 5). Under normal conditions, VAMP724 and VAMP726 showed PM and TGN localization (Figure S7A,B), but upon autophagy induction, a population of VAMP724 and VAMP726 colocalized with ATG8e (Figure 6). Furthermore, we have illustrated that VAMP724 and VAMP726 are closely associated with ATG9 (Figure 7 and Figure 8). The defected autophagic flux and ER-connected abnormal autophagosomes observed in this study are similar to those in ATG9-defected mutants [30]. Thus, VAMP724 and VAMP726 function in autophagosome formation in close proximity with ATG9. Future study will need to be carried out to examine and compare the autophagosome formation and the relative autophagic phenotypes among the vamp724-1 vamp726-1, atg9, and vamp724-1 vamp726-1 atg9 mutants, which will further illustrate the possible correlations between ATG9 and VAMP724 or VAMP726 in plant autophagy.
ATG9 is a six-TMD (transmembrane domain) protein which plays a key role in autophagosome formation. In yeast, Atg9-containing vesicles (30-60 nm in diameter) are derived from the Golgi apparatus and may shuttle between the Golgi and endosomes dependent on the TRAPPIII complex [36,37]. Atg9-containing vesicles nucleate on the PAS upon autophagic induction. After nucleation on the PAS, Atg9 is embedded in the phagophore outer membrane, and it may be recycled back into the cytoplasm for cargo recruitment by new Atg9-containing vesicles before autophagosome-vacuole fusion [36]. On the other hand, the mammalian ATG9 compartments cycle from the Golgi apparatus to endosomes dynamically [38]. Using correlative light electron microscopy (CLEM) and super-resolution imaging, ATG9-positive tubular-vesicular clusters are visualized on the RAB11-positive recycling endosomes or adjacent to the ATG13-positive ER subdomains upon starvation [39]. Recently, extensive studies in yeast and mammalian systems suggest that ATG9 is more of a scramblase of phospholipids than a transporter of lipids [40]. Instead, the yeast Atg2 and human ATG2A protein was shown to be the lipid transporter, which links the growing phagophores to membrane sources such as the ER [41-43]. It is suggested that Atg9-containing vesicles are not the membrane source, but rather the seeds of phagophore growth, as confirmed by a recent reconstitution study of yeast autophagosome nucleation [44]. In Arabidopsis, ATG9-containing vesicles also traffic through the early secretory pathway and closely associate with post-Golgi endosomes and may reside on motile compartments [30,45]. In Arabidopsis, ATG9-containing vesicles display transient interactions with the autophagosome membrane [30]. In this study, ATG9 was found to partially colocalize with autophagosome marker ATG8e, together with VAMP724 and VAMP726 in Arabidopsis protoplasts (Figure 7A), likely representing the transient interaction of ATG9-containing vesicles with PAS, and with the involvement of VAMP724 and VAMP726. In addition, ATG9 was detected on the TGN labeled by VAMP726 (Figure 8B), suggesting that Arabidopsis ATG9 residing on motile endosomal compartments and Golgi-endosomal system is a key route for the production of ATG9-containing vesicles as implied by previous findings [30,46]. Moreover, both ATG9 and VAMP726 labeled the edge of an expanding phagophore (Figure 8C-D), likely representing the remaining of the seed of autophagosome development. Previous studies have shown that, by using correlative light electron microscopy (CLEM) approaches, ATG9 localizes to vesicular-tubular clusters that are tightly associated with an ER subdomain in both mammalian cells [39] and yeast cells [47]. It will thus be interesting to use similar approaches to study the intercellular behavior of ATG9, VAMP724, and VAMP726 during autophagy in future study.
VAMP724 and VAMP726 may function similarly in Arabidopsis autophagy. In this study, double mutant vamp724-1 vamp726-1 showed enhanced sensitivity towards autophagy conditions compared to single mutant vamp724-1 or vamp726-1 (Figure 1 and 2), indicating the possible functional overlap of these two proteins. 70.8% identical in amino acid sequence, VAMP724 and VAMP726 belong to the VAMP72 SNARE family, which consists of 7 proteins: VAMP721 to VAMP727 [16]. It has been shown that VAMP721 and VAMP722, which share 96% identity in amino-acid sequence, function redundantly in the secretory pathway, as well as in pathogen resistance [48-51]. This further indicates the possibility that VAMP724 and VAMP726 may have a redundant function in autophagy in a similar manner. However, additional experiments are required to verify this possibility. The localization of the 7 SNAREs in the VAMP72 clade has been demonstrated [34] and our observations are consistent with previous results: PM plus endosome (VAMP721, VAMP722, VAMP724, VAMP725 and VAMP726), PVC/MVB (VAMP727) and ER (VAMP723). The subcellular targeting of the VAMP7 proteins in Arabidopsis is dependent on their longin domains as displayed by domain swap experiments [52]. As shown in this study, the autophagy-related phenotypes in the double mutant vamp724-1 vamp726-1 appear to be less severe as compared to the atg7-2 mutant (Figure 1). It is possible that, in the absence of VAMP724 and VAMP726, other SNAREs (possibly other VAMP72 SNAREs) may replace their function in autophagy conditions. It would thus be interesting to characterize these SNAREs for their possible involvement in autophagy in plants in future studies.
