Abstract
Transcription Regulatory Regions Database (TRRD) has been developed for accumulation of experimental information on the structure–function features of regulatory regions of eukaryotic genes. Each entry in TRRD corresponds to a particular gene and contains a description of structure–function features of its regulatory regions (transcription factor binding sites, promoters, enhancers, silencers, etc.) and gene expression regulation patterns. The current release, TRRD 4.2.5, comprises the description of 760 genes, 3403 expression patterns, and >4600 regulatory elements including 3604 transcription factor binding sites, 600 promoters and 152 enhancers. This information was obtained through annotation of 2537 scientific publications. TRRD 4.2.5 is available through the WWW at http://wwwmgs.bionet.nsc.ru/mgs/dbases/trrd4/
INTRODUCTION
Information on structure and function of long regulatory regions of eukaryotic genes, transcribed by RNA POL II, is accumulated in the TRRD database (1). Each entry of this database corresponds to a gene. The annotated part of an entry includes the structure–function description of gene regulatory regions composed by regulatory units (promoters, silencers, enhancers, etc.), individual transcription factor binding sites that constitute these regulatory units, and transcription factors that bind to these sites. In addition, the entry contains the gene expression patterns and references to original publications.
DATABASE DESCRIPTION
The format used in TRRD is based on the model of module-hierarchic organization of transcription regulatory regions of eukaryotic genes (Fig. 1). According to this model, the lowest level of regulatory region organization corresponds to transcription factor binding sites, the next level to composite elements (2) and the highest level to enhancers, silencers, core promoters and other types of regulatory units. The regulatory units listed above are components of transcription regulatory regions that represent the next hierarchic regulatory level and are located in 5′- and 3′-gene flanking regions and in introns. Description of a regulatory element of each level in addition to structure characteristics (element’s location and nucleotide sequence) may be supplemented with its functional features (effect on gene transcription activity, tissue- and stage-specificities, etc). Thus, the TRRD database allows both the structural and functional features of gene regulatory regions, which may contain various tissue- and stage-specific regulatory elements, alternative promoters (and the corresponding alternative transcription start sites), silencers, enhancers, etc., to be described. Finally, this allows the system of integral regulation of gene transcription to be described (Fig. 1). A more detailed description of the TRRD database format (release 4.1) has been previously published (1).
Figure 1.
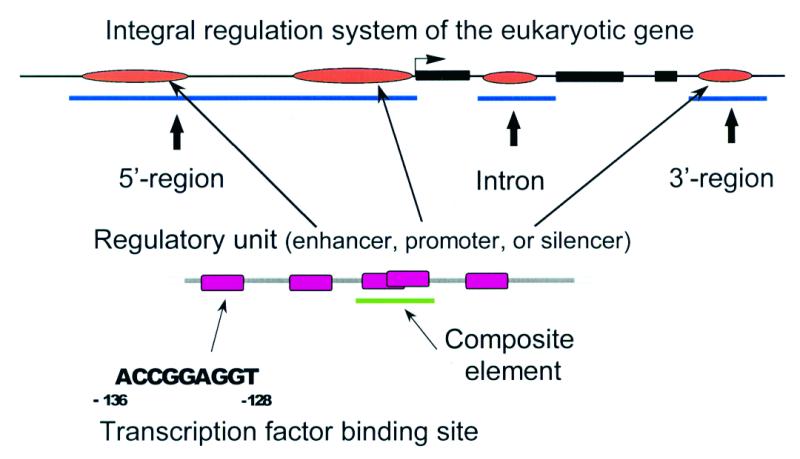
The model of structure–function organization of regulatory gene regions in eukaryotes, on which the TRRD database format is based.
The release number of the TRRD database consists of three digits divided by points, e.g., the current release number is 4.2.5. By adding the information without format modification, only the last digit is altered. Such updates occur 1–3 times per month. The second digit is altered when the format is slightly modified, for example, new fields are added into the database sections or the format of the fields is changed. The first digit of the release number is altered if the format globally changes, e.g., new informational blocks or tables are introduced.
The format of release TRRD 4.2.5 contains several additions as compared with release 4.1:
• A new field, NI, has been introduced to contain the site functionality index. This index is 1 where a site function has been demonstrated experimentally through studying the effects of deletions or mutations in the site on its binding with transcription factor and on the expression level of the reporter gene in plasmid constructions; otherwise the index is 2.
