Abstract
As the number of complete microbial genomes publicly available is still growing, the problem of annotation quality in these very large sequences remains unsolved. Indeed, the number of annotations associated with complete genomes is usually lower than those of the shorter entries encountered in the repository collections. Moreover, classical sequence database management systems have difficulties in handling entries of such size. In this context, the Enhanced Microbial Genomes Library (EMGLib) was developed to try to alleviate these problems. This library contains all the complete genomes from prokaryotes (bacteria and archaea) already sequenced and the yeast genome in GenBank format. The annotations are improved by the introduction of data on codon usage, gene orientation on the chromosome and gene families. It is possible to access EMGLib through two database systems set up on WWW servers: the PBIL server at http://pbil.univ-lyon1.fr/emglib/emglib.html and the MICADO server at http://locus.jouy.inra.fr/micado
INTRODUCTION
Since the obtention of the complete genome of Haemophilus influenzae Rd in 1995 (1), the number of complete microbial genomes sequenced has grown rapidly. From this date, sequence database management systems and analysis programs have had to deal with complete genomes and not only with genomic fragments. The main problem of these genomes is their relative lack of annotations, compared with the shorter entries found in the repository databases (2–4). This is due, of course, to the difference in scale and to the automation of sequence obtention, but also to an important deficit in biological information. Even if some efforts have been made in parallel to develop computer systems able to automate some parts of the annotation procedure (5), the level of information available for a single gene in a complete genome is usually lower than in shorter entries. Moreover, some database management systems, which were developed in the 1980s, are not able to handle sequences with a length >300 kb. Due to this fact, complete genomes are always split into smaller, overlapping entries of ~100 kb in the general databases.
In this context, we decided to develop the Enhanced Microbial Genome Library (EMGLib). This library contains all the prokaryotic genomes completely sequenced and the yeast genome with some improvements in their annotations. It is possible to access EMGLib through two databases set up on WWW servers that allow efficient queries on complete genome sequences. These two systems provide easy access to the sequences and their annotations but also to complementary data such as genetic maps.
DATABASE CONTENT
The sequences, except the one corresponding to the Bacillus subtilis genome, were taken from the genome division of GenBank (ftp://ncbi.nlm.nih.gov/genbank/genomes ). In the case of B.subtilis, we used the sequence from the NRSub database (6). Release 2.0 (September 1999) of EMGLib contains 39 entries from 22 complete bacterial and archaeal genomes, the complete genome of Saccharomyces cerevisiae, and chromosome II of Plasmodium falciparum. These 39 entries total 56 006 904 bp and they contain 48 092 protein genes, 1200 tRNA, 153 rRNA, 25 snRNA and 23 other miscellaneous RNA.
We performed some additions and corrections on the original GenBank genome entries. First, new identifiers are given for each genome (LOCUS field). Their names are based on the format xxxxxCG (for prokaryotes) or xxCHRnnnn (for eukaryotes). In the case of bacteria, xxxxx stands for an abbreviation of the systematic name of the organism (e.g., BACSUCG is the name of the B.subtilis genome entry). In the case of yeast and P.falciparum, nnnn stands for the chromosome number in roman numerals (e.g., SCCHRIX for chromosome IX). We also changed the GenBank accession numbers to our own numbers, which are based on the format CGnnnn.
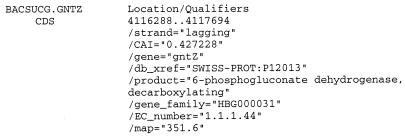
Features for CDS (coding DNA sequence) are completed with various information (Fig. 1). If the location of the replication origin and terminus are known or could be predicted (7), we add the orientation of the CDS on the chromosome (leading or lagging) under a ‘/strand’ qualifier. Then we add a Codon Adaptation Index (CAI) (8) value under a ‘/CAI’ qualifier and a cross-reference pointing to the corresponding entry in SWISS-PROT/TrEMBL (9) under a ‘/db_xref’ qualifier. We often rewrite or complete the ‘/product’ qualifier using data from SWISS-PROT. In the case of ORFs, we use ‘hypothetical protein’ for the product associated with these putative genes. When the encoded protein is an enzyme, we add its EC number, taken from the ENZYME database (10), in an ‘/EC_number’ qualifier. At last, when the gene is known to belong to a family defined in the HOBACGEN database (http://pbil.univ-lyon1.fr/databases/hobacgen.html ), we add the number of this family in a ‘/gene_family’ qualifier.