Our study also suggests the possible involvement of VAMP724 and VAMP726 in the interplay between the autophagy and endocytic pathways. Although the mechanism of interplay between the autophagosome and late endosome in yeast and mammalian cells is still elusive, evidence has shown that autophagosome formation as well as autophagic activity are severely affected upon malfunction of some regulators in the conventional MVB-vacuole/lysosome pathway [53]. Similarly, in Arabidopsis, dysfunction of ESCRT-III and related factors (e.g., VPS2.1 and AMSH3) results in defected autophagy pathway [54]. In yeast and animal cells, besides directly fusing with the vacuolar/lysosomal membrane, autophagosomes may also fuse with late endosomal compartments like MVBs to become an amphisome for expansion or maturation before their fusion with vacuole/lysosomes [53,55]. It has been reported that, in a cell line that is specialized towards exosome secretion, the mammalian SNARE protein VAMP3 mediates the fusion between autophagosome and MVB [56]. In addition, STX17 is also shown to mediate the fusion between autophagosome and late endosome [10]. In plants, the fusion between autophagosomes and late endosomal compartments is not well characterized [57,58]. However, hybrid structures between autophagosomes and MVBs have been observed in the free1 mutants, although the mechanism for their fusion is still unknown [29,59]. In this study, we have shown that VAMP724 and VAMP726 are PM- and TGN-localizing SNAREs (Figure S7A,B), with the same localization with VAMP721 and VAMP722. Additionally, it has been reported that VAMP724 interacts with the PM-localized Qa-SNARE SYP123, Qa-SNARE SYP121 and SYP132, which is similar to VAMP721 and VAMP722 [48,60]. In Petunia inflate, a homolog of VAMP726 showed a tip-focused localization in the growing pollen tube, suggesting its possible role in the recycling of endocytic vesicles in tip growth [61]. These indicate that VAMP724 and VAMP726 are involved in the conventional endocytic pathway. As shown in this study, depletion of VAMP724 and VAMP726 results in reduction of autophagy flux (Figure 2A-F) and the formation of tubular autophagic structures (Figure 2A,E, and 4A). Interestingly, connection between autophagosome and MVB was observed in the vamp724-1 vamp726-1 double mutant (Figure 5H-K). In addition, possible fusion between autophagosome and MVB was also observed in leaf protoplasts derived from the vamp724-1 vamp726-1 mutant plants (Figure S5). Thus, VAMP724 and VAMP726 may also play a role in the crosstalk between autophagy pathway and the endocytic pathway in plant, and it will be interesting to unravel the underlying mechanism in future studies.
Materials and methods
Plasmid construction
The YFP fusion constructs used for transient expression in protoplasts were created by cloning the PCR-amplified cDNA into the pBI221 backbone containing the constitutive 35S promoter [62] by restriction digestion. For the YFP-VAMP724 and YFP-VAMP726 transgenic plants, the full-length cDNA of VAMP724 or VAMP726 was amplified and cloned into the pBI121 backbone containing the ubiquitin (UBQ) promoter for construction of the YFP fusion [63]. Primers used for cloning are listed in Table S1.
Plant materials and growth conditions
All mutants and transgenic lines used in this study are in the Arabidopsis ecotype Columbia-0 background. To generate the transgenic plants, all the resulting constructs were introduced into Agrobacterium tumefaciens strain GV3101 and transformed into WT Arabidopsis Columbia-0 plants by the floral dip method [64]. The following previously published marker lines were crossed into the transgenic plants expressing YFP-VAMP724 and YFP-VAMP726 as described in the results section: the trans-Golgi marker (ST-RFP), PVC/late endosome marker (mCherry-RHA1), TGN marker line (VHA-a1-RFP), the autophagosome marker line (mCherry-ATG8e) and the transgenic line expressing ATG9-mCherry [30].
