Abstract
The current version of PLMItRNA has been realized to constitute a database for tRNA molecules and genes identified in the mitochondria of all green plants (Viridiplantae). It is the enlargement of a previous database originally restricted to seed plants [Ceci,L.R., Volpicella,M., Liuni,S., Volpetti,V., Licciulli,F. and Gallerani,R. (1999) Nucleic Acids Res., 27, 156–157]. PLMItRNA reports information and multialignments on 254 genes and 16 tRNA molecules detected in 25 higher plants (one bryophyta and 24 vascular plants) and seven green algae. PLMItRNA is accessible via the WWW at http://bio-WWW.ba.cnr.it:8000/srs6/
INTRODUCTION
In the past years we have developed a database for higher plant mitochondria tRNA molecules and genes aimed at facilitating the retrieval of information and sequences. The need for a database comes from the complex and non-homogeneous organization of tRNA genes and molecules in plant mitochondria (1). The database was originally restricted to seed plants (Angiosperms and Gymnosperms) whose mitochondria show the most complex organization. In fact, they contain active tRNA molecules coded by either mitochondrial genes, chloroplast genes heterogeneously transferred to the different mitochondrial genomes or nuclear genes (2).
The complete sequences of several higher plant and algae mitochondrial genomes [Arabidopsis thaliana (3), Chlamidomonas eugametos (4), Chlamidomonas reihnartdii (EMBL accession number U03843), Pedinomonas minor (EMBL accession number AF116775), Chlorogonium elongatum (5), Chondrus crispus (6), Prototheca wickerhamii (7), Marchantia polymorpha (8) and Cyanidioschyzon merolae (9)] reveal that, with the possible exception of P.wickerhamii, all the mitochondria surely need additional tRNAs to obtain a complete set of tRNAs. Moreover, specific studies on the import into M.polymorpha mitochondria of tRNA species from the cytoplasm, confirmed the import of the ‘necessary’ tRNA-Ile(AAU) (10) and tRNA-Thr(AGU) (11) species, but also revealed the unexpected import of two different tRNA-Val(AAC) molecules (12). A tRNA-Val(UAC) gene had been already identified in the M.polymorpha mitochondrial genome that would be sufficient to read all four codons GUN (8). These findings point out the impossibility of defining the complete set of tRNAs active in plant mitochondria until an exhaustive analysis on the import of tRNAs has been carried out.
In order to have as wide as possible an overview on the organization of tRNA molecules and genes in plant mitochondria, we thought it useful to include in the PLMItRNA database both information on tRNA genes and molecules in M.polymorpha mitochondria (encompassing, therefore, all the higher plants) and the numerous data coming from studies on green algae mitochondrial genomes. In this way the PLMItRNA database now includes all the information available for green plants (Viridiplantae).
Updating
The current version of the PLMItRNA database has been developed from a previous database for seed plant tRNA molecules and genes (1) by also including data for the bryophyta M.polymorpha and the green algae C.eugametos, C.reihnartdii, P.minor, Platimonas subcordiformis, C.elongatum, P.wickerhamii and Scenedesmus obliquus. It was updated in August 1999 by analyzing the whole EMBL database. All the sequences for tRNA genes or molecules detected in plant mitochondria were initially obtained through the SRS (Sequence Retrieval System) service at the European Bioinformatics Institute (EBI) WWW server: http://srs.ebi.ac.uk/ . A set of master sequences was then realised by choosing a single gene sequence for each tRNA species belonging to Viridiplantae. The master sequences were successively used to analyze the EMBL database (release 59) with the FastA program (13). The FastA analysis was performed directly at the http://www2.ebi.ac.uk/fasta3/ site of the EBI. Updating of PLMItRNA for the tRNA sequences has also been based on the analysis of the literature since many of their sequences have not been deposited in the nucleic acid databases. Literature references are reported in the relative entries.
DESCRIPTION OF THE DATABASE
PLMItRNA contains 270 entries for 254 genes and 16 tRNA sequences. The informative section of each entry is organized in several fields, which schematically report characteristics of the sequence. A query form (http://bio-www.ba.cnr.it:8000/srs6/ ) allows the retrieval of a list of specific entries by simply choosing among the different options available for each field. Combinations through Boolean operators are also possible. Fields and options are:
• Field ‘Molecule’ (options ‘DNA’ or ‘RNA’) refers to the primary source of the sequence. tRNA sequences, and cDNA sequences for which the corresponding gene was not sequenced have been cataloged as specific entries under the ‘RNA’ option. cDNA sequences of genes already sequenced have not been reported as single entries but are only mentioned in the ‘Note’ of the gene entry.