Figure 1.
Structure of the feature table for a protein gene from B.subtilis in EMGLib. Additional, non-standard qualifiers have been defined in a way to introduce specific information: ‘/strand’, for the strand location of the CDS (leading or lagging), ‘/CAI’, for the Codon Adaptation Index value, and ‘/gene_family’ for the accession number of the corresponding gene family in HOBACGEN, if any.
In the case of Escherichia coli, we added all the transcriptome data collected by Thieffry et al. (11). These data include operon structures, promoters and transcription start locations, –10 and –35 box scores, as well as the sequences of the promoters themselves.
DATABASE ACCESS
The easiest way to access EMGLib is through two databases installed on WWW servers that provide graphical interfaces and cover complementary aspects. The first one is hosted at the Bioinformatic Pole of Lyon (PBIL) and its URL is http://pbil.univ-lyon1.fr/emglib/emglib.html . Its main purpose is to provide a powerful retrieval system for gene sequences in a way to compose complex queries. Indeed, on this server EMGLib is indexed with the ACNUC sequence database management system (12). This system allows us to index all collections in EMBL, GenBank/DDBJ, SWISS-PROT/TrEMBL or NBRF/PIR (13) formats. Under ACNUC, each CDS or structural RNA gene can be seen as an independent sequence, and keywords corresponding to the contents of the different qualifiers are attached to these subsequences.
The home page of the server gives access to entry points allowing one to make simple or complex queries. Simple queries are made by keyword, sequence name and accession number. More sophisticated access is possible through WWW-Query, a general interface for the ACNUC databases installed on this server (14). WWW-Query permits the selection of sequences using different criteria like entry name, accession numbers, keywords, bibliographic references, dates of insertion in the bank, or the nature of the molecule sequenced (e.g., DNA, mRNA, tRNA, etc.). It is possible to combine many criteria using logical operators, and to use the results of previous queries to build new ones. Each kind of feature can be accessed and extracted as well as its flanking regions, and CDS can be translated into proteins. Different formats are available for extracting sequences including FASTA, MASE and GenBank. The server also gives access to various documents such as release notes, a history of the database, on-line documentation, etc.
It is also possible to install a local copy of the ACNUC version of EMGLib. This version is available at ftp://pbil.univ-lyon1.fr/pub/emglib and it requires the graphical retrieval system Query_win to run. This program is written in C and uses the Vibrant library, which is a part of the toolbox distributed by the NCBI. Binaries of Query_win are available through our server for UNIX workstations (Sun, DEC Alpha, IBM RISC, HP/UX, Silicon Graphics), and for all microcomputers under MacOS 7.x/8.x and Windows 95/98/NT 4.0 operating systems. Query_win integrates the same functionalities as the WWW-Query server plus more sophisticated options (like keywords and species browsing and keywords projections on sequences).
The second database is hosted at the INRA Microbial Genetics Laboratory, and its URL is http://locus.jouy.inra.fr/micado . MICADO is a relational database devoted to microbial genomes (15). On the WWW since 1994, it is now accessed 10 000 times a week. This horizontally integrates all DNA sequence information for archaea and bacteria, including complete genomes, with genetic maps of B.subtilis (16) and E.coli (17,18). As MICADO is the data repository of the functional analysis of B.subtilis unknown genes, a vertical integration of information is achieved for the bacterium. Physiological data concerning the disruption of 1200 genes is actually collected in the database by a European consortium of 17 laboratories (19), and linked to metabolic pathways classifications, DNA sequence and the genetic map. Information is searched through the WWW by text annotations (from DNA features to authors), sequence comparisons (BLAST and FASTA), browsing classification trees (metabolism and taxa), and finally, by navigating genome maps.
