Abstract
The Yeast Proteome Database (YPD™) has been for several years a resource for organized and accessible information about the proteins of Saccharomyces cerevisiae. We have now extended the YPD format to create a database containing complete proteome information about the model organism Caenorhabditis elegans (WormPD™). YPD and WormPD are designed for use not only by their respective research communities but also by the broader scientific community. In both databases, information gleaned from the literature is presented in a consistent, user-friendly Protein Report format: a single Web page presenting all available knowledge about a particular protein. Each Protein Report begins with a Title Line, a concise description of the function of that protein that is continually updated as curators review new literature. Properties and functions of the protein are presented in tabular form in the upper part of the Report, and free-text annotations organized by topic are presented in the lower part. Each Protein Report ends with a comprehensive reference list whose entries are linked to their MEDLINE abstracts. YPD and WormPD are seamlessly integrated, with extensive links between the species. They are freely accessible to academic users on the WWW at http://www.proteome.com/databases/index.html , and are available by subscription to corporate users.
INTRODUCTION
The genome and proteome of an organism do not correspond in a one-to-one fashion: one gene may give rise to multiple proteins by means of alternative splicing or post-translational modification, and its expression may be temporally or spatially regulated. Since proteins are the major players in most processes of living cells, knowledge of the proteome has great relevance to the study of the cell or organism in molecular detail. YPD™ and WormPD™ are proteome databases for Saccharomyces cerevisiae and Caenorhabditis elegans: organized collections of information about the complete sets of proteins of each of these model organisms. The protein-specific information in YPD and WormPD is derived from comprehensive and in-depth curation of the scientific literature. Curators read the full text of articles, select the key results, and record them as tabulated, searchable properties and also re-state them in clear, consistent language in free-text annotation lines. Full utilization of the scientific literature in this manner is impractical for an individual scientist studying more than a few proteins or interested in literature outside of his or her immediate field of expertise.
YPD and WormPD are useful in several different ways. For researchers interested in individual proteins, they present a summary of all available information about those proteins and a gateway to the original literature. For researchers investigating groups of proteins with common properties such as function or subcellular localization, the YPD and WormPD search capabilities allow the identification of all members of such groups, and also allow the searcher to specify whether properties are experimentally determined or predicted. Finally, for researchers engaged in global studies that generate new lists of proteins of interest, YPD and WormPD Title Lines lend meaning to such lists by providing succinct descriptions of the function of each protein.
YPD and WormPD have been developed as components, or ‘volumes’, of a larger relational database, the BioKnowledge™ Library. Within the BioKnowledge Library, natural connections exist between proteins of the same or different species based on sequence similarity, similarity of protein properties and similarities of function. These connections have allowed us to enrich the annotations for many unknown proteins of C.elegans by using information about their well-studied homologs in S.cerevisiae. As more volumes, representing more model organisms and humans, are entered into the BioKnowledge Library, many more connections and predictions of function will be possible.
NEW FEATURES OF YPD
The general features of YPD have been described in this issue of previous years (1), but it has recently undergone several significant improvements. The Protein Report format has been redesigned for greater clarity. The tabular information at the top of the Protein Report has been streamlined and reorganized to emphasize the most important properties of the protein, and a new section labeled ‘At-a-Glance’ provides a succinct summary of the significance of that protein to the cell. We have compiled, and continue to review, lists of subcellular localizations, cellular roles, biochemical functions, and other protein properties, with the aim of developing comprehensive sets of terms that will serve to classify proteins in any organism. This multi-dimensional classification system greatly increases the precision of searching for proteins with specific characteristics, and serves as an organizational tool for grouping proteins by involvement in cellular processes. In addition to changing the format of the tabular part of the Protein Report, the free-text annotations in the lower part of the page have been reorganized. Documentation of the Protein Report format is now available and easily accessible through links from the section titles.
The content of YPD has grown along with the body of scientific knowledge about yeast. By September 1999, curators had reviewed >12 500 articles to generate >90 000 curated annotation lines. Overall statistics on the characterization of the yeast proteome are presented in Table 1.
