Abstract
The objective of this work was to evaluate the antimicrobial resistant (AR) E. coli prevalence in recreational waters in Belgium and to assess the exposure risk for bathers. Nine stations were sampled during the 2021 bathing season. A total of 912 E. coli strains were isolated and tested by the disk diffusion method in accordance with EUCAST recommendations, including Extended-Spectrum Beta-Lactamase (ESBL) production. AR E. coli were counted at each bathing sites, 24% of strains were resistant to at least one antibiotic and 6% were Multi-Drug Resistant (MDR). A Multiple Antibiotic Resistance (MAR) index was calculated to compare the bathing sites. The Lesse river had the highest MAR index as well as the highest E. coli absolute abundance and the largest number of ESBL-producing E. coli. Conversely, the 3 lakes showed lower E. coli contamination levels and AR rates. A human health risk assessment of exposure to AR E. coli, based on the calculation of measured prevalence, was performed considering four different dose-response model scenarios. The human health risk (Pd) ranged from 10−9 to 0.183 (children). The exposure probabilities were low, except for scenario 3 (E. coli O157:H7), which is the most severe.
Keywords: Antibiotic resistance, Recreational water, Bathing water, Water quality, Wallonia, Human health exposure risk assessment
Highlights
-
•
Rivers contain higher E. coli absolute abundance and antimicrobial resistant (AR) E. coli than lakes.
-
•
Extended-Spectrum Beta-Lactamase-producing E. coli (ESBL-EC), found in all bathing areas, represent 3% of the isolated E. coli strains.
-
•
No correlation was observed between E. coli absolute abundance and Multiple Antibiotic Resistance (MAR) index.
-
•
The risk of exposure to bathers (Pd) varied from 10−9 to 0.183 (children).
1. Introduction
Antimicrobial resistance (AMR) is one of the greatest threats to health care in the world [1]. In the recent years, the “One Health” concept, recognizing, among others, the link between human, animal and environmental health, has become critically important [2]. The effect of clinically relevant antimicrobial resistance genes (ARGs) and antimicrobial resistant bacteria (ARB) that are released from anthropogenic sources, together with the excessive use of antibiotics in human or veterinary medicine and agriculture, is currently considered to be a critical environmental issue [3].
Bathing waters are monitored in Europe by the European Water Framework Directive (2006/7/EC), which classifies it as excellent, good, sufficient or poor based on thresholds for two fecal indicator organisms: Escherichia coli and intestinal enterococci. Although it is currently being revised, it is not intended to include an ARB monitoring despite the studies demonstrating the importance to monitor them [4,5].
Bathing waters are part of the environment that can be affected by the release of ARB through hospital or community wastewaters discharges and from intensive livestock production [6]. While conventional wastewater treatment plants (WWTPs) reduce the bacterial absolute abundance in water, they do not appreciably reduce the proportion of ARB [7] and pollute receiving waters with high levels of fecal indicator bacteria and ARGs [8]. Bathing waters contribute to the transmission of pathogenic agents including New Delhi Metallo-beta-lactamase (NDM) producing Enterobacteriaceae and ESBL-EC [9,10] and represent a putative reservoir and pathway for AR organisms spread [11,12]. Previous studies suggested that people with community-acquired urinary tract infection are more likely to be infected with an ESBL-EC if they bathed in freshwater within the last 12 months [13]. Furthermore, human intestinal colonization by AR commensal bacteria can persist for several months [14], and horizontal gene transfer of ARG on mobile genetic element can take place in vivo between ARB and transient or resident intestinal bacteria [15].
Though limited in number, previous epidemiological studies assessed the association between potential exposure to ARB through recreational aquatic activities and health endpoints such as gastrointestinal disease [16] and intestinal colonization with ESBL-EC bacteria [12]. For instance, higher rates of E. coli carrying Beta-lactamase encoding gene (blaCTX-M) colonization were observed in surfers (6.3%) than in non-surfers (1.5%) [12].
Several studies on AMR in bathing waters have been conducted in different countries [[17], [18], [19]] and a few of them estimate human health risks associated with AMR in recreational waters, with preliminary work focusing on exposure assessments [[20], [21], [22]]. Other studies proposed framework for the environmental surveillance of AR and its related parameters [[23], [24], [25]].
This study report, for the first time, the Belgian situation with a human health exposure risk assessment according to 4 scenarios following a new framework [26].
The aims of this study were (i) to assess the levels of AR E. coli in bathing sites in Belgium (ii) to study the resistance rate evolution during a bathing season (iii) to monitor the ESBL-EC prevalence and (iv) to determine the risk for human health.
2. Material and methods
2.1. Bathing stations
Nine areas of recreational waters were sampled from five watersheds in the Walloon Region of Belgium. The bathing stations were selected according to their different anthropic and demographic pressures and their nature: rivers (5 sampling zones), lakes (3 sampling zones) and canalized river (one sampling zone) (Fig. 1).
Fig. 1.
Sampling areas of the recreational waters in Belgium. Rivers (5); Lakes (3); Canalized river (1) (ArcMap).
2.2. Water sampling
Samples were collected monthly from April to September 2021 covering the bathing period.
Water samples were collected in 1-L sterile polyethylene bottles without any preservative, transported at 4 °C, stored at 5 ± 3 °C and analyzed within 24 h.
During each sampling, various field parameters were recorded. Firstly, the water physico-chemical parameters (temperature, dissolved oxygen, pH, conductivity and turbidity) were obtained and secondly, environmental observations were recorded (climate, number of animals, presence of bathers or kayaks, presence of waste or unusual smells).
