This report establishes a novel form of IEI due to DN RELA mutations as a cause of chronic mucocutaneous ulcerations with autoinflammatory and/or autoimmune manifestations. This also expands the clinical spectrum of human RELA-associated diseases.
Abstract
Inborn errors of the NF-κB pathways underlie various clinical phenotypes in humans. Heterozygous germline loss-of-expression and loss-of-function mutations in RELA underlie RELA haploinsufficiency, which results in TNF-dependent chronic mucocutaneous ulceration and autoimmune hematological disorders. We here report six patients from five families with additional autoinflammatory and autoimmune manifestations. These patients are heterozygous for RELA mutations, all of which are in the 3′ segment of the gene and create a premature stop codon. Truncated and loss-of-function RelA proteins are expressed in the patients’ cells and exert a dominant-negative effect. Enhanced expression of TLR7 and MYD88 mRNA in plasmacytoid dendritic cells (pDCs) and non-pDC myeloid cells results in enhanced TLR7-driven secretion of type I/III interferons (IFNs) and interferon-stimulated gene expression in patient-derived leukocytes. Dominant-negative mutations in RELA thus underlie a novel form of type I interferonopathy with systemic autoinflammatory and autoimmune manifestations due to excessive IFN production, probably triggered by otherwise non-pathogenic TLR ligands.
Introduction
Human inborn errors of immunity (IEIs) can underlie various infectious, autoinflammatory, autoimmune, allergic, and/or malignant phenotypes (Tangye et al., 2022). Inborn errors in the classical (canonical) or alternative (noncanonical) NF-κB pathway cause immunodeficiencies, except for complete deficiencies of I κ B kinase α (IKKα) or NF-κB essential modulator (NEMO), which are embryonic lethal mutations (Zhang et al., 2017). Most human IEIs affecting the classical NF-κB pathway impair both innate and adaptive immunity (Zhang et al., 2017). Some underlying clinical manifestations of IEIs are unrelated to hematopoiesis, such as anhidrotic ectodermal dysplasia (EDA), lymphedema, and osteopetrosis (Boisson et al., 2017). Inborn errors in the core classical NF-κB pathway were first described in 2001, with reports showing that X-linked recessive EDA with immunodeficiency (EDA-ID) is caused by hypomorphic mutations of IKBKG (encoding NEMO; Jain et al, 2001). Subsequently, autosomal dominant (AD) EDA-ID caused by hypermorphic mutations of the NFKBIA gene (encoding NF-κB inhibitor α) was observed (Courtois et al., 2003). These defects lead to a broad range of infections caused by multiple defects in innate and adaptive immunity (Casanova et al., 2011; Zhang et al., 2017; Courtois et al., 2003; Moriya et al., 2018). Other defects in the classical NF-κB pathway have since been reported, including autosomal recessive IKKβ deficiency (Pannicke et al., 2013; Mousallem et al., 2014; Nielsen et al., 2014), AD IKKβ gain-of-function (GOF; Cardinez et al., 2018), AD NF-κB1 deficiency (Fliegauf et al., 2015; Kaustio et al., 2017; Li et al. 2021), and autosomal recessive c-Rel deficiency (Lévy et al., 2021). Studies of these NF-κB–related disorders have improved our understanding of the clinical and immunological phenotypes seen in patients with inborn errors in the core components of the classical NF-κB pathway.
Recently, four related patients with RELA haploinsufficiency were described with TNF-dependent mucocutaneous ulcerations treatable with infliximab therapy (Badran et al., 2017). Subsequently, two additional affected families were reported: lymphoproliferation with autoimmune cytopenia due to RELA haploinsufficiency (Comrie et al., 2018) or Behçet’s disease associated with heterozygous mutation in the 3′ portion of the RELA gene (Adeeb et al., 2021; Lecerf et al., 2022). In the current study, we performed a multicenter survey investigating patients with heterozygous RELA variants.