The T-DNA insertional lines vamp724-1 (SAIL_569_E12), vamp724-2 (SALK_032219), vamp726-1 (SALK_082690), and vamp726-2 (SALK_147419) were obtained from the Arabidopsis Information Resource (TAIR) (https://www.arabidopsis.org/). The atg7-2 (GABI_655B06) [65] T-DNA insertional mutant was obtained from the Nottingham Arabidopsis Stock Centre. Genotyping was performed to obtain homozygous mutant lines according to instructions from Salk Institute Genomic Analysis Laboratory (http://signal.salk.edu). Homozygous double mutant vamp724-1 vamp726-1 was generated by crossing of the two single mutants vamp724-1 and vamp726-1, followed by genotyping selection of the F2 generation. All the primers used for genotyping are listed in Table S1. The 1300-UBQ-YFP-ATG8e construct [30] was used to generate various mutants expressing YFP-ATG8e.
For normal growth conditions, Arabidopsis seeds were sterilized and germinated on Murashige and Skoog (MS) [66] plates (full-strength MS salts [Sigma-Aldrich, M5524], 0.8% [w:v] phyto agar [PlantMedia, 40100072], and 1% [w:v] sucrose [ATCG Limited, S011], pH 5.7). The plates were kept at 4°C for 2 days, followed by incubation in the growth chamber at 22°C under a long-day (16 h light/8 h dark) photoperiod. Adult plants were grown on soil in a growth room at 22°C under a long-day (16 h light/8 h dark) cycle. The light intensity is ~150 μmol/m2/s.
Transient expression and confocal microscopy imaging
Transient expression in Arabidopsis PSBD protoplasts was performed as described previously [14,15,30]. Confocal fluorescence images were acquired 14 h after transformation using a Leica SP8 confocal microscope (Leica, Wetzlar, Germany) with a 63× water lens. A sequential acquisition was applied for observing fluorescent proteins. The Pearson–Spearman correlation for colocalization relationships was calculated using ImageJ software (Wayne Rasband, NIH, https://imagej.nih.gov/) and the PSC plug-in as described previously [67]. Images were processed using Adobe Photoshop software (https://www.adobe.com/) as described previously [68,69].
For transient expression analysis in leaf-derived protoplasts, mesophyll protoplasts preparation and plasmid transformation were performed as described previously [70]. Transformed protoplasts were cultured in dark for 16 h and were subsequently visualized by confocal microscopy.
FM 4-64 staining
For staining PM and endosomes, seedlings were washed with liquid MS medium and then stained with FM 4-64 (Invitrogen, T-3166) at a final concentration of 12 mM in liquid MS for 30 min. For tonoplast staining, seedlings were washed with liquid MS medium and then stained with FM 4-64 at a final concentration of 12 mM in liquid MS for 7 h.
Chemical treatments
For autophagic treatments, 4- or 5-day-old seedlings were transferred in liquid MS with methanol (1:100) as control or 100 μM BTH (Sigma-Aldrich, 32,820) for 6 h before observation or as indicated. Conc A (Santa Cruz Biotechnology, sc-202111) was used at a concentration of 0.5 μM.
RNA extraction and Real-time PCR
Total RNA was extracted from plate-grown plants by grinding the tissues in liquid nitrogen in the presence of TRIzol reagent (Invitrogen, 15596-026) according to the manufacturer’s instructions. First-strand cDNA was synthesized from 2 mg of total RNA using M-MLV reverse transcriptase (Promega, M170A) following the manufacturer’s protocols. The cDNA from WT or mutant seedlings was used as a template for real-time PCR. Gene specific primers are listed in Table S1. The mRNA expression level was normalized with reference gene ACTIN2.
Phenotypic assays
Etiolated hypocotyl elongation assay and hypocotyl length measurement was performed as described previously [25,26]. Briefly, seeds were sterilized and germinated on MS plates without sucrose (C-). The plates were kept at 4°C for 2 days, followed by incubation in dark for 7 days at 22°C. The length of hypocotyls was measured using ImageJ software.
