Abstract
Viperin is a broadly conserved radical SAM enzyme that synthesizes the antiviral nucleotide ddhCTP. In higher animals, viperin expression also accelerates the degradation of various cellular and viral proteins necessary for viral replication; however, the details of this process remain largely unknown. Here, we show that viperin activates a component of the protein ubiquitination machinery, which plays an important role in both protein degradation and immune signaling pathways. We demonstrate that viperin binds the E3 ubiquitin ligase, TRAF6, which catalyzes K63-linked ubiquitination associated with immune signaling pathways. Viperin activates ubiquitin transfer by TRAF6–2.5-fold and causes a significant increase in polyubiquitinated forms of TRAF6 that are important for mediating signal transduction. Our observations both imply a role for viperin as an agonist of immune signaling and suggest that viperin may activate other K48-linked E3-ligases involved in targeting proteins for proteasomal degradation.
Viperin (Virus Inhibitory Protein, Endoplasmic Reticulum-associated, Interferon iNducible), also denoted as cig5 and RSAD2 in humans,1 is strongly induced by type I interferons as part of the innate immune response to viral infection.2–4 Viperin is a member of the radical SAM enzyme superfamily and appears to be conserved in all 6 kingdoms of life,5–7 hinting at its ancient and ubiquitous role in combatting viral infection. Notably, viperin is one of the very few radical SAM enzymes found in higher animals.8 Viperin catalyzes the dehydration of CTP to form the antiviral nucleotide 3′-deoxy-3′,4′-didehydro-CTP (ddhCTP; Figure 1)9 through a radical mechanism initiated by reductive cleavage of SAM.7,10 The antiviral properties of this nucleotide against RNA viruses derive from its ability to act as a chain-terminating inhibitor of some, but not all, viral RNA-dependent RNA polymerases.9
Figure 1.
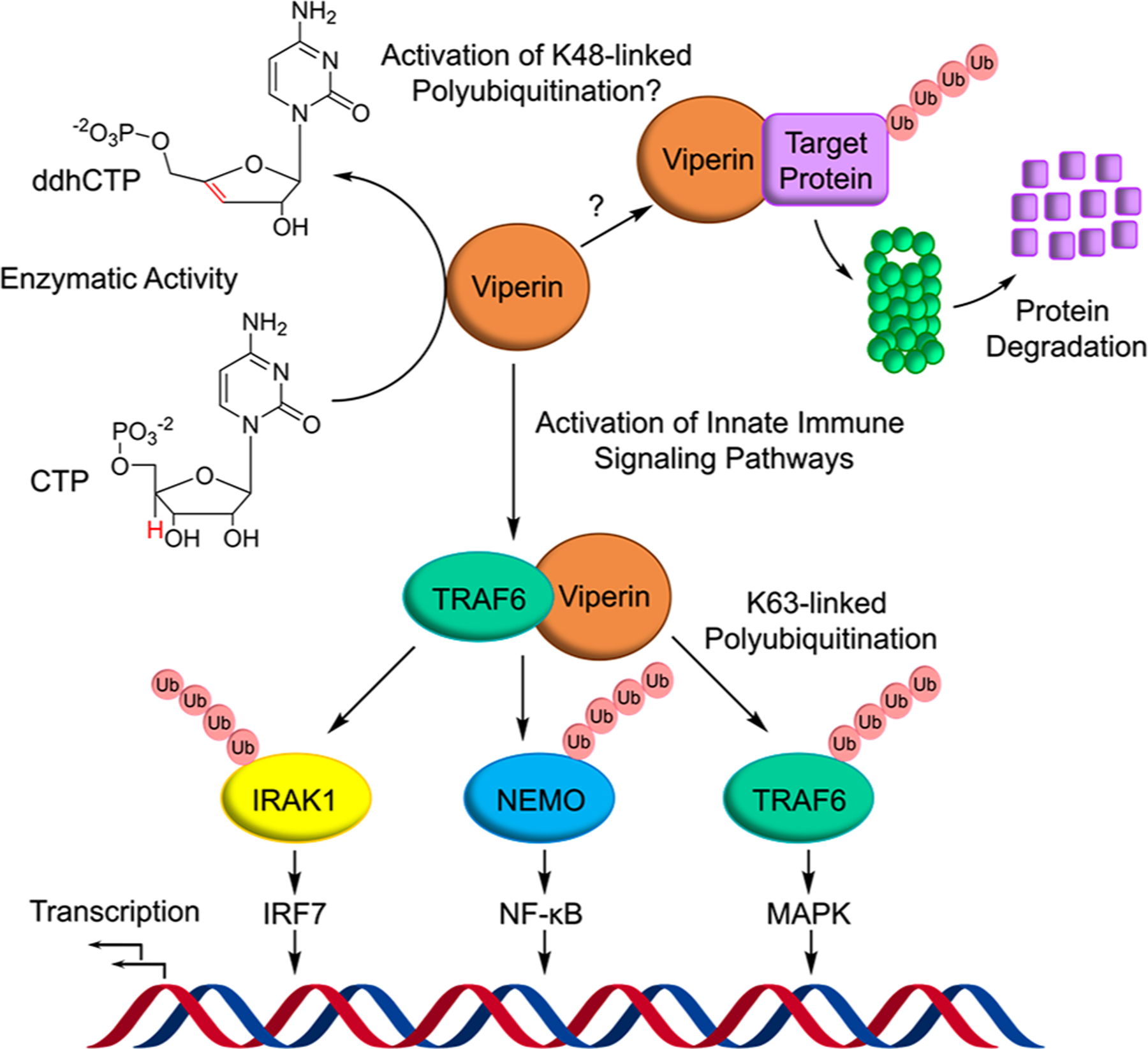
Overview of viperin’s interactions with the protein ubiquitination machinery and the E3-ligase, TRAF6.