Results
Identification of rare monoallelic RELA variants
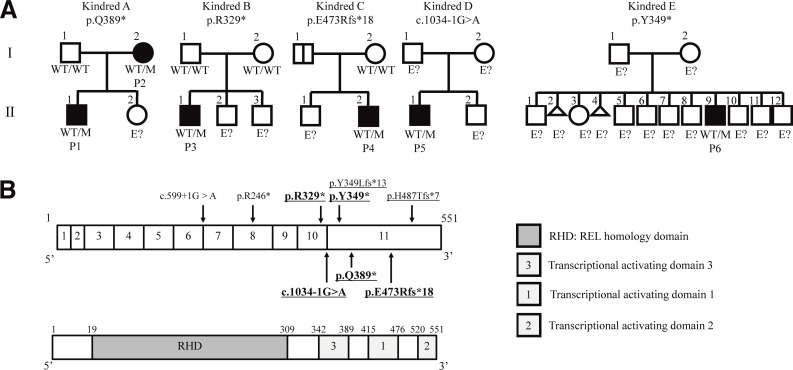
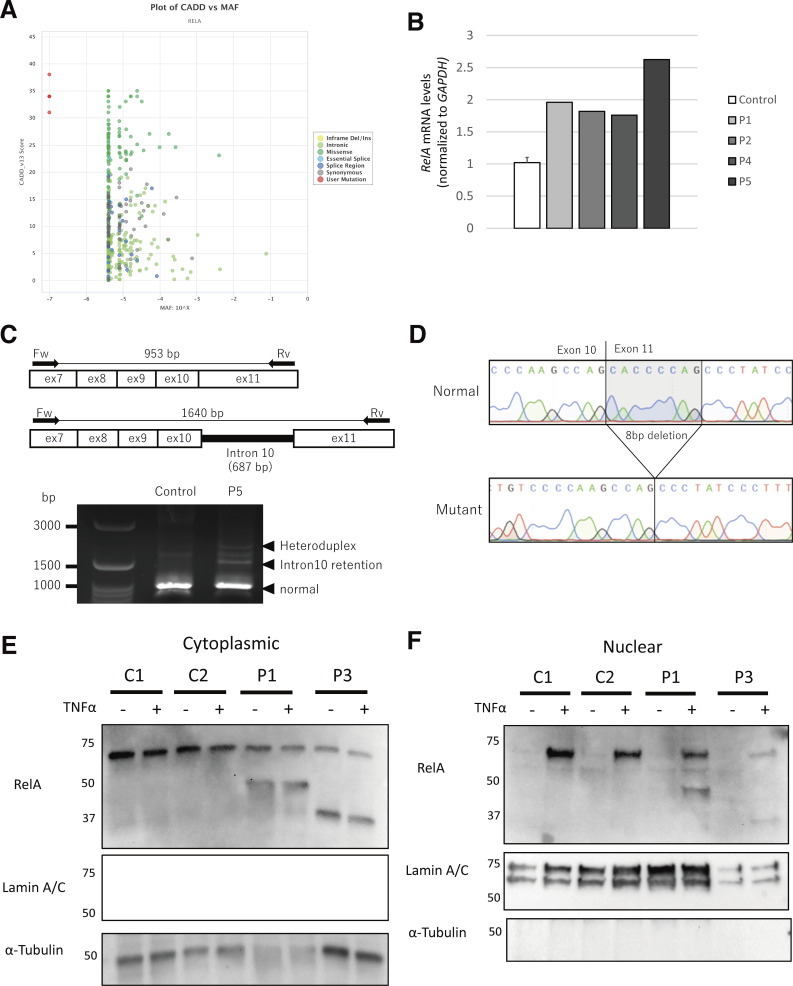
We identified six patients from five families with unreported heterozygous variants in RELA by multipanel genetic testing of primary immunodeficiency (patient 1 [P1] and P2; Table S4), a gene panel including nearly 400 primary immunodeficiency-related genes listed in IUIS 2019 (Tangye et al., 2021; P3/4/5), or whole-exome sequencing (P6; Fig. 1 A, Fig. S1 A, and Table S5). The list of candidate genes for patients in this study is shown in Table S6. All RELA mutations were confirmed by Sanger sequencing. P1 had a heterozygous nonsense mutation (NM_021975.4: c.1165C>T, p.Q389*) inherited from his mother (P2). P3 had a de novo heterozygous nonsense mutation (c.985C>T, p.R329*). P4 had a heterozygous frameshift mutation (c.1416dup, p.E473Rfs*18) inherited from her father, who was asymptomatic. P5 had a heterozygous essential splicing site mutation (c.1034-1G>A), the origin of which was undetermined due to the unavailability of parents’ and relatives’ DNA (Fig. 1 B). P6 had a heterozygous mutation (c.1047T>A, p.Y349*). In all of the patients tested (P1/2/4/5), RELA mRNA levels were normal or elevated compared with those in healthy controls, as shown by real-time quantitative PCR (RT-qPCR; Fig. S1 B). The expression of the mutant allele at the mRNA level was confirmed by cDNA sequencing in all of the patients except for P6, whose peripheral blood mononuclear cell (PBMCs) were not available. To determine the effect of the c.1034-1G>A mutation, we extracted total RNA from a blood sample and performed RT-PCR and TA cloning. Sanger sequencing of the cloned products showed two abnormal transcription products. One variant (P5-1) was a retention of intron 10 with a consequent stop gain at position 378, and the other (P5-2) was an 8 bp deletion (c.1035_1042del, p.P346Lfs*13; Fig. S1, C and D). We considered nonsense-mediated RNA decay unlikely because these mutations were located in the last exon or near the last exon junction. These mutations were not reported in the gnomAD v2.1.1 database (https://gnomad.broadinstitute.org/).