Nitrogen starvation assay was performed essentially as described previously [21-24]. Briefly, 6-day-old seedlings grown on nitrogen-rich (N+) MS plates under a long-day (16 h light/8 h dark) photoperiod were transferred for an additional 2 weeks to either nitrogen-rich (N+) MS liquid medium or nitrogen-deficient (N-) MS liquid medium. Chlorophyll contents were measured as described previously [71,72]. Briefly, chlorophyll of the seedling bundle was extracted by immersion in 3 mL of N,N-dimethylformamide (Sigma-Aldrich, D4254) for 48 h in the dark at 4°C. Absorbance was measured at 664 and 647 nm, and the total chlorophyll content was normalized to gram fresh weight per sample.
For detached leaf assay, 5th and 6th rosette leaves from 4-week-old soil-grown plants under a long-day (16 h light/8 h dark) photoperiod were detached and soaked in water. After 0 or 5 days of dark treatment, the leaves were photographed.
Protein extraction and immunoblot analysis
Protein isolation and blotting were performed as described previously [30,73,74]. Briefly, 5-d-old seedlings were ground in liquid nitrogen and extracted with the lysis buffer containing 25 mM Tris·HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% SDS, and 1× Complete Protease Inhibitor Cocktail (Roche, 11873580001). The total cell extracts were centrifuged at 20,000 × g for 30 min at 4°C. The supernatant protein samples were boiled in 1xSDS sample loading dye at 95°C and subjected to SDS-PAGE followed by immunoblotting. For immunoblot analysis, rabbit GFP antibodies (home-made) were used at a concentration of 4 μg/mL, and cFBPase (Agrisera, AS04043) was used at a dilution of 1:1,000. For quantification analysis, protein band intensities were quantified using ImageJ software, and all values were normalized according to the protein loading control before calculation.
ATG8 lipidation assay
An ATG8 lipidation assay was carried out essentially as described previously [20,30]. Briefly, 5-day-old seedlings were transferred in liquid MS with methanol (1:100) as control, or with 100 μM BTH and 0.5 μM Conc A for 6 h, respectively, followed by extraction in lysis buffer containing 25 mM Tris·HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, and 1× Complete Protease Inhibitor Cocktail (Roche, 11873580001). The total cell extracts were centrifuged at 500 × g for 5 min at 4°C. The supernatant was centrifuged at 100,000 × g for 30 min, and the membrane pellets were solubilized in an equal volume of lysis buffer with additional 1% Triton X-100 (Sigma-Aldrich, T8787). Protein samples were subjected to SDS-PAGE in the presence of 6 M urea (USB Corporation, 75826), followed by immunoblotting with anti-ATG8 (Agrisera, AS142769) antibodies at a dilution of 1:1,000.
TEM analysis and immunogold labeling
The general procedures for preparing TEM samples and ultrathin sectioning of samples have been described previously [30,59,74,75]. Briefly, for high-pressure freezing, root tips of 4- or 5-day-old seedlings were dissected and immediately frozen in a high-pressure freezing apparatus (HPM100, EM PACT2, Leica), with subsequent freeze substitution in dry acetone containing 0.1% uranyl acetate at -85°C. Infiltration with HM20, embedding, and UV polymerization were performed stepwise at -35°C. For immunolabeling, standard procedures were performed as described previously [30,59,74,75]. The rat anti-ATG8e [28] was used at 40 μg/mL, RFP antibodies (ChromoTek, 5F8) at 1:100, and calreticulin antibodies [32] at a dilution of 1:200. Gold-coupled secondary antibodies were used at 1:40 dilution. Sections were examined using a Hitachi H-7650 transmission electron microscope with a charge-coupled device camera operating at 80 kV (Hitachi High-Technologies).
Electron tomography, 3D reconstruction, and modeling
The general procedures for performing electron tomography have been described previously [30,75]. In brief, 300-nm-thick sections were cut and poststained with uranyl acetate and lead citrate. Electron tomography observations were performed with Tecnai F20 electron microscope (FEI Company) operated at 200 kV. For each grid, a tilt image stack from +60° to −60° with 1.5° increments was collected, and the second tilt image stack was collected by rotating the grid by 90°. Dual-axis tomograms were calculated from pairs of image stacks with the Etomo program of the IMOD software package (bio3d.colorado.edu). For model generation, the contours were drawn manually and meshed with the 3dmod program in the IMOD software package.