Table 1. Proportion of characterized proteins in the S.cerevisiae and C.elegans proteomes.
| S.cerevisiae | C.elegans | |
|---|---|---|
| Total number of known and predicted proteins |
6122 |
18 988 |
| Proteins characterized by genetics or biochemistry | 3199 (52% of total) | 1133 (6% of total) |
| Proteins known by similarity to characterized proteins | 975 (16% of total) | 12 638 (67% of total) |
| Proteins of unknown function | 1948 (32% of total) | 5217 (27% of total) |
These statistics are as of September 1999. For links to current statistics, see Supplementary Material.
INTRODUCTION OF WormPD
In July 1999, WormPD was introduced as the newest volume in our library of proteome databases. WormPD presents organized, comprehensive information for each of the nearly 19 000 proteins predicted from the recently completed C.elegans genome sequence (2). It utilizes the one-page-per-protein format familiar to YPD users, with the recent improvements described above. A major difference between C.elegans and yeast at the level of gene expression is that worm transcripts typically contain multiple introns and are often alternatively spliced, giving rise to several different gene products. This complexity necessitates the inclusion of more detailed transcript information and description of alternative protein forms in WormPD. If an alternative protein sequence has been documented in the literature or predicted by the genome sequencing project (2), this information is noted at the top of the Protein Report of the primary form. By following the link provided, the user can view an alignment of all alternative forms and access individual Protein Reports for each form. Annotations that are unique to a given protein form are distinguished from those applicable to all forms by boldface type.
By September 1999, >1400 articles had been reviewed to produce nearly 21 000 curated annotation lines in WormPD. Table 1 presents the extent of characterization of the C.elegans proteome at this time.
INTEGRATION OF YPD AND WormPD
Since YPD and WormPD are both part of the same underlying database, they are naturally integrated. The interface between the two appears seamless to the user, and navigation between them is as straightforward as navigation within each database. Every yeast or worm gene or protein name that appears within a Protein Report, in either database, is hyperlinked to its own respective Protein Report. Since the annotations are written in a standard style with a minimum of organism-specific terminology, users can easily browse both databases simultaneously. For example, a user who has identified an uncharacterized worm protein in an experimental screen can, with a single click, access all that is known from the scientific literature about the most closely related yeast protein. Such connections can allow the researcher to quickly assess whether or not to pursue an experimental lead. Furthermore, the information provided may suggest the design of future experiments.
Sequence similarity is at the heart of many connections between YPD and WormPD. It is of great interest to know whether a given protein is specific to one organism, is highly conserved, or is somewhere in between these two extremes. To facilitate such comparisons, each Protein Report in YPD and WormPD contains a section, ‘Related Proteins’, that lists similar proteins from S.cerevisiae, C.elegans, Drosophila melanogaster, Rattus norvegicus, Mus musculus, and Homo sapiens, as determined by biweekly BLAST analysis (3) refined by the Smith–Waterman algorithm (4). A pop-up window leads to a list of the number of matches in each organism, and the name of each organism is hyperlinked to the complete BLAST report. The BLAST reports represent similarity both graphically and as sequence alignments. S.cerevisiae and C.elegans gene names appearing within the BLAST reports are linked back to their respective Protein Reports. Thus it is simple and straightforward to find all proteins in both organisms with common sequence elements and examine what is known about their functions.
Having two comprehensively curated and interlinked model organism databases provides great power to comparative genomic analysis. With the complete genomic sequences of two eukaryotic organisms determined, it is now possible to compare both orthologous and divergent sets of proteins in order to identify proteins that are likely to provide core eukaryotic functions and those that may be specific to an individual species or family of organisms. Our analyses have suggested that ~41% of predicted yeast proteins and 19% of predicted worm proteins are conserved (with an expectation value of 10–10 or lower) between the two organisms. YPD and WormPD provide an information-rich background in which to expand such connections by tracing conserved pathways, protein associations and complexes across species lines.