Figure 1.
AD RELA deficiency in five families. (A) Pedigree of the five unrelated families showing familial segregation of the different RELA alleles. Generations are indicated by Roman numerals (I–II), and each individual is indicated by an Arabic numeral (1–12). Male and female individuals are represented by squares and circles, respectively. Affected patients are represented by closed black symbols, and asymptomatic carriers are indicated by a black vertical line. Individuals of unknown genotype are indicated by “E?” (B) Schematic representation of the RELA gene. Coding exons are numbered (1–11). The positions of the variants observed in the patients are indicated by arrows. Novel mutations in this study are shown in bold. DN mutations are underlined.
Figure S1.
Analysis of the RELA variants found in the patients. (A) A dot plot was created with PopViz-2 (Zhang et al., 2018). MAF: minor allele frequency. c.1416dupC is not shown because the CADD score was not available. (B) Total mRNA extracted from the whole blood of three controls and patients (P1/2/4/5) was subjected to RT-qPCR for the assessment of total RELA expression. Data are displayed as 2−ΔCt after normalization according to endogenous β-actin control gene expression. Mean ± SEM. N = 3. (C) RT-PCR products of control and P5 samples were separated by 3.0% agarose gel electrophoresis. We sequenced the three bands indicated by arrowheads. The normal size band in the P5 lane contained two sequences, and the extra band at ∼1,500 bp was retained by intron 10. The largest band was considered a heteroduplex. The PCR primers were 5′-CCTACTGTGTGACAAGGTGCAGA-3′ and 5′-CTCCTGAAAGGAGGCCATTG-3′. (D) The mutant sequence contained in the normal size band. Sequencing was performed by using the pCR2.1-TOPO vector (Thermo Fisher Scientific) with the purified band introduced. (E and F) Immunoblot analysis of the cytoplasmic (E) and nuclear (F) fractions primary fibroblasts stimulated with TNF for 30 min, including primary fibroblasts from two healthy controls (C1 and C2). P1 and P3 were subjected to immunoblot analysis with an anti-RELA antibody following induction with TNF for 30 min. Tubulin and lamin A/C were used as loading controls for the cytoplasmic and nuclear fractions, respectively. The results shown are representative of three independent experiments. Molecular weight units are given in kilodaltons. Source data are available for this figure: SourceData FS1.