Immunoprecipitation
Protein extraction and immunoprecipitation (IP) were performed as described previously [30,73,74]. In brief, transformed protoplasts (~107) were first diluted threefold with 250 mM NaCl and then harvested by centrifugation at 100 × g for 10 min. Total cell lysates of the harvested protoplasts were prepared in lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.5 mM EDTA, 0.4% Nonidet P-40 [USB Corporation, 19626], 5% glycerol, and 1×Complete Protease Inhibitor Cocktail), followed by centrifugation at 20,000 × g for 30 min at 4°C. The supernatant was incubated with GFP-Trap agarose beads (ChromoTek, gtma) for 4 h at 4°C in IP buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.5 mM EDTA, 0.2% Nonidet P-40, 5% glycerol, and 1×Complete Protease Inhibitor Cocktail) in a top to end rotator. After incubation, the beads were washed four times with ice-cold washing buffer (10 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.5 mM EDTA, 0.05% Nonidet P-40, and 5% glycerol) and boiled in SDS sample buffer. Samples were analyzed by SDS-PAGE and immunoblot using the anti-GFP (home-made) or anti-FLAG (Sigma-Aldrich, F1804) antibodies.
FRET analysis
FRET analysis was performed using the Leica SP8 confocal microscope as described previously [29,69]. Arabidopsis PSBD protoplasts were used to express the target proteins. Before FRET analysis, fixation was performed by incubation of protoplasts in 3% formaldehyde (Sigma-Aldrich, P-6148) for 15 min at room temperature, followed by two rounds of washing using PBS. Before and after photobleaching on the region of interest, the signal intensity of the donor and acceptor proteins was measured and FRET efficiency was calculated through the built-in SP8 algorithm.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 8 (GraphPad Software). Data were presented as mean ± SD (standard deviation) and statistical significance was analyzed by two-sided Student´s t-test (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). The colocalization relationship of two fluorescent signals was quantified using the PSC colocalization plug-in in the ImageJ program and results were presented by scatterplot images as previously described [67].
Accession numbers
The Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are as follows: VAMP721 (AT1G04750), VAMP722 (AT2G33120), VAMP723 (At2G33110), VAMP724 (AT4G15780), VAMP725 (AT2G32670), VAMP726 (AT1G04760), VAMP727 (AT3G54300), ATG8e (AT2G45170), ATG9 (AT2G31260), SH3P2 (AT4G34660), and FREE1 (AT1G20110).
Supplementary Material
Funding Statement
This work was supported by grants from the National Natural Science Foundation of China (91854201), the Research Grants Council of Hong Kong (AoE/M-05/12, CUHK 14100818, 14101219, C4033-19E, C4002-17G, C4002-20W, C4002-21EF, C2009-19G, and R4005-18), and The Chinese University of Hong Kong (CUHK) Research Committee and CAS-Croucher Funding Scheme for Joint Laboratories to L.J; the National Natural Science Foundation of China (32061160467 and 31870171) and Fok Ying-Tong Education Foundation for Young Teachers in the Higher Education Institutions of China (171014) to C.G.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary Material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15548627.2022.2127240.
References
- [1].Xie Z, Klionsky DJ.. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9(10):1102–9. [DOI] [PubMed] [Google Scholar]
- [2].Liu Y, Bassham DC.. Autophagy: pathways for self-eating in plant cells. Annu Rev Plant Biol. 2012;63:215–37. [DOI] [PubMed] [Google Scholar]
- [3].Morishita H, Mizushima N.. Diverse cellular roles of autophagy. Annu Rev Cell Dev Biol. 2019;35:453–475. [DOI] [PubMed] [Google Scholar]
- [4].Lipka V, Kwon C, Panstruga R.. SNARE-ware: the role of SNARE-domain proteins in plant biology. Annu Rev Cell Dev Biol. 2007;23:147–74. [DOI] [PubMed] [Google Scholar]
- [5].Nair U, Jotwani A, Geng J, et al. SNARE proteins are required for macroautophagy. Cell. 2011;146(2):290–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Darsow T, Rieder SE, Emr SD.. A multispecificity syntaxin homologue, Vam3p, essential for autophagic and biosynthetic protein transport to the vacuole. J Cell Biol. 1997;138(3):517–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Liu X, Klionsky DJ.. The Atg17-Atg31-Atg29 complex and Atg11 regulate autophagosome-vacuole fusion. Autophagy. 2016;12(5):894–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Bas L, Papinski D, Licheva M, et al. Reconstitution reveals Ykt6 as the autophagosomal SNARE in autophagosome-vacuole fusion. J Cell Biol. 2018;217(10):3656–3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Moreau K, Ravikumar B, Renna M, et al. Autophagosome precursor maturation requires homotypic fusion. Cell. 2011;146(2):303–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Itakura E, Kishi-Itakura C, Mizushima N.. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 2012;151(6):1256–69. [DOI] [PubMed] [Google Scholar]