2.3. E. coli absolute abundance
At first 10-fold dilutions of each sample were membrane filtered through 0.45-μm pore size filters (Millipore Corporation, USA) which were placed on Tryptone Bile X-glucuronide (TBX) (Bio-Rad, Marnes-la-Coquette, France) and incubated overnight at 37 °C to select the two optimal dilutions for Colony Forming Unit (CFU) counts, according to the ISO 8199:2018 norm (International Organization for Standardization, Switzerland).
The two optimal dilutions were membrane filtered a second time. Then, the filters were placed on TBX and on TBX supplemented with amoxicillin (AMX) at 8 mg l−1 corresponding to the minimum inhibitory concentration of AMX (Acros organics, New Jersey, USA) to select beta-lactam-resistant E. coli, and incubated overnight at 37 °C for E. coli absolute abundance enumeration and isolation [27].
2.4. E. coli isolation and confirmation
For each water sample, ten colonies of E. coli were randomly picked up from TBX and ten from TBX + AMX, inoculated on TBX medium to check the purity [27] and stored at 5 ± 3 °C until further used.
The confirmation of presumptive E. coli isolates was further tested for tryptophanase activity with Kovac’s reagent. Only indole positive isolates were considered for further analysis.
2.5. Antimicrobial susceptibility testing
For all strains isolated on TBX and TBX + AMX media, a susceptibility test was performed on Mueller-Hinton (MH) agar (Bio-Rad, Marnes-la-Coquette, France) using the disk diffusion assay and plates were incubated for 18 ± 2 h at 35 ± 1 °C according to the European Committee on Antimicrobials (EUCAST, 2020) [28]. E. coli ATCC 25922 (American Type Culture Collection, Manassas, VA, US) was included in each assay as negative control. The antimicrobial disks (Bio-Rad, Marnes-la-Coquette, France) were dispensed on MH agar with automatic disk dispenser (16-disks per plate) (Bio-Rad, Marnes-la-Coquette, France).
A total of 16 antibiotics selected based on sale volumes in human medicine (National Institute of Health and Disability Insurance of Belgium) and on their use (hospital, domestic or veterinary uses) were tested: ampicillin (AMP, content of the disk: 10 μg), amoxicillin/clavulanic acid (AMC, 20/10 μg), cefotaxim (COX, 5 μg), cefotaxim/clavulanic acid (CCO, 5/10 μg), ceftazidim (CZD, 10 μg), ceftazidim/clavulanic acid (CCZ, 10/10 μg), cefuroxime (CXM, 30 μg), ciprofloxacin (CIP, 5 μg), ertapenem (ETP, 10 μg), fosfomycin (FOS, 200 μg), gentamicin (CN, 10 μg), nitrofurantoin (NFE, 100 μg), meropenem (MEM, 10 μg), piperacillin/tazobactam (PTZ, 30/6 μg), sulfamethoxazole/trimethoprim (co-trimoxazole) (SXT, 23.75/1.25 μg), and tigecyclin (TGC, 15 μg).
Of these 16 antibiotics, cephalosporin discs without (COX, CZD) and with clavulanic acid (CCO, CCZ) were used for phenotypic confirmation of ESBL production in Enterobacterales according to the EUCAST technical guide.
2.6. Antimicrobial resistance rates
The AMX resistance rate was calculated by making the ratio between the numbers of CFU on TBX and on TBX + AMX. For the other antibiotics, the percentages of E. coli resistant to at least one, two or three antibiotics on TBX and TBX + AMX were calculated by making the ratio between the number of isolates respectively resistant to at least one, two or three antibiotics and the total of isolates. A Multi-Drug Resistant (MDR) strain is defined as being resistant to at least 3 antibiotics.
A Multiple Antimicrobial Resistant (MAR) index was calculated by sampling station. The MAR index of a sample was calculated with the following formula “a/(b*c)” where “a” is the aggregate antibiotic resistance score of all isolates from the sample (sum over all isolates of the number of antimicrobials to which each isolate is resistant), “b” is the number of tested antibiotics, and “c” is the number of isolates from the sample [29].
2.7. Statistical analysis
A Variance Analysis was performed with the Proc GLM of SAS software 9.4 program (SAS Institute, Cary, NC, USA) to study the impact of the localization and month of sampling on the E. coli absolute abundance and the MAR index. A logarithmic and a square root transformation were done on E. coli absolute abundance and MAR index respectively to normalize the data.
The Pearson correlation between E. coli absolute abundance and the MAR index was calculated with the Proc CORR of SAS software 9.4 program (SAS Institute, Cary, NC, USA).
2.8. Risk assessment
The daily exposure to AR E. coli probability (Pd) was performed based on the data obtained in this study following Tyagi & Kumar [26].
The input data were the concentration of E. coli (CFU/ml) and the number of isolates per sample on TBX, the number of isolates with at least one resistance and the number of non-resistant isolates.
The fvalue was calculated by antibiotic [26], but specific D-R models for each antibiotic were not available. In this study, fvalue, which ranged from 0 to 1, was defined as following:
| (1) |
The concentration of AR E. coli (CFU/ml) was calculated as following:
| AR E. coli concentration = fvalue × E. coli concentration | (2) |
The concentration of non-AR E. coli (CFU/ml) was calculated as following:
| non-AR E. coli concentration = (1 − fvalue) × E. coli concentration | (3) |
The number of AR and non-AR E. coli swallowed per exposure (N) during a dip [30] (Table 1) were measured as following:
| AR E. coli dose swallowed (N) = AR E. coli concentration × volume swallowed | (4) |
| non-AR E. coli dose swallowed (N) = non-AR E. coli concentration × volume swallowed | (5) |
Table 1.
Exposure parameters for bathers in freshwater: frequency of swimming per year, duration of swimming and volume swallowed per swimming event [30].
| Men | Women | Children | ||||
|---|---|---|---|---|---|---|
| Average | 95% CIa | Average | 95% CIa | Average | 95% CIa | |
| Frequency | 7 | 0–25 | 7 | 0–23 | 8 | 0–25 |
| Duration (min) | 54 | 7–200 | 54 | 6–222 | 79 | 12–270 |
| Volume swallowed (ml) | 27 | 0.016–140 | 18 | 0.022–86 | 37 | 0.14–170 |
CI: Confidence Interval.