In addition to synthesizing ddhCTP, viperin interacts with a wide range of cellular and viral proteins.11–16 This extensive network of protein–protein interactions remains poorly understood but constitutes an equally important aspect of the enzyme’s antiviral properties. In many cases, it appears that viperin exerts its antiviral effects by facilitating the degradation of cellular and viral proteins important for viral replication.11 A prevailing view is that viperin recruits the protein ubiquitination machinery to target proteins for proteasomal degradation (Figure 1).11,17 However, the evidence for viperin promoting protein ubiquitination is indirect and is based largely on studies using proteins transfected in mammalian cell lines.11,17 Thus, there’s uncertainty about which components of the ubiquitination system viperin interacts with or whether other proteins may be required for viperin to engage the ubiquitination system. Therefore, to better understand viperin’s role in promoting protein ubiquitination, we have examined viperin’s interaction with the E3 ubiquitin ligase, TRAF6.18,19
The ubiquitination machinery comprises 3 enzymes:20 E1 is responsible for the ATP-dependent activation of ubiquitin and transferring it as its C-terminal thioester to various ubiquitin conjugating enzymes (E2). E2 enzymes interact with a large set of E3 ubiquitin ligases, ~700 in humans, that recognize different protein targets for ubiquitination.21
Polyubiquitin chains may be constructed through isopeptide bonds to various ubiquitin lysine residues. K48-linked polyubiquitination marks proteins for degradation by the 26S proteasome,20,22 whereas Lys-63-linked polyubiquitination is important in activating various components of signal transduction pathways that trigger the immune response.23 E3 ligases, in particular, are highly regulated and may be activated or inhibited by a wide range of post-translational modifications and interactions with other proteins.24
TRAF6 (Tumor necrosis factor receptor-associated factor 6) is a member of the RING domain-containing E3 ligases.25 TRAF6 functions with the heterodimeric Ubc13/Uev1A E2-conjugating enzyme to synthesize K63-linked ubiquitin chains. TRAF6-mediated protein ubiquitination is central to several important signal transduction pathways,22,26 including activation of the NF-κB pathway and the MAPK signaling cascade. These pathways regulate such diverse biological processes as cell growth, oncogenesis, and immune and inflammatory responses.26 TRAF6 substrates include interleukin-1 receptor-associated kinases (IRAKs) and NF-κB essential modulator (NEMO).27,28 But TRAF6 also undergoes autoubiquitination specifically on Lys12429 and these polyubiquitinated forms serve to recruit and assemble downstream kinases and associated factors into signaling complexes that ultimately activate NF-κB and MAPK pathways.30,26
Recently, viperin was shown in cellullo to interact with TRAF6 to promote K63-linked polyubiquitination of interleukin receptor-associated kinase 1 (IRAK1) as part of innate immune signaling in the Toll-like receptor-7 and 9 (TLR-7/9) pathways.15,28 The viperin-TRAF6 interaction provides a unique opportunity to test whether viperin functions as an activator of protein ubiquitination in a well-defined biochemical system. We have reconstituted the TRAF6 autoubiquitination system in vitro using purified enzymes. This has allowed us to demonstrate that viperin does indeed activate the E3 ligase activity of TRAF6, leading to a significant increase in the amount of polyubiquitinated TRAF6 species formed.
Full-length TRAF6 is a multidomain protein that forms large oligomers in the cell and has proven refractory to expression in E. coli. Therefore, to reconstitute the ubiquitination system in vitro, we used a truncated TRAF6 construct comprising the RING and first 3 zinc-finger domains19 (designated TRAF6-N), which was previously shown to be functional and can be expressed and purified from E. coli.19 A human viperin construct lacking the first 50 residues of the ER-localizing N-terminal amphipathic helix, designated viperin-ΔN50, was expressed and purified from E. coli and the [4Fe-4S] cluster reconstituted as described previously.16 Preliminary pull-down experiments established that the truncated viperin-ΔN50 and TRAF6-N proteins form a stable complex with each other (Figure S1).