• Field ‘Gene structure’ (options ‘complete gene’ or ‘pseudo/truncated gene’) allows the user to distinguish between complete, potentially functional tRNA genes, and pseudo or truncated genes.
• Field ‘Genetic origin’ (options ‘Mitochondrial’, ‘Chloroplast’ or ‘Nuclear’) refers to the primary genetic origin of the sequence.
• Field ‘Plant’ has as an option every plant name (Latin or English) as listed in Table 1.
Table 1. Plants present in the PLMItRNA database.
| Latin name | English name | PLMItRNA code | |
|---|---|---|---|
| Bryophyta | |||
| Marchantiopsida | Marchantia polymorpha | Liverwort | M.p. |
| Magnoliophyta | |||
| Liliopsida | Arabidopsis thaliana | thale cress | A.t. |
| Beta vulgaris | sugar beet | B.v. | |
| Brassica napus | rapeseed | B.n. | |
| Brassica oleracea | cauliflower | B.o. | |
| Glycine max | soybean | G.m. | |
| Helianthus annuus | sunflower | H.a. | |
| Lupinus luteus | lupine | L.l. | |
| Lycopersicon esculentum | tomato | L.e. | |
| Oenothera berteriana | primrose | O.b. | |
| Petunia hybrida | petunia | P.h. | |
| Phaseolus vulgaris | bean | P.v. | |
| Pisum sativum | pea | P.s. | |
| Raphanus sativus | horseradish | R.s. | |
| Solanum tuberosum | potato | S.t. | |
| Vigna radiata | mung bean | V.r. | |
| Magnoliopsida | Lolium multiflorum | Italian ryegrass | L.m. |
| Lophopyrum elongatum | |||
| (Elytrigia elongata) | tall wheatgrass | Lo.e. | |
| Oryza sativa | rice | O.s. | |
| Secale cereale | rye | S.c. | |
| Sorghum bicolor | sorghum | S.b. | |
| Sorghum vulgare | sorghum | S.v. | |
| Triticum aestivum | wheat | T.a. | |
| Zea mays | maize | Z.m. | |
| Conipherophyta | |||
| Coniferopsida | Larix leptoeuropaea | larch | La.l. |
| Chlorophyta | |||
| Chlorophyceae | Chlamydomonas eugametos | C.e. | |
| Chlamydomonas reinhardtii | C.r. | ||
| Chlorogonium elongatum | Ch.e. | ||
| Prototheca wickerhamii | P.w. | ||
| Scenedesmus obliquus | S.o. | ||
| Pedinophyceae | Pedinomonas minor | P.m. | |
| Prasinophyceae | Platymonas subcordiformis | Pl.s. |
• Field ‘Plant group’ has as an option every plant group as listed in Table 2.
Table 2. Plant taxonomic groups present in the PLMItRNA database.