Different dose response (D-R) model scenarios were used to assess the probability of daily exposure (Pd) (Table 2). For the AR E. coli concentration, the 4 scenarios have been applied while for the non-AR E. coli concentration only scenario 1 has been applied since it corresponds to susceptible bacteria.
Table 2.
Assumed dose-response (D-R) models based on four considered scenarios to assess the human exposure risk to antibiotic resistant (AR) Escherichia coli [26].
| Scenarios | Host and response type | D-R model | Model parameters for infection endpoints |
|---|---|---|---|
| 1: AR E. coli D-R model is similar to non AR E. coli | Human, diarrhoea | Beta-poisson model |
α = 0.1705, β = 1.61 × 106 |
| 2: AR E. coli D-R model is similar to gentamicin resistant E. coli | Human, diarrhoea | Beta-poisson model |
α = 0.16, β = 1.41 × 106 |
| 3: AR E. coli D-R model is similar to most infectious/virulent bacteria (E. coli O157:H7/Shigella) | Human, severe diarrhoea | Beta-poisson model |
α = 0.277, β = 2.38 × 102 |
| 4: AR E. coli D-R model is similar to most persistent bacteria (Clostridium perfringens) | Human, vomiting, abdominal cramps and diarrhoea within 24 h | Exponential model |
r = 1.82 × 10−11 |
The annual exposure risk assessment of bathing water ingestion was given by:
| (6) |
with n, the number of bathing per year (Table 1).
To take into account the infectious potential of non-AR E. coli, an overall probability of exposure was calculated as following:
| (7) |
The gastrointestinal (GI) illness risk caused by E. coli ingestion was also given by the U.S. Environmental Protection Agency [31] as following:
| Risk of GI / 1000 persons = 9.4 × (log concentration E. coli in 100 ml) − 11.74 | (8) |
| GI probability / person = (9.4 × (log concentration E. coli in 100 ml) − 11.74) / 1000 | (9) |
3. Results
3.1. Water sampling
The nine stations were sampled monthly from April to September 2021, representing 54 samples and covering the bathing season in order to highlight the public exposure levels to AR E. coli.
3.2. E. coli absolute abundance
The samples from the Houyet station (Lesse river) contained the highest E. coli absolute abundance, with an average of 1807 CFU × (100 ml)−1 over the six months. Then came the lake of Neufchâteau and the Pont à Lesse station (Lesse river) with 1594 and 1574 CFU × (100 ml)−1 respectively (Table 3 and Fig. 2).
Table 3.
Average Escherichia coli absolute abundance (CFU/100 ml) per station and per month on the Tryptone Bile X-glucuronide (TBX) medium.
| a | SDb | SD/c | ||
|---|---|---|---|---|
| Sampling areas | Lake of Péronnes | 120 | 157 | 1.3 |
| Lake of Marlette | 255 | 394 | 1.5 | |
| Lake of Neufchâteau | 1594 | 1870 | 1.2 | |
| Jambes | 428 | 345 | 0.8 | |
| Bouillon | 590 | 663 | 1.1 | |
| Récréalle | 472 | 489 | 1.0 | |
| Chiny | 964 | 672 | 0.7 | |
| Houyet | 1807 | 1928 | 1.1 | |
| Pont à Lesse | 1574 | 1338 | 0.8 | |
| Months | April | 165 | 281 | 1.7 |
| May | 1912 | 1976 | 1.0 | |
| June | 350 | 2055 | 5.9 | |
| July | 1265 | 1673 | 1.3 | |
| August | 870 | 1790 | 2.1 | |
| September | 645 | 1882 | 2.9 |
Average E. coli absolute abundance.
Standard deviation.
Standard deviation divided by the average of E. coli absolute abundance.
Fig. 2.
E. coli absolute abundance measured by sampling station and/or month.
The samples from the lake of Péronnes were the least loaded with E. coli with an average of 120 CFU × (100 ml)−1 over the six months. All stations together, the highest average isolate numbers were counted in May and July with 1912 CFU and 1265 CFU × (100 ml)−1 respectively.
Statistical analysis showed that the localization and month effects were both highly significant on the E. coli absolute abundance (p < 0.001).
3.3. Antimicrobial resistance rates
Resistances were found against all antibiotics tested except for MEM which is a last resort molecule. The resistance rates measured by antibiotic and culture medium for the 912 isolated strains are presented in Fig. 3. On TBX medium, the highest resistance rates were found against AMP (22.1%), AMC (8.9%) and SXT (8.2%) and the lowest for CZD (0.4%) and TGC (0.2%). 23.8% of the E. coli strains were resistant to at least one antibiotic and 6.1% were multi-drug resistant.
Fig. 3.
Resistance rates based on antibiotic and culture medium for the 912 E. coli isolated strains (AMC: amoxicillin/clavulanic acid, AMP: ampicillin, CIP: ciprofloxacin, COX: cefotaxim, CXM: cefuroxime, CZD: ceftazidim, ETP: ertapenem, FOS: fosfomycin, CN: gentamicin, MEM: meropenem, NFE: nitrofurantoin, PTZ: piperacillin/tazobactam, SXT: sulfamethoxazole/trimethoprim, TGC: tigecyclin).