TRAF6 functions with the heterodimeric E2 ubiquitin-conjugating enzyme, Ubc13/Uev1A; this enzyme was expressed and purified from E. coli, as described previously.31 The complete ubiquitination system was then reconstituted using commercially obtained E1 and ubiquitin. A typical assay comprised 0.1 μM E1, 2 μM Ubc13, 2 μM Uev1A, 2 μM TRAF6-N, and 2 μM viperin-ΔN50 in 20 mM Tris-HCl buffer pH 7.5, 150 mM NaCl, 2 mM DTT, 2 mM ATP, and 5 mM MgCl2. Reactions were initiated by the addition of ubiquitin, 35 μM, and incubated at 37 °C. At various times, aliquots were removed and quenched by addition of SDS-PAGE loading buffer; samples were then analyzed by SDS-PAGE.
Initially, we examined the activity of TRAF6-N in the absence of viperin, with a typical experiment shown in Figure 2. Under these conditions, the formation of diubiquitin was clearly visible after 4 min and triubiquitin was visible as a faint band after 8 min. At longer times, the formation of polyubiquitin is evident as a faint smear of higher molecular weight material. Control experiments established that in the background, rate of ubiquitin ligation is negligible in the absence of TRAF6-N (Figure S2).
Figure 2.
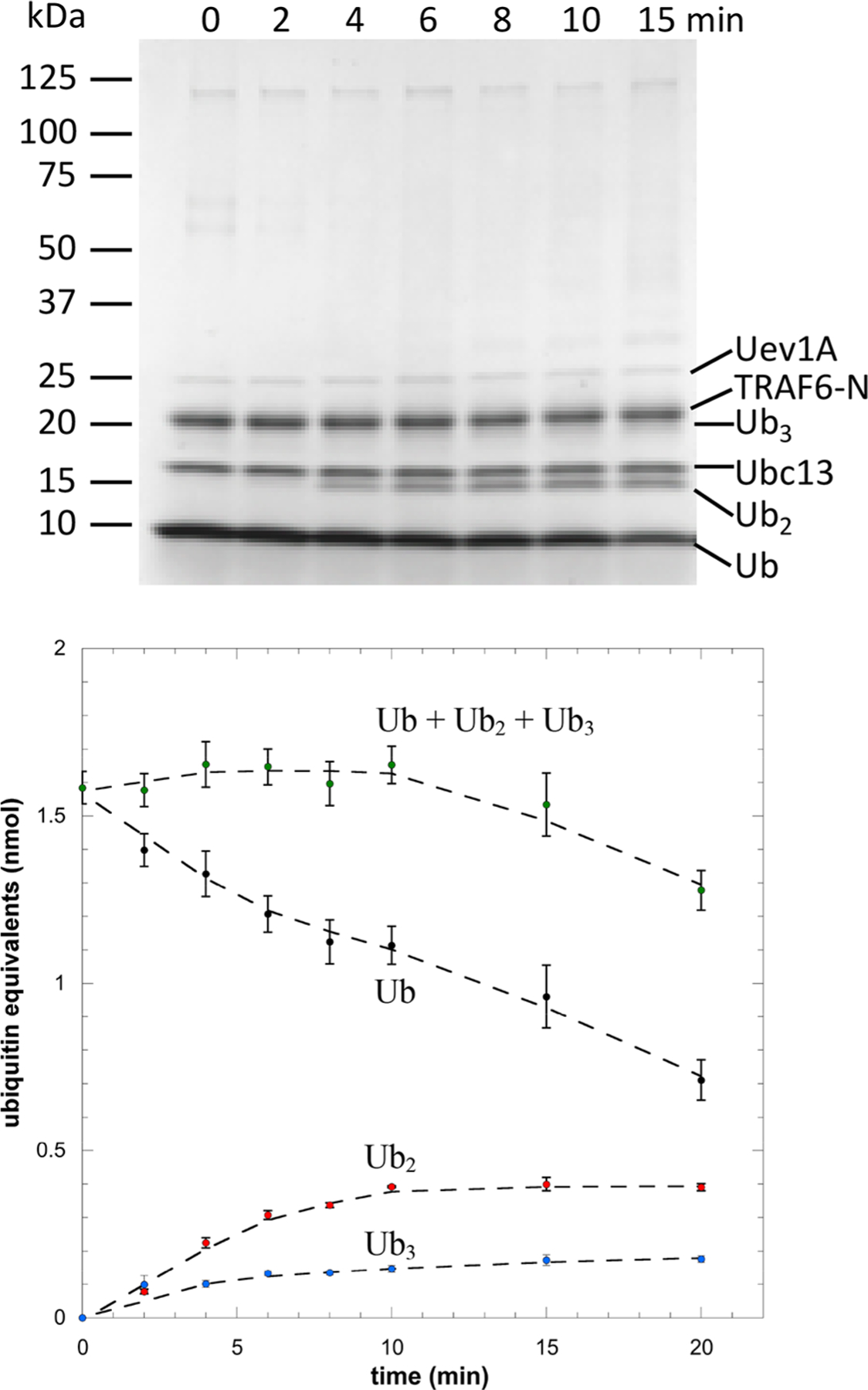
Kinetics of ubiquitin ligation catalyzed by TRAF6-N. Top: Representative Coomassie-stained gel showing consumption of ubiquitin and formation of ubiquitin oligomers. Bottom: Quantification of mono-, di- and triubiquitin; after 20 min, only a small fraction of the ubiquitin is converted to larger oligomers.