• Each TRRD entry contains the list of publications annotated to describe the corresponding gene. Description of each paper contains field AI reflecting its annotation state: 1, if the paper has been annotated; otherwise, 0.
• The letter A is added to the gene accession number (for example, the accession number 00274 of the gene for interferon-beta in release 4.1 is substituted with A00274 in release 4.2.5).
• The letter S is added to the site accession number (for example, accession number 2294 of ATP-2 binding site in release 4.1 is substituted with S2294 in release 4.2.5).
CONTENT OF THE CURRENT RELEASE
The current release, TRRD 4.2.5, contains the description of 760 genes, 1068 regulatory units, 3604 transcription factor binding sites and 3403 expression patterns. This information was extracted by annotating 2537 publications. The dynamics of information volume accretion in the TRRD database is shown in Figure 2.
Figure 2.
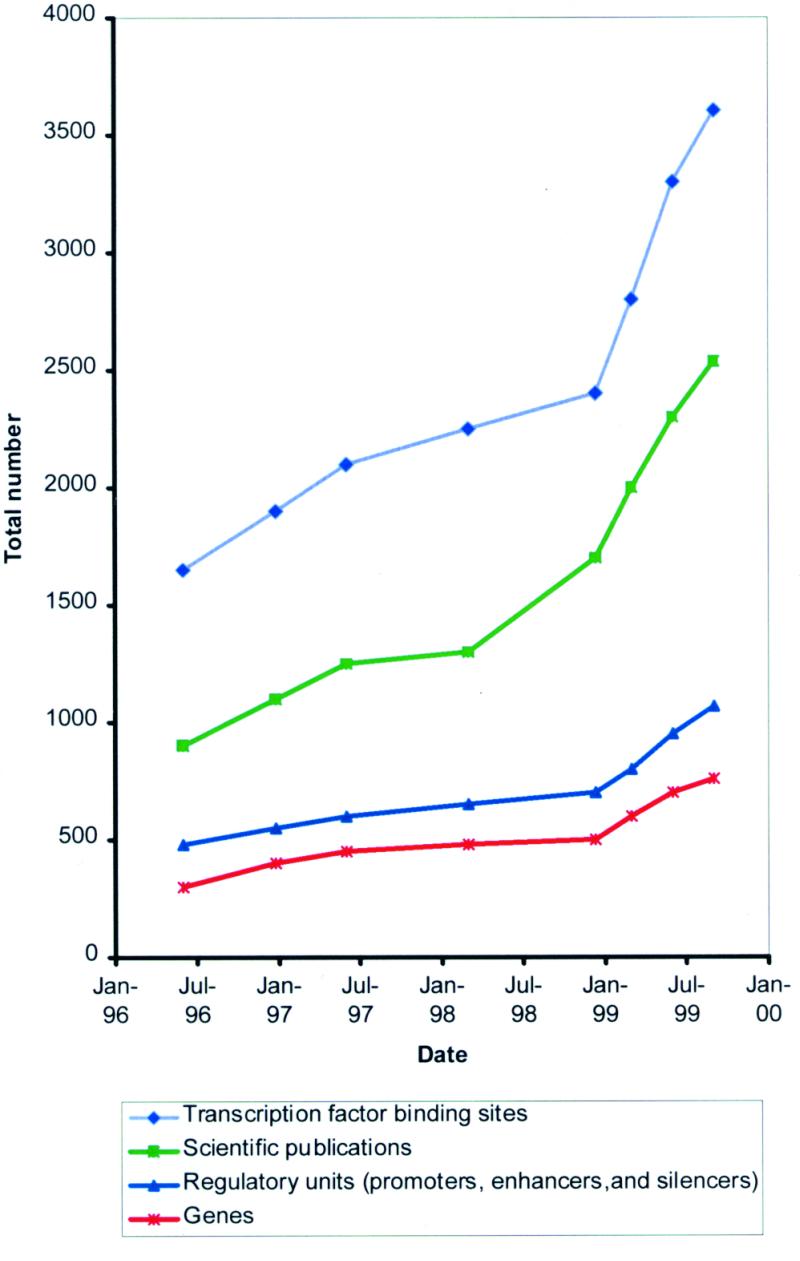
Dynamics of the TRRD database accretion (from 1996 to 1999).