3.4. Multiple antibiotic resistance (MAR) index
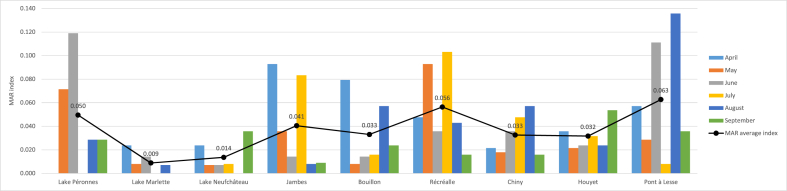
The MAR index was calculated for each of the 54 bathing water samples as well as globally per station and per month. The highest measured MAR index was 0.136 and corresponds to the Pont à Lesse sample from August (Fig. 4). A MAR index greater than 0.2 means that the high risk source of contamination is where antibiotics are frequently used [40]. The highest multi-drug resistant strain (resistant to six antibiotics) isolated in this study came from this sample. Four samples out of the 54 had a null MAR index, i.e. no resistance to the 14 tested antibiotics. These were sampled from the three lakes.
Fig. 4.
MAR index calculated by sampling station and month and average MAR index by station.
Overall, over the six months of sampling, the Pont à Lesse station showed the highest MAR index (0.063) and the highest percentages of multi-drug resistant strains (14.3%). Then, it was followed by the station of Récréalle for which the MAR index reached 0.056.
The lakes showed the lowest MAR index with 0.009 (Lake of La Marlette) and 0.014 (Lake of Neufchâteau).
Variations in MAR index, and thus antibiotic resistance, within the same station during the bathing season were observed (Fig. 4). The localization effect on the MAR index was slightly significant (p < 0.05) due to the lower MAR index values for the lakes of La Marlette. Removing this localization from the statistical analysis led to an absence of significant difference between localizations.
The MAR index of the different stations did not identically vary within the same month. Overall, at the nine bathing stations sampled, the MAR index was highest in April (0.049), followed by August (0.041) and June (0.040). September samples showed the lowest one with a value of 0.026. However, the effect of month was not significant (p = 0.47) on it.
The correlation analysis between the MAR index and the E. coli absolute abundance showed no significant correlation (p = 0.56) meaning that the E. coli absolute abundance was not directly related to the ARB presence.
The Lake of Péronnes had a higher overall MAR index than the other two lakes (0.050). The higher percentages observed in the lake of Péronnes compared to the two other lakes may be partially explained by the lower strain numbers that could be isolated from this sampling point over the entire campaign and from both environments (n = 43 on TBX and n = 26 on TBX + AMX). E. coli enumeration and isolation could not be performed in April for this lake.
3.5. ESBL-EC prevalence
During this campaign, ESBL-EC were found at every bathing point and in 33% (18/54) of the samples.
The two stations located on the Lesse, at Pont à Lesse and Houyet, were the most affected with 14 ESBL-EC detected. This corresponds to 50% of the strain detections with an ESBL-EC phenotype (28 in total). Only one ESBL-EC strain was found for each lakes and in Bouillon area. The highest number of ESBL-EC strains was found in July with 13 detections and the lowest in June.
Overall, these ESBL-EC strains represent 3% of the strains isolated during this study (28 out of 912 isolated strains).
3.6. Health exposure risk assessment
There is currently no dose-response model to quantify the risk of infection due to ARB swallowed by humans. Therefore, several scenarios were tested for the AR E. coli concentration (Table 2). The fact that ARB are potentially more dangerous than non-resistant bacteria was taken into account by applying the first scenario for the non-AR E. coli concentration and combining it with the 4 scenarios tested for the AR E. coli concentration.
Pd, Pa, and Poverall obtained are shown in Table 4A, Table 4B, Table 4C, Table 4D.
Table 4A.
Daily human exposure probabilities (PdAR) calculated based on four scenarios for AR E. coli.
| PdAR |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Scenario 1a |
Scenario 2b |
Scenario 3c |
Scenario 4d |
|||||||||
| Men | Women | Children | Men | Women | Children | Men | Women | Children | Men | Women | Children | |
| Mean | 6.2E-06 | 4.1E-06 | 8.5E-06 | 6.6E-06 | 4.4E-06 | 9.1E-06 | 0.047 | 0.035 | 0.060 | 1.1E-09 | 7.1E-10 | 1.5E-09 |
| Median | 2.4E-06 | 1.6E-06 | 3.3E-06 | 2.6E-06 | 1.7E-06 | 3.6E-06 | 0.025 | 0.017 | 0.034 | 4.2E-10 | 2.8E-10 | 5.7E-10 |
| Min | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Max | 5.9E-05 | 3.9E-05 | 8.0E-05 | 6.3E-05 | 4.2E-05 | 8.6E-05 | 0.283 | 0.229 | 0.328 | 1.0E-08 | 6.7E-09 | 1.4E-08 |
| P 95 | 2.5E-05 | 1.7E-05 | 3.5E-05 | 2.7E-05 | 1.8E-05 | 3.7E-05 | 0.176 | 0.133 | 0.214 | 4.4E-09 | 2.9E-09 | 6.0E-09 |
Table 4B.
PdnonAR calculated based on the scenario 1 for non-AR E. coli.
| PdnonAR |
|||
|---|---|---|---|
| Scenario 1a | |||
| Men | Women | Children | |
| Mean | 1.9E-05 | 1.3E-05 | 2.6E-05 |
| Median | 8.4E-06 | 5.6E-06 | 1.2E-05 |
| Min | 1.1E-07 | 7.1E-08 | 1.5E-07 |
| Max | 1.2E-04 | 8.1E-05 | 1.7E-04 |
| P 95 | 7.0E-05 | 4.7E-05 | 9.6E-05 |
Table 4C.