- [11].Matsui T, Jiang P, Nakano S, et al. Autophagosomal YKT6 is required for fusion with lysosomes independently of syntaxin 17. J Cell Biol. 2018;217(8):2633–2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Surpin M, Zheng H, Morita MT, et al. The VTI family of SNARE proteins is necessary for plant viability and mediates different protein transport pathways. Plant Cell. 2003;15(12):2885–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zouhar J, Rojo E, Bassham DC.. AtVPS45 is a positive regulator of the SYP41/SYP61/VTI12 SNARE complex involved in trafficking of vacuolar cargo. Plant Physiol. 2009;149(4):1668–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Miao Y, Jiang L.. Transient expression of fluorescent fusion proteins in protoplasts of suspension cultured cells. Nat Protoc. 2007;2(10):2348–53. [DOI] [PubMed] [Google Scholar]
- [15].Gao C, Yu CK, Qu S, et al. The Golgi-localized Arabidopsis endomembrane protein12 contains both endoplasmic reticulum export and Golgi retention signals at its C terminus. Plant Cell. 2012;24(5):2086–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Sanderfoot A. Increases in the number of SNARE genes parallels the rise of multicellularity among the green plants. Plant Physiol. 2007;144(1):6–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Oyama T, Shimura Y, Okada K.. The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Gene Dev. 1997;11(22):2983–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Yang C, Shen WJ, Yang LM, et al. HY5-HDA9 module transcriptionally regulates plant autophagy in response to light-to-dark conversion and nitrogen starvation. Mol Plant. 2020;13(3):515–531. [DOI] [PubMed] [Google Scholar]
- [19].Thompson AR, Doelling JH, Suttangkakul A, et al. Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 2005;138(4):2097–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Chung T, Phillips AR, Vierstra RD.. ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A and ATG12B loci. Plant J. 2010;62(3):483–93. [DOI] [PubMed] [Google Scholar]
- [21].Phillips AR, Suttangkakul A, Vierstra RD.. The ATG12-conjugating enzyme ATG10 Is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics. 2008;178(3):1339–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Suttangkakul A, Li F, Chung T, et al. The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell. 2011;23(10):3761–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Li F, Chung T, Vierstra RD.. AUTOPHAGY-RELATED11 plays a critical role in general autophagy- and senescence-induced mitophagy in Arabidopsis. Plant Cell. 2014;26(2):788–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Lin Y, Zeng Y, Zhu Y, et al. Plant Rho GTPase signaling promotes autophagy. Mol Plant. 2021;14(6):905–920. [DOI] [PubMed] [Google Scholar]
- [25].Avin-Wittenberg T, Bajdzienko K, Wittenberg G, et al. Global analysis of the role of autophagy in cellular metabolism and energy homeostasis in Arabidopsis seedlings under carbon starvation. Plant Cell. 2015;27(2):306–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Huang X, Zheng C, Liu F, et al. Genetic analyses of the Arabidopsis ATG1 kinase complex reveal both kinase-dependent and independent autophagic routes during fixed-carbon starvation. Plant Cell. 2019;31(12):2973–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Yoshimoto K, Hanaoka H, Sato S, et al. Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell. 2004;16(11):2967–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zhuang X, Wang H, Lam SK, et al. A BAR-domain protein SH3P2, which binds to phosphatidylinositol 3-phosphate and ATG8, regulates autophagosome formation in Arabidopsis. Plant Cell. 2013;25(11):4596–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Gao C, Zhuang X, Cui Y, et al. Dual roles of an Arabidopsis ESCRT component FREE1 in regulating vacuolar protein transport and autophagic degradation. Proc Natl Acad Sci U S A. 2015;112(6):1886–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Zhuang X, Chung KP, Cui Y, et al. ATG9 regulates autophagosome progression from the endoplasmic reticulum in Arabidopsis. Proc Natl Acad Sci U S A. 2017;114(3):E426–E435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Le Bars R, Marion J, Le Borgne R, et al. ATG5 defines a phagophore domain connected to the endoplasmic reticulum during autophagosome formation in plants. Nat Commun. 2014;5:4121. [DOI] [PubMed] [Google Scholar]