RELA variants result in the expression of truncated RelA proteins
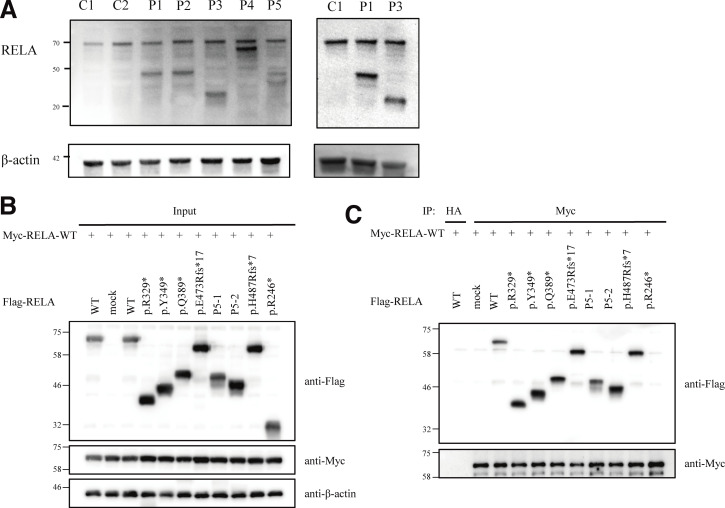
We examined the molecular and functional consequences of RELA variants in the patients identified. We established patient-derived B lymphoblastoid cell lines and primary fibroblasts derived from skin biopsy samples from P1 and P3. We detected WT RelA protein in cells from controls, and truncated RelA proteins in all of the patient-derived cells, using an anti–N-terminal RelA antibody (Fig. 2 A). Truncated proteins of 43.6, 37.3, 53.5, and 40 kD were detected in samples from P1, P2, P3, P4, and P6, respectively. The sample from P5 expressed two splicing variants at ∼40.5 and 42.5 kD. Besides, both WT and truncated RelA protein showed nuclear translocation in fibroblasts stimulated with TNF (Fig. S1, E and F). Next, we knocked out the RELA gene in HEK293 cells using the CRISPR/Cas9 system and transfected these cells with a pCMV4 plasmid with cDNA corresponding to the WT or mutant RELA variants. All variants, as well as a p.R246* variant that causes RELA haploinsufficiency (Comrie et al., 2018) and a previously reported uncharacterized variant (p.H487Tfs*7; Adeeb et al., 2021), expressed truncated RelA proteins with estimated molecular weights similar to those seen in the patients’ samples (Fig. 2 B).
Figure 2.
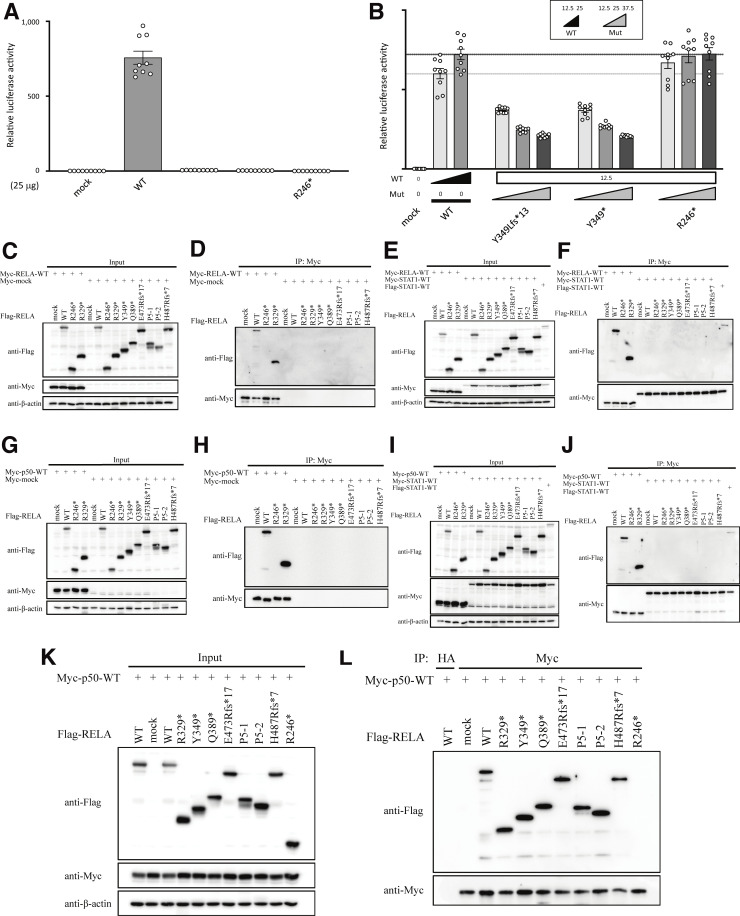
Effect of the pathogenic RELA variants on the expression and dimerization of RelA protein. (A) Immunoblot analysis of RelA protein levels in lymphoblastoid cell line and primary fibroblasts from controls and patients with AD RELA deficiency. The results shown are representative of three independent experiments. Molecular weight units were given in kilodaltons. C1 and C2 mean healthy controls 1 and 2. (B) Immunoblot analysis of RelA protein levels in total protein extracts of HEK293 cells transfected with anti–c-Myc, anti-Flag, or anti–β-actin antibody. (C) Whole-cell lysate immunoprecipitated with anti-HA antibody was used as an experimental negative control. Whole cell lysates were immunoprecipitated with anti–c-Myc antibody and then immunoblotted with anti-Flag or anti-Myc antibody. The p.R246* mutant, which causes RELA haploinsufficiency, impaired binding to WT RelA, whereas the other mutants exhibited normal binding to WT RelA. Two independent experiments were performed to confirm the results. Source data are available for this figure: SourceData F2.