As an example, using the information presented in YPD and WormPD Protein Reports we have been able to identify likely components of the C.elegans mitochondrial translocation apparatus (see Supplementary Material). While not studied directly in C.elegans, the protein machinery involved in the import of proteins into the mitochondria, a process essential to life, has been well-studied in yeast (5–8). Through our analyses, we have found that most, but not all, essential yeast proteins involved in this process have recognizably conserved counterparts in C.elegans. In all cases of apparent conservation the similarities are surprisingly weak, with expectation values for most >10–12. This finding is consistent with our observation that mitochondrial proteins, as a group, are more highly diverged between yeast and worm than are proteins found in most other subcellular compartments (Fig. 1).
Figure 1.
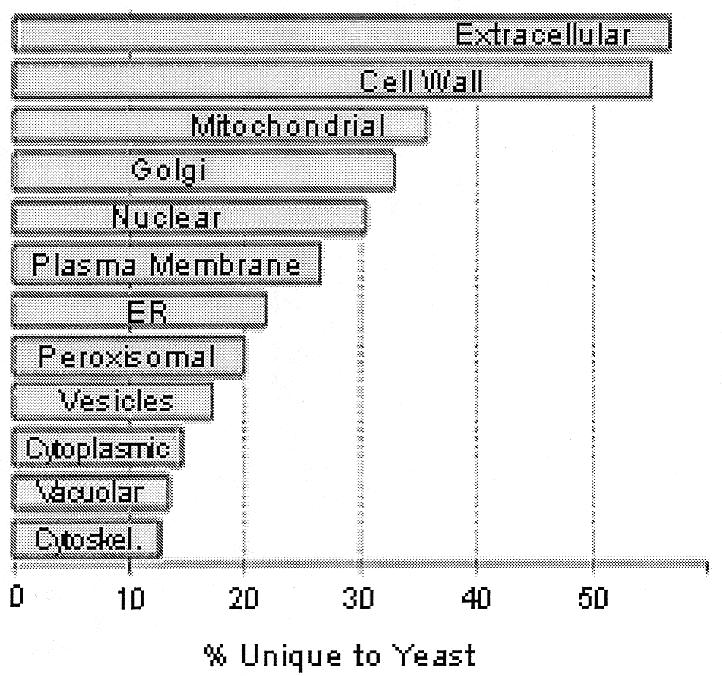
Conservation of proteins between S.cerevisiae and C.elegans, sorted by major subcellular localization. For each group, the percentage of all proteins of that group without a match in the worm proteome (E value of 10–6 or greater) is represented.
ENRICHING ANNOTATION OF UNKNOWN PROTEINS
As the first multicellular organism whose entire genomic sequence is known (2), C.elegans serves as a model for the interpretation of this wealth of information. Nearly 18 000 predicted worm proteins have not yet been characterized by focused experimental projects. We have provided an initial level of characterization for ~12 500 of these proteins through curated similarity comparisons. As discussed above, each predicted yeast or worm protein is compared to all other yeast, worm, fly, rat, mouse and human proteins using BLAST similarity analysis (3). Such analyses and additional sources of information (including our annotated databases) are evaluated by trained curators and provide the basis for annotation of the as-yet-uncharacterized proteins. Using guidelines we have developed to assess the significance of various levels of similarity, our staff of curators has defined protein families and made intelligent predictions for each of the experimentally uncharacterized worm proteins with significant similarity to characterized proteins. The level and extent of similarity are stated in the Title Line, and for those proteins highly similar to known proteins, we have predicted properties such as biochemical function, cellular role, subcellular localization and molecular environment when possible. All predicted properties are clearly distinguished from experimentally demonstrated properties.
FUNCTIONAL GENOMICS
We continue to focus on making functional genomic data available to our users as well as providing the context to help make sense of them. YPD currently provides access to transcription profiles derived from DNA microarray hybridization experiments. As of September 1999, the results of 36 such experiments, which measure the relative expression of each gene under a variety of experimental conditions, are now incorporated into YPD. Users gain access to these data by clicking on a link in the Gene Expression section of the upper part of the Protein Report. Such data will be added to WormPD in the near future.