Quantification of the bands due to mono-, di-, and triubiquitin by imaging of Coomassie-stained gels (Figure 2) allowed the consumption of ubiquitin ligation to be quantified and the amount of ubiquitin incorporated into high molecular weight oligomers to be estimated. For the first 10 min of the reaction, the concentrations of di- and triubiquitin increased and then plateaued. In contrast, the concentration of monoubiquitin steadily decreased as more ubiquitin was incorporated into high molecular weight oligomers. After 20 min, ~80% of the ubiquitin was accounted for by mono-, di-, and triubiquitin, with only ~20% converted to high molecular weight oligomers.
We then repeated the reaction with the addition of viperin-ΔN50 in a 1:1 ratio with TRAF6-N (2 μM of each enzyme). The addition of viperin markedly altered the kinetics of ubiquitination (Figure 3). In this case, there was an initial rapid increase in the amount of diubiquitin formed, which then decayed to a steady state level. Notably, ubiquitin was converted to high molecular weight species much more rapidly when viperin was bound to TRAF6-N. High molecular weight ubiquitin oligomers accounted for ~70% of the ubiquitin pool while mono-, di- and triubiquitin comprised only ~30%.
Figure 3.
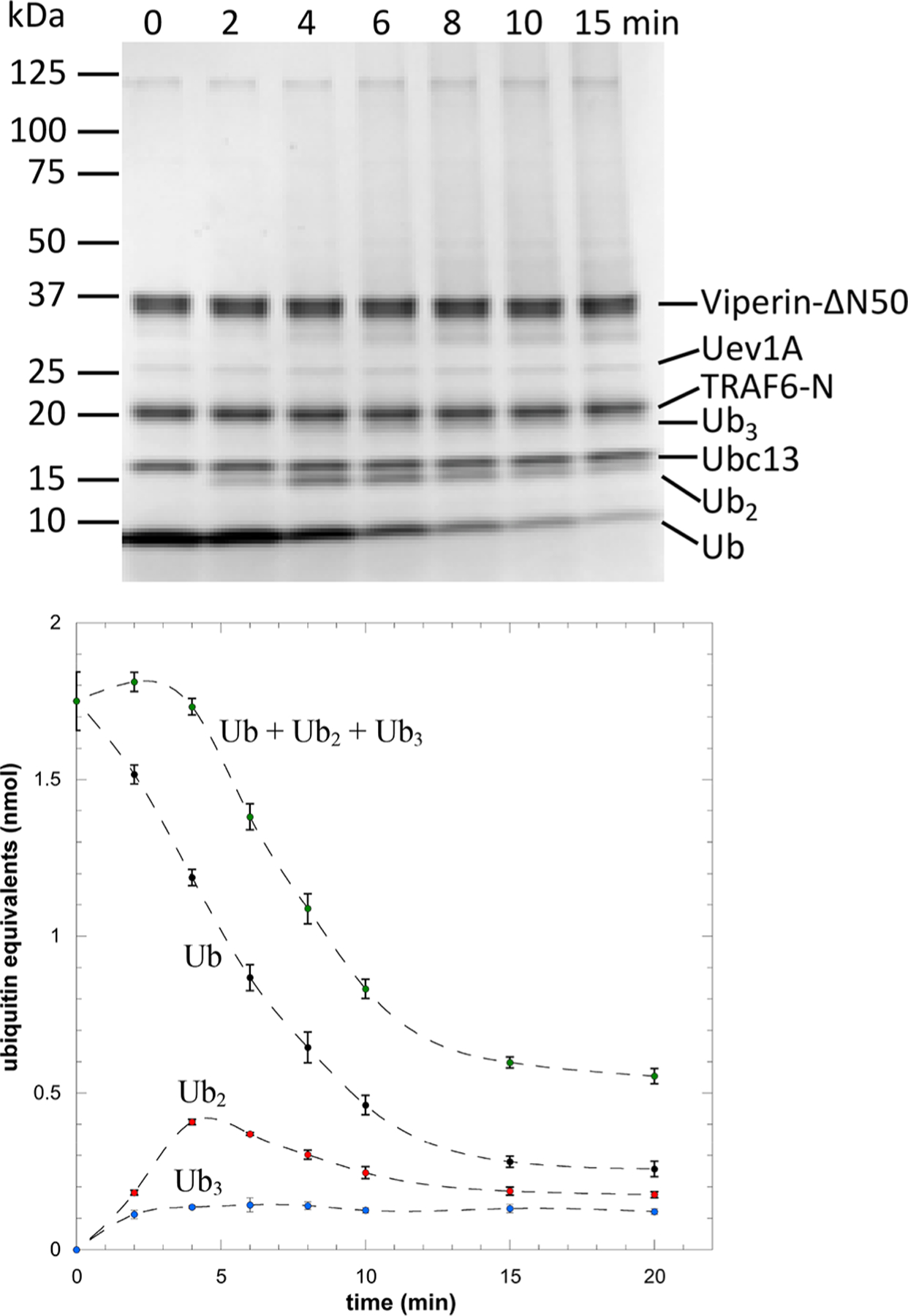
Activation of TRAF6-N by viperin. Top: Representative Coomassie-stained gel showing consumption of ubiquitin and formation of ubiquitin oligomers, note the smear of high Mr species at longer times. Bottom: Quantification of mono-, di-, and triubiquitin; these oligomers are rapidly depleted as they are converted to higher Mr species. (Experiments were performed with 1:1 molar ratio of TRAF6-N to viperin.)