Truncated RelA mutants do not have transcriptional activity
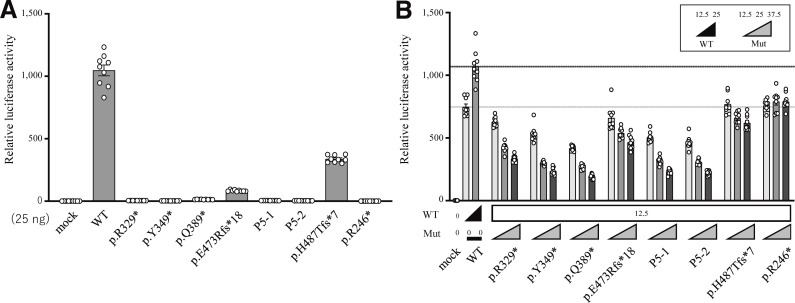
RelA protein consists of a REL homology domain at the N-terminal region and transcriptional activating domains at the C-terminal region. Therefore, we hypothesized that the expressed truncated proteins do not have a proper transcriptional activity. To this end, we established a dual-luciferase reporter assay to assess the ability of each mutant to activate NF-κB–dependent gene expression. Specifically, we cotransfected RELA KO HEK293 cells with plasmids encoding either WT or mutant RelA proteins or EV (empty vector), a plasmid encoding firefly luciferase under the control of five NF-κB responsive elements, and a plasmid encoding renila luciferase as an internal control. The assay revealed that four RELA variants (p.R329*, p.Y349*, p.Q389*, and the two mutant isoforms [P5-1 and P5-2] produced due to c.1034-1G>A), as well as p.R246* and p.Y349Lfs*13, are complete loss-of-function (LOF; Fig. 3 A and Fig. S2 A). On the other hand, P4’s variant (p.E473Rfs*18) was hypomorphic, similar to a known hypomorphic variant p.H487Tfs*7. Overall, all five variants identified in the current study were LOF or hypomorphic and, therefore, likely pathogenic.
Figure 3.
NF-κB reporter assay. (A) 25 ng plasmids encoding WT or mutant RelA protein was used. The p.E473Rfs*18 and p.H487Tfs*7 RELA variants were hypomorphic, having ∼8.0 and 32.3% residual activity, respectively, whereas the other five mutants, including p.R246*, were classified as LOF. (B) The total amount of expression vector, containing WT (12.5 or 25 ng) and mutant (12.5, 25, or 37.5 ng) RELA, was adjusted to 50 ng by supplementation with EV. All of the mutants, except for p.R246* mutants, which cause RELA haploinsufficiency, showed a dose-dependent negative effect against WT RelA. The mean ± SEM of three independent experiments is shown.
Figure S2.
Functional characterization of the RelA mutants in an overexpression system. (A and B) NF-κB reporter assay. 25 ng of WT or mutant RELA was used. The p.Y349Lfs* RELA was considered LOF. (B) The total amount of expression vector containing WT (12.5 or 25 ng) and mutant (12.5, 25, or 37.5 ng) RELA was adjusted to 50 ng by supplementation with EV. The p.Y349Lfs* RELA, as well as the p.Y349* mutant, showed dose-dependent negative effects against WT RELA. (C–J) Optimization of co-immunoprecipitation conditions. (C, E, G, and I) Immunoblot analysis to assess expression levels of Flag-RELA and Myc-tagged protein (RELA, p50, STAT1 or mock) in total protein extracts of HEK293 cell transfectants. (D, F, H, and J) Whole-cell lysates were immunoprecipitated with anti–c-Myc antibody and then immunoblotted with anti-Flag, anti-Myc, or anti–β-actin antibody. WT and mutant Flag-RELA did not bind to Myc-mock (D and H) or Myc-STAT1 (F and J). Under the same conditions, the p.R246* mutant which caused RELA haploinsufficiency impaired binding to WT RELA, whereas WT and the p.R329* mutant presented normal binding to WT RELA (D and F). Similarly, the p.R246* mutant impaired binding to WT p50, whereas WT and the p.R329* mutant presented normal binding to WT p50 (H and J). To confirm the result, two independent experiments were performed. (K and L) Dimerization between RELA and p50. (K) Immunoblot analysis to assess the expression levels of Flag-RELA and Myc-p50 protein in total protein extracts of HEK293 cells transfectants. (L) Extracts immunoprecipitated with anti-HA antibody are shown as experimental negative controls. Whole-cell lysates were co-immunoprecipitated with anti–c-Myc antibody and immunoblotted with anti-Flag, anti-Myc, or anti–β-actin antibody. The p.R246* mutant, which causes RELA haploinsufficiency, exhibited impaired binding to WT p50, whereas other mutants bound to WT p50. Two independent experiments were performed to confirm the result. Source data are available for this figure: SourceData FS2.
