Summary
Gastruloids are 3D structures generated from pluripotent stem cells recapitulating fundamental principles of embryonic pattern formation. Using single-cell genomic analysis, we provide a resource mapping cell states and types during gastruloid development and compare them with the in vivo embryo. We developed a high-throughput handling and imaging pipeline to spatially monitor symmetry breaking during gastruloid development and report an early spatial variability in pluripotency determining a binary response to Wnt activation. Although cells in the gastruloid-core revert to pluripotency, peripheral cells become primitive streak-like. These two populations subsequently break radial symmetry and initiate axial elongation. By performing a compound screen, perturbing thousands of gastruloids, we derive a phenotypic landscape and infer networks of genetic interactions. Finally, using a dual Wnt modulation, we improve the formation of anterior structures in the existing gastruloid model. This work provides a resource to understand how gastruloids develop and generate complex patterns in vitro.
Keywords: gastruloids, symmetry breaking, cell states, pluripotency, embryoids, imaging, screening
Graphical abstract
Highlights
-
•
Spatial map of cell-type emergence in gastruloids at single-cell resolution
-
•
Phenotypic compound screen reveals functional modules underlying gastruloid formation
-
•
Spatial and temporal variabilities in the pluripotency state determines binary Wnt responses
-
•
Dual Wnt modulation improves the representation of anterior structures in gastruloids
Suppinger et al. employ scRNA-seq, an image-based trajectory and a phenotypic compound screen, to provide a resource characterizing murine gastruloid formation. Focusing on the early stages of gastruloid development, they show that variability in pluripotency states determines Wnt response and symmetry breaking. They, then, use dual Wnt modulation enriching for anterior structures.
Introduction
The metazoan body undergoes an essential phase of development called gastrulation, during which substantial morphological changes establish all major body axes. During the gastrula stage, cells spatially organize and differentiate into the three germ layers, which subsequently give rise to all organs and specialized cell types. In recent years, pluripotent stem cell-derived in vitro embryoids mimicking aspects of embryonic development, including gastrulation,1,2,3,4,5,6,7,8,9 have been developed. Among them are mouse gastruloids that recapitulate aspects such as axial organization and germ layer specification in a reductionist manner without extraembryonic tissues.10,11,12 Variations of the gastruloid protocol have also enabled the initiation of organogenesis,13,14,15 and with modified conditions, the generation of anterior neural derivatives.16,17,18,19 Because gastruloids are highly scalable and amenable to a variety of perturbations ranging from genetic manipulations to chimerism approaches,20 they are gaining popularity. Starting from ∼300 mESCs, the addition of a Wnt signaling agonist between 48 and 72 h of development induces a symmetry-breaking event resulting in elongated gastruloids exhibiting expression of the mesodermal marker Brachyury (Bra, T) at the posterior pole.8,21 Although it is known that this process relies on Wnt and Nodal signaling,10 the precise cellular behavior causing symmetry breaking, namely differentiation into distinct specialized lineages and their axial organization, starting from a uniform cell population in a homogeneous environment, remains largely unknown.
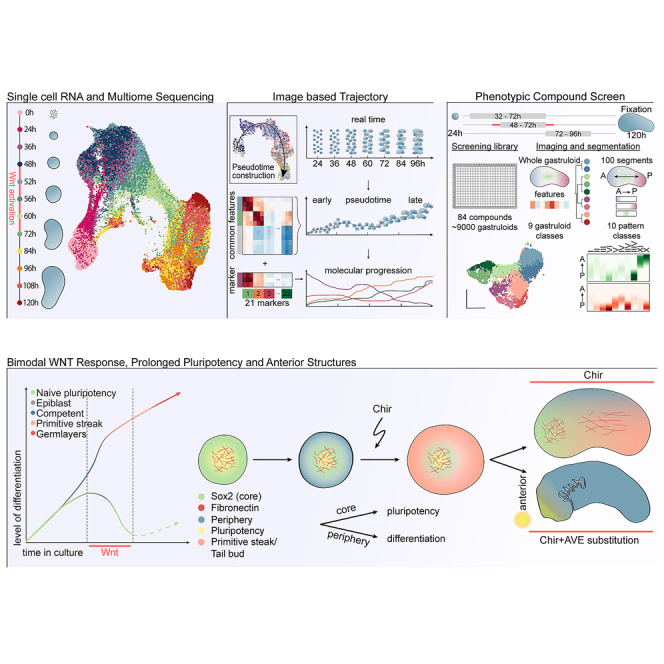
In this resource, we use single-cell RNA sequencing (scRNA-seq) and a high-content imaging platform to profile gastruloid formation from aggregation to elongation in tens of thousands of gastruloids. We identify important events regulating symmetry breaking: (1) a temporal difference in pluripotency exit between cells in the gastruloid core and periphery, (2) a differential response to Wnt activation between 48 and 72 h, with the gastruloid core reverting to a pluripotent state, whereas the periphery starts a primitive-streak-like genetic program, and (3) a subsequent radial symmetry breaking localizing the two populations at opposing poles along an anterior-posterior (AP) axis. We further performed a time-dependent compound screen that uncovered regulatory modules controlling the three main steps of gastruloid development and provide insights into the signaling mechanisms regulating gastruloid formation and cell fate determination. Finally, we further characterized screening hits and used them to develop a dual Wnt modulation approach generating gastruloids with an improved representation of anterior foregut and neural structures.
Results
Time course of gastruloid development
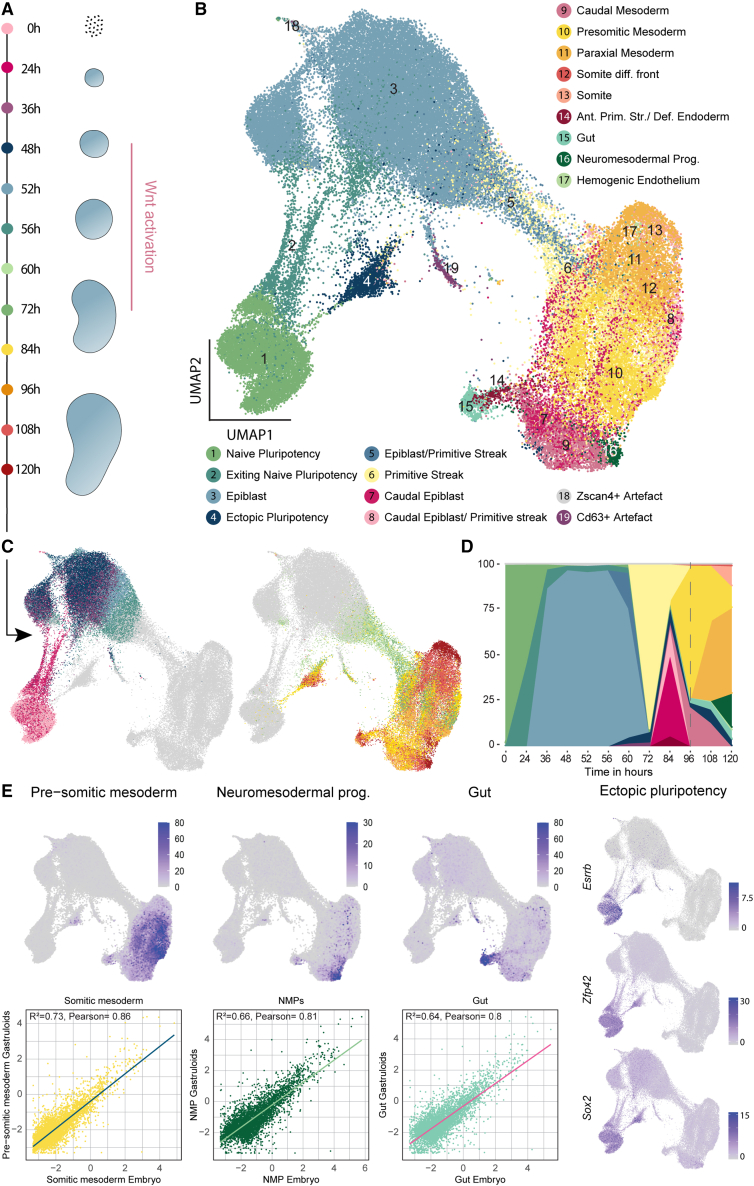
To study gastruloid development, we generated gastruloids as described by Beccari et al.11 (see STAR Methods) and performed scRNA-seq time course experiments with sampling from 0 to 120 h (Figure 1A). To identify gastruloid cell states, we clustered single-cell transcriptomes globally (Figure S1A) and from individual time points (Figure S1B) and used cluster alignment tool (CAT)22 to compare the clusters with annotated cell types from a published in vivo dataset21,23 (see STAR Methods). For most gastruloid clusters, the analysis resulted in single or strong matches to a particular embryonic cell type (Tables S1, S2, and S3). Based on the results from this analysis and marker gene expression, we annotated the cells generating a comprehensive atlas of gastruloid development (Figures 1B and S1C; Tables S4 and S5). Cells originated as naive pluripotent cells and were exiting this state during the first 24 h (Figure 1C). At 36 h, the cells resided in a broad epiblast state until 48 h when they received Wnt activation. During this activation, between 60 and 72 h, most of the cells started differentiating via a primitive streak-like state. At later time points, between 84 and 120 h, most of the cells fully committed to the three germ layers (Figure 1D). As expected, the gastruloids had an underrepresentation of anterior structures and rostral neuronal fates8,10 with a clear population of neuro-mesodermal progenitors (NMPs). We also saw the emergence of the definitive endoderm lineage, which further differentiated into the gut. Mesoderm was the most diverse lineage including cells with pre-somitic mesoderm (PSM), somite, and paraxial mesodermal identity. We report high similarities between gastruloid cell types and their respective in vivo counterparts (Figure 1E). Surprisingly, during Wnt activation, some cells (cluster 4 in Figure 1B; see temporal dynamics in Figures 1C and 1D) reverted to a population we term ectopic pluripotency (EP), as it displayed strong similarities with naive ES cells and expressed pluripotency markers such as Sox2, Esrrb, and Zfp42 (Figures 1E and S1C).24,25,26
Figure 1.
scRNA-seq time course of gastruloid development
(A) Scheme of gastruloid formation between 0 and 120 h, including sampling time points for scRNA-seq. Pink bar: Wnt activation.
(B) Uniform Manifold Approximation and Projection (UMAP) of scRNA-seq time course experiments including cell type annotations. Somite diff. front, somite differentiation front; Ant. Prim. Str., anterior primitive streak; Def. endoderm, definitive endoderm; NMPs, neuro-mesodermal progenitors.
(C) UMAP of single-cell transcriptomes highlighting sampling time points. Legend: (A).
(D) Ribbon plot showing changes in cell type composition over time. Dashed line: 96 h.
(E) UMAPs showing the aggregated expression of marker genes for the NMPs (Hes3, Hoxb9, Cdx4, and Epha5), gut (Sox17, Foxa2, Cer1, Krt8, Krt18, Shh, and Gsc), and pre-somitic mesoderm (Hes7, Aldh1a2, Dll1, Tbx6, Cyp26a1, and Hoxb1) (top). Scatter plots and inferred linear regression for conservation analysis between gastruloid and embryonic cells. Scatter plot: log2 mean expression of genes in gastruloid and embryonic cell types. Expression UMAPs for pluripotency genes Esrrb, Zfp42, and Sox2 (right).
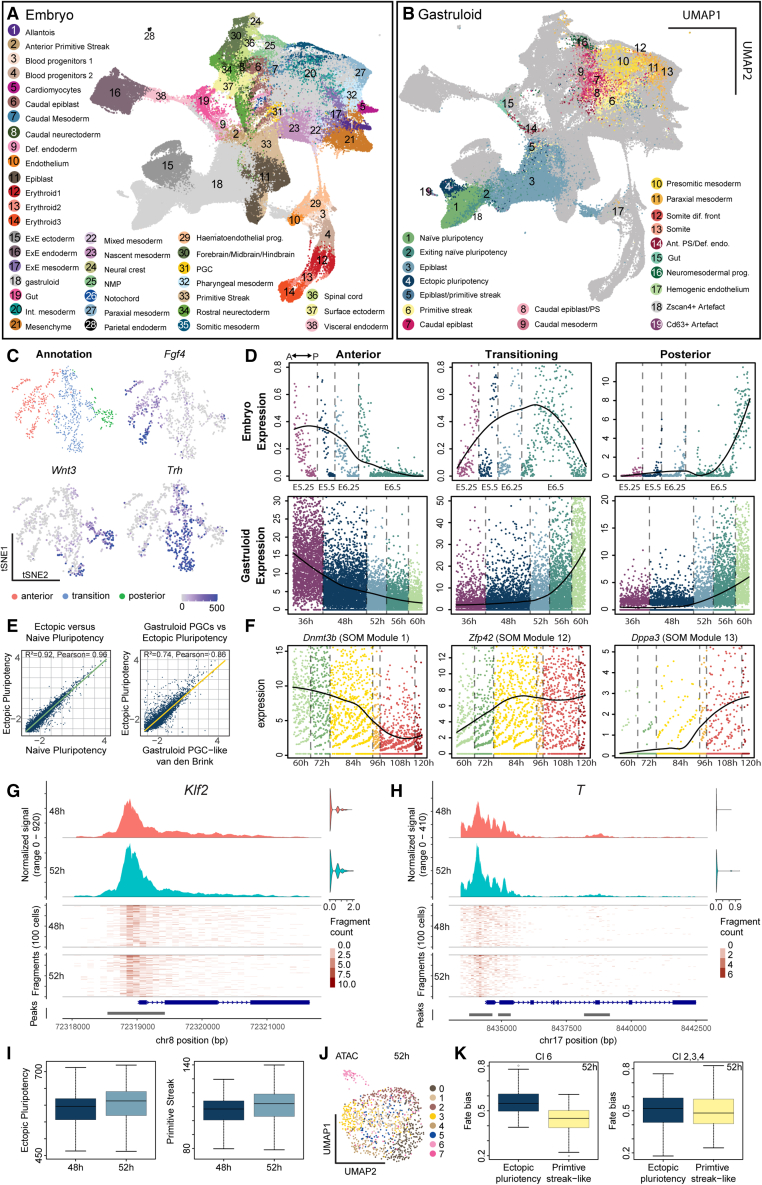
To systematically compare gastruloids with in vivo embryonic development, we integrated and co-embedded gastruloid and embryonic cells23 (Figures 2A, 2B, S2A, and S2B). The gastruloid cell types from time points after Wnt activation (>72 h) mostly co-clustered with their in vivo counterparts (Figure S2C). In contrast with these strong similarities, the cell types from earlier time points did not co-cluster as prominently, likely because the time points sampled in the reference dataset (E.6.5–E8.5) were not equivalent to early gastruloid time points. To further characterize our epiblast population, we compared it with an in vivo dataset that identified anterior, transition, and posterior states in early post-implantation epiblast and captured the acquisition of primitive streak propensity from E5.25 to E6.5 (Figures 2C and S2E).27 Post-implantation epiblast cells formed a continuum with a major axis of cellular state variability (see t-distributed stochastic neighbor embedding, t-SNE 2) corresponding to the AP axis (respective markers: Fgf4, Trh, and Wnt3) (Figures 2C and S2D). Gene signatures for the three epiblast states (Table S6) allowed us to generate temporal expression maps (Figures 2D and S2G) and showed that epiblast cells in gastruloids change from an anterior-like epiblast state at 36 h (pre-Wnt pulse) to a mixed transitioning and posterior-like state on Wnt activation (56–60 h).
Figure 2.
In vivo comparison and characterization of epiblast and pluripotency states
(A) UMAP of co-embedded gastruloid and embryonic cells highlighting embryonic cell types from Pijuan-Sala et al.23
(B) UMAP of co-embedded gastruloid and embryonic cells highlighting gastruloid cell types. PS, primitive streak; dif., differentiation; Ant. PS/Def. endo., anterior primitive streak/definitive endoderm.
(C) t-SNE map of single-cell transcriptomes from Cheng et al.27 highlighting three embryonic epiblast states and expression maps of Fgf4, Trh, and Wnt3.
(D) Temporal gene expression maps of anterior, transition, and posterior gene signatures for embryonic and gastruloid epiblast cells. y axis: normalized expression.
(E) Scatter plot and inferred linear regression comparing ectopic and naive pluripotency populations (left) and PGC-like15 populations (right). Scatter plot: mean expression of individual genes in ectopic and naive pluripotency populations or PGC-like populations.
(F) Temporal gene expression maps of Dppa3, Zfp42, Dnmt3b, of EP cells. y axis: normalized expression. Self-organizing map (SOM) modules (see Figure S2F).
(G) Coverage plot of chromatin accessibility for Klf2 and 1,000 bp upstream region from transcription start site (TSS) at 48 and 52 h. Right: multiome RNA expression of Klf2 in the same cells. Top: averaged frequency of DNA fragments within the genomic region. Middle: frequency of fragments within the genomic region for single cells. Lower: arrows indicate transcriptional direction. Bottom: peak coordinates within genome region.
(H) Coverage plot of chromatin accessibility for T and 1,000 bp upstream region from the TSS containing the promoter.
(I) Boxplots: aggregated gene activity scores for the EP and primitive streak-like state.
(J) UMAP of the scATAC-seq data-modality from the multiome highlighting clusters. Time point: 52 h.
(K) Boxplots of fate bias analysis at 52 h for multiome clusters toward EP and primitive steak-like populations.
At the same time, we report the emergence of the EP at 60 h. This population was very similar to naive pluripotent cells and a population of cells found in a published dataset (primordial germ cell [PGC]-like in van den Brink et al.15) (Figure 2E). Dynamic analysis of the EP (Figure S2F; Table S6) (see STAR Methods) revealed early, intermediate, and late gene expression modules with epiblast markers, such as Dnmt3b and the transitioning epiblast marker Trh gradually decreasing over time (Figure 2F). Conversely, the expression of pluripotency genes like Zfp42 increases gradually over time. In later time points (>84 h), a subset of EP starts to upregulate the PGC marker Dppa3, (PGCs marker). Interestingly, the CAT analysis shows that early EP had only a few matches, whereas the later EP matched to numerous distinct and mature cell types (Figure S1B). This indicates that EP started homogeneously and then acquired more heterogeneity over time, likely due to increasing complexity in the tissue context. At 120 h, one match of the EP was PGC, suggesting some similarities to in vivo PGCs (Figure S2H). However, we did not find EP co-clustering with in vivo PGCs (Figure 2A) which suggested that a subset of late EP might have had the potential to acquire but did not fully commit to a PGC identity at the assessed time points. Overall, we report a good resemblance between gastruloids with their in vivo counterparts. Nonetheless, we observed two phenomena, which were aberrant from in vivo gastrulation, namely the emergence of a mixed transitioning and posterior epiblast state and the existence of an EP population.