| 1. | Viridiplantae (green plants) |
| 1.1. | Streptophyta |
| 1.1.1. | Embryophyta (higher plants) |
| 1.1.1.1. | Bryophyta |
| 1.1.1.1.1. | Marchantiopsida |
| 1.1.1.2. | Trachephyta (vascular plants) |
| 1.1.1.2.1. | Spermatophyta (seed plants) |
| 1.1.1.2.1.1. | Conipherophyta |
| 1.1.1.2.1.1.1. | Coniferopsida |
| 1.1.1.2.1.2. | Magnoliophyta = Angiospermae (angiosperms, flowering plants) |
| 1.1.1.2.1.2.1. | Liliopsida = Monocotyledonae (monocots, monocotyledons) |
| 1.1.1.2.1.2.2. | Magnoliopsida = Dicotyledonae (dicots, dicotyledons) |
| 1.2. | Chlorophyta (green algae) |
| 1.2.1. | Chlorophyceae |
| 1.2.2. | Pedinophyceae |
| 1.2.3. | Prasinophyceae |
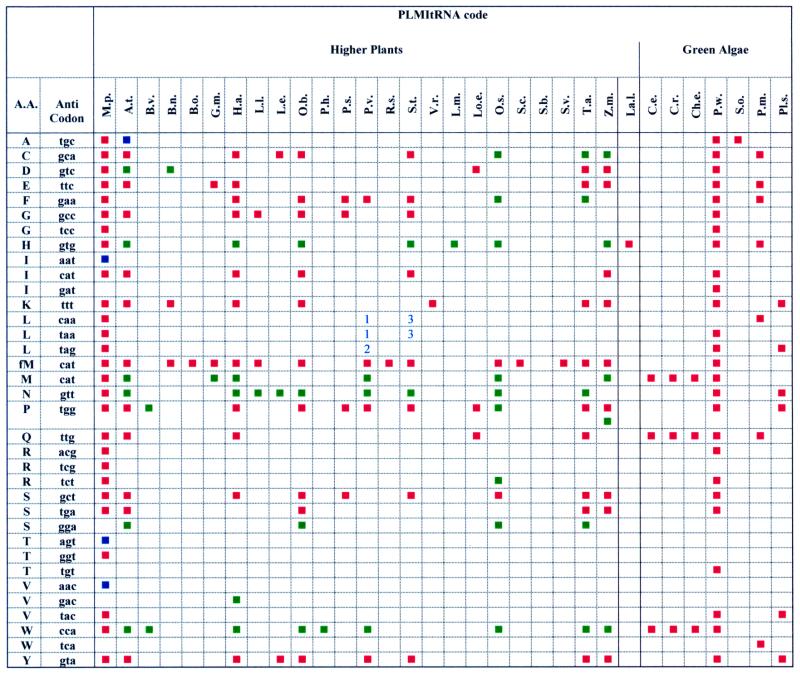
• Fields ‘Aminoacid’ and ‘Anticodon’ allow to choose every aminoacid or anticodon deduced from Table 3.
Table 3. Genes coding for mitochondrial tRNAs in Viridiplantae (only complete genes have been reported).
Red square, mitochondrial gene.
Blue square, nuclear gene.
Green square, chloroplast gene.
1, Two imported tRNA-Leu(NAA) have been found in P.vulgaris (14,15).
2, An imported tRNA-Leu(NAG) has been found in P.vulgaris (16).
3, An imported tRNA-Leu(NAA) has been found in S.tuberosum (17).
• Field ‘Transcription’ has as options the symbol ‘+’ (the gene is transcribed), ‘–’ (the gene is not transcribed) and ‘NS’ (transcription not studied).
• Field ‘Link’ allows retrieval of specific entries through their EMBL database accession number. A direct link to the EMBL database through the reported accession numbers is also possible.
• Field ‘Notes’ reports explicative notes, written including standard sentences to allow retrieval of entries by specific keywords. Topic keywords are ‘not described’ (genes not described in the feature field of the primary database accession number), ‘not available’ (sequences detected in the literature, but not present in nucleic acid libraries), ‘cDNA’ and ‘RNA editing’. It is also possible to select entries by indicating specific keywords in the ‘All text’ field of the query form.
Each entry also has a multialignment section where all the homologous sequences available for green plant mitochondria have been aligned. Sequences have been reported in the same order as the relative plant appears in the systematic classification adopted in the ACNUC retrieval program (18) (also see Table 1).
In the multialignments, sequences have been subdivided into the classical tRNA domains. Nucleotide positions have been numbered from 0 to 76. Positions of nucleotides which are not always present (0, 17, 17a, 20a, 20b, nucleotides in the extra-arm, 47 and 74–76) are indicated by dashes. Each sequence is also preceded by some informative characters: the ID number, the initials of the Latin name of the plant species and the letter D or R for DNA or RNA sequences respectively.
When possible, tRNA sequences have been aligned with the corresponding gene sequences. Symbols of modified nucleotides in the tRNA sequences are according to Sprinzl et al. (19) with the exception of the Å character that stands for either i6A or ms2 i6A modified nucleotides, and for the £ character which represents a lysidine-like hypermodified nucleotide.
