Abstract
The RESID Database contains supplemental information on post-translational modifications for the standardized annotations appearing in the PIR-International Protein Sequence Database. The RESID Database includes: systematic and frequently observed alternate names, Chemical Abstracts Service registry numbers, atomic formulas and weights, enzyme activities, indicators for N-terminal, C-terminal or peptide chain cross-link modifications, keywords, literature citations with database cross-references, structural diagrams and molecular models. Since 1995 updates of the RESID Database have appeared as often as weekly, and full releases appear quarterly. The database is freely accessible through the PIR Web site http://pir.georgetown.edu/pirwww/dbinfo/resid.html and by FTP.
INTRODUCTION
As part of the effort of the Protein Information Resource (PIR) to produce the PIR-International Protein Sequence Database (1) as a comprehensive, accurate, precise and consistent resource, the PIR maintains a number of auxiliary annotation databases. The RESID Database is the auxiliary database of modified amino acid residues annotated as features in the Protein Sequence Database. This database was designed:
1) to document standardized features annotations for covalent binding sites, modified sites and cross-links, and to provide appropriate keywords and other annotations to accompany those features,
2) to furnish more detailed chemical information than is possible in the Protein Sequence Database, and to enable annotators to recognize when authors are using synonymous descriptions of previously described features,
3) to provide an adaptable mechanism for calculating the molecular weights of modified proteins and their peptide fragments, and
4) to be accessible through the Internet and multi-database access programs with useful search capabilities and display of chemical structures.
During each update, all feature records in the Protein Sequence Database are automatically checked for syntax and vocabulary using the standard records in the latest version of the RESID Database. These procedures ensure accuracy and consistency in annotation, and through the use of automated scripts help propagate annotation revisions quickly throughout Protein Sequence Database entries (2). RESID is the only publicly available, human- and computer-readable database comprehensively documenting protein structural modifications. It provides scientists interested in protein sequence and structure with visual display and molecular modeling information for a comprehensive collection of protein post-translational modifications.
DATABASE DESCRIPTION
The database includes entries for the 22 alpha-amino acids currently known to be genetically encoded, including N-formyl methionine and selenocysteine, three ambiguous ‘residues’ represented in the standard single-letter code, and more than 245 other residues either predicted or observed in proteins arising through natural, post-translational modification of encoded amino acids. Included are several molecular structures that are known not to exist, but are encountered or referred to in the literature or other databases.
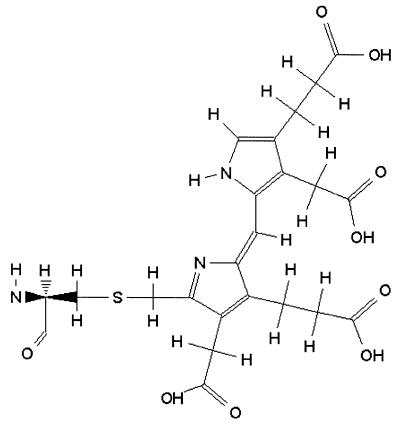
Information in the RESID Database entries includes: dates for database entry and modification of text and structure; a systematic chemical name and Chemical Abstracts Service registry number for the free residue; frequently observed alternate names; the atomic formula and weight; original amino acids with difference formulas and weights; enzyme activities producing the modification; indicators for N-terminal, C-terminal or peptide chain cross-link modifications; how the modification is represented in feature tables in the Protein Sequence Database and the keywords associated with it; and literature citations with database cross-references. The RESID Database maintains dynamic links with the PIR-International Protein Sequence and NRL_3D databases, and concurrent cross-references to Chemical Abstracts (CAS), to the MEDLINE citation database, and to the Protein Data Bank (PDB). (CAS Registry Numbers are copyrighted by the American Chemical Society and used with permission of the Chemical Abstracts Service of the American Chemical Society.) The RESID Database provides a means for calculating both chemical-average and monoisotopic molecular weights for modified peptides in order to facilitate their identification by mass-spectroscopy (3,4). Structural diagrams are presented in GIF format, and molecular models in PDB format are provided for use with widely available WWW display programs such as RasMol (5). A sample entry in the RESID Database is presented in Table 1.
Table 1. RESID Database sample entry.
Release 20.00 of the database contains >270 entries, an increase of ~10% annually. During the fall of 1999, molecular models for RESID Database entries began to be introduced. Also during 1999, several enhancements were made in WWW access for the database. Additional search capabilities were provided for users to search lists of modifications arising from particular encoded amino acids, and to search for modifications based on either the molecular weight of the modified residue or the difference in the molecular weight produced by the modification.
AVAILABILITY AND ACCESS
The RESID Database is updated as often as weekly and is released quarterly with the PIR-International Protein Sequence Database through Internet distribution. Database entries may be retrieved through WEB search at http://pir.georgetown.edu/pirwww/search/textresid.html by entry code or other unique identifier, by name, citation, or keyword text search, by molecular weight search, or from selection lists based on encoded amino acids. Active links to RESID Database entries are also provided through the feature table of Protein Sequence Database entries.
The database textual annotations, structural diagrams and molecular models are copyrighted. The RESID Database is distributed free with no license required.
SUBMISSIONS AND REVISIONS
Most new entries for the RESID Database are generated as a result of annotation and re-evaluation of sequences in the PIR-International Protein Sequence Database. The author invites the submission of information for new entries or for the revision of existing entries. Those wishing to submit material may do so using the electronic submission form at http://pir.georgetown.edu/pirwww/otherinfo/subinfo.html or by Email directed to the author’s attention at pirmail@nbrf.georgetown.edu . New database entries are assigned unique access codes which may be cited in publications. It is appreciated if authors referring to the RESID Database cite this article or the introductory announcement (6).
Acknowledgments
ACKNOWLEDGEMENTS
The author acknowledges Mr David J. Miller for his help as research assistant, and Dr Bruce C. Orcutt and Mr Joseph Janda for their support in maintaining the WWW pages. This publication and enhancement of the RESID database are supported by NSF grant DBI-9808414. PIR is a registered mark of the NBRF.
REFERENCES
- 1.Barker W.C., Garavelli,J.S., Huang,H.Z., McGarvey,P.B., Orcutt,B.C., Srinivasarao,G.Y., Xiao,C.L., Yeh,L.S., Ledley,R.S., Janda,J.F., Mewes,H.W., Pfeiffer,F., Tsugita,A. and Wu,C. (2000) Nucleic Acids Res., 28, 41–44 (this issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garavelli J.S., Miller,D.J. and Srinivasarao,G.Y. (1999) Protein Sci., 8 (Suppl. 1), 75 (abstract 107).10210185 [Google Scholar]
- 3.Biemann K. and Scoble,H. (1987) Science, 237, 992–998. [DOI] [PubMed] [Google Scholar]
- 4.Takao T., Yoshino,K., Suzuki,N. and Shimonishi,Y. (1990) Biomed. Environ. Mass Spectrom., 19, 705–712. [DOI] [PubMed] [Google Scholar]
- 5.Sayle R.A. and White,E.J.M. (1995) Trends Biochem. Sci., 20, 374–376. [DOI] [PubMed] [Google Scholar]
- 6.Garavelli J.S. (1993) Protein Sci., 2 (Suppl. 1), 133 (abstract 450). [Google Scholar]