To further characterize the EP population during Wnt activation, we performed multiome sequencing (scRNA-seq + scATAC-seq) on gastruloids sampled at 48 and 52 h. Although the cells were in an epiblast state, the promoter regions of naive pluripotency genes such as Klf2 and Klf4 as well as those of primitive streak genes like T were accessible. However, the expression of these genes was not detected (Figures 2G, 2H, S2I, and S2J). Gene activity scores for single cells based on promoter and gene body accessibilities (see STAR Methods) revealed gene accessibility for naive, EP, and primitive streak signatures (Figures 2I and S2K). Interestingly, although there was no significant difference in the gene activity for the naive signature, we detected an increase in gene activity at 52 h for the EP and primitive streak-like signatures. Unsupervised clustering using both multiome-modalities (see STAR Methods) revealed several clusters at 52 h (e.g., cluster 6), (Figures 2J and S2L). Interestingly, the percentage of cells at 52 h in cluster 6 was 7.5%, which was similar to the fraction of cells annotated as EP at 60 h (5.6%) in the scRNA-seq data. Cell fate bias analysis toward the EP and primitive streak-like populations showed that cluster 6 had a higher fate bias toward EP compared with the primitive streak-like population (Figure 2K). These data suggest that on Wnt activation, there is a differential response to Wnt in epiblast cells, which drives a binary fate response: EP and primitive streak-like.
Spatial cell-type organization during gastruloid development
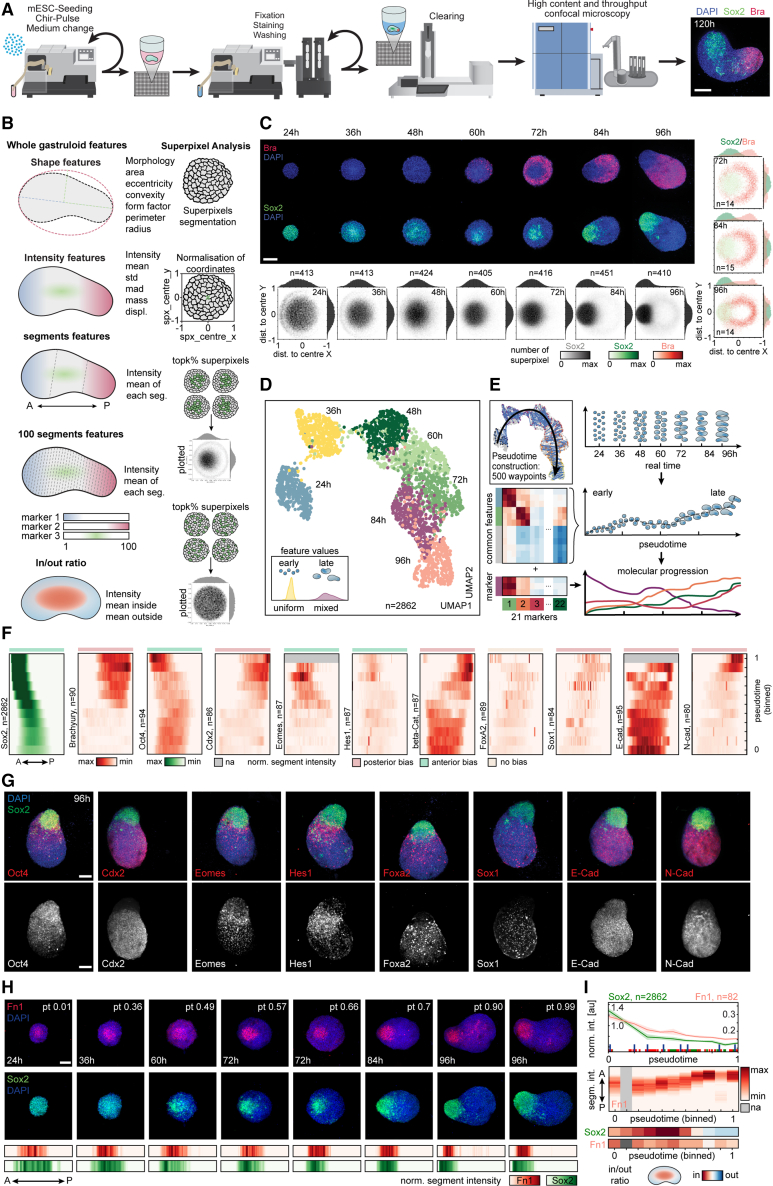
To study their spatial organization, we established an automated handling procedure and a pipeline for high-throughput culture, compound and genomic perturbations, immunofluorescence staining, sample clearing, and high-content imaging of tens of thousands of gastruloids (Figure 3A). This approach and some aspects described here28 allowed us to increase the elongation efficiency from the previously reported 70%8 to 100% (Figure S3A). Gastruloid images were then automatically segmented and processed with a custom workflow extracting features at multiple levels as illustrated in Figure 3B (see STAR Methods).
Figure 3.
Mapping spatial cell-type emergence with image-based trajectory
(A) Scheme illustrating automatized handling workflow. Right: representative image of gastruloids fixed at 120 h. Maximum intensity projection (MIP) of z stack: DAPI and antibody stainings for Sox2 and Bra. Scale bars, 150 μm.
(B) Scheme illustrating extracted features and super pixel analysis.
(C) Representative images at indicated time points (middle z plane of a z stack showing DAPI, Bra, and Sox2. Scale bars, 100 μm. Hexbin plots: mean normalized superpixel intensities of Sox2. Kernel density plots: distribution of intensities along the x and y axes; dist. to center, normalized distance of superpixel to object center (bottom). Co-expression hexbin plots: expression of Sox2 and Bra. Kernel density plots: distribution of intensities along x and y axes (right). Sample numbers (n) are indicated.
(D) UMAP plots of n = 2,862 gastruloids color-coded by time points. Bottom left: scheme illustrates increasing heterogeneity in later time points.
(E) Inferred pseudotime (top left) and scheme of pseudotime ordering, trajectory inference, and molecular progression.
(F) Heatmaps depicting distribution of stainings from the anterior (left) to the posterior (right) pole along pseudotime (progressing from bottom to top). n, number of gastruloids. Anterior bias (light green), posterior bias (light red), and unbiased markers (light yellow).
(G) Representative images at 96 h. (MIPs of z stack showing DAPI and antibody stainings for Oct4, Cdx2, Eomes, Hes1, Foxa2, Sox1, E-Cad and N-Cad, and Sox2). Scale bars, 100 μm.
(H) Top: representative images at indicated pseudotime and sampling time points. Middle z plane: DAPI and antibody stainings for Fn1 and Sox2. Scale bars, 100 μm. Bottom: heatmaps depicting distribution of Fn1 and Sox2 staining from anterior (left) to posterior (right).
(I) Top: line plots of mean staining intensity for Sox2 and Fn1 (n = 82). Blue bars: individual gastruloids shown in (H). Middle: heatmaps depicting Fn1 distribution from anterior (top) to posterior (bottom) along pseudotime. Bottom: heatmap depicting Sox2 and Fn1 in/out ratio measured on the middle z plane.
We established that radial symmetry breaking, axial elongation, as well as the majority of cell types are formed within the first 96 h (Figure 1D). We therefore performed time course experiments starting from 24 to 96 h with fixation intervals of 12 h and stained for Bra, monitoring tail bud and mesodermal induction,8,10 and for Sox2, expressed in naive pluripotent cells, epiblast, NMPs, and neural progenitors in vivo.29,30,31 The choice of Sox2 allowed us to follow the cell type annotations 1–4 and 16 and is continuously expressed throughout gastruloid development. Making use of the extracted features, we further created “meta-gastruloids” showing Sox2 and Bra expression patterns in an average gastruloid representation (Figure 3C). Gastruloids from ESCs grown in S/L exhibit Bra expression before 48 h,10,21 although starting from cells in the S/L/2i medium, Bra protein expression started only at 60 h and displayed a salt-and-pepper yet peripheral pattern. Interestingly, the initially uniform Sox2 staining developed into a heterogeneous pattern at 36 h, forming a gradient with high levels in the gastruloid core. The Chir pulse converted this gradient into a binary pattern with only former Sox2-low regions expressing Bra and a persistent Sox2-positive core population of Bra-negative cells in the center. Spatial variability in Sox2 expression therefore preceded the induction of Bra expression. Remarkably, the Sox2-positive and Bra-positive populations exhibited an increasing bias toward opposing poles starting at 72 h, marking the initiation of radial symmetry breaking and axial organization, culminating in the translocation of the Sox2-positive cell population from the core to the anterior tip and the establishment of a primary body axis.
We then used a panel of antibodies to profile the expression of 21 cell types, adhesion, and signaling activity markers (see STAR Methods). In each case, we co-stained one marker from the panel with DAPI (4′,6-diamidino-2-phenylindole) and Sox2 as a fiducial marker. Gastruloids do not show a perfectly synchronous developmental progression (Figures 3D and S3B; see STAR Methods). To gain developmental resolution, we inferred an image-based gastruloid trajectory.32,33 This pseudotime trajectory enabled us to correlate expression patterns along gastruloid formation (Figures 3E, S3C, and S3D) at the whole gastruloid (Figure S3E) as well as at the segment and inside/outside level (Figures 3F, S3F, and S3G). Patterning maps aligned to the trajectory showed a robust formation of the AP axis, as evidenced by the progressive anterior and posterior localization of polarized markers (Figures 3F and 3G). Of note, the anterior localization of Sox2-positive cells was not marking a rostral neural identity, as we saw segment-level colocalization with pluripotency markers (e.g., Oct4). As shown by scRNA-seq, the only population that expressed a pluripotency signature at 96 h was the EP population. Thus, we used anterior Sox2-positive cells to mark the EP state in post-Wnt pulse gastruloids. From here onward, we refer to the EP population also as the “gastruloid core.”
Other markers such as the mesodermal and epithelial-to-mesenchymal transition (EMT) regulator Eomes34 and Hes1 formed an anteriorly biased band pattern but did not fully reach the anterior pole. Although Hes1 suggested Notch activity near the Sox2 core, we saw that Wnt and Nodal activity (β-catenin and pSmad2 antibodies, respectively) was posteriorly polarized. We observed N-cadherin expression at the posterior, consistent with mesoderm specification, whereas E-cadherin was globally expressed. This segment-level co-expression suggested incomplete EMT at 96 h. We also observed the expression of transcription factors suggesting the emergence of the endodermal (Foxa2) and neural lineage (Sox1). Remarkably, the ECM component Fibronectin 1 (Fn1) showed similar behaviors as the Sox2 core (Figures 3H and 3I). Fn1 already defined a domain at the core 24 h post-seeding and continuously overlapped with Sox2 expression throughout morphogenesis.
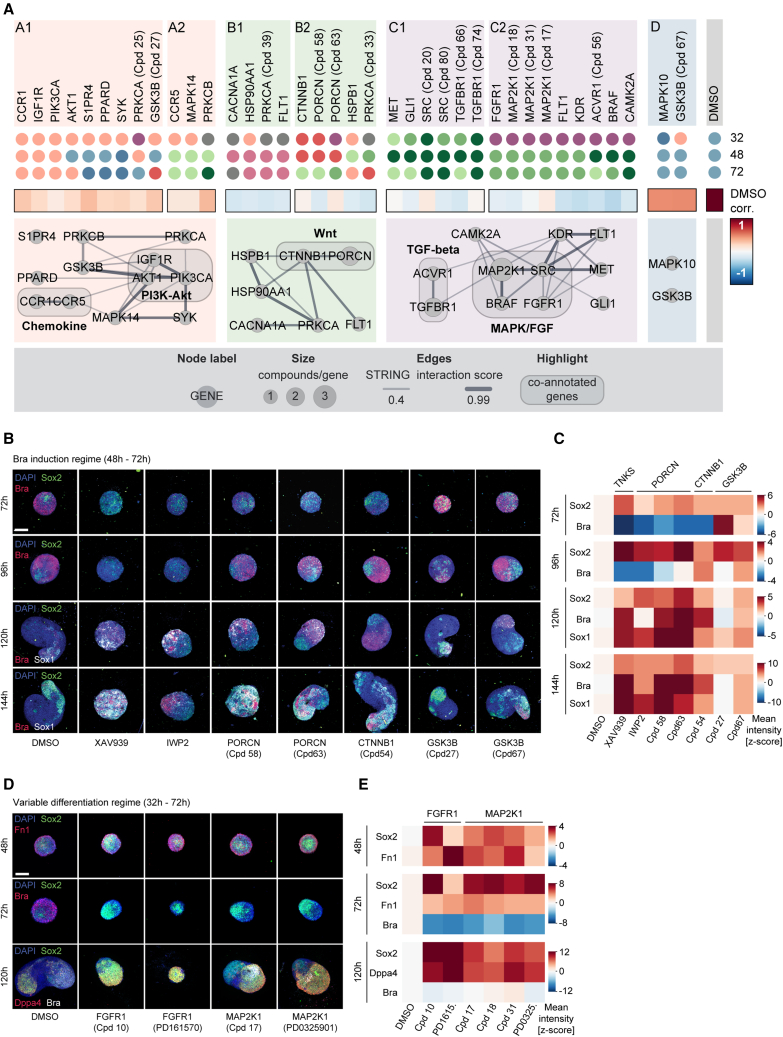
Molecular regulators and regime-dependent phenotypic differences
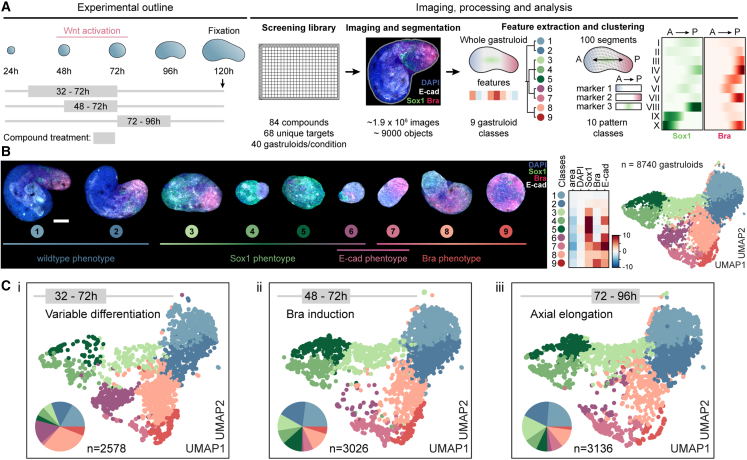
Our analysis revealed a three-step process of symmetry breaking: (1) establishment of cellular variability in Sox2 levels as a possible consequence of differentiation progression, (2) a binary response to Wnt activation, and the formation of two cell populations whose organization ultimately culminates in (3) radial symmetry breaking and elongation. To systematically identify molecular regulators of each step, we designed an image-based compound screen (Figure 4A). The screening library consisted of 84 compounds (Cpd) (Figure S4A; Table S7) selected from a pre-screening of 200 small molecules (Figure S4B). The library was annotated with 68 unique primary targets. Compound treatment was performed in three separate regimes: from 32 to 72 h (“variable differentiation”), 48 to 72 h (“Bra induction”), or 72 to 96 h (“axial elongation”). In all treatment regimes, gastruloids were fixed at 120 h and stained for markers of mesoderm (Bra),35 neuroectoderm (Sox1),36 epiblast or endoderm (E-cadherin),12 and DAPI. 40 gastruloids per condition and regime were then imaged, analyzed, and quality controlled (Figure S4C) and used to generate a multivariate feature set on the whole gastruloid and a segment level of ∼9,000 gastruloids.
Figure 4.
Image-based compound screen and phenotypic landscape
(A) Scheme of experimental outline, image processing, and analysis of the screen.
(B) Left: representative maximum intensity projection images of whole gastruloid phenotypes. Stainings: DAPI, Sox1, Brachyury, and E-cadherin, Scale bars, 100 μm. Right: UMAP plot color-coded by whole gastruloid class. Data points: individual gastruloids, n = 8,740. Heatmap: mean values of indicated features for each class, Z score normalized.
(C) UMAP plots and pie charts color-coded by whole gastruloid classes from indicated treatment regimens (left to right: 32–72 h, 48–72 h, and 72–96 h).
To generate a phenotypic landscape of gastruloid development, we grouped gastruloids by phenotypic similarity by separately clustering37,38 whole gastruloid and segment features (Figures S4D and S4E). Each gastruloid was thus unambiguously assigned to a whole gastruloid and to an AP-pattern class. At the whole gastruloid level, we observed nine phenotypes ranging over four major groups (Figure 4B): wild-type phenotype (classes 1–2) to which the majority (97.6%) of control gastruloids was assigned; Sox1-enriched (classes 3–5) with an increased or exclusive expression of the neuroectoderm marker, indicating failure to produce primitive streak-derived cell lineages; E-cadherin-enriched, exhibiting increased expression of the epithelial marker either together with Sox1 (class 6) or Bra (class 7) expression; and Bra-enriched (classes 8 and 9) with an increased or exclusive expression of Bra (Figure 4B). At the segment level, we detected 10 pattern classes, among them, classes observed in control conditions (classes I–IV, “wild-type” classes) and those that occurred mostly under perturbation (classes VI–X, “perturbed” classes) (Figure S4E). The latter included gastruloids with an increased polarized expression of Bra (class VII) or Sox1 (classes VIII and IX), localization of Bra to the center (class VI), or expression of the two markers on opposing poles (class X). We then highlighted gastruloids from each treatment regime separately (Figure 4C) and inferred a network of functional annotations and color-coded the nodes by the most frequently assigned phenotype for each treatment regime (see STAR Methods). Each phenotypic class was detected in all three temporal regimes. However, their ratios differed significantly, especially between the earliest regime and the latter two. Although perturbation during the establishment of “variable differentiation” (32–72 h) favored classes 8 (light pink) and 6 (purple), the abundance of Sox1-enriched phenotypes (classes 3–5, shades of green) increased when perturbing in the latter two regimes (Figure S4F).