Control experiments established that TRAF6-N activation is specific to viperin, as proteins such as bovine serum albumin had no effect on TRAF6-N activity (Figure S2). Furthermore, the Fe–S cluster of viperin appears to be important for TRAF6 activation, as a viperin-ΔN50–C83A mutant that is unable to bind the Fe–S cluster did not activate TRAF6-N (Figure S3). These results clearly demonstrate that viperin activates TRAF6-N and promotes the formation of longer polyubiquitin chains that are considered to be important mediators of signaling in the MAPK and NF-kB pathways.26,30
TRAF6 is known to autoubiquitinate Lys124,30 a process that is important for its role in signal transduction.30 To examine whether viperin promotes TRAF6 autoubiquitination, we probed gels with antibodies against ubiquitin and the N-terminal domain of TRAF6 (Figure 4). Immunoblotting with antiubiquitin antibody confirmed identity of the di- and triubiquitin bands and, as expected, strongly stained the high molecular weight material evident in Coomassie-stained gels. The high molecular weight material also cross-reacted with anti-TRAF6 antibodies demonstrating that it represents autoubiquitinated forms of TRAF6-N. When probed with antiviperin antibodies, only the viperin band was cross-reactive. This result demonstrates that although viperin promotes TRAF6 polyubiquitination, it is not itself a substrate for ubiquitination.
Figure 4.
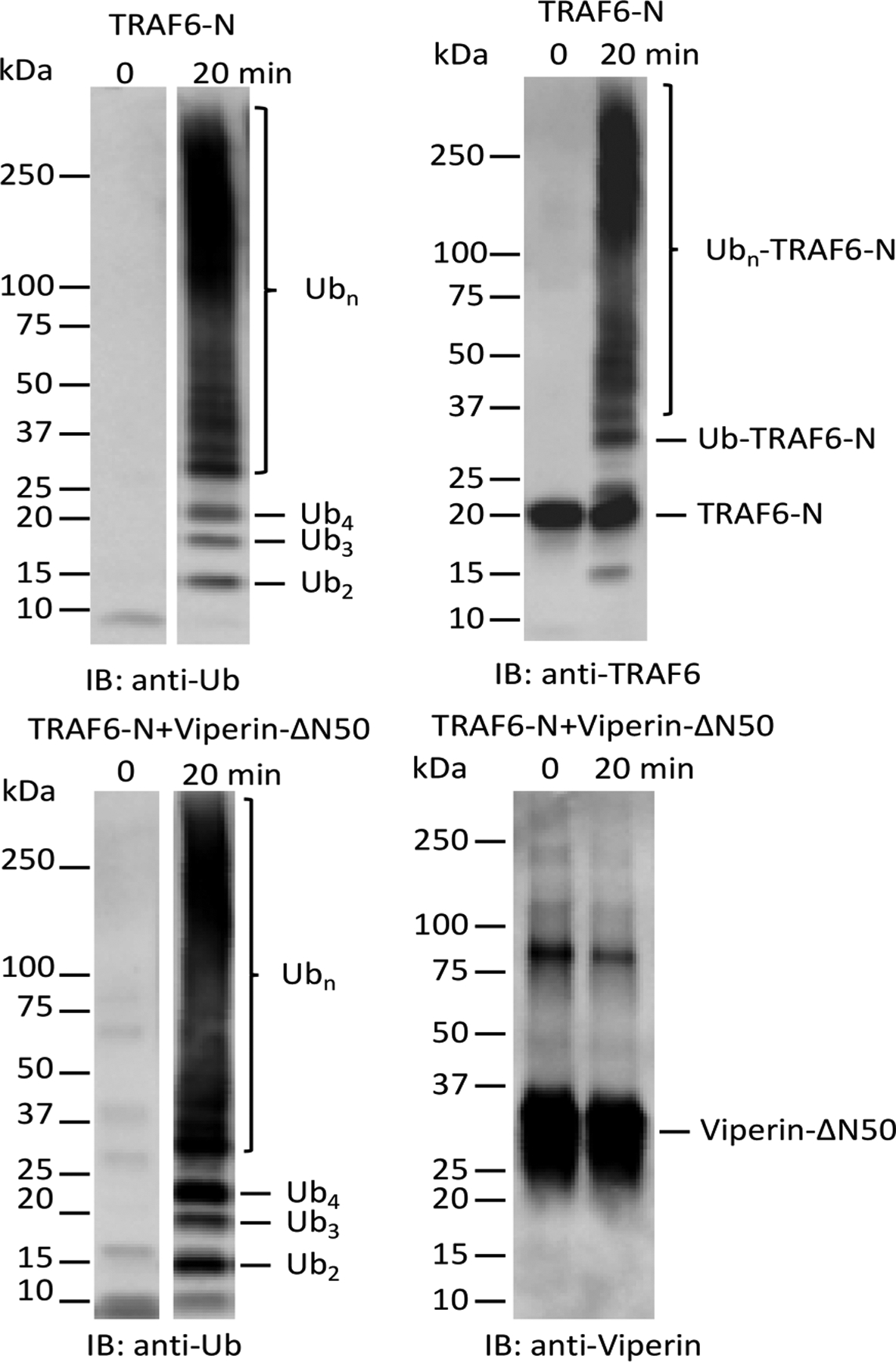
Immunoblot analysis of ubiquitination reactions. Top: Staining for ubiquitin (left) and TRAF6 (right) in reactions containing TRAF6-N. Bottom: Staining for ubiquitin (left) and viperin (right) in reactions containing TRAF6-N and viperin. (Note: the polyclonal antiubiquitin antibody used in staining recognizes monoubiquitin very poorly; both t = 0 and t = 20 min lanes contain similar amounts of ubiquitin.).
Although the time course for ubiquitin ligation is complex, at early time points, the major reaction catalyzed by TRAF6-N is the formation of diubiquitin