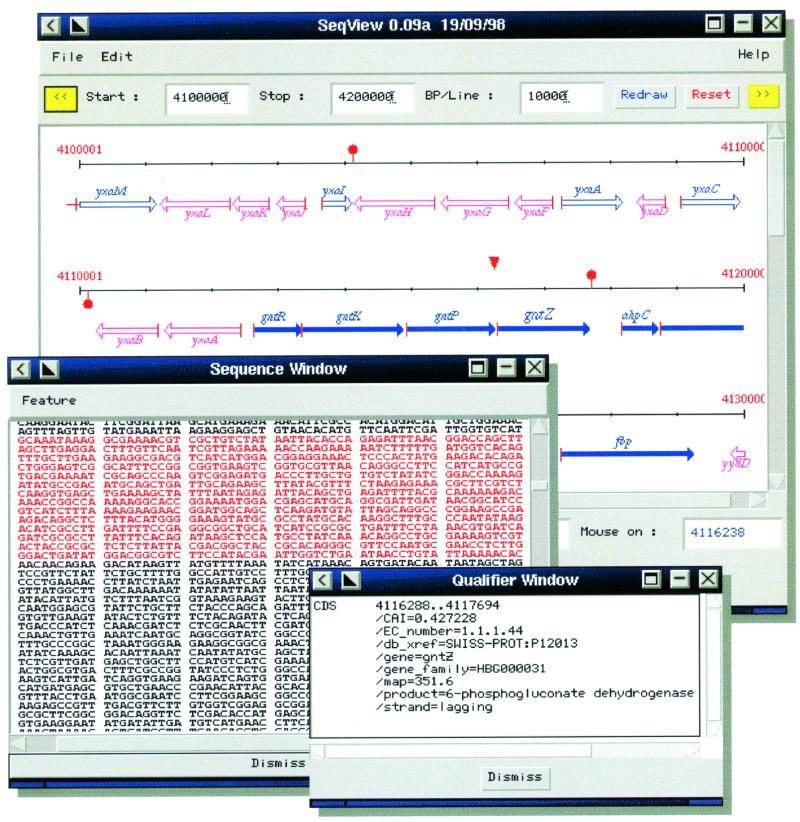
A graphical navigation on genome maps has been programmed to offer selective information retrieval on large-scale data sets, thus providing global overview and easy access to genome information. The maps have hierarchical relationships; they display chromosomal information at different scales, from the chromosome and the genetic map to featured physical maps, and finally, text of the sequences and their annotations (Fig. 2). Two versions of the graphical interface to MICADO are now accessible on WWW, the most complete and reliable in Perl 5 language, running on the WWW server and, derived from this Perl version, a new one in Java (20). The latter, running on the client side, brings more interactivity to users, and allows multi-windowing, with synchronized browsing of map representations at different scales. The Java client communicates with the database through an object server in C++ and uses the CORBA application interface protocol. This new standard is relevant to our growing needs, especially for future extensions. It is providing an interesting alternative to physical integration, by allowing us to establish virtual federations of shared databases.
Figure 2.
Viewing microbial genome information with the version of the interface in Java and CORBA. Here is shown a featured physical map of the chromosome of B.subtilis with the text of its DNA sequence, and scrolling is synchronized between the two windows. Also, a qualifier window displays the characteristics of a chosen feature, and is dynamically updated when the cursor is moved to another feature in the window of the physical map.
PERSPECTIVES
In the short term we want to continue the process of annotation improvement by adding protein sequence data such as molecular weight, isoelectric point value or subcellular location prediction. Next we want to introduce comparative genomics data taken from both the HOBACGEN and COLIPAGE (http://www-colipage.igmors.u-psud.fr ) databases. Indeed, the data provided by these two systems are complementary. While HOBACGEN contains homology data obtained on the whole sequences, COLIPAGE has assembled a lot of information about the constitutive segments of homology (modules) in paralogous proteins (21,22). Thanks to these data it would be possible to check gene names and the functions associated with their products, as there are often historical assignations that have been proven to be wrong in the light of the complete genome obtention. Also, it would be possible to assign functions to hypothetical proteins as the proportion of these proteins in completely sequenced bacterial genomes is sometimes very high (~30%).
Acknowledgments
ACKNOWLEDGEMENT
This work was supported by a grant from the PRFMMIP (Projet de Recherche Fondamentale pour les Maladies Microbiennes Infectieuses et Pathogènes).