Regulatory modules of gastruloid development
To systematically uncover regulatory modules, we combined the abundance of the whole gastruloid and segment classes in the three regimes into a phenotypic signature (57-feature vector for each compound, Figures S5A–S5C). This revealed 4 regulatory modules that were divided into categories grouping compounds with similar phenotypic effects over time (Figure 5A). We selected hits by significance and robustness (see STAR Methods and Figures S5D and S5E) for a final hitlist of 38 compounds (Table S7). These were predominantly assigned to modules A–C that included gastruloids with delayed development (module A), increased Bra expression (module B), or increased Sox1 expression (module C) (see Figures S5E and S5F).
Figure 5.
Regulatory modules of gastruloid development
(A) Top: dot plots color-coded by most frequent whole gastruloid phenotypes for 38 hit compounds. Compound names: targets with multiple compound coverage. Middle: heatmap correlation of phenotypic signature to control. Bottom: functional interactions network—STRING database for similarity clusters. Nodes: targets of compounds.
(B) Representative images of gastruloids treated with Wnt pathway compounds corresponding to (C). MIPs of z stacks: DAPI and antibody stainings. Compound concentrations: 5 μM. Scale bars: 150 μm.
(C) Heatmaps of Z-scored mean intensity of Sox2, Sox1 and Bra of compound treated gastruloids at 48 h, fixed at indicated time points. n = minimum 25 gastruloids per time point and condition.
(D) Representative images of gastruloids from (E). Middle z plane (48 and 72 h) or MIP (120 h) of z stack, showing DAPI and antibody stainings. Treatment was performed from 32 to 72 h. Scale bars, 150 μm.
(E) Heatmap of Z-scored mean intensity of Sox2, Fn1, Bra, and Dppa4. n = minimum 20 gastruloids per time point and condition.
Module A contained compounds that produced gastruloids with minor phenotypes indicated by high correlations to DMSO controls. Indeed, the perturbation of targets in module A1 such as Akt1, Igf1r, or Pik3ca resulted in gastruloids with a slight increase in Bra expression (class 8) that exhibited a delay but not a full developmental failure, as gastruloids at 120 h resembled wild-type gastruloids at an earlier time point (96 h). Inhibition of Ccr5, Mapk14, or Prkcb during or after the Chir pulse (module A2) produced gastruloids of class 3 (light green) with both Sox1 and Bra-positive domains.
Module B contained compounds that had an increased Bra expression with either elongated (module B1, class 7, and dusky pink) or almost spherical morphology (module B2, class 9, and red). Unexpectedly, inhibition of Ctnnb1 (β-catenin) and Porcn (porcupine O-acyltransferase), members of the Wnt signaling pathway, produced spherical, Bra-increased gastruloids (class 9) when treated before or during the Chir pulse and elongated, Sox1/Bra double-positive gastruloids (class 3) when treated later. To understand the counterintuitive emergence of Bra-positive phenotypes, we performed follow-up experiments for Wnt pathway-related hits including additional compounds (IWP2 and XAV939) (Figures 5B, 5C, S5G, and S5H). Gastruloids were treated from 48 to 72 h or from 72 to 96 h and fixed every 24 h after treatment up to 144 h and stained for Sox1, Sox2, and Bra. Wnt-agonistic treatments (Gsk3b inhibitors) caused an increase in Bra at early time points but resulted in mild phenotypes with only limited effect by 120 and 144 h. This suggests that Wnt overexposure does not cause an increase in Bra expression at the expense of Sox1 at later time points but rather that cell ratios found in late gastruloids are not strongly dependent on the dose of Wnt activation during the Chir pulse. Inhibitors of β-catenin (Cpd54), tankyrase (XAV939), and porcupine (IWP2, Cpd58, and Cpd63), on the other hand, had very drastic effects. All compounds except Cpd54 caused AP axis failure. When treated between 48 and 72 h, gastruloids had a strongly reduced Bra expression (72 h), which resulted in increased levels of Bra, Sox1, and Sox2 at 120 and 144 h. The delayed expression of Bra after the Chir pulse suggested that endogenous Wnt activity is sufficient to cause Bra induction. Thus, endogenously secreted Wnt ligands and their gradients play an important role in primary axial elongation.10 Here, we detected an expanded Sox1- and Sox2-positive territory during Ctnnb1 inhibition and formation of rosette-like structures in XAV939 treatment (Figures S5G–S5I).
Module C mainly contained Sox1-enriched gastruloids. Lack of Bra expression implied the absence of inductive signals and failure of mesodermal differentiation, which consequently skewed development toward neural differentiation.39 This suggests that calcium, hedgehog, mitogen-activated protein kinase (MAPK), and TGF-β signaling control induction and/or maturation of mesodermal cell types. Although perturbation of Tgfbr1, Src, Gli1, and Met (module C1) produced Sox1-enriched gastruloids (classes 3–5) irrespective of the treatment regime, the inhibition of Map2k1, Fgfr1, Braf, or Acvr resulted in the phenotypic class 6 when perturbed before the Chir pulse. Intriguingly, class 6 gastruloids exhibited the most severe defect with a nearly complete absence of the mesoderm. The phenotypic signature of MAPK/fibroblast growth factor (FGF) signaling inhibition underlined its importance throughout gastruloid development: although its inhibition at later time points impeded mesoderm and favors neural induction, at earlier time points, it reduced differentiation in general. This suggests that a perturbation during the variable differentiation states (32–72 h) is incompatible with subsequent development. To better understand the role of MAPK/FGF signaling in pluripotency exit, we analyzed the core in screening hits related to class 6 and FGF-mediated MAPK signaling. After treating gastruloids between 32 and 72 h and fixing at 48, 72, and 120 h, we stained for Bra, Sox2, Fn1, and the pluripotency marker Dppa4 (Figures 5D and 5E). Inhibition of FGF receptors as well as their downstream target MAP2K1 resulted in an expansion of the Sox2 and Dppa4-positive cells as well as an increase in Fn1 at the expense of Bra expression and axial elongation. Inhibition of MAPK/FGF signaling in combination with Wnt activation maintained naive pluripotency40 and under the inhibition of FGF-mediated MAPK signaling, pluripotency is maintained for longer preventing gastruloids to enter a state that is competent for Bra induction on Wnt activation.
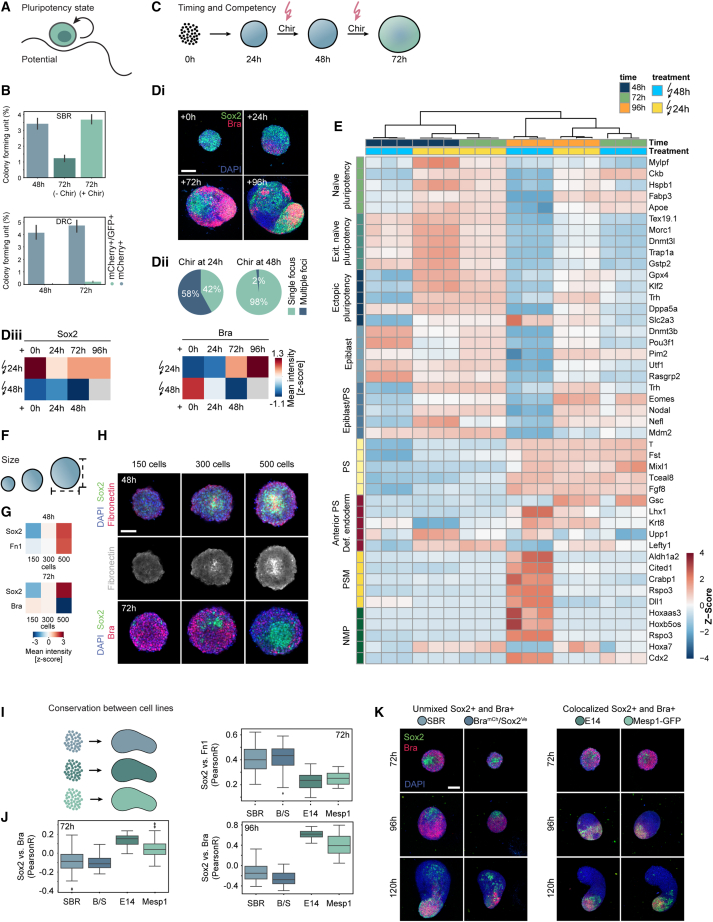
Cell-state heterogeneity in early gastruloids and characterization of the gastruloid core
We then addressed five aspects of the EP core population: the pluripotency state, the timing of differentiation competency, the role of Fn1, gastruloid size dependence, and the reproducibility between multiple cell lines. To verify the presence of a pluripotent subpopulation, we performed clonogenicity assays (Figures 6A and 6B), which select for naive pluripotent stem cells in the N2B27/2i medium.41,42 When dissociated into single cells, gastruloids showed a colony forming unit of ∼3.5% at 48 h with an increase after Wnt pulse. Cloning efficiency of ESCs is usually below 50%.41 Given that EP cells made up for ∼5.6% of all sequenced cells at 72 h, the efficiency exceeded this clonogenicity score. The colony formation was Chir-dependent since the lack of Wnt activation caused a decrease <1.5%. The EPs maintained naive pluripotency, since PGCs and embryonic germ cells cannot be maintained in N2B27/2i in the absence of leukemia inhibitory factor (LIF).43 The same assay was performed on a miR-290-mCherry/miR-302-eGFP reporter line.10 We sorted cells and assessed different pluripotency states with miR-290-mCherry expressed in E3.5–E6.5 embryos and miR-302-eGFP expressed from E5.5 to E8.0.44 In early gastruloids (48–72 h), we found cells corresponding to pre-implantation (mCherry+/GFP−) and early post-implantation epiblast (mCherry+/GFP+). Displaying a continuum, cells shifted toward a double-positive post-implantation epiblast state. By 72 h, ∼9.3% of the sorted cells remained mCherry single positive (Figure S6A). As expected, colony-forming efficiency was dependent on the pluripotency state, and we saw an enrichment for that state after the Wnt pulse.
Figure 6.
Core characterization
(A) Scheme illustrating the stem cell pluripotency state.
(B) Top bar plots: colony formation efficiency for SBR gastruloids at 48 and 72 h with or without Wnt activation (−Chir, Chir). Bottom barplots: colony formation efficiency for single cells sorted from DRC gastruloids at 48 and 72 h. Blue bars: clonogenicity of mCherry+. Green bars: efficiency for double-positive cells. Clonogenicity score: fraction of seeded cells forming a colony in %. Error bars show standard deviation for 12 wells/condition.
(C) Scheme showing different Wnt activation timings.
(D) Di: representative images of gastruloid pulsed at different time points. (n = 334 at +0 h, n = 328 at +24 h, n = 299 at +72 h, n = 231 at +96 h.) Middle z plane (+0, +2, and +72 h) or MIP (+96 h). DAPI and antibody stainings for Sox2 and Bra. Scale bars, 150 μm. Dii: quantification of Chir treatment at 24 h (144 h quantification) or 48 h (120 h quantification). (n = 1,192 treated at 24 h, n = 807 treated at 48 h.) Diii: heatmaps: Z scored mean intensity of Sox2 and Bra.
(E) Bulk RNA sequencing (in triplicates) of gastruloids collected at 48, 72, and 96 h pulsed at indicated time points. Heatmap: Z scored expression levels of top 5 differentially expressed genes for cell-type annotations obtained from scRNA-seq. NMPs, neuro-mesodermal progenitors; PSM, pre-somitic mesoderm; PS, primitive streak; Def. endoderm, definitive endoderm; Exit. Naive pluripotency, exiting naive pluripotency.
(F) Scheme showing size regulation.
(G) Heatmaps: Z scored mean intensities corresponding to (H). (n = 170 gastruloids [150 cells], n = 239 gastruloids [300 cells], n = 215 gastruloids [500 cells]).
(H) Representative images at indicated cell number and time points. Middle z plane of z stack: DAPI and antibody stainings for Sox2, Fn1, and Bra. Scale bars, 150 μm.
(I) Scheme showing gastruloids seeded from different cell lines.
(J) Boxplots: comparisons of Pearson correlations of marker intensities (Sox2, Bra, and Fn1) stained in different cell lines (SBR, B/S [BramCh/Sox2Ve], E14, and Mesp1-GFP) at indicated time points. n = minimum 80 gastruloids per cell line and time point.
(K) Images corresponding to (J) of gastruloids from different cell lines (SBR, B/S [BramCh/Sox2Ve], E14, and Mesp1-GFP) fixed at 72, 96, and 120 h and stained for Bra, Sox2, and DAPI. Middle z plane for 72 h time point and MIPs for the other time points. Scale bars, 150 μm.
To assess the effects of different levels of differentiation competency, we performed Wnt pulses at different time points (Figures 6C and 6Di–6Diii). An early pulse showed an increase in Sox2 expression and a delayed Bra expression onset. At 144 h post-seeding, the prematurely pulsed gastruloids also had multiple Bra+ foci instead of a single tail bud. We also performed bulk RNA-sequencing of prematurely pulsed gastruloids (24 h) and assessed gene signatures obtained from scRNA-seq (Figures 6E and S6B). We saw that gastruloids pulsed at 24 h showed an upregulation of genes associated with EP, naive, and exiting pluripotency signatures. Interestingly, gastruloids that received an early pulse showed a downregulation of the epiblast signature at 48 h suggesting that cells in early gastruloids did not maintain an epiblast identity longer but responded with an EP signature. Late primitive streak and anterior primitive streak/definitive endoderm signatures showed lower expression levels at 72 h when pulsed prematurely. Genes associated with PSM or NMPs were only expressed in 96 h gastruloids when pulsed at 48 h.
To understand the importance of Sox2 levels when gastruloids receive Wnt activation, we successfully performed siRNA knockdown (KD) experiments (Figure S6C), adding siRNA during aggregation. Sox2 KD had a minor effect on pluripotency markers Oct4 and Dppa4, however, caused an increased expression of Nanog at 48 h, which can act as an early primitive streak marker.45 Sox2 KD also resulted in an increase of Bra expression by 72 h and failed axial elongation by 120 h suggesting that increased levels of differentiation in early gastruloids before Wnt activation lead to failure of efficient axial elongation, which was in line with previous studies.21,28 Another plausible explanation for the failure of axial elongation is the depletion of Sox2-dependent lineages such as NMP cells, which are important for in vivo axial elongation.46
Fn1 colocalized with the Sox2-positive core and was expressed by naive pluripotent and core cells (Figures S2D and S6E).47 To understand if Fn1 plays a functional role in core maintenance, we treated gastruloids with RGD-peptide (minimal integrin binding motif48 preventing cellular attachment to Fn1 and thus inhibiting downstream signaling49,50,51). RGD treatment increased Sox2 expression and decreased Bra expression (Figure S6F). Inhibition of focal adhesion kinase (FAK) caused an analogous but stronger effect (Figure S6G) with complete failure of axial elongation. This suggests that deposited Fn1 may keep the level and extent of pluripotency within certain boundaries.
A crucial aspect of gastruloid development8 that could have an effect on the core is the initial seeding cell number. Accordingly, seeding gastruloids with different cell numbers (150, 300, and 500) resulted in a size-dependent relative expansion of the core as well as the Fn1 expression (Figures 6F and 6H). Gastruloids generated from 150 cells showed impaired Fn1 and Sox2 core formation, whereas gastruloids generated from 500 cells showed an expanded core. In gastruloids generated from 500 cells, the peripheral cells also showed reduced Bra expression at 72 h. Fn1 secretion is stimulated under hypoxia, which could also explain the cell number-dependent increase of Fn1 expression.52
Finally, we explored if the core is a unique feature of gastruloids generated from the Sox1-GFP::Brachyury-mCherry (SBR) cells and their parental line CGR8.53 We tested additional cell lines (Figures 6I–6K) using a BramCh/Sox2Ve reporter line,14 E14, and a Mesp1 reporter line.54 Although SBR and BramCh/Sox2Ve showed a tight clustering of Sox2 cells, E14 and Mesp1-GFP gastruloids did not have the same level of organization. We also stained for Fn1 (Figure S6H), which was only expressed in SBR and BramCh/Sox2Ve gastruloids. In all cell lines, we observed variability of Sox2 expression and a differential Wnt response, with some cells maintaining high levels of Sox2 after the Wnt pulse. The separation between Sox2- and Bra-positive cells did not happen as efficiently in E14 and Mesp1-GFP gastruloids. At 96 h, E14 and Mesp1-GFP gastruloids had a larger NMP population indicated by the co-expression of Sox2 and Bra.55 At 120 h only SBR, CGR8 (not shown) and BramCh/Sox2Ve gastruloids have anteriorly localized Sox2 pluripotent cells. It is possible that there are convergent mechanisms of gastruloid formation56 that might also be dependent on cell line-specific differences.57
The window of competency to differentiation in gastruloids is dependent on multiple factors such as pluripotency state, aggregate size, and time of Wnt activation, as well as cell line-specific aspects. Careful assessment of these aspects is necessary to generate developmentally meaningful cell types in gastruloids.
Dual WNT modulation causes anterior structures
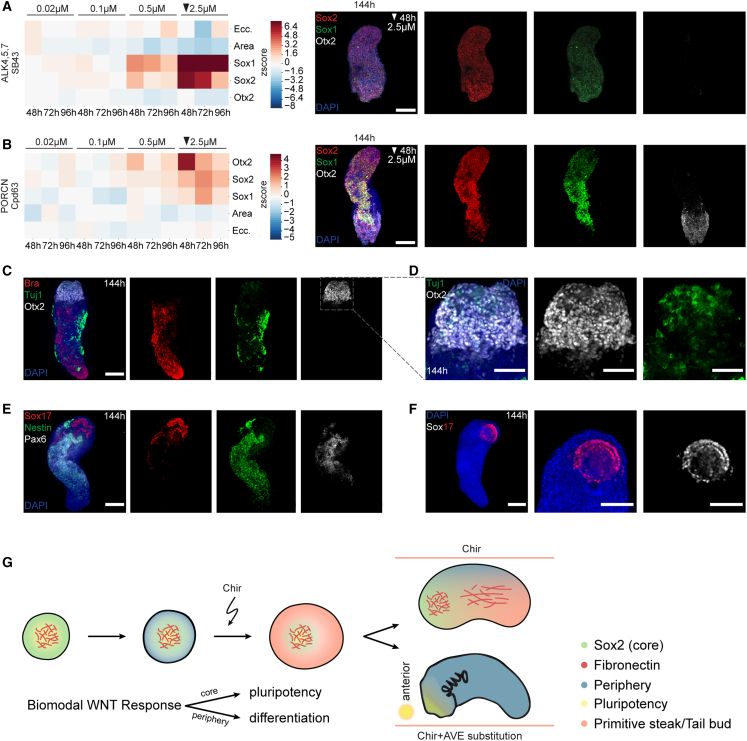
We then hypothesized that the core population and surrounding cells might be an opportunity to reach a better representation of anterior embryonic identities in gastruloids. To successfully form anterior parts of the in vivo gastrula, respective cells are situated in a region that is shielded from Wnt and Nodal activation by local inhibitors (Dkk1, Lefty1, and Cer1) secreted by the anterior visceral endoderm (AVE).58 Screening hits that could restrict caudalizing gradients and phenocopy the effect of the AVE are thus compounds targeting the Wnt pathway and the TGF-b superfamily (especially Nodal signaling).