Truncated RelA mutants are dominant-negative (DN)
To further characterize the functional significance of the RELA mutations, we next asked whether these RelA mutants exert DN effects on the WT protein. Using the NF-κB reporter assay, we assessed the impact of increasing the concentrations of the plasmids encoding various RelA mutants while maintaining the concentration of the plasmid encoding the WT RelA protein. Intriguingly, all five mutants, as well as the previously reported but uncharacterized mutants p.H487Tfs*7 and p.Y349Lfs*13 (Adeeb et al., 2021; Lecerf et al., 2022), exerted a DN effect in a dose-dependent manner compared with the p.R246* mutant that was used as a control for RELA haploinsufficiency (Fig. 3 B and Fig. S2 B; Comrie et al., 2018). To clarify the molecular mechanism of RELA DN mutations, we next assessed the formation of RelA-RelA homodimer (Fig. 2, B and C; and Fig. S2, C–F) and RelA-p50 heterodimer (Fig. S2, G–L). Immunoprecipitation assay showed that DN RelA mutants form dimers with WT RelA protein. On the other hand, p.R246* RelA mutant, which causes haploinsufficiency, did not form a dimer with WT RelA protein. Additionally, we performed pull-down assays with appropriate controls using Myc-empty (Myc-EV) or Myc-STAT1 vectors as bait (Fig. S2, D, F, H, and J). Neither WT nor mutant Flag-RelA bound to Myc-STAT1 and Myc-EV. This result suggests that the pull-down assay actually depends on Myc-p50 or Myc-RelA and that R246* RelA cannot bind to WT RelA or WT p50. Overall, the patients described in this study have RELA DN mutations, which likely underlie clinical manifestations distinct from patients with RELA haploinsufficiency.
Patients’ leukocytes display an enhanced type I IFN signature
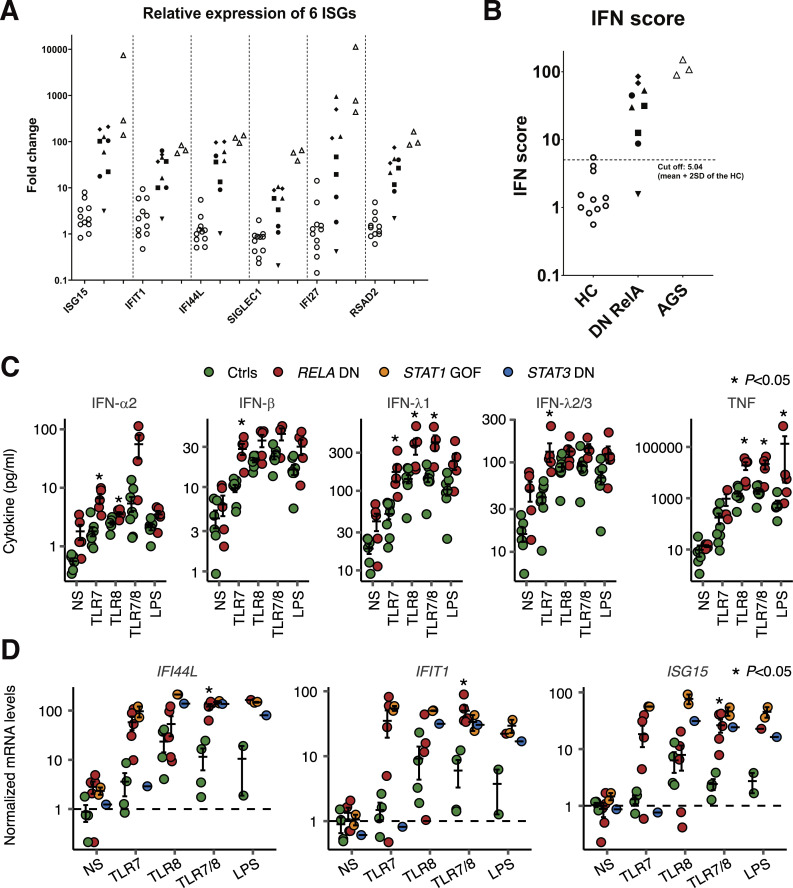
We unexpectedly found that the whole-blood samples of patients with RELA DN mutations contained elevated mRNA levels of six IFN-stimulated genes (ISGs) compared to healthy controls, similar to the patients with genetically confirmed Aicardi-Goutières syndrome type 1 (AGS1; Fig. 4, A and B). We then measured the IFN-α2 concentration in the serum by ELISA. The sera from P2, P5, and one healthy control showed detectable levels of IFN-α2 (minimum detectable concentration: <4 pg/ml), whereas the sera from P1/3/4 and four healthy controls showed undetectable levels of IFN-α2 (Fig. S3 A). On the other hand, all of the patients’ serum samples, but none of the five healthy controls, contained detectable levels of TNF (the limit of quantification: <6.23 pg/ml; Fig. S3 B). We reasoned that the instability of type I IFNs in the peripheral blood of the patients hindered their robust detection, and we therefore hypothesized that the patients’ leukocytes produce excessive amounts of IFNs. To test this hypothesis, we cultured cryopreserved PBMCs from P2/3/5 and healthy