- [32].Crofts AJ, Leborgne-Castel N, Hillmer S, et al. Saturation of the endoplasmic reticulum retention machinery reveals anterograde bulk flow. Plant Cell. 1999;11(11):2233–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Hayashi-Nishino M, Fujita N, Noda T, et al. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol. 2009;11(12):1433–7. [DOI] [PubMed] [Google Scholar]
- [34].Uemura T, Ueda T, Ohniwa RL, et al. Systematic analysis of SNARE molecules in Arabidopsis: dissection of the post-Golgi network in plant cells. Cell Struct Funct. 2004;29(2):49–65. [DOI] [PubMed] [Google Scholar]
- [35].Gomez-Sanchez R, Rose J, Guimaraes R, et al. Atg9 establishes Atg2-dependent contact sites between the endoplasmic reticulum and phagophores. J Cell Biol. 2018;217(8):2743–2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Yamamoto H, Kakuta S, Watanabe TM, et al. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J Cell Biol. 2012;198(2):219–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Shirahama-Noda K, Kira S, Yoshimori T, et al. TRAPPIII is responsible for vesicular transport from early endosomes to Golgi, facilitating Atg9 cycling in autophagy. J Cell Sci. 2013;126(Pt 21):4963–73. [DOI] [PubMed] [Google Scholar]
- [38].Orsi A, Razi M, Dooley HC, et al. Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, are required for autophagy. Mol Biol Cell. 2012;23(10):1860–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Karanasios E, Walker SA, Okkenhaug H, et al. Autophagy initiation by ULK complex assembly on ER tubulovesicular regions marked by ATG9 vesicles. Nat Commun. 2016;7:12420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Chang C, Jensen LE, Hurley JH.. Autophagosome biogenesis comes out of the black box. Nat Cell Biol. 2021;23(5):450–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Maeda S, Otomo C, Otomo T.. The autophagic membrane tether ATG2A transfers lipids between membranes. Elife. 2019;8: e45777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Osawa T, Kotani T, Kawaoka T, et al. Atg2 mediates direct lipid transfer between membranes for autophagosome formation. Nat Struct Mol Biol. 2019;26(4):281–288. [DOI] [PubMed] [Google Scholar]
- [43].Valverde DP, Yu S, Boggavarapu V, et al. ATG2 transports lipids to promote autophagosome biogenesis. The J Cell Biol. 2019;218(6):1787–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Sawa-Makarska J, Baumann V, Coudevylle N, et al. Reconstitution of autophagosome nucleation defines Atg9 vesicles as seeds for membrane formation. Science. 2020;369(6508):eaaz7714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Zhuang X, Chung KP, Luo M, et al. Autophagosome biogenesis and the endoplasmic reticulum: a plant perspective. Trends Plant Sci. 2018;23(8):677–692. [DOI] [PubMed] [Google Scholar]
- [46].Davis S, Wang J, Ferro-Novick S.. Crosstalk between the secretory and autophagy pathways regulates autophagosome formation. Dev Cell. 2017;41(1):23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Mailler E, Guardia CM, Bai X, et al. The autophagy protein ATG9A enables lipid mobilization from lipid droplets. Nat Commun. 2021;12(1):6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Kwon C, Neu C, Pajonk S, et al. Co-option of a default secretory pathway for plant immune responses. Nature. 2008;451(7180):835–40. [DOI] [PubMed] [Google Scholar]
- [49].Zhang L, Zhang H, Liu P, et al. Arabidopsis R-SNARE proteins VAMP721 and VAMP722 are required for cell plate formation. PloS One. 2011;6(10):e26129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Zhang B, Karnik R, Wang Y, et al. The Arabidopsis R-SNARE VAMP721 Interacts with KAT1 and KC1 K+ Channels to Moderate K+ Current at the Plasma Membrane. Plant Cell. 2015;27(6):1697–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Yun HS, Kwaaitaal M, Kato N, et al. Requirement of vesicle-associated membrane protein 721 and 722 for sustained growth during immune responses in Arabidopsis. Mol Cells. 2013;35(6):481–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Uemura T, Sato MH, Takeyasu K.. The longin domain regulates subcellular targeting of VAMP7 in Arabidopsis thaliana. FEBS lett. 2005;579(13):2842–6. [DOI] [PubMed] [Google Scholar]