For the TGF-b superfamily inhibition, we performed treatments (Cpd56 [ALK2i], Cpd66 [TGFRi], Cpd74 [TGFRi], and SB43 [ALK4,5,7i]), with different concentrations, in 24 h treatment windows starting at 48, 72, or 96 h, fixed at 144 h and stained for Sox1, Sox2, and Otx2, which marks rostral neurectoderm.59,60,61 Otx2 is also associated with the foregut and anterior foregut and thus also marks anterior endodermal derivatives.62,63 TGF-b superfamily inhibitors caused a strong increase in Sox1 and Sox2 levels with a drastic expansion of neural lineages, although having only a minor effect on elongation (Figures 7A and S7A–S7D). All conditions, however, did not show an increased Otx2 expression suggesting the absence of anterior neural and endodermal structures.
Figure 7.
Dual Wnt modulation for anterior neuronal structures
(A) Left: heatmap of Z scored area, eccentricity, and mean intensity of Otx2, Sox2, and Sox1 of SB43 treatment fixed at 144 h. Per time point and condition 24 gastruloids were treated. Arrow: condition of representative images. Right: representative images. MIP (144 h) of z stack, DAPI, and antibody stainings. Scale bars, 200 μm.
(B) Left: heatmap of Z scored mean intensity of Otx2, Sox2, Sox1, area, and eccentricity of Cpd 63 treatment fixed at 144 h. n = 24 gastruloids were treated per time point and condition. Arrow: condition of representative images. Right: representative images. MIP (144 h) of z stack, DAPI and antibody stainings. Scale bars, 200 μm.
(C) Representative image of 2.5 μM Cpd63 at 48 h treatment with 2.5 μM Cpd63 at 48 h. MIP (144 h) of z stack, DAPI, and antibody stainings. Scale bars, 200 μm.
(D) Region of interest (ROI) of (F) (dashed square). MIP (144 h) of z stack, nuclear staining (DAPI), and antibody stainings. Scale bars, 150 μm.
(E) Representative image of 2.5 μM Cpd63 treated at 48 h. MIP (144 h) of z stack, DAPI, and antibody stainings. Scale bars, 200 μm.
(F) Representative image of 2.5 μM Cpd63 treatment at 48 h. MIP (144 h) of z stack, DAPI, and antibody staining. Left: scale bars, 200 μm. Right: scale bars, 150 μm.
(G) Scheme of symmetry breaking in gastruloids (left). Below: binary response to Wnt activation. Right: top gastruloid shows patterns found in unperturbed gastruloids. Bottom: potential phenotype with limited posterior Wnt gradients. Curved arrow: Wnt activation. AVE, anterior visceral endoderm.
For the Wnt pathway, we utilized the porcupine inhibitors (IWP2, Cpd58, and Cpd63) as they affect Wnt secretion and the formation of endogenous AP-gradients. We also used a Ctnnb1 inhibitor (Cpd54) as it previously caused an increase in Sox1 and Sox2 while maintaining the AP axis. Each Wnt inhibitor showed a dose-dependent upregulation of the three markers (Sox1, Sox2, and Otx2), with a striking anterior localization of Otx2 (Figures 7B and S7E–S7H). In the most promising condition (2.5 μM Cpd63 administered at 48 h), gastruloids displayed neuronal maturation, as indicated by laterally and anterior Tuj1 expression and long cell protrusions (Figures 7C and 7D). We also observed Pax6- and Nestin-positive cells localized between the AP poles with a potential spinal cord identity (Figure 7E).64,65 In some cases, anterior Tuj1-positive cells were in proximity to a bi-layered Otx2 and Sox17 double-positive ring (Figure 7F). The morphology of this structure is reminiscent of endoderm compartments in 168-h gastruloids previously annotated as anterior foregut.12 Our results are also in line with previous conclusions that gastruloids develop endodermal progenitors that do not transition through EMT or a Bra-positive state.
Discussion
In this study, we used single-cell genomics and developed automated culture and imaging approaches to map the spatial unfolding of cell states and types during gastruloid development. We focused on the early time points to understand pluripotency exit, epiblast states, and germ layer commitment. We defined three events that characterize symmetry breaking and patterning in these gastruloids: (1) ES cells display a spatial heterogeneity of pluripotency exit and a heterogeneous time of differentiation in 3D, (2) which causes a binary response to the Wnt activation driving the cells into two distinct cell populations (EP core cells and peripheral primitive-streak-like cells), (3) leading to radial symmetry breaking, morphological changes, and axial elongation (Figure 7G). Drawing on the scalability of gastruloids, we performed a high-content screen targeting each of these events separately. We uncovered regulatory modules that orchestrate symmetry breaking and identified several screening hits involving pathways that have yet to be investigated. We finally used the gained insights to perform a dual Wnt modulation, generating gastruloids with additional anterior neural and endodermal structures (Figure 7G).
Within 36 h, Sox2 and other pluripotency-related transcription factors exhibited a graded expression from the center to the periphery, reflecting a state continuum from the naive ground state to primed pluripotency. The sources of the cell-to-cell variability, how such graded expression patterns are established, and how other signaling pathways like bone morphogenetic protein (BMP) and Nodal contribute to radial patterning, as reported in human 2D gastruloids,66,67,68 remain to be determined. As for the consequences of the variability in cellular states, it has been shown that the state of a cell determines its response to external stimuli69 and variability in a population is a prerequisite for symmetry breaking and functional diversification.33,70 Although initial cell states form a continuum, Wnt activation results in a binary cell fate decision with upregulation of an EP or mesodermal program. Sox2 is known to inhibit mesodermal differentiation,71,72 and it has recently been shown that the level of Sox2 expression dictates the response to Wnt: high levels of Sox2 activate pluripotency genes, whereas low levels induce mesoderm identities.73 However, what ultimately determines the threshold between one or the other response in gastruloids remains unclear and likely is not solely dependent on Sox2 levels alone. For instance, our scATAC-seq data suggest that lineage specification may also involve early changes in the chromatin state.
The localization of a naive pluripotent population in the core of gastruloids and the perseverance of that state was unexpected and does not recapitulate known in vivo development.74 Accordingly, conclusions on developmental mechanisms for this particular population of cells should not directly be extrapolated to in vivo systems.21 Nevertheless, dual Wnt modulation enabled the development of gastruloids with an AP axis including anterior Otx2-positive cells (likely neuroecto and endodermal structures).
In this study, we provide both a systematic description of the gastruloid phenotypic landscape and its response to perturbations and a toolbox of methods to quantitatively describe emerging patterns in a time-resolved manner. Although other embryoid systems recapitulate the morphology and cell-type composition of the early1,5,6,75 and peri gastrulation embryo76,77,78 more faithfully, gastruloids still combine high cell-type complexity with high formation efficiency and precision of scalable patterning79 enabling studies of cellular mechanisms in diverse contexts.13,14,15
Limitations of the study
Although gastruloids recapitulate aspects of the mouse embryo, they do not necessarily employ the same mechanisms as their in vivo counterpart.11,21 However, observations made in gastruloids give insight into how cells coordinate their behavior to achieve symmetry breaking in a uniform environment in vitro. Furthermore, it has been shown that the pluripotency state80 as well as the genetic background57 greatly affects the propensity of a cell for differentiation. These cell line-specific differences also became clear in our work. So far, gastruloids have been generated from various cell lines cultured in distinct conditions.11,14,15,81 Therefore, although gastruloids from all culture conditions form similar morphological structures, their differentiation path, and exact cellular composition may vary, even with the same cell line and culture conditions.56
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-AKT2 (phospho S474), (used in gastruloid trajectory) | Abcam | Cat # ab38513; RRID: AB_867564 |
| Rabbit monoclonal anti-Activin A Receptor Type IB/ALK-4, (used in gastruloid trajectory) | Abcam | Cat # ab109300; RRID: AB_10860328 |
| Mouse monoclonal anti-β-catenin, (used in gastruloid trajectory) | BD Biosciences | Cat # 610154; RRID: AB_397555 |
| Rabbit monoclonal anti-Brachyury, (used in gastruloid trajectory) | Abcam | Cat # ab209665; RRID: AB_2750925 |
| Rabbit monoclonal anti-Cdx2, (used in gastruloid trajectory) | Abcam | Cat # ab76541; RRID: AB_1523334 |
| Goat polyclonal anti-Dppa4 | R&D Systems | Cat # AF3730; RRID: AB_2094166 |
| Mouse monoclonal anti-E-cadherin, (used in gastruloid trajectory) | BD Biosciences | Cat # 610181; RRID: AB_397580 |
| Rabbit monoclonal anti-EGFR (phospho Y1068), (used in gastruloid trajectory) | Abcam | Cat # ab40815; RRID: AB_732110 |
| Rabbit polyclonal anti-Fibronectin, (used in gastruloid trajectory) | Merck | Cat # F3648; RRID: AB_476976 |
| Rabbit monoclonal anti-FoxA2, (used in gastruloid trajectory) | Abcam | Cat # ab108422; RRID: AB_11157157 |
| Rabbit monoclonal anti-Gata6, (used in gastruloid trajectory) | Cell Signaling Technology | Cat # 5851S; RRID: AB_10705521 |
| Rabbit monoclonal anti-Hes1, (used in gastruloid trajectory) | Cell Signaling Technology | Cat # 11988S; RRID: AB_2728766 |
| Rabbit monoclonal anti-Ki67, (used in gastruloid trajectory) | Abcam | Cat # ab16667; RRID: AB_302459 |
| Mouse monoclonal anti-N-cadherin, (used in gastruloid trajectory) | BD Biosciences | Cat # 610920; RRID: AB_2077527 |
| Rabbit polyclonal anti-Nanog | Abcam | Cat # ab80892; RRID: AB_2150114 |
| Mouse monoclonal anti-Nestin | Millipore | Cat # MAB353; RRID: AB_94911 |
| Mouse monoclonal anti-Oct3/4, (used in gastruloid trajectory) | BD Biosciences | Cat # 611203; RRID: AB_398737 |
| Rabbit polyclonal anti-Otx1/2 | Abcam | Cat # ab21990; RRID: AB_776930 |
| Goat polyclonal anti-Otx2 | RnD | Cat# AF1979; RRID:AB_2157172 |
| Mouse monoclonal anti-p44/42 MAPK (Erk1/2), (used in gastruloid trajectory) | Cell Signaling Technology | Cat # 4696S; RRID: AB_390780 |
| Rabbit monoclonal anti-Pax6 | Abcam | Cat# ab195045; RRID:AB_2750924 |
| Rabbit polyclonal anti-Phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204), (used in gastruloid trajectory) | Cell Signaling Technology | Cat # 9101S; RRID: AB_331646 |
| Rabbit polyclonal anti-Phospho-NPM (Thr95), (used in gastruloid trajectory) | Cell Signaling Technology | Cat # 3517S; RRID: AB_2155177 |
| Rabbit polyclonal anti-Phospho-Smad2 (Ser465/467), (used in gastruloid trajectory) | Thermo Fisher Scientific | Cat # 44-244G; RRID: AB_2533614 |
| Goat polyclonal anti-Sox1, (used in gastruloid trajectory) | Cell Signaling Technology | Cat # 4194; RRID: AB_1904140 |
| Rabbit monoclonal anti-Sox2 | Cell Signaling Technology | Cat # 23064S; RRID: AB_2714146 |
| Rat monoclonal anti-Sox2, (used in gastruloid trajectory) | Thermo Fisher Scientific | Cat # 14-9811-82; RRID: AB_11219471 |
| Rabbit monoclonal anti-Tbr2/Eomes, (used in gastruloid trajectory) | Abcam | Cat # ab183991; RRID: AB_2721040 |
| Mouse monoclonal anti-Tubulin beta-3 | BioLegend | Cat# 801201; RRID:AB_2313773 |
| Donkey anti-rabbit Alexa Fluor 488 | Thermo Fisher Scientific | Cat # A-21206; RRID:AB_2535792 |
| Donkey anti-rabbit Alexa Fluor 488 Plus | Thermo Fisher Scientific | Cat # A32790; RRID:AB_2762833 |
| Donkey anti-rat Alexa Fluor 488 | Thermo Fisher Scientific | Cat # A-21208; RRID:AB_2535794 |
| Donkey anti-rabbit Alexa Fluor 568 | Thermo Fisher Scientific | Cat # A10042; RRID:AB_2534017 |
| Donkey anti-mouse Alexa Fluor 568 | Thermo Fisher Scientific | Cat # A10037; RRID:AB_2534013 |
| Donkey anti-rabbit Alexa Fluor 647 | Thermo Fisher Scientific | Cat # A-31573; RRID:AB_2536183 |
| Donkey anti-rabbit Alexa Fluor 647 Plus | Thermo Fisher Scientific | Cat # A32795; RRID:AB_2762835 |
| Donkey anti-mouse Alexa Fluor 647 | Thermo Fisher Scientific | Cat A-31571; RRID:AB_162542 |
| Donkey anti-goat Alexa Fluor 647 | Thermo Fisher Scientific | Cat # A-21447; RRID:AB_2535864 |
| Donkey anti-goat Alexa Fluor 647 | Abcam | Cat # ab150135; RRID:AB_2687955 |
| Chemicals, peptides, and recombinant proteins | ||
| CHIR99021 | STEMCELL Technologies | Cat # 72054 |
| PD0325901 | STEMCELL Technologies | Cat # 72182 |
| Mouse Recombinant LIF | STEMCELL Technologies | Cat # 78056 |
| SB431542 | Tocris | Cat # 1614 |
| BMS605541 | Tocris | Cat # 6069 |
| IWP2 | Tocris | Cat # 3533 |
| SU5402 | STEMCELL Technologies | Cat # 73914 |
| XAV939 | STEMCELL Technologies | Cat # 72672 |
| Screening library (see Table S7) | Novartis | N/A |
| PD161570 | Novartis | N/A |
| Sorafenib | Novartis | N/A |
| RGD-(Arg-Gly-Asp)-peptide | Selleck Chemicals | Cat # S8008 |
| Focal Adhesion Kinase Inhibitor III | Merk | Cat # 5.04045 |
| Deposited data | ||
| Single cell RNA sequencing and multiome data | This paper | GEO: GSE229513 |
| Bulk RNA-sequencing data | This paper | GEO: GSE229386 |
| Experimental model/ Cell lines | ||
| BramCh/Sox2Ve T::H2B-mCherry,Sox2::H2B-Venus |
Laboratory of Jesse V. Veenvliet | N/A |
| CGR8 | Laboratory of Matthias Lutolf | 129P2 |
| E14 | Laboratory of Matthias Lutolf | 129P2 |
| Mesp1-GFP | Laboratory of Matthias Lutolf, originating from: Laboratory of Cédric Blanpain | N/A |
| miR-290-mCherry/mir-302-eGFP, DRC | Laboratory of Matthias Lutolf, originating from the Laboratory of Robert Blelloch | N/A |
| Sox1-GFP::Brachyury-mCherry (SBR) | Laboratory of Matthias Lutolf originating from: Laboratory of David Suter | based on CGR8 129P2 |
| 129sv/ev | Experiments using this cell line were performed in the Laboratory of Denis Duboule. | CMTI-1, Embryomax |
| Oligonucleotides | ||
| AllStars Negative Control siRNA | Qiagen | Cat #SI03650318 |
| Mm_Sox2_4 FlexiTube siRNA |
Qiagen | Cat #SI01429596 |
| Mm_Sox2_3 FlexiTube siRNA |
Qiagen | Cat #SI01429589 |
| Mm_Fn1_1 FlexiTube siRNA |
Qiagen | Cat #SI01004059 |
| Mm_Fn1_2 FlexiTube siRNA |
Qiagen | Cat # 1027415, SI01004066 |
| Software and algorithms | ||
| Fiji/ImageJ | N/A | https://imagej.net/Fiji |
| FlowJo™ | BD Life Sciences | https://www.flowjo.com |
| Python | Python Software Foundation | https://www.python.org |
| R | R Project | https://www.r-project.org |
| Gastruloid feature extraction pipeline | This paper https://github.com/fmi-basel/gliberal-gastruloid-2023-methods |
https://doi.org/10.5281/zenodo.7858557 |
| Other | ||
| 384-well black/clear round bottom ultra-low attachment spheroid microplates | Corning | Cat # 4516 |
| EL406 washer dispenser | BioTek Instruments | N/A |
| CyBio SELMA 384/25 μl | Analytik Jena AG | N/A |
| Integra Assist Plus | Integra | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Prisca Liberali (prisca.liberali@fmi.ch).
Materials availability
This study did not generate new unique reagents. Non commercial small molecules and compounds can not be shared as they were a gift from Novartis.
Experimental model and subject details
Cell lines
mESCs were cultured at 37°C and 5% CO2 on gelatin-coated tissue culture plates/flasks in GMEM (Merck) supplemented with 10% embryonic stem cell qualified FBS (Gibco), GlutaMAX (Gibco), sodium pyruvate (Gibco), EmbryoMAX MEM NEAA (Merck), β-mercaptoethanol, or N2B27 (see below) supplemented with 3 μM CHIR99021 (Chir) (Stem Cell Technologies), 1 μM PD0305901 (Stem Cell Technologies) and 0.01 μg/ml LIF (Stem Cell Technologies). Cells were passaged every other day with Accutase (Merck) and maintained in culture for at least two passages post-thawing prior to experimental use. Cells were routinely tested for mycoplasma. If not stated otherwise, Sox1-GFP::Brachyury-mCherry cells53 were used.