The ubiquitin-charged E2 functions as a substrate for TRAF6-N, which is then rapidly replenished through the action of E1 so that the steady state concentration of E2 ~ Ub remains constant. This simplification allowed us to quantify the ubiquitin ligase activity of TRAF6-N and compare its activity when complexed with viperin. Preliminary experiments established that the rate of ubiquitin consumption was linear with TRAF6-N concentration (Figure S4). Under these conditions, the apparent turnover number for diubiquitin formation by TRAF6-N was kapp = 0.47 ± 0.06 min−1, whereas in the presence of viperin, kapp = 1.25 ± 0.08 min−1 representing a ~ 2.5-fold rate enhancement (Figure S5).
TRAF6 is one of the better studied members of this class E3 ligases, in part due to the important role it plays in NF-kB and MAPK signaling.22,26 However, to our knowledge, quantitative measurements of rate at which TRAF6 catalyzes ubiquitin transfer have not been previously been reported. The kinetics of ubiquitin transfer catalyzed by various other E3 ligases have been quite extensively investigated, with kcat ranging from several per second, e.g., the RING-E3 ligase SCFCdc432 and HECT-E3 ligase E6AP,33 to several per minute, e.g., the RING-E3 ligase San1.34 Compared with those of these E3 ligases, the rate of ubiquitin ligation catalyzed by TRAF6-N is relatively slow, but we note that ubiquitination rates are also dependent on the protein substrate and may accelerate as the polyubiquitin chain is extended.32 Furthermore, TRAF6-N lacks the C-terminal TRAF domain through which TRAF6 binds many of its protein substrates and which may also influence the ligase activity of the enzyme.
A role for viperin in immune signaling was initially suggested through studies on TRAF6-catalyzed polyubiquitination of IRAK1 in mouse cell-lines lacking viperin.28 More recently, our studies in HEK 293T cells demonstrated that cotransfection of viperin with TRAF6 significantly increased the polyubiquitination of IRAK1.15 However, these studies left open the possibility that viperin activated TRAF6 indirectly through additional unknown factor(s). Reconstituting the ubiquitination system in vitro with purified enzymes has allowed us to unambiguously demonstrate viperin’s role in activating TRAF6. Viperin both speeds up the rate of ubiquitin consumption and increases the formation of high molecular weight autoubiquitinated forms of TRAF6 that mediate downstream signaling. Although the ~2.5-fold activation of TRAF6 by viperin is relatively modest, this level of amplification may be appropriate to modulating transcription of the various genes needed to establish the antiviral response.