- [53].Lamb CA, Dooley HC, Tooze SA.. Endocytosis and autophagy: Shared machinery for degradation. BioEssays. 2013;35(1):34–45. [DOI] [PubMed] [Google Scholar]
- [54].Kalinowska K, Isono E.. All roads lead to the vacuole-autophagic transport as part of the endomembrane trafficking network in plants. J Exp Bot. 2018;69(6):1313–1324. [DOI] [PubMed] [Google Scholar]
- [55].Muller M, Schmidt O, Angelova M, et al. The coordinated action of the MVB pathway and autophagy ensures cell survival during starvation. Elife. 2015;4:e07736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Fader CM, Sanchez DG, Mestre MB, et al. TI-VAMP/VAMP7 and VAMP3/cellubrevin: two v-SNARE proteins involved in specific steps of the autophagy/multivesicular body pathways. Biochim Biophys Acta. 2009;1793(12):1901–16. [DOI] [PubMed] [Google Scholar]
- [57].Zhuang X, Cui Y, Gao C, et al. Endocytic and autophagic pathways crosstalk in plants. Curr Opin Plant Biol. 2015;28:39–47. [DOI] [PubMed] [Google Scholar]
- [58].Cui Y, He Y, Cao W, et al. The multivesicular body and autophagosome pathways in plants. Front Plant Sci. 2018;9:1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Gao C, Luo M, Zhao Q, et al. A unique plant ESCRT component, FREE1, regulates multivesicular body protein sorting and plant growth. Curr Biol. 2014;24(21):2556–63. [DOI] [PubMed] [Google Scholar]
- [60].Ichikawa M, Hirano T, Enami K, et al. Syntaxin of plant proteins SYP123 and SYP132 mediate root hair tip growth in Arabidopsis thaliana. Plant Cell Physiol. 2014;55(4):790–800. [DOI] [PubMed] [Google Scholar]
- [61].Guo F, McCubbin AG.. The pollen-specific R-SNARE/longin PiVAMP726 mediates fusion of endo- and exocytic compartments in pollen tube tip growth. J Exp Bot. 2012;63(8):3083–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Woo CH, Gao C, Yu P, et al. Conserved function of the lysine-based KXD/E motif in Golgi retention for endomembrane proteins among different organisms. Mol Biol Cell. 2015;26(23):4280–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Lam SK, Siu CL, Hillmer S, et al. Rice SCAMP1 defines clathrin-coated, trans-golgi-located tubular-vesicular structures as an early endosome in tobacco BY-2 cells. Plant Cell. 2007;19(1):296–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Clough SJ, Bent AF.. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–43. [DOI] [PubMed] [Google Scholar]
- [65].Hofius D, Schultz-Larsen T, Joensen J, et al. Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell. 2009;137(4):773–83. [DOI] [PubMed] [Google Scholar]
- [66].Murashige T, Skoog F.. A revised medium for rapid growth and bio assays with tobacco fissue cultures. Physiol Plant. 1962;15(3):473–497. [Google Scholar]
- [67].French AP, Mills S, Swarup R, et al. Colocalization of fluorescent markers in confocal microscope images of plant cells. Nat Protoc. 2008;3(4):619–28. [DOI] [PubMed] [Google Scholar]
- [68].Jiang L, Rogers JC.. Integral membrane protein sorting to vacuoles in plant cells: evidence for two pathways. J Cell Biol. 1998;143(5):1183–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Chung KP, Zeng Y, Li Y, et al. Signal motif-dependent ER export of the Qc-SNARE BET12 interacts with MEMB12 and affects PR1 trafficking in Arabidopsis. J Cell Sci. 2018;29;131(2):jcs202838. [DOI] [PubMed] [Google Scholar]
- [70].Yoo SD, Cho YH, Sheen J.. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc. 2007;2(7):1565–72. [DOI] [PubMed] [Google Scholar]
- [71].Xiao S, Gao W, Chen QF, et al. Overexpression of Arabidopsis acyl-CoA binding protein ACBP3 promotes starvation-induced and age-dependent leaf senescence. Plant Cell. 2010;22(5):1463–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Qi H, Xia FN, Xie LJ, et al. TRAF family proteins regulate autophagy dynamics by modulating AUTOPHAGY PROTEIN6 stability in Arabidopsis. Plant Cell. 2017;29(4):890–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Shen J, Zhao Q, Wang X, et al. A plant Bro1 domain protein BRAF regulates multivesicular body biogenesis and membrane protein homeostasis. Nat Commun. 2018;9(1):3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Zhao Q, Shen J, Gao C, et al. RST1 is a FREE1 suppressor that negatively regulates vacuolar trafficking in Arabidopsis. Plant Cell. 2019;31(9):2152–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Cui Y, Cao W, He Y, et al. A whole-cell electron tomography model of vacuole biogenesis in Arabidopsis root cells. Nat Plants. 2019;5(1):95–105. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