Method details
Automated gastruloid culture
The original gastruloid protocol81 was modified as follows: mESCs were detached from tissue culture plates with Accutase, collected with DMEM/F-12 (Gibco) supplemented with 0.1% bovine serum albumin (Gibco), centrifuged and washed once with N2B27 medium. N2B27 medium contained DMEM/F-12 and Neurobasal medium (Gibco) supplemented with N2 (homemade), B-27 serum-free supplement (Gibco), GlutaMAX, HEPES (Sigma) and β-mercaptoethanol. Cells were resuspended in N2B27 medium and the cell concentration was determined using the TC20 cell counter (Bio-Rad). A cell suspension containing the required cell number in N2B27 medium (300 cells/well and additional dead volume) was prepared and 20 μl per well were seeded into black, ultra-low attachment, round-bottom 384-well plates (4516, Corning) using the EL406 liquid handling robot (BioTek Instruments). All following medium changes were performed with the EL406 liquid handling robot. At 48h, 75 μl N2B27 supplemented with 3 μM Chir was added. Afterwards, medium (75 μl) was replaced every 24h with the same volume of fresh N2B27 medium until gastruloids where fixed.
Sample preparation, immunofluorescence, and imaging
All steps were performed using the EL406 liquid handling robot at room temperature (RT) if not indicated otherwise. Gastruloids were fixed at indicated time points with 4% PFA for 30min and washed 6 times with PBS. Gastruloids were permeabilised with 1% Triton X-100 for 1h and washed 6 times with 0.1% BSA, then blocked with 3% donkey serum (Sigma) for 1h. Primary and secondary antibodies were diluted in 3% donkey serum with 0.1% Triton X-100. Cell nuclei were stained with 0.2 μg/ml DAPI (Invitrogen) during the secondary antibody incubation. Antibody incubation was performed shaking overnight at 4°C. On the next day, washing was performed 6 times with PBS for 15min. After the secondary antibody wash, gastruloids were washed 6 times with ddH2O. Refractive index matching was performed with ScaleS482 (gastruloid screen) or FOCM83 (all other experiments) using the Assist Plus (Integra Biosciences) pipetting robot.
Following primary antibodies were used: rabbit-anti pAKT2 (Akt signalling) (1:500, Abcam), rabbit-anti Activin A Receptor Type IB (ALK4) (1:500, Abcam), mouse-anti β-catenin (Wnt signalling effector) (1:500, BD Biosciences), rabbit-anti Brachyury (early mesoderm and PS marker) (1:500, Abcam), rabbit-anti Cdx2 (posterior marker) (1:500, Abcam), goat-anti Dppa4 (pluripotency marker) (1:500, R&D Systems), mouse-anti E-cadherin (marker used for epithelial identity and endoderm) (1:500, BD Biosciences), rabbit-anti EGFR (phosphor Y1068) (EGF signalling) (1:500, Abcam), rabbit- anti Fibronection (ECM protein) (1:500, Merck), rabbit-anti FoxA2 (Endoderm) (1:500, Abcam), rabbit-anti Gata6 (Cardiac mesoderm) (1:500, Cell Signaling Technology), rabbit-anti Hes1 (Notch signalling target) (1:500, Cell Signaling Technology), rabbit-anti Ki67 (proliferating cells) (1:500, Abcam), mouse-anti N-cadherin (mesenchymal marker) (1:500, BD Biosciences), rabbit-anti Nanog (naïve pluripotency) (1:500, Abcam), mouse-anti Nestin (neural intermediate filament) (1:500, Millipore), mouse-anti Oct3/4 (pluripotency) (1:500, BD Biosciences), rabbit-anti Otx1/2 (pluripotency, foregut, anterior neuectoderm) (1:500, Abcam), goat-anti Otx2 (1:500, RnD), mouse-anti p44/42 MAPK (Erk1/2) (MAPK (FGF) signalling) (1:500, Cell Signaling Technology), rabbit-anti Pax6 (neurectoderm) (1:500, Abcam), rabbit-anti Phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) (MAPK (FGF) signalling) (1:500, Cell Signaling Technology), rabbit-anti Phospho Smad2 (Ser465/467) (Active TGFβ signalling) (1:500, Thermo Fisher Scientific), rabbit-anti Sox1 (neurectoderm) (1:500, Cell Signaling Technology), rat-anti Sox2 (neurectoderm, pluripotency) (1:500, Thermo Fisher Scientific), rabbit-anti Sox2 (1:500, Cell Signaling Technology), rabbit-anti Tbr2/Eomes (primitive streak) (1:500, Abcam) and anti-mouse Tuj1 (neurectoderm) (1:500, BioLegend).
Following secondary antibodies were used: donkey anti-rabbit Alexa Fluor 488 (1:500, Thermo Fisher Scientific), donkey anti-rabbit Alexa Fluor 488 Plus (1:500, Thermo Fisher Scientific), donkey anti-rat Alexa Fluor 488 (1:500, Thermo Fisher Scientific), donkey anti-rabbit Alexa Fluor 568 (1:500, Thermo Fisher Scientific), donkey anti-mouse Alexa Fluor 568 (1:500, Thermo Fisher Scientific), donkey anti-rabbit Alexa Fluor 647 (1:500, Thermo Fisher Scientific), donkey anti-rabbit Alexa Fluor 647 Plus (1:500, Thermo Fisher Scientific), donkey anti-mouse Alexa Fluor 647 (1:500, Thermo Fisher Scientific), and donkey anti-goat Alexa Fluor 647 (1:500, Abcam).
High-throughput imaging was performed with the automated spinning disk microscope CellVoyager 7000S (Yokogawa), an enhanced CSU-W1 spinning disk (Microlens-enhanced dual Nipkow disk confocal scanner), a 10x Olympus objective and a Neo sCMOS camera (Andor, 2560 x 2160 pixels). Z-planes were acquired in 3, 5 or 10 μm z-steps.
Image-based time course
Gastruloids were fixed every 12h from 24h to 96h and stained as described above. Each gastruloid was stained with DAPI, an antibody against Sox2, and one additional antibody (for full list of co-stained antibodies see Figure S3E and key resources table). Per timepoint, 16 gastruloids were stained with the same antibody combination.
Single-cell RNA sequencing
Gastruloids were generated as previously described81 Briefly, 300 mESC were plated in 40 μl N2B27 into ultra-low attachment, round-bottom 96-well plates (7007, Corning). After 48h, 150 ml of N2B27 supplemented with 3 μM Chir were added to each well. After 72 h, medium was changed with N2B27. Gastruloids were collected at 24h (240 gastruloids), 36h (96 gastruloids), 48h (2x48 gastruloids), 52h (48 gastruloids), 56h (48 gastruloids), 60h (48 gastruloids), 72h (24 gastruloids) and (2x 24 gastruloids), 96h (24 gastruloids), 108h (16 gastruloids) and 120h (16 gastruloids). Gastruloids were transferred into a tube, spun and medium was removed. Gastruloids were dissociated by incubating with Accutase for 5 min at 37°C with intermediate mechanical dissociation (pipetting). After spinning with DMEM/F-12 with 0.1% BSA, cells were resuspended in PBS, passed through a cell strainer with 30 μm pore size and dead cells were stained with DRAQ7 dye (Thermo Fisher Scientific). Per sample, 12,000 alive cells (for the 24h time point, only 7,500 cells were obtained) were sorted by FACS (Becton Dickinson FACS Aria cell sort or Becton Dickinson Influx cell sorter). Cellular suspensions were loaded on a 10x Genomics Chromium Single Cell instrument to generate single cell GEMs. Single cell RNAseq libraries were prepared using the 10x Genomics Single Cell 3` Gel Bead and Library Kit according to CG000183 Single Cell 3`Reagent Kit v3 User Guide_RevA. GEMRT was performed in a Bio-Rad PTC-200 Thermal Cycler with 0.2ml PCR Tube Strips (Eppendorf P/N 0030 124.359): 53 °C for 45min, 85 °C for 5min; held at 4 °C. After RT, GEMs were broken and the single strand cDNA was cleaned up with DynaBeads® MyOneTM Silane Beads (Life Technologies P/N 37002D). cDNA was amplified using a Bio-Rad PTC-200 Thermal cycler with 0.2ml PCR Tube Strips (Eppendorf P/N 0030 124.359): 98 °C for 3min; cycled 11x: 98 °C for 15 s, 63 °C for 20 s, and 72 °C for 1min; 72 °C for 1min; held at 4 °C. Amplified cDNA product was cleaned up with the SPRIselect Reagent Kit (0.6X SPRI). Indexed sequencing libraries were constructed using the reagents in the Chromium Single Cell 3` library kit V3 ( 10x Genomics P/N1000078), following these steps: 1) Fragmentation, End Repair and A-Tailing; 2) Post Fragmentation, End Repair & A-Tailing Double Sided Size Selection with SPRIselect Reagent Kit (0.6X SPRI and 0.8X SPRI); 3) adaptor ligation; 4) post-ligation cleanups with SPRIselect (0.8X SPRI); 5) sample index PCR using the Chromium Multiplex kit (10x Genomics P/N-120262); 6) Post Sample Index Double Sided Size Selection- with SPRIselect Reagent Kit (0.6X SPRI and 0.8X SPRI). The barcode sequencing libraries were quantified using a Qubit 2.0 with a Qubit TM dsDNA HS Assay Kit (Invitrogen P/N Q32854) and the quality of the libraries were assessed on a 2100 Bioanalyzer from Agilent using an Agilent High Sensitivity DNA kit (Agilent P/N 5067-4626). Sequencing libraries were loaded at 1.6pM on an Illumina Nextseq500 with 75cycle kits using the following read length: 28 cycles Read1, 8 cycles i7 Index and 56 cycles Read2. The CellRanger suite (1.3.0) was used to generate the aggregated gene expression matrix from the BCL files generated by the sequencer based on the mm10 Cell Ranger mouse genome annotation files.
Multiome Sequencing
For 10x Multiome experiments, single nuclei were extracted from dissociated gastruloids. Gastruloids were generated as previously described.81 Briefly, 300 mESC were plated in 40 μl N2B27 into ultra-low attachment, round-bottom 96-well plates (7007, Corning). After 48h, 150 ml of N2B27 supplemented with 3 μM Chir were added to each well. Gastruloids were collected at 48h (384 gastruloids) and 52h (288 gastruloids) were collected. Gastruloids were transferred into a tube, spun and medium was removed. Gastruloids were dissociated by incubating with Accutase for 4min at 37°C with intermediate mechanical dissociation. After spinning with DMEM/F-12 with 0.1% BSA, cells were resuspended in Freezing medium (90% Serum medium, 10% DMSO (Sigma, 276855)), cell concentration was determined and 180,000 cells per vial were frozen and stored at -80°C for later continuation of the sample preparation. After thawing, cells were resuspended in PBS 0.8% BSA and washed twice in PBS 0.04% BSA. Cells were then resuspended in Lysis Buffer containing 10 mM Tris-HCl, pH 7.4 (Sigma-Aldrich, T2194), 10 mM Sodium Chloride (Sigma-Aldrich, 59222C), 3 mM Magnesium Chloride (Sigma-Aldrich, M1028), 0.01% Tween-20 (Bio-Rad, 1662404), 0.01% NP40 Substitute (Sigma-Aldrich, 74385), 0.005% Digitonin (Sigma-Aldrich D141-100MG), 1% BSA (Miltenyi Biotec, 130-091-376), 1 mM DTT (Sigma-Aldrich, 646563), Protector RNase inhibitor (Sigma-Aldrich 3335402001) and nuclease-free water (Ambion, AM9937). The suspension was incubated for 5min on ice. The reaction was stopped with Wash Buffer (10 mM Tris-HCl, pH 7.4, 10 mM Sodium Chloride, 3 mM Magnesium Chloride, 1% BSA, 0.1% Tween-20, 1 mM DTT, 1 U/μl RNase inhibitor, and nuclease-free water) and span down for 5min at 500g at 4°C. The pellet was resuspended in Wash buffer and again span down for 5 min at 500g at 4°C. The pellet was resuspended in 1X Diluted Nuclei Buffer consisting of 1X Nuclei Buffer (stock of 20X, 10x Genomics, 2000207), 1 mM DTT, 1 U/μl RNase inhibitor and nuclease-free water and nuclei concentration was determined. Nuclei were span down for 5min at 500g at 4°C and resuspended in the respective volume of 1X Diluted Nuclei Buffer according to the 10x Multiome user guide for theoretically targeting 5,000 nuclei.
Single cell Multiome experiments were performed using the 10x Chromium Single Cell Multiome ATAC + Gene Expression (GEX) kit (1000283) following manufacturer’s instructions.
Briefly, nuclei suspensions were incubated in a transposition mix where open chromatin DNA was preferentially fragmented and adapter sequences were added to the ends of the DNA fragments.
Afterwards, nuclei were mixed with reverse transcription mix, and gel beads and oil were loaded onto a 10x microfluidic chip to be co-encapsulated into nanodroplets, forming GEMs. Inside each GEM containing a nucleus, for GEX, first strand cDNA synthesis occurred, where each mRNA was tagged with a UMI and a barcode unique for each nucleus and a 30nt poly (dT). In the same partition, for ATAC, transposed DNA was tagged with P5 adaptor followed by 16nt barcode and a spacer. Subsequently, the reaction was quenched. The droplets were broken, pooled fractions recovered and purified using Dynabeads MyOne Silane. Barcoded transposed DNA and barcoded full-length cDNA from polyadenylated mRNA was amplified to fill gaps and generate sufficient amounts for library generation. The pre-amplified product was used for generation of both GEX and ATAC libraries. Single-cell gene expression libraries were generated using fragmentation, end repair, A-tailing and double-sided size selection using SPRIselect. P5 and P7 adaptor sequences were ligated and was further amplified with the number of PCR cycles depending on the number of nuclei loaded. Individual sample indices provided as a Dual Index Plate TT Set A (10x Genomics, 3000431) were used during amplification to enable pooling and subsequent demultiplexing of multiple libraries.
For ATAC libraries construction, P7 adapter sequences and sample indices from a Sample Index Plate N, Set A (10x Genomics, 3000427) were added to the pre-amplified product. Quantification and quality control of libraries was performed using High Sensitivity DNA assays on an Agilent Bioanalyzer. GEX expression libraries were sequenced on Illumina Novaseq. ATAC libraries were sequenced on Illumina Nextseq using custom sequencing read lengths 50-(8)i-(16)i-49.
Image-based screening assay
A total of 30 plates were prepared as described above. The screen was split into three assays which were performed in parallel and differed in the time point of compound library addition. The compound library (kind gift from Novartis) was composed of 84 compounds (Table S7) in form of 10 mM DMSO stocks in 384-well diamond-bottom plates. It contained 29 wells of vehicle control (DMSO). Compound treatment was performed at three different time points: from 32h-72h, from 48h-72h and from 72h-96h. Compounds were added in respective volumes from a 200 μM intermediate compound library dilution in N2B27 using the SELMA 384 automated pipettor (Analytik Jena AG) to achieve a final concentration of 5 μM. Medium changes were performed as described above. Plates were fixed and prepared for imaging at 120h.
Compound treatment of gastruloids
Compound treatments apart from the screening assay were performed as follows: If gastruloids were treated with compounds before 48h, a 2x concentrated solution in N2B27 was prepared and 20 μl were added to the gastruloids. If gastruloids were treated with compounds at or after 48h, the compound was added during the medium change. Gastruloids treated with the same dilution of DMSO were used as controls.
Additional compounds for the validation experiments (core perturbation) were selected to target either the same target or other component of the same signalling pathway as the compounds from the screening library to confirm or refute the observed phenotype. Compounds used in this experiment included: inhibitors SB 431542 (Tocris, Cat# 1614); FGFR1 inhibitors compound 10 (PD-166866, gift from Novartis), PD-161570 (Sigma Aldrich, Cat# PZ0109); MAP2K1 inhibitors compounds 17, 18, 31 (gifts from Novartis), and PD0325901 (STEMCELL Technologies, Cat# 72182). Compounds were used in the same concentrations as in the screen. For the broad Wnt pathway follow up experiments the following compounds: XAV939 (STEMCELL Technologies, Cat# 72672), IWP2 (Tocris, Cat#3533), inhibitors of Porcn (compounds 58 and 63, gifts from Novartis), inhibitor of Ctnnb1 (compound 54, gift from Novartis) and inhibitors of Gsk3b (compounds 27 and 67, gifts from Novartis) were added either at 48h or 72h of gastruloid differentiation for 24h. A concentration of 5μM was used. DMSO used for controls was used at the same volume as resuspended compounds. Gastruloids were fixed at 72h, 96h, 120h and 144h and processed with immunofluorescence as mentioned above.
Bulk RNA sequencing – sample preparation
We analyzed three independent batches of gastruloids for each condition. For each batch of bulk RNA samples, 96 gastruloids were grown as described above from mESCs (Embryomax 129sv/ev). Control samples received a pulse of Chir between 48h and 72h AA; Early samples received a pulse of Chir between 24h and 48h AA. Gastruloids were collected on ice washed twice with PBS and snap froze in liquid nitrogen. RNA extraction was performed using RNeasy (Qiagen) columns according to manufacturer recommendation and on-columns DNAse treatment was performed. RNA quality was assessed on a Tapestation TS4200 with all RNA showing Quality Number above 9.5. Library preparation was performed by the EPFL Gene expression core facility, using "Illumina stranded mRNA ligation" (ISML) prep starting from 1000ng of RNA, according to Illumina protocol 1000000124518 v01. Libraries were quantified by qubit DNA HS and profile analysis was done on TapeStation TS4200. Libraries were sequenced on HiSeq 4000 Illumina, with paired end 75bp.
Clonogenicity assay
12 well tissue culture plates were coated with Laminin (0.01μg/ml in PBS) by incubating them O/N at 37°C in a tissue culture incubator. SBR gastruloids were grown in round-bottom 96-well plates (7007, Corning) until harvesting at 48h and 72h. At 48h control gastruloids were pulsed with N2B27 medium supplemented with 3μM Chir as described above. Gastruloids were transferred into a tube and spun to remove medium. Gastruloids were then incubated for 5min in Accutase for dissociation. After resuspension in DMEM/F-12 supplemented with 0.1% BSA, cells were sedimented via centrifugation. The obtained pellet was resuspended in PBS and, passed through a cell strainer with 30 μm pore size. 14000 alive cells were sorted by FACS (Becton Dickinson Influx cell sorter) and eventually plated at a concentration of 1000 cells per well in N2B27/2i. One 12 well plate was used per condition.