REFERENCES
- 1.Fraser C.M., Gocayne,J.D., White,O., Adams,M.D., Clayton,R.A., Fleischmann,R.D., Bult,C.J., Kerlavage,A.R., Sutton,G., Kelley,J.M. et al. (1995) Science, 270, 397–403. [DOI] [PubMed] [Google Scholar]
- 2.Benson D.A., Boguski,M.S., Lipman,D.J., Ostell,J., Ouellette,B.F.F., Rapp,B.A. and Wheeler,D.L. (1999) Nucleic Acids Res., 27, 12–17. Updated article in this issue: Nucleic Acids Res. (2000), 28, 15–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stoesser G., Tuli,M.A., Lopez,R. and Sterk,P. (1999) Nucleic Acids Res., 27, 18–24. Updated article in this issue: Nucleic Acids Res. (2000), 28, 19–23.9847133 [Google Scholar]
- 4.Sugawara H., Miyazaki,S., Gojobori,T. and Tateno,Y. (1999) Nucleic Acids Res., 27, 25–28. Updated article in this issue: Nucleic Acids Res. (2000), 28, 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Médigue C., Rechenmann,F., Danchin,A. and Viari,A. (1999) Bioinformatics, 15, 2–15. [DOI] [PubMed] [Google Scholar]
- 6.Perrière G., Gouy,M. and Gojobori,T. (1998) Nucleic Acids Res., 26, 60–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lobry J.R. (1996) Science, 272, 745–746. [DOI] [PubMed] [Google Scholar]
- 8.Sharp P.M. and Li,W.-H. (1987) Nucleic Acids Res., 15, 1281–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bairoch A. and Apweiler,R. (1999) Nucleic Acids Res., 27, 49–54. Updated article in this issue: Nucleic Acids Res. (2000), 28, 45–48.9847139 [Google Scholar]
- 10.Bairoch A. (1999) Nucleic Acids Res., 27, 310–311. Updated article in this issue: Nucleic Acids Res. (2000), 28, 380–382.9847212 [Google Scholar]
- 11.Thieffry D., Salgado,H., Huerta,A.M. and Collado-Vides,J. (1998) Bioinformatics, 14, 391–400. [DOI] [PubMed] [Google Scholar]
- 12.Gouy M., Gautier,C., Attimonelli,M., Lanave,C. and di Paola,G. (1985) Comput. Appl. Biosci., 1, 167–172. [DOI] [PubMed] [Google Scholar]
- 13.Barker W.C., Garavelli,J.S., Haft,D.H., Hunt,L.T., Marzec,C.R., Orcutt,B.C., Srinivasarao,G.Y., Yeh,L.S.L., Ledley,R.S., Mewes,H.W., Pfeifer,F. and Tsugita,A. (1998) Nucleic Acids Res., 26, 27–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perrière G. and Gouy,M. (1996) Biochimie, 78, 364–369. [DOI] [PubMed] [Google Scholar]
- 15.Biaudet V., Samson,F. and Bessières,P. (1997) Comput. Appl. Biosci., 13, 431–438. [DOI] [PubMed] [Google Scholar]
- 16.Biaudet V., Samson,F., Anagnostopoulos,C., Ehrlich,S.D. and Bessières,P. (1996) Microbiology, 142, 2669–2729. [DOI] [PubMed] [Google Scholar]
- 17.Wahl R., Rice,P., Rice,C.M. and Kröger,M. (1994) Nucleic Acids Res., 22, 3450–3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rudd K.E. (1996) Trends Genet., 12, 156–157. [DOI] [PubMed] [Google Scholar]
- 19.Harwood C.R. and Wipat,A. (1996) FEBS Lett., 389, 84–87. [DOI] [PubMed] [Google Scholar]
- 20.Samson F., Biaudet,V. and Bessières,P. (1998) In Robinson,A.J. (ed.), Abstracts of the Objects in Bioinformatics 1998 Conference. EMBL-EBI, Hinxton, Cambridge, UK, p. 19.
- 21.Riley M. and Labedan,B. (1997) J. Mol. Biol., 269, 1–12. [DOI] [PubMed] [Google Scholar]
- 22.Labedan B. and Riley,M. (1999) In Charlebois,R.L. (ed.), Organization of the Prokaryotic Genome. ASM Press, Washington, DC, pp. 311–329.