Overall exposure probabilities (Poverall).
| Poverall | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Scenario 1a (AR) + scenario 1a (non-AR) | Scenario 2b (AR) + scenario 1a (non-AR) | Scenario 3c (AR) + scenario 1a (non-AR) | Scenario 4d (AR) + scenario 1a (non-AR) | ||||||||
| Men | Women | Children | Men | Women | Children | Men | Women | Children | Men | Women | Children |
| 2,5E-05 | 1,7E-05 | 3,5E-05 | 2,6E-05 | 1,7E-05 | 3,5E-05 | 0.047 | 0.035 | 0.060 | 1,9E-05 | 1,3E-05 | 2,6E-05 |
Table 4D.
Annual overall exposure probabilities (Poverall annual).
| Poverall annual | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Scenario 1a (AR) + scenario 1a (non-AR) | Scenario 2b (AR) + scenario 1a (non-AR) | Scenario 3c (AR) + scenario 1a (non-AR) | Scenario 4d (AR) + scenario 1a (non-AR) | ||||||||
| Men | Women | Children | Men | Women | Children | Men | Women | Children | Men | Women | Children |
| 1.8E-04 | 1.2E-04 | 2.8E-04 | 1.8E-04 | 1.2E-04 | 2.8E-04 | 0.238 | 0.185 | 0.308 | 1.3E-04 | 8.9E-05 | 2.1E-04 |
Scenario 1: non-AR E. coli D-R model.
Scenario 2: gentamicin-resistant E. coli model.
Scenario 3: AR E. coli D-R model similar to most infectious/virulent bacteria (E. coli O157:H7/Shigella).
Scenario 4: AR E. coli D-R model is similar to most persistent bacteria (Clostridium perfringens).
For scenario 1, Pd varied from 0 to 5.9 × 10−5 (men), 0 to 3.9 × 10−5 (women), and 0 to 8.0 × 10−5 (children).
For scenario 2, Pd ranged from 0 to 6.3 × 10−5 (men), 0 to 4.2 × 10−5 (women), and 0 to 8.6 × 10−5 (children).
For scenario 3, Pd ranged from 0 to 0.283 (men), 0 to 0.229 (women), and 0 to 0.328 (children).
For scenario 4, Pd ranged from 0 to 1.0 × 10−8 (men), 0 to 6.7 × 10−9 (women) and 0 to 1.4 × 10−8 (children).
The GI probability for a person during a dip (Formula (9)) calculated from the E. coli concentrations measured from the 54 samples followed a normal distribution (mean = 0.0122, SD = 0.0064) meaning that there is more than 1/100 chance of developing a gastrointestinal disorder based on measured E. coli levels.
4. Discussion
This study focuses on nine bathing sites in the Walloon Region of Belgium collected monthly for 6 months in 2021. The 2021 bathing season was exceptional in regard to the spring heavy rains and summer huge floods [32]. Fecal indicator concentration and AR tend to increase during precipitations, with greater concentrations occurring at higher rainfall intensity [33,34]. Thus, the E. coli levels observed during the 2021 bathing monitoring were three to ten times higher than in 2020 (ISSeP, unpublished data). In the future, the expected consequences of climate change include increased frequency of extreme rainfall, longer periods of drought, and higher stream temperatures, which increase the vulnerability of aquatic environments to bacterial contamination, including ARB [35].
Even if increasing the number of samples would allow a better assessment of the AR E. coli prevalence, the results obtained in this study are close to those of other recent studies in the same geographical area. Indeed, in 2019 in Belgium, resistance rates of 26.4% for AMP, 15.9% for AMC and 14.6% for SXT were observed in freshwater and hospital effluents [36]. These were slightly higher than the resistance rates found in this study (AMP (22.1%)-AMC (8.9%)-SXT (8.2%)) but included hospital effluent known for their high rate of MDR E. coli [37]. Furthermore, the percentages of AR E. coli in the Netherlands were lower during the bathing season than during the winter season [38], which could explain the higher resistance rates measured in 2019 because a sampling campaign was conducted in winter.
The prevalence measured in the Seine River watershed in France were 42% of E. coli strains resistant to at least one antibiotic and 35% resistant to at least two antibiotics [39]. These higher resistances in the Seine watershed were probably related to the high population density of the Parisian area. The MAR index measured in the Seine (0.075) is also higher than the average MAR index measured in this study, which suggests the good quality of the water at the bathing stations [41].
Out of the 912 E. coli isolated, 28 were identified as ESBL-producer (3% of the isolated strains) and 33% (18/54) of the samples were concerned. This is quite similar to Norway, where a study comparing ESBL-EC from clinical, recreational water and wastewater samples found that 40% (8/20 occasions) of the recreational water samples contained ESBL-EC, representing all sites [9]. Exposure to ESBL-EC through bathing is likely, if recreational waters are located downstream of WWTPs or livestock farms [20]. In England and Wales, 0.12% of the E. coli detected in coastal surface waters were resistant to third generation cephalosporins (3GCs). Despite this low prevalence, the authors conclude that there is still an identifiable human exposure risk for recreational water users [4].
The ultimate objective of this work was to be able to estimate the risk incurred by bathers. The potential human health risks caused by exposition to ARB and/or ARGs present in aquatic environment have not yet been completely evaluated because specific information such as dose-response curves and exposure assessment data needed for a quantitative microbial risk assessment (QMRA) are scare [[40], [41], [42]]. The development of suitable dose-response models for the ingestion of AR vs non AR E. coli, specifically by antibiotic or antibiotic class would allow a more accurate risk evaluation for human health.
For the exposure assessment step of the QMRA process, it is necessary to quantify the ARBs or ARGs absorbed by an individual through various routes such as ingestion, inhalation or contact during recreational activities [42]. Although AR bacteria and ARG involved in skin infections are present in recreational waters [43], only the risk of exposure through ingestion was considered in this study.