DRC gastruloids were generated and harvested as described above. Both conditions collected at 72h were Chir pulsed as described above. For both timepoints of collection (48h and 72h) 8000 mCherry-positive and 8000 mCherry and GFP double positive cells were sorted and plated into N2B27/2i at a concentration of 1000 cells per well of a 12 well plate.
Assays were terminated after 7 days and colonies were counted manually using a light microscope.
Flow cytometry results from sorts of 48h and 72h DRC gastruloids were assessed using FlowJo.
siRNA knockdown experiment
When cells were seeded to differentiate gastruloids for the knock down experiments a reverse transfection of siRNAs was performed. The cell suspension generated for gastruloid seeding already included the transfection mix. The transfection mix was prepared in the following way: siRNAs pairs (Mm_Sox2_3 and Mm_Sox2_4, Qiagen Cat#SI01429589 and Cat#SI01429596 respectively) and (Mm_Fn1_1 and Mm_Fn1_2, Qiagen Cat#SI01004059 and Cat#SI01004066 respectively) were diluted in Opti-MEM I (Gibco Ref# 31985-047). The negative control RNA (Qiagen Cat#SI03650318) was diluted in Opti-MEM I to a matching total amount of siRNA. Next 1μl/100μl of Lipofectamine RNAiMAX (Invitrogen, Cat# 13778-150) was added. The transfection mix was then gently mixed and incubated at room temperature for 25min. After incubation the cell suspension in N2B27 was added (83.15% Cell suspension in N2B27, 16.85% transfection mix) to reach a final siRNA concentration of 10nM per clone. After seeding medium changes were performed according to the protocol described above. Gastruloids were fixed at 48h, 72h and 120h and processed for immunofluorescence as described above.
RGD peptide experiment
Gastruloids were either seeded with a final concentration of 1mg/ml RGD peptide or H2O (control). At 48h gastruloids were pulsed with Chir as described above. In the treatment regime in addition to Chir this medium was also supplemented with 1mg/ml RGD peptide (Selleck Chemicals Cat# S8008). Gastruloids were fixed at 72h and processed for immunofluorescence as described above.
Double modulation of the Wnt pathway
Dose finding experiments were performed for the inhibitor of Ctnnb1 (Compound 54) and the Porcn inhibitors Compounds 58, 63 and IWP2. As wells as for the TGFb superfamily using the following compounds: inhibitors of TGFR (compounds 66 and 74, gifts from Novartis), inhibitor of ALK2 (compound 56, gift from Novartis) and SB431542 (Tocris Cat# 1614) inhibiting ALK4, 5 and 7. Compounds were used in the working concentrations 0.02, 0.1, 0.5 and 2.5μM and were added either at 48h, 72h or 96h and kept for 24h. Gastruloids treated with the same dilution of DMSO were used as controls. Treated gastruloids were fixed at 144h and processed with immunofluorescence as describe above. Extracted features were subsequently z-scored using the DMSO controls corresponding to the relevant compound dilution and treatment timing.
Quantification and statistical analysis
Image analysis and extraction of features
Gastruloids segmentation on MIPs
Maximum intensity projections (MIPs) were generated from each acquired z-stack. Gastruloids were automatically segmented from the DAPI channel using a fully convolutional neural network (FCN) on manually curated threshold-based segmentation masks. The network was based on a RDCNet backbone with a single class segmentation head and was trained in TensorFlow, using a soft-Jaccard loss.84 Most wells contained a single gastruloid and when more than one was present, only the largest one was considered for further analysis.
Skeleton extraction
The medial axis skeleton was computed from the gastruloid mask (set of all points having more than one closest point on the object's boundary, i.e. ridges of the distance transform). To avoid small spurious branches, the distance transform was smoothed with a Gaussian kernel prior to extracting the ridges. Large, curled gastruloids were split along their folding line by subtracting a separator map from the distance transform prior to extracting the ridges. In case of the gastruloid screen analysis, the separator map was predicted from all four channels (DAPI, Sox1, Bra and E-cadherin) by a second FCN trained on manual annotations using a mean squared error loss.
Gastruloid shape model
A regular rectangular grid was deformed to the shape of the mask and was used to measure its length/width as well as measure intensity profile along its length. First, gastruloid's “corners" were determined by extrapolating the skeleton until the object boundaries. For skeletons with side branches, the two ends resulting in the longest path were used. Points along the contour that are equidistant from the ends of the original and extrapolated skeletons were used as corners. Equidistant points were placed along the contour between each corner, to define a mapping for the edges of a regular grid. The rest of the grid was mapped by thin plate interpolation. Finally, the grid orientation along the length of the gastruloid was normalised so that the mean intensity of the Bra channel over the second half of the grid was always larger than over the first half.
Shading occlusions
Precipitates from the clearing solutions sometimes partially occluded the optical path, resulting in shaded regions. A shading mask was estimated by re-thresholding the DAPI channel at 30% of the difference between background intensity (10th percentile over background) and the object intensity (90th percentile over foreground). Shaded areas were excluded from intensity measurements and samples with more than 20% of their area shaded were completely discarded.
Fluorescent debris
On occasions, debris from the liquid dispenser ended up in the wells, resulting in bright fluorescent debris with very distinctive shape and texture. Binary masks of those debris were predicted by a FCN trained on manual annotations. Similarly to shading occlusions, affected regions were excluded from intensity measurements.
Whole gastruloids feature extraction
For each segmented gastruloids, a set of morphological and intensity-based features were extracted from MIPs. Intensity features (mean, standard deviation, quantiles) were computed on image (for quality control), gastruloid, coarse and fine segment levels. Morphological features included perimeter, area, eccentricity, convexity, form factor, mean radius, length, and width, estimated from the mapped grid and number of skeleton branches.
Segment feature extraction
The grid mapped to the gastruloid was used to generate coarse (posterior/middle/anterior) and fine (100 steps) segments along its length. The anterior-posterior orientation was then determined by finding the centre of mass of either the Sox2 signal (corresponding to the anterior pole, used in the trajectory analysis) or the Bra signal (corresponding to the posterior pole, used in the screen analysis), see also “Gastruloid shape model”. Hence, the intensity profile of the obtained line faithfully recapitulated the changes in readout intensity going from the anterior to posterior side of the gastruloid. Measured intensity values were normalised for each gastruloid by dividing the mean intensity of each segment over the sum of mean segment intensities of the gastruloid.
Inside/outside feature extraction
In order to analyse markers localisation in the interior of the gastruloid, features were also extracted from the middle slice of the z-stack. The middle slice was taken as the z-plane having the largest mean intensity over the DAPI channel. The binary mask extracted from the MIP was also refined by re-thresholding the DAPI channel at 30% of the difference between background intensity (10th percentile over background) and the object intensity (90th percentile over foreground). To quantify radial signal localisation, the refined mask was partitioned into inside/outside regions with a separation at 50% of the maximum distance transform value. The in/out ratio represents the ratio of measured intensity between the two regions.
Superpixel feature extraction
For fine-graine localisation, the mask was partitioned into superpixels calculated with the SLIC (Simple Linear Iterative Clustering) method, initialised with uniform regions of approximately the size of a single cell (300 pixel). Superpixels were used as an approximation to single cells to determine (1) the localisation of the highest expressing superpixels and (2) the number of Sox2-high expressing cells. For the former, polar coordinates of superpixels were normalised by the maximum radius of the gastruloid. Superpixels were then ranked according to their mean intensity value and their angle normalised with respect to the mean angle of the 25% top-k pixels. Finally, a hexbin histogram of the top-k superpixels in normalised coordinates was built to represent a “mean” gastruloid at each timepoint and under different growth conditions. For the latter, the upper quartile of the mean intensity of superpixels of control gastruloids was used as threshold to distinguish Sox2 high and low values and the percentage of Sox2 high superpixels over total was calculated.
List of extracted features
Whole gastruloid features: Area, Centroid 0, Centroid 1, Convex area, Convex perimeter, Convexity, Eccentricity, Form factor, L3 segment n area, L3 segment n median average deviation of intensity, L3 segment n mean intensity, L3 segment n quantile 0 0.000, L3 segment n quantile 0 0.250, L3 segment n quantile 0 0.500, L3 segment n quantile 0 0.750, L3 segment n quantile 1 0.000, L3 segment n standard deviation of intensity, Major axis length, Max radius, Mean radius, Median average deviation of intensity, Median radius, Mass displacement, Mean intensity, Minor axis length, Perimeter, Quantile 0 0.000, Quantile 0 0.001, Quantile 0 0.250, Quantile 0 0.500, Quantile 0 0.750, Quantile 0 0.999, Quantile 1 0.000, Standard deviation of intensity, Solidity, Weighted centroid 0, Weighted centroid 1.
100 Segment features: Area, Median average deviation of intensity, Median radius, Quantile 0 0.000, Quantile 0 0.250, Quantile 0 0.500, Quantile 0 0.750, Quantile 1 0.000, Standard deviation of intensity. In/Out features: Inside area, Inside mean intensity, Outside area, Outside mean intensity.
Time course analysis and trajectory inference
Trajectory of gastruloid development
To align gastruloids according to their developmental states, we generated a pseudotime serving as the axis of molecular progression. For pseudotime inference we used Palantir32 (python implementation, as included in the scanpy package), a trajectory inference algorithm originally developed for single cell sequencing and mass cytometry data.
Trajectory inference comprised the following steps:
-
1.
z-score normalisation of the extracted features: Features were first normalised using z-score normalisation to reduce batch effect. Datapoints were grouped by experimental batches (2 in total). Subsequently, data was again z-score normalised whereby the 24h timepoint was used as reference.
-
2.
Quantile-based filtering of the input data: datapoints outside of the 0.2 percentile–99.8 percentile range for each of the 23 input features (common feature set) were discarded as outliers, pruning 303 objects.
-
3.
Pseudotime calculation: we considered gastruloid development as a stochastic process where gastruloids develop over time by a series of steps through a low dimensional phenotypic manifold. To this end, original 23-dimensional data was used to calculate 20 principal components that then served for calculating diffusion components, representing the dimension-reduced manifold.
-
4.
Iterative pseudotime construction: Random datapoints from 24h and 96h timepoints were selected as staring and terminal points of the trajectory, respectively. Pseudotime inference was based on 500 waypoints and performed for 100 iterations to generate an averaged pseudotime.
-
5.
Pseudotime resampling: Since pseudotime ordering is inferred from molecular events and depends on the features selected to describe them, large distances in feature space are captured by the inferred pseudotime, resulting in “warping” of the pseudotime that can result in empty pseudotime intervals (Figure S3). To address this issue, we resampled the pseudotime, removing intervals that lacked datapoints, thus obtaining a continuous trajectory.
Mapping individual readouts to pseudotime
Having aligned the individual gastruloids to the pseudotime-based developmental progression, we then assessed the dynamics of 21 antibody-based readouts (see key resources table) along the pseudotime. To compensate for staining and imaging variation, we normalised the measured fluorescence intensity to that of the nuclear counterstain (DAPI). Mean values for each readout were then calculated for 11 bins of pseudotime (0 to 1, binned in 0.1 intervals) for visualising changes along the pseudotime. For assessing gradient dynamics along pseudotime, mean values for each segment were calculated per pseudotime interval (binning performed as described in previous section, for the intensity analysis in segments, see section “Segment feature extraction”).
Gastruloid screen analysis
Filtering of sparse conditions
For image-based screening, conditions where fewer than 5 gastruloids from less than 3 individual plates were detected were discarded from the analysis, with the final dataset including 9311 gastruloids. In other assays, the threshold level for sparse conditions was assessed on an assay-to-assay basis.
For the segment workflow, same quality control criteria have been applied, hence we started from 9157 gastruloids. Additional filters for the segment workflow included discarding all gastruloids that did not have signal in Channel 2 or 3 above baseline in any of the 100 segments per channel. Final object count for the segment analysis was thus 8724.
Gastruloid phenotypic analysis
In the gastruloid screening assay, extracted features were normalised using z-score normalisation within respective assay plates. Phenotypic clustering was carried out using the entire dataset (whole gastruloid and segment dataset used separately) using the software package PhenoGraph (python implementation; part of scanpy.external package). For the whole gastruloid features, the feature set defined in Figure S4 was used. For pattern class analysis, a 200-element array composed of the normalised Sox1 and Bra intensities of segments was used. Quality control was performed on the gastruloid area, fluorescent debris, and shading occlusions as described above.
To determine the appropriate number of neighbours for the clustering, PhenoGraph analysis was performed for community sizes from 10 to 100 in increments of 10. Data was subsequently analysed and visualised using ClusTree,85 an R package (https://github.com/lazappi/clustree) to select the optimal resolution for clustering.
Combined phenotypic space
To incorporate the information on both the whole gastruloid and pattern phenotypes in all three treatment regimes, the abundance of both the whole gastruloid and pattern class types was calculated as fraction of total gastruloids measured for each screened condition. The resulting 57-element array was used to describe every condition in multivariate space and used for clustering by compound-level similarity, dimensionality reduction, and assessing DMSO similarity (Figures 5 and S5). DMSO similarity was defined as the correlation coefficient of the 57-element phenotypic signature between the tested condition and that of the DMSO control.
Annotation term network analysis
To non-ambiguously annotate targets of the compound library with functional annotation terms, we first assigned the Kyoto Encyclopedia of Genes and Genomes (KEGG) terms to the target genes of the compounds. To resolve remaining ambiguous annotations (for example, when well-defined classes of compounds such as PKC inhibitors were assigned to much broader annotation terms), custom annotation terms were introduced to improve interpretability of the analysis (Table S7). Term-term interactions (kappa-scores) were then calculated for the canonical KEGG terms using the Cytoscape3 plugin ClueGO.
Hit selection
Individual treatment conditions were ranked by reproducibility of the resulting phenotypic effect. To this end, we defined a reproducibility score (see Figure S5). In brief, the reproducibility score described how homogeneous the observed phenotype was and how robustly it was observed in replicates of the same condition, penalising conditions with more pleiotropic phenotypes. Conditions with a reproducibility score of more than 1.0 and DMSO similarity (see previous section) below 0.5 were selected as hits of the screen.
Interaction Networks
To visualise known interactions present in the STRING database, annotated target genes of the compounds were used as input network nodes and interactions with STRING combined score above 0.4 were retrieved as network edges. Genes co-annotated with functional annotation terms (Table S7) were highlighted for visualisation.
scRNA-seq analysis
Reference and quantification of transcript abundance
Genome sequence and transcript annotation was obtained from Gencode Mouse release M24 (GRCm38 primary assembly and annotation gtf file from ftp://ftp.ebi.ac.uk/pub/databases/gencode/Gencode_mouse/release_M24), and the coordinates of transcripts and introns were extracted using R (https://r-project.org, version 3.6) and the getFeatureRanges function from the eisaR package (version 0.9 available from https://github.com/fmicompbio/eisaR/tree/bbd8787d7e4b87d158a276af0787226530867b88) with arguments featureType = c("spliced", "intron"), intronType = "separate", flankLength = 0, joinOverlappingIntrons = FALSE. A transcript-to-gene map was created using the getTx2Gene function from eisaR, which links transcripts to genes, and introns to distinct genes for simultaneous quantification (see below). Sequences were extracted from the genome using extractTranscriptSeqs from the GenomicFeatures package (version 1.38.2, )86 and indexed with Salmon (version 1.1.087) with arguments -k 23 –gencode and using the genome as a decoy. Reads were then quantified using Salmon/Alevin (version 1.1.088) with parameters -l ISR --chromiumV3 and the transcript-to-gene map created above.
Quality control and filtering
Technical quality of single cell experiments, cell barcode identification and quantification were assessed using the Bioconductor package alevinQC (version 1.4.0, https://doi.org/doi:10.18129/B9.bioc.alevinQC). Salmon/Alevin counts from all twelve timepoints were imported into R (version 4.0.2) using the tximeta package (version 1.6.389) and stored in a SingleCellExperiment container for downstream analysis. The number of detected genes per cell and the fraction of counts in genes encoded on the mitochondrion (chrM) were calculated using the addPerCellQC function from the scater package (version 1.16.290), and cells with more than 1800 detected genes and less than 10% mitochondrial counts were retained. Gms, mitochondrial, and ribosomal protein genes (genes associated with the GeneOntology term "structural constituent of ribosome", GO:0003735, based on the org.Mm.eg.db package, version 3.11.4) were removed, resulting in a final analysis set of 27027 genes and 71005 cells.
Single-cell RNA sequencing data analysis
For batch effect correction, cells were assigned to one of two respective batches, and SCTransform-based normalization and variance stabilization91 were performed followed by unsupervised identification of anchors between pairs of datasets, canonical correlation analysis (CCA) and integration92,93 using Seurat (version 4.1.1). Clustering (Louvain clustering of a constructed KNN graph based on the Euclidean distance in PCA space) and visualization were then performed using Seurat (version 4.1.1).
CAT analysis and cell type annotation
Gastruloid cells from each timepoint were clustered separately using Seurat. Cells from the Pijuan-Sala et al.23 mouse embryonic dataset and the corresponding cell type annotation were used as the target reference for the CAT22 analysis. Raw counts for the gastruloid (for each timepoint separately) and embryonic datasets were normalised in scanpy94 based on total counts with a target sum of 10000 transcripts/cells and then log transformed. The CAT analysis was performed on clusters for each timepoint separately to match the gastruloid clusters to cell types. We would like to note that we employ both manual and CAT analysis-based annotations. Our results of the CAT analysis are dependent on the in vivo reference (Pijuan-Sala et al.).23 Thus, the CAT-annotations -obtained for the gastruloid cells can be affected by limitations and possible annotation inaccuracies of this in vivo reference. For that reason, we invested major efforts to perform manual annotations and comparisons with additional datasets.15,27
Conservation analysis
The cells corresponding to a given cell type (gut, pre-somitic mesoderm or NMPs) were selected from the gastruloid and Pijuan-Sala et al.23 embryonic datasets and their raw gene expression counts were merged to form a single gene expression matrix. RaceID3 (version 0.2.3)95 was then used for normalisation and filtering using the default parameters. The means of the normalised counts for the genes were calculated for each of the embryonic and gastruloid cell type populations. The means were then plotted in a scatter plot and a linear regression model was inferred using ggplot2 and ggpmisc packages. Pearson correlation analysis was performed between the embryonic and gastruloid cell type populations using the means.