We also aim to help researchers bring meaning to their large-scale functional genomics experiments in both yeast and worm by providing access to a current list of YPD and WormPD Title Lines which include protein properties, functions and roles. As described above, every yeast and worm protein is defined by a single line summarizing its function. Title Lines are constantly updated to reflect new experimental findings and up-to-date BLAST analyses. ‘Hit lists’ generated by functional genomics experiments (for example, groups of co-regulated genes) are much more immediately informative when they are presented with informative and up-to-date Title Lines. We encourage the use of YPD and WormPD Title Lines for annotation of functional genomic data, and these Title Lines can be used with permission on other non-commercial web sites or in publications.
FUTURE DIRECTIONS
YPD and WormPD are nearing complete curation of the yeast and worm literature respectively. Both YPD and WormPD will be maintained as comprehensively curated databases. We continue to seek ways to improve our existing databases through addition of new features and expansion of older features, such as the continued incorporation of functional genomics data. At the same time, we are working to develop new databases that will be beneficial to life science researchers in diverse fields of study. We have recently begun a Candida albicans proteome database (CalPD™) and plan to expand it into a series of volumes for the BioKnowledge Library (the Fungal Knowledge Collection) representing more fungal model organisms and pathogens. In the future our work will extend to proteome databases concerning higher eukaryotes, including humans.
CONTACTING YPD AND WormPD
We appreciate feedback from our users concerning new data submission, additions, clarifications and corrections. Personal communications will be cited as such. Functional genomic datasets (both for yeast and worms) are especially welcomed, and users with functional genomics websites are encouraged to link to our site. Any correspondence, including requests for YPD and WormPD spreadsheets, should be directed to ypd@proteome.com , wormpd@proteome.com , or by mail to the address of the authors.
CITING YPD AND WormPD
Authors wishing to make use of the information provided by YPD or WormPD should cite this article as a general reference for access to and content of YPD and WormPD.
SUPPLEMENTARY MATERIAL
The following material is available as supplementary material via NAR Online:
• View current yeast proteome statistics.
• View current worm proteome statistics.
• View a sample YPD Protein Report.
• View a sample WormPD Protein Report.
• Identification of conserved C.elegans mitochondrial import machinery using information presented within YPD and WormPD Protein Reports.
Acknowledgments
ACKNOWLEDGEMENTS
We thank the staff of the Saccharomyces Genome Database (http://genome-www.stanford.edu/Saccharomyces ), the Munich Information Centre for Protein Sequences (http://www.mips.biochem.mpg.de ), and a C.elegans Database (http://www.sanger.ac.uk/Projects/C_elegans/webace_front_end.shtml and http://stein.cshl.org/elegans ) for their help and cooperation. We appreciate the advice of many C.elegans researchers during the development of WormPD, particularly Leon Avery, Richard Durbin, Michael Hengartner, Jonathan Hodgkin, Barbara Meyer, John Spieth, Lincoln Stein, Paul Sternberg and Jean Thierry-Mieg. We wholeheartedly thank the members of two groups of fundamental importance to these efforts: the yeast and worm research communities, and the curators of YPD and WormPD. The development of YPD has been partially funded by a Phase II SBIR grant from the National Institute of General and Medical Sciences (R44 GM54110-02).
REFERENCES
- 1.Hodges P.E., McKee,A.H.Z., Davis,B.P., Payne,W.E. and Garrels,J.I. (1999) Nucleic Acids Res., 27, 69–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.The C.elegans Sequencing Consortium (1998) Science, 282, 2012–2018.9851916 [Google Scholar]
- 3.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waterman M.S. (1995) Introduction to Computational Biology: Maps, Sequences and Genomes. Chapman & Hall, London, UK.
- 5.Leuenberger D., Bally,N.A., Schatz,G. and Koehler,C.M. (1999) EMBO J., 18, 4816–4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schatz G. (1996) J. Biol. Chem., 271, 31763–31766. [DOI] [PubMed] [Google Scholar]
- 7.Pfanner N., Craig,E.A. and Honlinger,A. (1997) Annu. Rev. Cell. Dev. Biol., 13, 25–51. [DOI] [PubMed] [Google Scholar]
- 8.Rassow J., Dekker,P.J.T., van Wilpe,S., Meijer,M. and Soll,J. (1999) J. Mol. Biol., 286, 105–120. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