An estimate of the daily (Pd), annual (Pa) and overall (Poverall) probability of exposure to ARB in Belgian bathing waters was calculated. Based on the measured ARB prevalence and the volume of water potentially swallowed, the ARB dose potentially swallowed by humans was calculated. In order to assess the human health risks, the absolute abundance of ARB present in a particular aquatic sample is essential [44]. Assuming that, the behaviour of Belgians is similar to that of the neighboring Dutch and swallowed volumes were extrapolated from the study by Schets et al. (2011) [30]. However, the frequency and duration of bathing can also be influenced by local factors such as climatic and cultural aspects. Four dose-response model scenarios were tested. Scenario 3, which was recommended by Tyagi and Kumar (2021) because it currently represents the fittest model for AR E. coli, given the lack of epidemiological/experimentally observed data, was the most severe one. With this scenario, the overall probability of exposure (Poverall) was on average 0.047 for men, 0.035 for women and 0.060 for children. Taking into account the annual bathing frequency (Table 1), Poverall annual increased to 0.238 for men, 0.185 for women and 0.308 for children. The scenario therefore strongly influenced the results. The probabilities (Pd, Pa) obtained with the other 3 scenarios were lower (scenario 2 > scenario 1 > scenario 4) (Table 4A, Table 4B, Table 4C, Table 4D). Conversely, the probabilities obtained by Tyagi and Kumar (2021) based on literature data coming mainly from developing countries were high (99th percentile ∼ 1 for the 4 scenarios). This is due to the low ARB prevalence found in Belgium compared to other developing countries.
In central Italy, human exposure to AR E. coli by recreational activities performed in bathing sites located in close proximity to WWTPs were predicted to be between 0.45 and 345.09 CFU/100 ml [21]. Therefore, even though information on the exact quantity of ARB necessary to colonize or infect the humans is not available, it will be reasonable to assume that an extremely low dose of ARB can pose human health risks [42].
5. Conclusions
This study shows, for the first time in Belgium, that an antibiotic resistance monitoring within freshwaters seems important in terms of public health.
The exposure risk of bathers currently appears to be limited in recreational waters in Belgium. However, it is important to maintain global efforts to control antibiotic resistance. Indeed, ESBL-EC have been found in good classified bathing areas and the MAR index fluctuates strongly between samples independently of the E. coli absolute abundance.
In application of the precautionary principle, the non-detection of a given ARB and ARG, based on state-of-the-art methodologies, cannot be interpreted as an absence of risk. Due to the large number of ARB, ARGs and antibiotics present in aquatics environments, quantification of each bacteria, gene or antibiotic concentration is time, cost and labor intensive. A solution would be to choose a panel of commonly accepted indicators, such as ESBL-EC but other approaches such as metagenomics could also be envisaged as monitoring tool in a near future.
Author contribution statement
Leslie Crettels: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Léa Champon: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Damien Thiry: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Nadine Burlion: Conceived and designed the experiments.
Elisa Delrée: Performed the experiments.
Claude Saegerman: Contributed reagents, materials, analysis tools or data; analyzed and interpreted the data.
Data availability statement
The authors do not have permission to share data.
Additional information
No additional information is available for this paper.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Leslie Crettels reports financial support was provided by Walloon Public Service (SPW), "Antibiobug 2 " project, ENVIeS plan (Action I-4-4).
References
- 1.WHO . 2019. Ten Threats to Global Health in 2019.https://www.who.int/vietnam/news/feature-stories/detail/ten-threats-to-global-health-in-2019 Geneva, Switzerland. [Google Scholar]
- 2.White A., Hughes J.M. Critical importance of a one health approach to antimicrobial resistance. EcoHealth. 2019;16:404–409. doi: 10.1007/s10393-019-01415-5. [DOI] [PubMed] [Google Scholar]
- 3.Berendonk T.U., Manaia C.M., Merlin C., Fatta-Kassinos D., Cytryn E., Walsh F., Bürgmann H., Sørum H., Norström M., Pons M.-N., Kreuzinger N., Huovinen P., Stefani S., Schwartz T., Kisand V., Baquero F., Martinez J.L. Tackling antibiotic resistance: the environmental framework. Nat. Rev. Microbiol. 2015;13:310–317. doi: 10.1038/nrmicro3439. [DOI] [PubMed] [Google Scholar]
- 4.Leonard A.F.C., Zhang L., Balfour A.J., Garside R., Gaze W.H. Human recreational exposure to antibiotic resistant bacteria in coastal bathing waters. Environ. Int. 2015;82:92–100. doi: 10.1016/j.envint.2015.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Herrig I., Fleischmann S., Regnery J., Wesp J., Reifferscheid G., Manz W. Prevalence and seasonal dynamics of blaCTX-M antibiotic resistance genes and fecal indicator organisms in the lower Lahn River, Germany. PLoS One. 2020;15 doi: 10.1371/journal.pone.0232289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hassoun-Kheir N., Stabholz Y., Kreft J.-U., de la Cruz R., Romalde J.L., Nesme J., Sørensen S.J., Smets B.F., Graham D., Paul M. Comparison of antibiotic-resistant bacteria and antibiotic resistance genes abundance in hospital and community wastewater: a systematic review. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140804. [DOI] [PubMed] [Google Scholar]