Integration of gastruloid and embryonic datasets
For the integration of gastruloid and embryonic scRNA-seq datasets, single-cell trasncriptomes of gastruloid cells and embryonic cells were merged to create a single-gene expression matrix. Three batches were created. Gastruloid cells were assigned to one of two respective batches for batch effect correction and embryonic cells from Pijuan-Sala et al.23 were assigned to a third batch. SCTransform-based normalization and variance stabilization were then performed followed by CCA and integration using Seurat (version 4.1.1). Clustering and visualization were then performed on the integrated dataset using Seurat.
Analysis of embryonic epiblast data
Single-cell transcriptomes from the embryonic epiblast data of Cheng et al.27 (GSE109071) were clustered and visualized using Seurat. Epiblast subpopulations were annotated based on the original annotation from Cheng et al.27
Differential gene expression analysis
Differential gene expression analysis between cells from different origins (mouse embryo versus gastruloids) or timepoints was performed using the diffexpnb function from the RaceID3 package.95 First, negative binomial distributions reflecting the gene expression variability within each subgroup were inferred on the basis of the background model for the expected transcript count variability computed by RaceID3. Using these distributions, a P-value for the observed difference in transcript counts between the two subgroups was calculated and corrected for multiple testing using the Benjamini-Hochberg method as described.96
Cell ordering and generation of SOM
For the Cheng et al.27 embryonic epiblast dataset (GSE109071), cells were ordered in chronological order of the sampled timepoints, and cells from each timepoint were ordered in ascending order along the anterior-posterior axis based on the coordinates of the first t-SNE dimension, that is the cells’ x-axis coordinates from the t-SNE. Gastruloid epiblast cells and ectopic pluripotent cells were ordered in chronological order of the sampled timepoints. For the ectopic pluripotency population, the SOM was generated using the FateID package based on the chronological ordering of the timepoints95 and the temporally variable genes inferred from the differential gene expression analysis as described above. Only genes with more than two counts after size normalization in at least 10 cells were included for the SOM analysis. In brief, smooth profiles were derived by applying local regression on normalized transcript counts after ordering cells. A one-dimensional SOM with 144 nodes was computed on these profiles after z-transformation. Neighbouring nodes were merged if the Pearson’s correlation coefficient of the average profiles of these nodes exceeded 0.85. The remaining aggregated nodes represent the gene modules shown in the SOM figures.
Generation of temporal gene expression profiles
After ordering cells, smooth temporal gene expression profiles were derived by applying local regression on normalized transcript counts using the FateID package. Gene signatures for the epiblast subpopulations (anterior, transitioning, and posterior) were assembled based on gene expression data from Cheng et al.27
scMultiome - ATAC and RNA expression analysis
Alignment, processing, and counting of both ATAC and RNA molecules were performed using Cell Ranger ARC. The mouse mm10 genome was used a reference. For the scATAC-seq data, gene annotations were extracted from EnsDb.Mmusculus.v79. Cells from the 48h and 52h samples were filtered using total counts from both RNA and ATAC-seq datasets; cells that had greater than 1000 ATAC-seq counts/cells and 1000 transcript counts/cell were kept, resulting in a dataset of 1317 cells from the 48h sample and a dataset of 1096 cells from the 52h sample. The scATAC-seq data was normalized by performing term frequency-inverse document frequency (TF-IDF) normalization.97 For the scRNA-seq data SCTransform-based normalization was performed in Seurat (version 4.1.1). Clustering of cells and visualization were performed using Seurat (version 4.1.1). For the scATAC-seq modality, after normalisation, top features were identified and partial singular value decomposition was applied; dimensionality reduction was performed using iterative latent semantic indexing followed by the generation of a UMAP on the first 50 dimensions from the iterative latent semantic indexing reduction while discarding the first dimension associated with sequencing depth variability. For the snRNA-seq modality, after normalisation, the UMAP was generated based on the first 50 dimensions from the PCA. Weighted nearest neighbor clustering was performed on RNA+ATAC-seq data in single cells. Visualizations for gene accessibility profiles were performed using Signac (version 1.9.0) and Seurat (version 4.1.1). For inferring gene activity in single cells, gene coordinates were extracted and then extended to include the 2 kb region upstream of the transcription start site (region containing the promoter). The number of fragments for each cell that map to each of these regions were counted and aggregated. Gene activities were then log normalized using the median total count of gene activities as a scale factor. Gene activities were inferred in single cells for single genes that belong to a particular cell type/state signature. The gene activities for genes belonging to a particular signature were then aggregated.
Fate bias analysis
The single-cell transcriptomes from the 52h multiome data were integrated with the single-cell transcriptomes of the ectopic pluripotency and primitive-streak like populations from the scRNA-seq data. Cells were assigned to one of three respective batches, and SCTransform-based normalization and variance stabilization91 were performed followed by unsupervised identification of anchors between pairs of datasets, canonical correlation analysis (CCA) and integration92,93 using Seurat (version 4.1.1). FateID was used to infer the cell fate probabilities.95 The ectopic pluripotency and primitive-streak like populations were assigned as the target states. After quantifying the cell fate probabilities or fate bias in single cells, the fate bias of the multiome clusters towards the ectopic pluripotency and primitive streak-like populations were quantified.
Bulk-RNA-sequencing analysis
Raw RNA-seq reads were trimmed to remove Nextera adapters or bad quality bases (Cutadapt v4.0 -a CTGTCTCTTATACACATCTCCGAGCCCACGAGAC -A CTGTCTCTTATACACATCTGACGCTGCCGACGA -q 30 -m 15).98 Mapping was done from filtered reads on the mouse genome mm10 with STAR version 2.7.8a99 with the gencode.vM24.primary assembly annotations and ENCODE parameters. FPKM values were obtained by Cufflinks version 2.2.1100,101 with options --no-effective-length-correction -b 'mm10.fa' --multiread-correct --library-type fr-firststrand --mask-file 'chrM_mm10.gtf' --max-bundle-length 10000000 --max-bundle-frags 1000000 (where chrM_mm10.gtf contains a transcript on each strand of the whole chrM). PCA, correlation matrices and clustering were performed on log2(1 + FPKM) values of the 2000 most variant genes. Visualization was done in R v4.2.1, Heatmaps were generated with the Pheatmap package (v1.0.12) using log2(1 + FPKM) values scaled for each gene. Samples were clustered using Pearson correlation from the visualized genes. No samples were excluded from the analysis. The sample correlation matrix was obtained using the 2000 most variable genes using the "ward.D2" clustering method on the spearman inter-sample correlations.
Acknowledgments
We thank D. Vischi and E. Tagliavini for IT support; S. Smallwood and S. Aluri for sequencing; F.J. Theis for input on scRNA-seq analysis; M. Kahnwald, N. Repina, and L. Gelman for input about imaging, experimental design, and analysis; D. Suter for providing cells; and D. Duboule, J. Betschinger, A. Schier, C. Tsiairis, M. Turco, and laboratory members for scientific discussions and feedback on the manuscript. Funding: European Research Council under the European Union’s Horizon 2020 research and innovation programme (grant agreement number 758617), SNSF Sinergia grant (CRSII5_189956), and Human Frontier Science Program (LT000032/2019-L).
Author contributions
P.L., S.S., and M.Z. conceived the study and designed the experiments. M.Z. established the gastruloid culture and initiated this project. M.Z. performed the transcriptomics, image-based screen, and validation experiments. S.S. performed the time course/trajectory, validation, core perturbation, and core anteriorization experiments. N.A. analyzed and interpreted the scRNA-seq and multiome sequencing data. N.A. and S.S. performed the cell type annotation and contributed to the interpretation of the scRNA-seq data. I.L. performed image analysis on the time course and screen and M.Z. and S.S. on all other experiments. R.O. designed the gastruloid feature extraction pipeline. C.A. helped with experiments and performed the multiome sample preparation. M.B.S. contributed to the processing of the scRNA-seq and multiome sequencing data. G.P. contributed to the scRNA-seq data analysis. H.K. performed fluorescent cell sorting experiments. A.M. performed the Chir timing bulk RNA-seq experiment and analysis. M.P.L. and S.V. contributed to study design and discussion of results. S.S., M.Z., N.A., I.L., and P.L. wrote the paper.
Declaration of interests
M.P.L. is an employee of Roche and has equity in Roche.
Inclusion and diversity
One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in their field of research or within their geographical location. One or more of the authors of this paper self-identifies as a gender minority in their field of research. One or more of the authors of this paper self-identifies as a member of the LGBTQIA+ community. We support inclusive, diverse, and equitable conduct of research.
Published: May 19, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stem.2023.04.018.
Supplemental information
Data and code availability
-
•
The single cell RNA-seq, multiome, and bulk RNA-seq datasets generated during this study are available at NCBI GEO (https://www.ncbi.nlm.nih.gov/geo/). The accession numbers are listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request. Any additional data reported in this paper will be shared by the lead contact upon request.
-
•
The original code has been deposited on Github and is publicly available. See key resources table for the link and DOI.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Rivron N.C., Frias-Aldeguer J., Vrij E.J., Boisset J.-C., Korving J., Vivié J., Truckenmüller R.K., van Oudenaarden A., van Blitterswijk C.A., Geijsen N. Blastocyst-like structures generated solely from stem cells. Nature. 2018;557:106–111. doi: 10.1038/s41586-018-0051-0. [DOI] [PubMed] [Google Scholar]
- 2.Yu L., Wei Y., Duan J., Schmitz D.A., Sakurai M., Wang L., Wang K., Zhao S., Hon G.C., Wu J. Blastocyst-like structures generated from human pluripotent stem cells. Nature. 2021;591:620–626. doi: 10.1038/s41586-021-03356-y. [DOI] [PubMed] [Google Scholar]
- 3.Zheng Y., Xue X., Shao Y., Wang S., Esfahani S.N., Li Z., Muncie J.M., Lakins J.N., Weaver V.M., Gumucio D.L., Fu J. Controlled modelling of human epiblast and amnion development using stem cells. Nature. 2019;573:421–425. doi: 10.1038/s41586-019-1535-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Warmflash A., Sorre B., Etoc F., Siggia E.D., Brivanlou A.H. A method to recapitulate early embryonic spatial patterning in human embryonic stem cells. Nat. Methods. 2014;11:847–854. doi: 10.1038/nmeth.3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sozen B., Amadei G., Cox A., Wang R., Na E., Czukiewska S., Chappell L., Voet T., Michel G., Jing N., et al. Self-assembly of embryonic and two extra- embryonic stem cell types into gastrulating embryo-like structures. Nat. Cell Biol. 2018;20:979–989. doi: 10.1038/s41556-018-0147-7. [DOI] [PubMed] [Google Scholar]
- 6.Sozen B., Cox A.L., De Jonghe J., Bao M., Hollfelder F., Glover D.M., Zernicka-Goetz M. Self-organization of mouse stem cells into an extended potential blastoid. Dev. Cell. 2019;51:698–712.e8. doi: 10.1016/j.devcel.2019.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shao Y., Taniguchi K., Townshend R.F., Miki T., Gumucio D.L., Fu J. A pluripotent stem cell-based model for post-implantation human amniotic sac development. Nat. Commun. 2017;8 doi: 10.1038/s41467-017-00236-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van den Brink S.C., Baillie-Johnson P., Balayo T., Hadjantonakis A.K., Nowotschin S., Turner D.A., Martinez Arias A. Symmetry breaking, germ layer specification and axial organisation in aggregates of mouse embryonic stem cells. Development. 2014;141:4231–4242. doi: 10.1242/dev.113001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moris N., Anlas K., van den Brink S.C., Alemany A., der, J. S.x.F., Ghimire S., Balayo T., van Oudenaarden A., Arias A.M. An in vitro model of early anteroposterior organization during human development. Nature. 2020;582:410–415. doi: 10.1038/s41586-020-2383-9. [DOI] [PubMed] [Google Scholar]
- 10.Turner D.A., Girgin M., Alonso-Crisostomo L., Trivedi V., Baillie-Johnson P., Glodowski C.R., Hayward P.C., Collignon J., Gustavsen C., Serup P., et al. Anteroposterior polarity and elongation in the absence of extra-embryonic tissues and of spatially localised signalling in gastruloids: mammalian embryonic organoids. Development. 2017;144:3894–3906. doi: 10.1242/dev.150391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beccari L., Moris N., Girgin M., Turner D.A., Baillie-Johnson P., Cossy A.C., Lutolf M.P., Duboule D., Arias A.M. Multi-axial self-organization properties of mouse embryonic stem cells into gastruloids. Nature. 2018;562:272–276. doi: 10.1038/s41586-018-0578-0. [DOI] [PubMed] [Google Scholar]
- 12.Vianello S., Rosa V.S., Lutolf M.P. In vitro endoderm emergence and self-organisation in the absence of extraembryonic tissues and embryonic architecture. Preprint at bioRxiv. 2021 doi: 10.1101/2020.06.07.138883. [DOI] [Google Scholar]
- 13.Rossi G., Broguiere N., Miyamoto M., Boni A., Guiet R., Girgin M., Kelly R.G., Kwon C., Lutolf M.P. Capturing cardiogenesis in gastruloids. Stem Cells. 2020;28:230–240.e6. doi: 10.1016/j.stem.2020.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Veenvliet J.V., Bolondi A., Kretzmer H., Haut L., Scholze-Wittler M., Schifferl D., Koch F., Guignard L., Kumar A.S., Pustet M., et al. Mouse embryonic stem cells self-organize into trunk-like structures with neural tube and somites. Science. 2020;370:eaba4937–eaba4939. doi: 10.1126/science.aba4937. [DOI] [PubMed] [Google Scholar]
- 15.van den Brink S.C., Alemany A., van Batenburg V., Moris N., Blotenburg M., Vivié J.V., Baillie-Johnson P., Nichols J., Sonnen K.F., Arias A.M., van Oudenaarden A. Single-cell and spatial transcriptomics reveal somitogenesis in gastruloids. Nature. 2020;582:405–409. doi: 10.1038/s41586-020-2024-3. [DOI] [PubMed] [Google Scholar]
- 16.Girgin M.U., Broguiere N., Hoehnel S., Brandenberg N., Mercier B., Arias A.M., Lutolf M.P. Bioengineered embryoids mimic post-implantation development in vitro. Nat. Commun. 2021;12 doi: 10.1038/s41467-021-25237-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Girgin M.U., Broguiere N., Mattolini L., Lutolf M.P. Gastruloids generated without exogenous Wnt activation develop anterior neural tissues. Stem Cell Rep. 2021;16:1143–1155. doi: 10.1016/j.stemcr.2021.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu P.-F., Borges R.M., Fillatre J., De Oliveira-Melo M., Cheng T., Thisse B., Thisse C. Construction of a mammalian embryo model from stem cells organized by a morphogen signalling centre. Nat. Commun. 2021;12 doi: 10.1038/s41467-021-23653-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bérenger-Currias N.M., Mircea M., Adegeest E., Van Den Berg P.R., Feliksik M., Hochane M., Idema T., Tans S.J., Semrau S. A gastruloid model of the interaction between embryonic and extra-embryonic cell types. J. Tissue Eng. 2022;13 doi: 10.1177/20417314221103042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wehmeyer A.E., Schüle K.M., Conrad A., Schröder C.M., Probst S., Arnold S.J. Chimeric 3D gastruloids – a versatile tool for studies of mammalian peri-gastrulation development. Development. 2022;149 doi: 10.1242/dev.200812. [DOI] [PubMed] [Google Scholar]
- 21.Anlas K., Gritti N., Oriola D., Arató K., Nakaki F., Le Lim J., Sharpe J., Trivedi V. Dynamics of anteroposterior axis establishment in a mammalian embryo-like system. Preprint at bioRxiv. 2021 doi: 10.1101/2021.02.24.432766. [DOI] [Google Scholar]
- 22.Rothová M.M., Nielsen A.V., Proks M., Wong Y.F., Riveiro A.R., Linneberg-Agerholm M., David E., Amit I., Trusina A., Brickman J.M. Identification of the central intermediate in the extra-embryonic to embryonic endoderm transition through single-cell transcriptomics. Nat. Cell Biol. 2022;24:833–844. doi: 10.1038/s41556-022-00923-x. [DOI] [PubMed] [Google Scholar]
- 23.Pijuan-Sala B., Griffiths J.A., Guibentif C., Hiscock T.W., Jawaid W., Calero-Nieto F.J., Mulas C., Ibarra-Soria X., Tyser R.C.V., Ho D.L.L., et al. A single-cell molecular map of mouse gastrulation and early organogenesis. Nature. 2019;566:490–495. doi: 10.1038/s41586-019-0933-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Masui S., Nakatake Y., Toyooka Y., Shimosato D., Yagi R., Takahashi K., Okochi H., Okuda A., Matoba R., Sharov A.A., et al. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat. Cell Biol. 2007;9:625–635. doi: 10.1038/ncb1589. [DOI] [PubMed] [Google Scholar]
- 25.Festuccia N., Osorno R., Halbritter F., Karwacki-Neisius V., Navarro P., Colby D., Wong F., Yates A., Tomlinson S.R., Chambers I. Esrrb is a direct Nanog target gene that can substitute for Nanog function in pluripotent cells. Stem Cells. 2012;11:477–490. doi: 10.1016/j.stem.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi W., Wang H., Pan G., Geng Y., Guo Y., Pei D. Regulation of the pluripotency marker Rex-1 by Nanog and Sox2. J. Biol. Chem. 2006;281:23319–23325. doi: 10.1074/jbc.M601811200. [DOI] [PubMed] [Google Scholar]
- 27.Cheng S., Pei Y., He L., Peng G., Reinius B., Tam P.P.L., Jing N., Deng Q. Single-cell RNA-seq reveals cellular heterogeneity of pluripotency transition and X chromosome dynamics during early mouse development. Cell Rep. 2019;26:2593–2607.e3. doi: 10.1016/j.celrep.2019.02.031. [DOI] [PubMed] [Google Scholar]