Development of TRRD now is directed to description of individual functional gene systems (3–8). Listed in Table 1 are the main functional sections of the TRRD database, with the numbers of genes, regulatory units and sites contained in each section.
Table 1. Functional sections of the TRRD database.
| TRRD section | Genes | Regulatory units | Sites |
|---|---|---|---|
| Genes of Lipid Metabolism (LM-TRRD) | 77 | 124 | 495 |
| Endocrine System Transcription Regulatory Regions Database (ES-TRRD) | 60 | 90 | 232 |
| Cell Cycle-Dependent Genes | 51 | 84 | 213 |
| Glucocorticoid-Controlled Genes | 39 | 68 | 328 |
| Erythroid-Specific Regulated Genes (ESRG-TRRD) | 51 | 103 | 442 |
| Plant Genes (PLANT-TRRD) | 95 | 96 | 307 |
| Heat Shock-Induced Genes (HS-TRRD) | 66 | 59 | 168 |
| Interferon-Inducible Genes (IIG-TRRD) | 87 | 122 | 379 |
| Others | 234 | 421 | 1015 |
STANDARDIZATION OF INFORMATION INPUT
The program TRRD-INPUT (1) is used to standardize the information in TRRD. The controlled vocabularies for database sections, supported by this program, have been considerably expanded. The vocabularies describing organs, tissues and cells where the genes described in TRRD are expressed have been developed such that they are now united and organized hierarchically (Fig. 3). This hierarchy is used for modification of queries (generalized or specific) and for realization of associated searches in TRRD.
Figure 3.
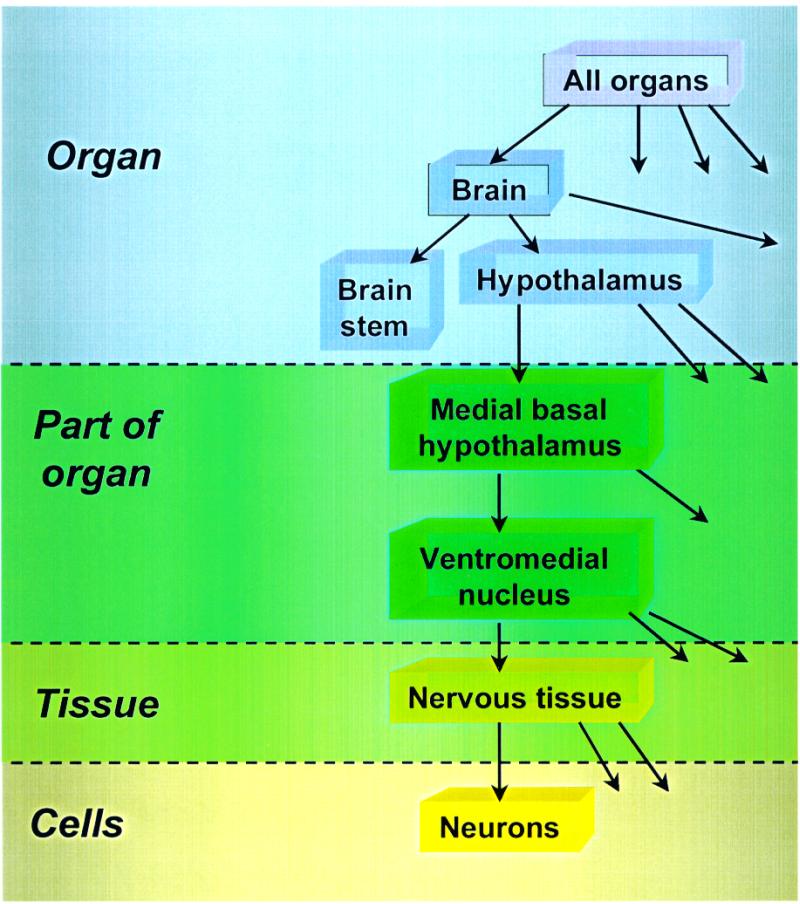
Hierarchical organization of controlled vocabularies of morphological terms in the TRRD database (using ventromedial nucleus neurons as example).