- 7.Blaak H., Lynch G., Italiaander R., Hamidjaja R.A., Schets F.M., de Roda Husman A.M. Multidrug-resistant and extended spectrum beta-lactamase-producing Escherichia coli in Dutch surface water and wastewater. PLoS One. 2015;10 doi: 10.1371/journal.pone.0127752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reynolds L.J., Sala-Comorera L., Martin N.A., Nolan T.M., Stephens J.H., Gitto A., O’Hare G.M.P., O’Sullivan J.J., Meijer W.G. Correlation between antimicrobial resistance and faecal contamination in small urban streams and bathing waters. Sci. Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.140242. [DOI] [PubMed] [Google Scholar]
- 9.Jørgensen S.B., Søraas A.V., Arnesen L.S., Leegaard T.M., Sundsfjord A., Jenum P.A. A comparison of extended spectrum β-lactamase producing Escherichia coli from clinical, recreational water and wastewater samples associated in time and location. PLoS One. 2017;12 doi: 10.1371/journal.pone.0186576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mahon B.M., Brehony C., McGrath E., Killeen J., Cormican M., Hickey P., Keane S., Hanahoe B., Dolan A., Morris D. Indistinguishable NDM-producing Escherichia coli isolated from recreational waters, sewage, and a clinical specimen in Ireland, 2016 to 2017. Euro Surveill. 2017;22 doi: 10.2807/1560-7917.ES.2017.22.15.30513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farrell M.L., Joyce A., Duane S., Fitzhenry K., Hooban B., Burke L.P., Morris D. Evaluating the potential for exposure to organisms of public health concern in naturally occurring bathing waters in Europe: a scoping review. Water Res. 2021;206 doi: 10.1016/j.watres.2021.117711. [DOI] [PubMed] [Google Scholar]
- 12.Leonard A.F.C., Zhang L., Balfour A.J., Garside R., Hawkey P.M., Murray A.K., Ukoumunne O.C., Gaze W.H. Exposure to and colonisation by antibiotic-resistant E. coli in UK coastal water users: environmental surveillance, exposure assessment, and epidemiological study (Beach Bum Survey) Environ. Int. 2018;114:326–333. doi: 10.1016/j.envint.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 13.Søraas A., Sundsfjord A., Sandven I., Brunborg C., Jenum P.A. Risk factors for community-acquired urinary tract infections caused by ESBL-producing Enterobacteriaceae – a case–control study in a low prevalence country. PLoS One. 2013;8 doi: 10.1371/journal.pone.0069581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kennedy K., Collignon P. Colonisation with Escherichia coli resistant to “critically important” antibiotics: a high risk for international travellers. Eur. J. Clin. Microbiol. Infect. Dis. 2010;29:1501–1506. doi: 10.1007/s10096-010-1031-y. [DOI] [PubMed] [Google Scholar]
- 15.Blake D.P., Hillman K., Fenlon D.R., Low J.C. Transfer of antibiotic resistance between commensal and pathogenic members of the Enterobacteriaceae under ileal conditions. J. Appl. Microbiol. 2003;95:428–436. doi: 10.1046/j.1365-2672.2003.01988.x. [DOI] [PubMed] [Google Scholar]
- 16.Griffith J.F., Weisberg S.B., Arnold B.F., Cao Y., Schiff K.C., Colford J.M. Epidemiologic evaluation of multiple alternate microbial water quality monitoring indicators at three California beaches. Water Res. 2016;94:371–381. doi: 10.1016/j.watres.2016.02.036. [DOI] [PubMed] [Google Scholar]
- 17.Döhla M., Sib E., Dericks B., Grobe S., Behringer K., Frechen M., Simon K., Färber H., Lenz F., Parcina M., Skutlarek D., Voigt A., Felder C., Exner M., Schmithausen R.M. Assessment of the prevalence of antibiotic-resistant bacteria and the concentration of antibiotics in EU bathing waters in western Germany. Expo Health. 2020;12:323–334. doi: 10.1007/s12403-019-00313-z. [DOI] [Google Scholar]
- 18.Di Cesare A., Eckert E.M., Teruggi A., Fontaneto D., Bertoni R., Callieri C., Corno G. Constitutive presence of antibiotic resistance genes within the bacterial community of a large subalpine lake. Mol. Ecol. 2015;24:3888–3900. doi: 10.1111/mec.13293. [DOI] [PubMed] [Google Scholar]
- 19.Hooban B., Fitzhenry K., Cahill N., Joyce A., O’ Connor L., Bray J.E., Brisse S., Passet V., Abbas Syed R., Cormican M., Morris D. A point prevalence survey of antibiotic resistance in the Irish environment, 2018–2019. Environ. Int. 2021;152 doi: 10.1016/j.envint.2021.106466. [DOI] [PubMed] [Google Scholar]
- 20.Schijven J.F., Blaak H., Schets F.M., de Roda Husman A.M. Fate of extended-spectrum β-lactamase-producing Escherichia coli from faecal sources in surface water and probability of human exposure through swimming. Environ. Sci. Technol. 2015;49:11825–11833. doi: 10.1021/acs.est.5b01888. [DOI] [PubMed] [Google Scholar]
- 21.O’Flaherty E., Solimini A., Pantanella F., Cummins E. The potential human exposure to antibiotic resistant-Escherichia coli through recreational water. Sci. Total Environ. 2019;650:786–795. doi: 10.1016/j.scitotenv.2018.09.018. [DOI] [PubMed] [Google Scholar]
- 22.Nappier S.P., Liguori K., Ichida A.M., Stewart J.R., Jones K.R. Antibiotic resistance in recreational waters: state of the science. IJERPH. 2020;17:8034. doi: 10.3390/ijerph17218034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huijbers P.M.C., Flach C.-F., Larsson D.G.J. A conceptual framework for the environmental surveillance of antibiotics and antibiotic resistance. Environ. Int. 2019;130 doi: 10.1016/j.envint.2019.05.074. [DOI] [PubMed] [Google Scholar]