- 28.Cermola F., D’Aniello C., Tatè R., De Cesare D., Martinez-Arias A., Minchiotti G., Patriarca E.J. Gastruloid development competence discriminates different states of pluripotency. Stem Cell Rep. 2021;16:354–369. doi: 10.1016/j.stemcr.2020.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koch F., Scholze M., Wittler L., Schifferl D., Sudheer S., Grote P., Timmermann B., Macura K., Herrmann B.G. Antagonistic activities of Sox2 and brachyury control the fate choice of neuro-mesodermal progenitors. Dev. Cell. 2017;42:514–526.e7. doi: 10.1016/j.devcel.2017.07.021. [DOI] [PubMed] [Google Scholar]
- 30.Bergsland M., Ramsköld D., Zaouter C., Klum S., Sandberg R., Muhr J. Sequentially acting Sox transcription factors in neural lineage development. Genes Dev. 2011;25:2453–2464. doi: 10.1101/gad.176008.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Avilion A.A., Nicolis S.K., Pevny L.H., Perez L., Vivian N., Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Amp Dev. 2002;17:126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Setty M., Kiseliovas V., Levine J., Gayoso A., Mazutis L., Pe’er D. Characterization of cell fate probabilities in single-cell data with Palantir. Nat. Biotechnol. 2019;37:451–460. doi: 10.1038/s41587-019-0068-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Serra D., Mayr U., Boni A., Lukonin I., Rempfler M., Challet Meylan L.C., Stadler M.B., Strnad P., Papasaikas P., Vischi D., et al. Self-organization and symmetry breaking in intestinal organoid development. Nature. 2019;569:66–72. doi: 10.1038/s41586-019-1146-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arnold S.J., Hofmann U.K., Bikoff E.K., Robertson E.J. Pivotal roles for eomesodermin during axis formation, epithelium-to-mesenchyme transition and endoderm specification in the mouse. Development. 2008;135:501–511. doi: 10.1242/dev.014357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herrmann B.G. Expression pattern of the brachyury gene in whole mount Twis/Twis mutant embryos. Development. 1991;113:913–917. doi: 10.1242/dev.113.3.913. [DOI] [PubMed] [Google Scholar]
- 36.Pevny L.H., Sockanathan S., Placzek M., Lovell-Badge R. A role for Sox1 in neural determination. Development. 1998;125:1967–1978. doi: 10.1242/dev.125.10.1967. [DOI] [PubMed] [Google Scholar]
- 37.Levine J.H., Simonds E.F., Bendall S.C., Davis K.L., Amir E.-a.D., Tadmor M.D., Litvin O., Fienberg H.G., Jager A., Zunder E.R., et al. Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell. 2015;162:184–197. doi: 10.1016/j.cell.2015.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lukonin I., Serra D., Challet Meylan L.C., Volkmann K., Baaten J., Zhao R., Meeusen S., Colman K., Maurer F., Stadler M.B., et al. Phenotypic landscape of intestinal organoid regeneration. Nature. 2020;586:275–280. doi: 10.1038/s41586-020-2776-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hemmati-Brivanlou A., Melton D. Vertebrate embryonic cells will become nerve cells unless told otherwise. Cell. 1997;88:13–17. doi: 10.1016/S0092-8674(00)81853-X. [DOI] [PubMed] [Google Scholar]
- 40.Ying Q.L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wray J., Kalkan T., Smith A.G. The ground state of pluripotency. Biochem. Soc. Trans. 2010;38:1027–1032. doi: 10.1042/BST0381027. [DOI] [PubMed] [Google Scholar]
- 42.Betschinger J., Nichols J., Dietmann S., Corrin P.D., Paddison P.J., Smith A. Exit from pluripotency is gated by intracellular redistribution of the bHLH transcription factor Tfe3. Cell. 2013;153:335–347. doi: 10.1016/j.cell.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leitch H.G., Nichols J., Humphreys P., Mulas C., Martello G., Lee C., Jones K., Surani M.A., Smith A. Rebuilding pluripotency from primordial germ cells. Stem Cell Rep. 2013;1:66–78. doi: 10.1016/j.stemcr.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parchem R.J., Ye J., Judson R.L., LaRussa M.F., Krishnakumar R., Blelloch A., Oldham M.C., Blelloch R. Two miRNA clusters reveal alternative paths in late-stage reprogramming. Cell Stem Cell. 2014;14:617–631. doi: 10.1016/j.stem.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hart A.H., Hartley L., Ibrahim M., Robb L. Identification, cloning and expression analysis of the pluripotency promoting Nanog genes in mouse and human. Dev. Dyn. 2004;230:187–198. doi: 10.1002/dvdy.20034. [DOI] [PubMed] [Google Scholar]
- 46.Sambasivan R., Steventon B. Neuromesodermal progenitors: A basis for robust axial patterning in development and evolution. Front. Cell Dev. Biol. 2020;8 doi: 10.3389/fcell.2020.607516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hunt G.C., Singh P., Schwarzbauer J.E. Endogenous production of fibronectin is required for self-renewal of cultured mouse embryonic stem cells. Exp. Cell Res. 2012;318:1820–1831. doi: 10.1016/j.yexcr.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kapp T.G., Rechenmacher F., Neubauer S., Maltsev O.V., Cavalcanti-Adam E.A., Zarka R., Reuning U., Notni J., Wester H.-J., Mas-Moruno C., et al. A comprehensive evaluation of the activity and selectivity profile of ligands for RGD-binding integrins. Sci. Rep. 2017;7 doi: 10.1038/srep39805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ruoslahti E., Pierschbacher M.D. New perspectives in cell adhesion: RGD and integrins. Science. 1987;238:491–497. doi: 10.1126/science.2821619. [DOI] [PubMed] [Google Scholar]
- 50.Desrochers L.M., Bordeleau F., Reinhart-King C.A., Cerione R.A., Antonyak M.A. Microvesicles provide a mechanism for intercellular communication by embryonic stem cells during embryo implantation. Nat. Commun. 2016;7 doi: 10.1038/ncomms11958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hur Y.H., Feng S., Wilson K.F., Cerione R.A., Antonyak M.A. Embryonic stem-cell-derived extracellular vesicles maintain ESC stemness by activating FAK. Dev. Cell. 2020;56:277–291.e6. doi: 10.1016/j.devcel.2020.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee S.H., Lee Y.J., Han H.J. Role of hypoxia-induced fibronectin-integrin β1 expression in embryonic stem cell proliferation and migration: involvement of PI3K/Akt and FAK. J. Cell. Physiol. 2011;226:484–493. doi: 10.1002/jcp.22358. [DOI] [PubMed] [Google Scholar]
- 53.Deluz C., Friman E.T., Strebinger D., Benke A., Raccaud M., Callegari A., Leleu M., Manley S., Suter D.M. A role for mitotic bookmarking of SOX2 in pluripotency and differentiation. Genes Dev. 2016;30:2538–2550. doi: 10.1101/gad.289256.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bondue A., Tännler S., Chiapparo G., Chabab S., Ramialison M., Paulissen C., Beck B., Harvey R., Blanpain C. Defining the earliest step of cardiovascular progenitor specification during embryonic stem cell differentiation. J. Cell Biol. 2011;192:751–765. doi: 10.1083/jcb.201007063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wymeersch F.J., Huang Y., Blin G., Cambray N., Wilkie R., Wong F.C., Wilson V. Position-dependent plasticity of distinct progenitor types in the primitive streak. eLife. 2016;5 doi: 10.7554/eLife.10042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rosen L.U., Stapel L.C., Argelaguet R., Barker C.G., Yang A., Reik W., Marioni J.C. Inter-gastruloid heterogeneity revealed by single cell transcriptomics time course: implications for organoid based perturbation studies. Preprint at bioRxiv. 2022 doi: 10.1101/2022.09.27.509783. [DOI] [Google Scholar]
- 57.Ortmann D., Brown S., Czechanski A., Aydin S., Muraro D., Huang Y., Tomaz R.A., Osnato A., Canu G., Wesley B.T., et al. Naive pluripotent stem cells exhibit phenotypic variability that is driven by genetic variation. Stem Cells. 2020;27:470–481.e6. doi: 10.1016/j.stem.2020.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stower M.J., Srinivas S. Heading forwards: anterior visceral endoderm migration in patterning the mouse embryo. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014;369 doi: 10.1098/rstb.2013.0546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ang S.L., Jin O., Rhinn M., Daigle N., Stevenson L., Rossant J. A targeted mouse Otx2 mutation leads to severe defects in gastrulation and formation of axial mesoderm and to deletion of rostral brain. Development. 1996;122:243–252. doi: 10.1242/dev.122.1.243. [DOI] [PubMed] [Google Scholar]
- 60.Acampora D., Mazan S., Lallemand Y., Avantaggiato V., Maury M., Simeone A., Brûlet P. Forebrain and midbrain regions are deleted in Otx2−/− mutants due to a defective anterior neuroectoderm specification during gastrulation. Development. 1995;121:3279–3290. doi: 10.1242/dev.121.10.3279. [DOI] [PubMed] [Google Scholar]
- 61.Matsuo I., Kuratani S., Kimura C., Takeda N., Aizawa S. Mouse Otx2 functions in the formation and patterning of rostral head. Genes Dev. 1995;9:2646–2658. doi: 10.1101/gad.9.21.2646. [DOI] [PubMed] [Google Scholar]
- 62.Vianello S.D., Lutolf M. In vitro endoderm emergence and self-organisation in the absence of extraembryonic tissues and embryonic architecture. Preprint at bioRxiv. 2020 doi: 10.1101/2020.06.07.138883. [DOI] [Google Scholar]
- 63.Nowotschin S., Setty M., Kuo Y.-Y., Liu V., Garg V., Sharma R., Simon C.S., Saiz N., Gardner R., Boutet S.C., et al. The emergent landscape of the mouse gut endoderm at single-cell resolution. Nature. 2019;569:361–367. doi: 10.1038/s41586-019-1127-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Osumi N., Shinohara H., Numayama-Tsuruta K., Maekawa M. Concise review: Pax6 transcription factor contributes to both embryonic and adult neurogenesis as a multifunctional regulator. Stem Cells. 2008;26:1663–1672. doi: 10.1634/stemcells.2007-0884. [DOI] [PubMed] [Google Scholar]
- 65.Bel-Vialar S., Medevielle F., Pituello F. The on/off of Pax6 controls the tempo of neuronal differentiation in the developing spinal cord. Dev. Biol. 2007;305:659–673. doi: 10.1016/j.ydbio.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 66.Chhabra S., Liu L., Goh R., Kong X., Warmflash A. Dissecting the dynamics of signaling events in the BMP, WNT, and NODAL cascade during self-organized fate patterning in human gastruloids. PLoS Biol. 2019;17 doi: 10.1371/journal.pbio.3000498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Etoc F., Metzger J., Ruzo A., Kirst C., Yoney A., Ozair M.Z., Brivanlou A.H., Siggia E.D. A balance between secreted inhibitors and edge sensing controls gastruloid self-organization. Dev. Cell. 2016;39:302–315. doi: 10.1016/j.devcel.2016.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu L., Nemashkalo A., Rezende L., Jung J.Y., Chhabra S., Guerra M.C., Heemskerk I., Warmflash A. Nodal is a short-range morphogen with activity that spreads through a relay mechanism in human gastruloids. Nat. Commun. 2022;13 doi: 10.1038/s41467-022-28149-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kramer B.A., Sarabia del Castillo J., Pelkmans L. Multimodal perception links cellular state to decision-making in single cells. Science. 2022;377:642–648. doi: 10.1126/science.abf4062. [DOI] [PubMed] [Google Scholar]
- 70.Xavier da Silveira Dos Santos A., Liberali P. From single cells to tissue self-organization. FEBS Journal. 2018;18 doi: 10.1111/febs.14694. 483–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kinney B.A., Al Anber A., Row R.H., Tseng Y.-J., Weidmann M.D., Knaut H., Martin B.L. Sox2 and canonical Wnt signaling interact to activate a developmental checkpoint coordinating morphogenesis with mesoderm fate acquisition. Cell Rep. 2020;33:108311. doi: 10.1016/j.celrep.2020.108311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gouti M., Delile J., Stamataki D., Wymeersch F.J., Huang Y., Kleinjung J., Wilson V., Briscoe J. A gene regulatory network balances neural and mesoderm specification during vertebrate trunk development. Dev. Cell. 2017;41:243–261.e7. doi: 10.1016/j.devcel.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Blassberg R., Patel H., Watson T., Gouti M., Metzis V., Delás M.J., Briscoe J. Sox2 levels configure the WNT response of epiblast progenitors responsible for vertebrate body formation. 2020;17 doi: 10.1101/2020.12.29.424684. 3165–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Posfai E., Schell J.P., Janiszewski A., Rovic I., Murray A., Bradshaw B., Yamakawa T., Pardon T., El Bakkali M., Talon I., et al. Evaluating totipotency using criteria of increasing stringency. Nat. Cell Biol. 2021;23:49–60. doi: 10.1038/s41556-020-00609-2. [DOI] [PubMed] [Google Scholar]
- 75.Yanagida A., Spindlow D., Nichols J., Dattani A., Smith A., Guo G. Naive stem cell blastocyst model captures human embryo lineage segregation. Cell Stem Cell. 2021;28:1016–1022.e4. doi: 10.1016/j.stem.2021.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kagawa H., Javali A., Khoei H.H., Sommer T.M., Sestini G., Novatchkova M., Scholte op Reimer Y., Castel G., Bruneau A., Maenhoudt N., et al. Human blastoids model blastocyst development and implantation. Nature. 2022;601:600–605. doi: 10.1038/s41586-021-04267-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tarazi S., Aguilera-Castrejon A., Joubran C., Ghanem N., Ashouokhi S., Roncato F., Wildschutz E., Haddad M., Oldak B., Gomez-Cesar E., et al. Post-gastrulation synthetic embryos generated ex utero from mouse naive ESCs. Cell. 2022;185:3290–3306.e25. doi: 10.1016/j.cell.2022.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Amadei G., Handford C.E., Qiu C., De Jonghe J., Greenfeld H., Tran M., Martin B.K., Chen D.Y., Aguilera-Castrejon A., Hanna J.H., et al. Embryo model completes gastrulation to neurulation and organogenesis. Nature. 2022;610:143–153. doi: 10.1038/s41586-022-05246-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Merle M., Friedman L., Chureau C., Gregor T. Precise and scalable self-organization in mammalian pseudo-embryos. Preprint at arXiv. 2023 doi: 10.48550/arXiv.2303.17522. [DOI] [PubMed] [Google Scholar]
- 80.Ghimire S., Van der Jeught M., Neupane J., Roost M.S., Anckaert J., Popovic M., Van Nieuwerburgh F., Mestdagh P., Vandesompele J., Deforce D., et al. Comparative analysis of naive, primed and ground state pluripotency in mouse embryonic stem cells originating from the same genetic background. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-24051-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Baillie-Johnson P., van den Brink S.C., Balayo T., Turner D.A., Martinez Arias A. Generation of aggregates of mouse embryonic stem cells that show symmetry breaking, polarization and emergent collective behaviour in vitro. J. Vis. Exp. 2015 doi: 10.3791/53252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hama H., Hioki H., Namiki K., Hoshida T., Kurokawa H., Ishidate F., Kaneko T., Akagi T., Saito T., Saido T., et al. ScaleS: an optical clearing palette for biological imaging. Nat. Neurosci. 2015;18:1518–1529. doi: 10.1038/nn.4107. [DOI] [PubMed] [Google Scholar]
- 83.Zhu X., Huang L., Zheng Y., Song Y., Xu Q., Wang J., Si K., Duan S., Gong W. Ultrafast optical clearing method for three-dimensional imaging with cellular resolution. Proc. Natl. Acad. Sci. USA. 2019;116:11480–11489. doi: 10.1073/pnas.1819583116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ortiz R., de Medeiros G., Peters A.H.F.M., Liberali P., Rempfler M. Springer International Publishing; 2020. RDCNet: Instance Segmentation with a Minimalist Recurrent Residual Network. Held in Cham; pp. 434–443. [Google Scholar]
- 85.Zappia L., Oshlack A. Clustering trees: a visualization for evaluating clusterings at multiple resolutions. GigaScience. 2018;7:1–9. doi: 10.1093/gigascience/giy083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lawrence M., Huber W., Pagès H., Aboyoun P., Carlson M., Gentleman R., Morgan M.T., Carey V.J. Software for computing and annotating genomic ranges. PLoS Comp. Biol. 2013;9 doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Patro R., Duggal G., Love M.I., Irizarry R.A., Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Srivastava A., Malik L., Smith T., Sudbery I., Patro R. Alevin efficiently estimates accurate gene abundances from dscRNA-seq data. Genome Biol. 2019;20 doi: 10.1186/s13059-019-1670-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Love M.I., Soneson C., Hickey P.F., Johnson L.K., Pierce N.T., Shepherd L., Morgan M., Patro R. Tximeta: reference sequence checksums for provenance identification in RNA-seq. PLoS Comp. Biol. 2020;16 doi: 10.1371/journal.pcbi.1007664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.McCarthy D.J., Campbell K.R., Lun A.T., Wills Q.F. Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics. 2017;33:1179–1186. doi: 10.1093/bioinformatics/btw777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hafemeister C., Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019;20 doi: 10.1186/s13059-019-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wolf F.A., Angerer P., Theis F.J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19 doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Herman J.S., Sagar, Grün D. FateID infers cell fate bias in multipotent progenitors from single-cell RNA-seq data. Nat. Methods. 2018;15:379–386. doi: 10.1038/nmeth.4662. [DOI] [PubMed] [Google Scholar]
- 96.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11 doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cusanovich D.A., Daza R., Adey A., Pliner H.A., Christiansen L., Gunderson K.L., Steemers F.J., Trapnell C., Shendure J. Multiplex single-cell profiling of chromatin accessibility by combinatorial cellular indexing. Science. 2015;348:910–914. doi: 10.1126/science.aab1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17 doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 99.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., Van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Roberts A., Pimentel H., Trapnell C., Pachter L. Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics. 2011;27:2325–2329. doi: 10.1093/bioinformatics/btr355. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The single cell RNA-seq, multiome, and bulk RNA-seq datasets generated during this study are available at NCBI GEO (https://www.ncbi.nlm.nih.gov/geo/). The accession numbers are listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request. Any additional data reported in this paper will be shared by the lead contact upon request.
-
•
The original code has been deposited on Github and is publicly available. See key resources table for the link and DOI.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.