TRRD IMPLEMENTATION IN SRS AND LINKAGE TO OTHER INFORMATION RESOURCES
TRRD is maintained under SRS to provide an Internet access to the database. Implementation in SRS distributes the contents of the TRRD flat-file corresponding to an entry between the five interconnected tables (1): (i) TRRDGENES, general description of genes; (ii) TRRDEXP, description of the gene expression patterns; (iii) TRRDSITES, description of transcription factor binding sites; (iv) TRRDFACTORS, description of transcription factors; and (v) TRRDBIB, references. Such a distribution represents the information on expression regulation of each gene in a convenient manner.
The TRRD database is a constituent module of the GeneExpress system (9). The hyperlinks between the TRRD tables and informational resources of the GeneExpress system and other molecular–biological information resources are shown in Table 2. The links contained in different TRRD database tables allow the user to gain additional information on the sites and regulatory regions available in the other modules of GeneExpress. For example, from the field WW in TRRDSITES (Table 2), it is possible to activate the software for binding site recognition using the program RGSiteScan (http://wwwmgs.bionet.nsc.ru/Programs/yura/rgscan1.html ) (10).
Table 2. Links from TRRD tables to other information resources.
| TRRD table | Database/System | Number of links |
|---|---|---|
| TRRDSITES | ACTIVITYa | 282 |
| SAMPLESa | 1489 | |
| RGSiteScana | 633 | |
| EMBL/GenBank | 3363 | |
| TRANSFAC | 625 | |
| TRRDGENES | SWISS-PROT | 571 |
| EMBL/GenBank | 803 | |
| COMPEL | 101 | |
| EpoDB/GERD | 66 | |
| GeneNeta | 62 | |
| TRANSFAC | 299 | |
| Others | 199 | |
| TRRDFACTORS | TRANSFAC | 1331 |
| TRRDBIB | MEDLINE | 2443 |
aGeneExpress resources.
Field DR in TRRDSITES provides links to the ACTIVITY and SAMPLES databases. The ACTIVITY database contains experimental data on activity of transcription factor binding sites (11). The SAMPLES database contains samples of transcription factor binding sites and other regulatory regions of DNA and RNA molecules (12). As a result, it is possible to navigate via hyperlinks to the other programming modules and databases of the GeneExpress system from any TRRD database entry. It provides the user with the ability to carry out an integrated analysis of the expression regulation of any gene described in TRRD.
In addition, the TRRD database is linked to other relevant databases, such as SWISS-PROT (13), EMBL/GenBank (14,15), TRANSFAC (16), COMPEL (2), EpoDB (17), GeneNet (18) and others (Table 2).
AVAILABILITY
TRRD 4.2.5 is available through the WWW at http://wwwmgs.bionet.nsc.ru/mgs/dbases/trrd4/ . A list of mirror sites is given at http://wwwmgs.bionet.nsc.ru/mgs/links/mirrors.html . TRRD flat-files are available on a collaborative basis. TRRD cannot be included into other databases without explicit permission of the authors. All rights reserved. The administrator of TRRD, Nikolay A. Kolchanov, can be contacted by Email: kol@bionet. nsc.ru . Please send comments, corrections and requests for additional information by Email or Fax (+7 3832 331278). Users are asked to refer to this paper and the previous publication (1) in reporting results obtained through TRRD application.
Acknowledgments
ACKNOWLEDGEMENTS
The authors are grateful to I. V. Lokhova for assistance in bibliographic searching, to G. B. Chirikova for translation of the paper into English, and to G. V. Orlova for helpful comments. The work was supported by the Russian Foundation for Basic Research (grant nos. 97-04-49740, 98-04-49479, 98-07-91078), Russian Human Genome Program, Ministry of Science and Technology of Russian Federation, Integrated Program of the Siberian Department of the Russian Academy of Sciences and the National Institutes of Health, USA (grant No. 5-R01-RR04026-09).