- 24.Mortimer M., Winchell A., Holden P.A. Evaluation of frameworks proposed as protective of antimicrobial resistance propagation in the environment. Environ. Int. 2020;144 doi: 10.1016/j.envint.2020.106053. [DOI] [PubMed] [Google Scholar]
- 25.Jampani M., Gothwal R., Mateo-Sagasta J., Langan S. Water quality modelling framework for evaluating antibiotic resistance in aquatic environments. J. Hazard. Mater. Lett. 2022;3 doi: 10.1016/j.hazl.2022.100056. [DOI] [PubMed] [Google Scholar]
- 26.Tyagi N., Kumar A. Evaluation of recreational risks due to exposure of antibiotic-resistance bacteria from environmental water: a proposed framework. J. Environ. Manag. 2021;279 doi: 10.1016/j.jenvman.2020.111626. [DOI] [PubMed] [Google Scholar]
- 27.Bessa L.J., Barbosa-Vasconcelos A., Mendes Â., Vaz-Pires P., Martins da Costa P. High prevalence of multidrug-resistant Escherichia coli and Enterococcus spp. in river water, upstream and downstream of a wastewater treatment plant. J. Water Health. 2014;12:426–435. doi: 10.2166/wh.2014.160. [DOI] [PubMed] [Google Scholar]
- 28.Matuschek E., Brown D.F.J., Kahlmeter G. Development of the EUCAST disk diffusion antimicrobial susceptibility testing method and its implementation in routine microbiology laboratories. Clin. Microbiol. Infection. 2014;20:O255–O266. doi: 10.1111/1469-0691.12373. [DOI] [PubMed] [Google Scholar]
- 29.Krumperman P.H. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foodst. Appl. Environ. Microbiol. 1983;46:6. doi: 10.1128/aem.46.1.165-170.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schets F.M., Schijven J.F., de Roda Husman A.M. Exposure assessment for swimmers in bathing waters and swimming pools. Water Res. 2011;45:2392–2400. doi: 10.1016/j.watres.2011.01.025. [DOI] [PubMed] [Google Scholar]
- 31.U.S. Environmental Protection Agency . 1986. Bacteriological Ambient Water Quality Criteria for Marine and Fresh Recreational Waters - 1986. U.S. [Google Scholar]
- 32.Cornwall W. Europe’s deadly floods leave scientists stunned. Science. 2021;373:372–373. doi: 10.1126/science.373.6553.372. [DOI] [PubMed] [Google Scholar]
- 33.Cho K.H., Cha S.M., Kang J.-H., Lee S.W., Park Y., Kim J.-W., Kim J.H. Meteorological effects on the levels of fecal indicator bacteria in an urban stream: a modeling approach. Water Res. 2010;44:2189–2202. doi: 10.1016/j.watres.2009.12.051. [DOI] [PubMed] [Google Scholar]
- 34.Kubera Ł. Spread patterns of antibiotic resistance in faecal indicator bacteria contaminating an urbanized section of the brda river. Microb. Ecol. 2021;81:592–600. doi: 10.1007/s00248-020-01624-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anses . 2020. Antibiorésistance et environnement. État et causes possibles de la contamination des milieux en France par les antibiotiques, les bactéries résistantes aux antibiotiques et les supports génétiques de la résistance aux antibiotiques. Avis de l’Anses. Rapport d’expertise collective. [Google Scholar]
- 36.Crettels L., Burlion N., Breyer R., Mainil J., Servais P., Korfer J., Thiry D. Antimicrobial resistance of Escherichia coli isolated from freshwaters and hospital effluents in Belgium. Lett. Appl. Microbiol. 2022;74:411–418. doi: 10.1111/lam.13625. [DOI] [PubMed] [Google Scholar]
- 37.Gaşpar C.-M., Cziszter L.T., Lăzărescu C.F., Ţibru I., Pentea M., Butnariu M. Antibiotic resistance among Escherichia coli isolates from hospital wastewater compared to community wastewater. Water. 2021;13:3449. doi: 10.3390/w13233449. [DOI] [Google Scholar]
- 38.Blaak H., van Rooijen S.R., Schuijt M.S., Docters van Leeuwen A.E., Italiaander R., van den Berg H.H.J.L., Lodder-Verschoor F., Schets F.M., de Roda Husman A.M. National Institute for Public Health and the Environment. Ministry of Health, Welfare and Sport; 2011. Prevalence of Antibiotic Resistant Bacteria in the Rivers Meuse, Rhine, and New Meuse. [Google Scholar]
- 39.Passerat J., Tamtam F., Le Bot B., Eurin J., Chevreuil M., Servais P. Rejets hospitaliers d’antibiotiques et de bactéries fécales antibiorésistantes dans les rivières du bassin de la Seine. Eur. J. Water Qual. 2010;41:1–13. doi: 10.1051/water/2010004. [DOI] [Google Scholar]
- 40.Davis R., Brown P.D. Microbiology Society; 2016. Multiple Antibiotic Resistance Index, Fitness and Virulence Potential in Respiratory Pseudomonas aeruginosa from Jamaica. (accessed April 14, 2023) [DOI] [PubMed] [Google Scholar]
- 41.Servais P., Passerat J. Antimicrobial resistance of fecal bacteria in waters of the Seine river watershed (France) Sci. Total Environ. 2009;408:365–372. doi: 10.1016/j.scitotenv.2009.09.042. [DOI] [PubMed] [Google Scholar]
- 42.Manaia C.M. Assessing the risk of antibiotic resistance transmission from the environment to humans: non-direct proportionality between abundance and risk. Trends Microbiol. 2017;25:173–181. doi: 10.1016/j.tim.2016.11.014. [DOI] [PubMed] [Google Scholar]
- 43.Santiago-Rodriguez T.M., Rivera J.I., Coradin M., Toranzos G.A. Antibiotic-resistance and virulence genes in Enterococcus isolated from tropical recreational waters. J. Water Health. 2013;11:387–396. doi: 10.2166/wh.2013.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Amarasiri M., Sano D., Suzuki S. Understanding human health risks caused by antibiotic resistant bacteria (ARB) and antibiotic resistance genes (ARG) in water environments: current knowledge and questions to be answered. Crit. Rev. Environ. Sci. Technol. 2020;50:2016–2059. doi: 10.1080/10643389.2019.1692611. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors do not have permission to share data.