DATABASE AVAILABILITY AND CITATION
Access to the PLMItRNA database is possible by WWW at the address http://bio-www.ba.cnr.it:8000/srs6/ . Entries from the database can be retrieved by either a standard ‘All text’ based query form or by an extended query form where the different fields and options can be specified.
The collection of sequences present in PLMItRNA is also available for database searching at the address http://bigarea.area.ba.cnr.it:8000/BioWWW/fasta.htm . The FastA output file is designed in order to allow links with the PLMItRNA entries.
Users of the database should cite the present publication as a reference. Comments, corrections and new entries are welcome.
REFERENCES
- 1.Ceci L.R., Volpicella,M., Liuni,S., Volpetti,V., Licciulli,F. and Gallerani R. (1999) Nucleic Acids Res., 27, 156–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dietrich A., Small,I., Cosset,A., Weil,J.H. and Maréchal-Drouard,L. (1996) Biochimie, 78, 518–529. [DOI] [PubMed] [Google Scholar]
- 3.Unseld M., Marienfeld,J.R., Brandt,P. and Brennicke,A. (1997) Nature Genet., 15, 57–61. [DOI] [PubMed] [Google Scholar]
- 4.Denovan-Wright E.M., Nedelcu,A.M. and Lee,R.W. (1998) Plant Mol. Biol., 36, 285–295. [DOI] [PubMed] [Google Scholar]
- 5.Kroymann J. and Zetsche,K. (1998) J. Mol. Evol., 47, 431–440. [DOI] [PubMed] [Google Scholar]
- 6.Leblanc C., Boyen,C., Richard,O., Bonnard,G., Grienenberger,J.M. and Kloareg,B. (1995) J. Mol. Biol., 250, 484–495. [DOI] [PubMed] [Google Scholar]
- 7.Wolff G., Plante,I., Lang,B.F., Kuck,U. and Burger,G. (1994) J. Mol. Biol., 237, 75–86. [DOI] [PubMed] [Google Scholar]
- 8.Oda K., Yamato,K., Ohta,E., Nakamura,Y., Takemura,M., Nozato,N., Akashi,K., Kanegae,T., Ogura,Y., Kohochi,T. and Ohyama,K. (1992) J. Mol. Biol., 223, 1–7. [DOI] [PubMed] [Google Scholar]
- 9.Ohta E., Sato,N. and Kuroiwa,T. (1998) Nucleic Acids Res., 26, 5190–5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akashi K., Sakurai,K., Hirayama,J., Fukuzawa,H. and Ohyama,K. (1996) Curr. Genet., 30, 181–185. [DOI] [PubMed] [Google Scholar]
- 11.Akashi K., Hirayama,J., Takenaka,M., Yamaoka,S., Suyama,Y., Fukuzawa,H. and Ohyama,K. (1997) Biochim. Biophys. Acta, 1350, 262–266. [DOI] [PubMed] [Google Scholar]
- 12.Akashi K., Takenaka,M., Yamaoka,S., Suyama,Y., Fukuzawa,H. and Ohyama,K. (1998) Nucleic Acids Res., 26, 2168–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pearson W.R. and Lipman,D.J. (1988) Proc. Natl Acad. Sci. USA, 85, 2444–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Green G.A., Marechal,L., Weil,J.H. and Guillemaut,P. (1987) Plant Mol. Biol., 10, 13–19. [DOI] [PubMed] [Google Scholar]
- 15.Marechal-Drouard L. and Guillemaut,P. (1988) Nucleic Acids Res., 16, 11812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marechal-Drouard L., Weil,J.H. and Guillemaut,P. (1988) Nucleic Acids Res., 16, 4777–4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marechal-Drouard L., Neuburger,M., Guillemaut,P., Douce,R., Weil,J.H. and Dietrich,A. (1990) FEBS Lett., 262, 170–172. [DOI] [PubMed] [Google Scholar]
- 18.Gouy M., Gautier,C., Attimonelli,M., Lanave,C. and Di Paola,G. (1985) Comput. Appl. Biosci., 1, 167–172. [DOI] [PubMed] [Google Scholar]
- 19.Sprinzl M., Horn,C., Brown,M., Ioudovitch,A. and Steinberg,S. (1998) Nucleic Acids Res., 26, 148–153. [DOI] [PMC free article] [PubMed] [Google Scholar]