REFERENCES
- 1.Kolchanov N.A., Ananko,E.A., Podkolodnaya,O.A., Ignatieva,E.V., Stepanenko,I.L., Kel-Margoulis,O.V., Kel,A.E., Merkulova,T.I., Goryachkovskaya,T.N., Busygina,T.V., Kolpakov,F.A., Podkolodny,N.L., Naumochkin,A.N. and Romashchenko,A.G. (1999) Nucleic Acids Res., 27, 303–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kel O.V., Kel,A.E., Romaschenko,A.G., Wingender,E. and Kolchanov,N.A. (1997) Mol. Biol. (Mosk.), 31, 498–512. [Google Scholar]
- 3.Ananko E.A., Bazhan,S.I., Belova,O.E. and Kel,A.E. (1997) Mol. Biol. (Mosk.), 31, 592–604. [Google Scholar]
- 4.Podkolodnaya O.A. and Stepanenko,I.L. (1997) Mol. Biol. (Mosk.), 31, 562–574. [Google Scholar]
- 5.Ignateva E.V., Merkulova,T.I., Vishnevskii,O.V. and Kel,A.E. (1997) Mol. Biol. (Mosk.), 31, 575–591. [PubMed] [Google Scholar]
- 6.Merkulova T.I., Merkulov,V.M. and Mitina,R.L. (1997) Mol. Biol. (Mosk.), 31, 605–615. [Google Scholar]
- 7.Kel O.V. and Kel,A.E. (1997) Mol. Biol. (Mosk.), 31, 548–561. [Google Scholar]
- 8.Goryachkovskaya T.N., Ananko,E.A. and Peltek,S.E. (1998) Proceedings of the First International Conference on Bioinformatics of Genome Regulation and Structure, (BGRS ’98), ICG, Novosibirsk, Russia, Vol. I, pp. 50–53.
- 9.Kolchanov N.A., Ponomarenko,M.P., Frolov,A.S., Ananko,E.A., Kolpakov,F.A., Ignatieva,E.V., Podkolodnaya,O.A., Goryachkovskaya,T.N., Stepanenko,I.L., Merkulova,T.I., et al. (1999) Bioinformatics, 15, 669–686. [DOI] [PubMed] [Google Scholar]
- 10.Kondrakhin Yu.V., Babenko,V.N., Milanesi,L., Lavryushev,C.V. and Kolchanov,N.A. (1998) Bioinformatics, 14, 95–104. [Google Scholar]
- 11.Ponomarenko M.P., Ponomarenko,J.V., Frolov,A.S., Podkolodny,N.L., Savinkova,L.K., Kolchanov,N.A. and Overton,G.C. (1999) Bioinformatics, 15, 687–703. [DOI] [PubMed] [Google Scholar]
- 12.Vorobiev D.G., Ponomarenko,J.V. and Podkolodnaya,O.A (1998) Proceedings of the First International Conference on Bioinformatics of Genome Regulation and Structure, (BGRS ’98), ICG, Novosibirsk, Russia, Vol. I, pp. 58–61.
- 13.Bairoch A. and Apweiler,R. (1999) Nucleic Acids Res., 27, 49–54. Updated article in this issue: Nucleic Acids Res. (2000), 28, 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stoesser G., Tuli,M.A., Lopez,R. and Sterk,P. (1999) Nucleic Acids Res., 27, 18–24. Updated article in this issue: Nucleic Acids Res. (2000), 28, 19–23.9847133 [Google Scholar]
- 15.Benson D.A., Boguski,M.S., Lipman,D.J., Ostell,J., Ouellette,B.F., Rapp,B.A. and Wheeler,D.L. (1999) Nucleic Acids Res., 27, 12–17. Updated article in this issue: Nucleic Acids Res. (2000), 28, 15–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heinemeyer T., Chen,X., Karas,H., Kel,A.E., Kel,O.V., Liebich,I., Meinhardt,T., Reuter,I., Schacherer,F. and Wingender,E. (1999) Nucleic Acids Res., 27, 318–322. Updated article in this issue: Nucleic Acids Res. (2000), 28, 316–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stoeckert C.J. Jr, Salas,F., Brunk,B. and Overton,G.C. (1999) Nucleic Acids Res., 27, 200–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolpakov F.A., Ananko,E.A., Kolesov,G.B. and Kolchanov,N.A. (1998) Bioinformatics, 14, 529–537. [DOI] [PubMed] [Google Scholar]