Here, we have shown that viperin interacts with the N-terminal RING-domain of TRAF6, whereas it is known that the C-terminal TRAF domain (lacking in TRAF6-N) mediates TRAF6’s interactions with most other protein substrates.24 This modular arrangement suggests that activation of TRAF6 E3-ligase activity by viperin may enhance polyubiquitination of other target proteins, which would be consistent with our observation that viperin stimulates TRAF6-catalyzed polyubiquitination of IRAK1.15 These observations provide further support the idea, for which there is extensive but indirect evidence in the literature, that viperin, more broadly, activates other K48-linked E3 ligases to increase proteasomal degradation of specific proteins in response to viral infection.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH grant GM 093088 to E.N.G.M. We thank Prof. Hao Wu (Harvard University) and Prof. Catherine Day (University of Otago) for their kind gifts of expression vectors for TRAF6-N and Ubc13/Uev1A, respectively.
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.1c01045.
Experimental methods and supporting figures (PDF)
Complete contact information is available at: https://pubs.acs.org/10.1021/jacs.1c01045
Notes
The authors declare no competing financial interest.
Contributor Information
Ayesha M. Patel, Department of Chemistry, University of Michigan, Ann Arbor, Michigan 48109, United States
E. Neil G. Marsh, Department of Chemistry, University of Michigan, Ann Arbor, Michigan 48109, United States
REFERENCES
- (1).Chin KC; Cresswell P Viperin (cig5), an IFN-inducible antiviral protein directly induced by human cytomegalovirus. Proc. Natl. Acad. Sci. U. S. A 2001, 98, 15125–15130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Crosse KM; Monson EA; Beard MR; Helbig KJ Interferon-Stimulated Genes as Enhancers of Antiviral Innate Immune Signaling. J. Innate Immun 2018, 10, 85–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Helbig KJ; Beard MR The role of viperin in the innate antiviral response. J. Mol. Biol 2014, 426, 1210–1219. [DOI] [PubMed] [Google Scholar]
- (4).Lindqvist R; Överby AK The Role of Viperin in Antiflavivirus Responses. DNA Cell Biol. 2018, 37, 725–730. [DOI] [PubMed] [Google Scholar]
- (5).Fenwick MK; Li Y; Cresswell P; Modis Y; Ealick SE Structural studies of viperin, an antiviral radical SAM enzyme. Proc. Natl. Acad. Sci. U S A 2017, 114, 6806–6811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Chakravarti A; Selvadurai K; Shahoei R; Lee H; Fatma S; Tajkhorshid E; Huang RH Reconstitution and substrate specificity for isopentenyl pyrophosphate of the antiviral radical SAM enzyme viperin. J. Biol. Chem 2018, 293, 14122–14133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Fenwick MK; Su D; Dong M; Lin H; Ealick SE Structural Basis of the Substrate Selectivity of Viperin. Biochemistry 2020, 59, 652–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Landgraf BJ; McCarthy EL; Booker SJ Radical S-Adenosylmethionine Enzymes in Human Health and Disease. Annu. Rev. Biochem 2016, 85, 485–514. [DOI] [PubMed] [Google Scholar]
- (9).Gizzi AS; Grove TL; Arnold JJ; Jose J; Jangra RK; Garforth SJ; Du Q; Cahill SM; Dulyaninova NG; Love JD; Chandran K; Bresnick AR; Cameron CE; Almo SC A naturally occurring antiviral ribonucleotide encoded by the human genome. Nature 2018, 558, 610–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Ebrahimi KH; Rowbotham JS; McCullagh J; James WS Mechanism of Diol Dehydration by a Promiscuous Radical-SAM Enzyme Homologue of the Antiviral Enzyme Viperin (RSAD2). ChemBioChem. 2020, 21, 1–9. [DOI] [PubMed] [Google Scholar]
- (11).Ghosh S; Marsh ENG Viperin: an ancient radical SAM enzyme finds its place in modern cellular metabolism and innate immunity. J. Biol. Chem 2020, 295, 11513–11528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Ghosh S; Patel AM; Grunkemeyer TJ; Dumbrepatil AB; Zegalia K; Kennedy RT; Marsh ENG Interactions between Viperin, Vesicle-Associated Membrane Protein A, and Hepatitis C Virus Protein NS5A Modulate Viperin Activity and NS5A Degradation. Biochemistry 2020, 59, 780–789. [DOI] [PubMed] [Google Scholar]
- (13).Dumbrepatil AB; Zegalia KA; Sajja K; Kennedy RT; Marsh ENG Targeting viperin to the mitochondrion inhibits the thiolase activity of the trifunctional enzyme complex. J. Biol. Chem 2020, 295, 2839–2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Crosse KM; Monson EA; Dumbrepatil AB; Smith M; Tseng Y-Y; Van der Hoek KH; Revill PA; Tscharke DC; Marsh ENG; Beard MR; Helbig KJ, Viperin binds STING and enhances the type-I interferon response following dsDNA detection. Immunol. Cell Biol 2020, DOI: 10.1111/imcb.12420, 1–19. [DOI] [PubMed] [Google Scholar]
- (15).Dumbrepatil AB; Ghosh S; Zegalia KA; Malec PA; Hoff JD; Kennedy RT; Marsh ENG Viperin interacts with the kinase IRAK1 and the E3 ubiquitin ligase TRAF6, coupling innate immune signaling to antiviral ribonucleotide synthesis. J. Biol. Chem 2019, 294, 6888–6898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Makins C; Ghosh S; Roman-Melendez GD; Malec PA; Kennedy RT; Marsh ENG Does Viperin Function as a Radical S-Adenosyl-l-methionine-dependent Enzyme in Regulating Farnesyl-pyrophosphate Synthase Expression and Activity? J. Biol. Chem 2016, 291, 26806–26815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Panayiotou C; Lindqvist R; Kurhade C; Vonderstein K; Pasto J; Edlund K; Upadhyay AS; Overby AK Viperin Restricts Zika Virus and Tick-Borne Encephalitis Virus Replication by Targeting NS3 for Proteasomal Degradation. J. Virology 2018, 92, e02054–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Fu TM; Shen C; Li QB; Zhang PF; Wu H Mechanism of ubiquitin transfer promoted by TRAF6. Proc. Natl. Acad. Sci. U. S. A 2018, 115, 1783–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Yin Q; Lin SC; Lamothe B; Lu M; Lo YC; Hura G; Zheng LX; Rich RL; Campos AD; Myszka DG; Lenardo MJ; Darnay BG; Wu H E2 interaction and dimerization in the crystal structure of TRAF6. Nat. Struct. Mol. Biol 2009, 16, 658–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Pickart CM; Rose IA Functional-Heterogeneity of Ubiquitin Carrier Proteins. J. Biol. Chem 1985, 260, 1573–1581. [PubMed] [Google Scholar]
- (21).Komander D The emerging complexity of protein ubiquitination. Biochem. Soc. Trans 2009, 37, 937–953. [DOI] [PubMed] [Google Scholar]
- (22).Park Y; Jin HS; Aki D; Lee J; Liu YC The Ubiquitin System in Immune Regulation. Adv. Immunol 2014, 124, 17–66. [DOI] [PubMed] [Google Scholar]
- (23).Bhoj VG; Chen ZJ Ubiquitylation in innate and adaptive immunity. Nature 2009, 458, 430–437. [DOI] [PubMed] [Google Scholar]
- (24).Hu H; Sun SC Ubiquitin signaling in immune responses. Cell Res. 2016, 26, 457–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Metzger MB; Pruneda JN; Klevit RE; Weissman AM RING-type E3 ligases: Master manipulators of E2 ubiquitinconjugating enzymes and ubiquitination. Biochim. Biophys. Acta, Mol. Cell Res 2014, 1843, 47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Oeckinghaus A; Hayden MS; Ghosh S Crosstalk in NF-kappa B signaling pathways. Nat. Immunol 2011, 12, 695–708. [DOI] [PubMed] [Google Scholar]
- (27).Ordureau A; Smith H; Windheim M; Peggie M; Carrick E; Morrice N; Cohen P The IRAK-catalysed activation of the E3 ligase function of Pellino isoforms induces the Lys63-linked polyubiquitination of IRAK1. Biochem. J 2008, 409, 43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Saitoh T; Satoh T; Yamamoto N; Uematsu S; Takeuchi O; Kawai T; Akira S Antiviral protein Viperin promotes Toll-like receptor 7- and Toll-like receptor 9-mediated type I interferon production in plasmacytoid dendritic cells. Immunity 2011, 34, 352–63. [DOI] [PubMed] [Google Scholar]
- (29).Lamothe B; Besse A; Campos AD; Webster WK; Wu H; Darnay BG Site-specific Lys-63-linked tumor necrosis factor receptor-associated factor 6 auto-ubiquitination is a critical determinant of I kappa B kinase activation. J. Biol. Chem 2007, 282, 4102–4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Shi JH; Sun SC, Tumor Necrosis Factor Receptor-Associated Factor Regulation of Nuclear Factor kappa B and Mitogen-Activated Protein Kinase Pathways. Front. Immunol 2018, 9. article 1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Middleton AJ; Budhidarmo R; Das A; Zhu JY; Foglizzo M; Mace PD; Day CL, The activity of TRAF RING homo- and heterodimers is regulated by zinc finger 1. Nat. Commun 2017, 8, article 1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Pierce NW; Kleiger G; Shan SO; Deshaies RJ Detection of sequential polyubiquitylation on a millisecond timescale. Nature 2009, 462, 615–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Purbeck C; Eletr ZM; Kuhlman B Kinetics of the Transfer of Ubiquitin from UbcH7 to E6AP. Biochemistry 2010, 49, 1361–1363. [DOI] [PubMed] [Google Scholar]
- (34).Ibarra R; Sandoval D; Fredrickson EK; Gardner RG; Kleiger G The San1 Ubiquitin Ligase Functions Preferentially with Ubiquitin-conjugating Enzyme Ubc1 during Protein Quality Control. J. Biol. Chem 2016, 291, 18778–18790. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