controls for 24 h with or without TLR7, TLR8, TLR7/8 agonists, or LPS. We measured secreted cytokines and mRNA in total leukocytes via multiplex ELISA and RT-qPCR, respectively. We found that the patients’ leukocytes secreted significantly high levels of IFN-α2, IFN-β, IFN-λ1, and IFN-λ2/3 in response to a TLR7 agonist (Fig. 4 C). We also observed significantly high levels of cytokine secretion by other TLR agonists (IFN-α2, IFN-λ1, and TNF by a TLR8 agonist; IFN-λ1 and TNF by a TLR7/8 agonist; and TNF by LPS) (Fig. 4 C). Moreover, the patients’ leukocytes displayed excessive induction of IFI44L, IFIT1, and ISG15 when stimulated with a TLR7 agonist, TLR7/8 agonist, or LPS (Fig. 4 D), similar to patients with STAT1 GOF or STAT3 DN mutations as a disease control with an enhanced type I IFN signature (Goel et al., 2021; Kaleviste et al., 2019; Scott et al., 2021). In contrast, TLR8 agonist-induced upregulation of ISGs was largely normal (Fig. 4 D). Thus, leukocytes with RELA DN mutations display type I IFN signatures in vivo, probably because of TLR7-dependent excessive type I/III IFN production.
Figure 4.
RELA DN mutations underlie type I interferonopathy. (A and B) Relative expression of (A) six ISGs and (B) IFN scores of 11 healthy controls, five patients with DN RELA mutations, and three patients with AGS were evaluated by RT-qPCR. The relative abundance of each transcript was normalized to the expression level of β-actin, and the results are shown relative to a single calibrator. The experiment was performed in triplicate. The median of the relative quantification of the six ISGs was used to calculate the IFN score for each patient. IFN scores greater than +2 SD of the average of 11 healthy controls (5.04) were designated as positive. For each DN RELA patient except P3, multiple blood draws were taken at regular follow-up appointments in the absence of obvious signs of infection or fever. For P3, only one sample, taken just before bone marrow transplant (under conditioning), was available. For disease controls, data from three AGS patients (two genetically diagnosed: IFIH1 [p.R779H/WT] and IFIH1 [p.R720Q/WT], and one clinically diagnosed) was used. (C and D) PBMC stimulation assay. PBMCs of P2/3/5, patients with STAT1 GOF or STAT3 DN mutations, and healthy donors were stimulated with TLR7, TLR8, TLR7/8 agonists, or LPS for 24 h. (C) Secreted cytokines as measured by a LEGENDplex assay. NS, not stimulated. (D) Gene expression as determined by qPCR. GUSB was used as an internal control. In C and D, results from two experiments were compiled. Statistical significance of the difference between RELA DN patients and healthy controls was determined by two-tailed Wilcoxon’s rank sum test with FDR adjustment. Two independent experiments were performed to confirm the results. Bars represent the mean and SEM.
Figure S3.
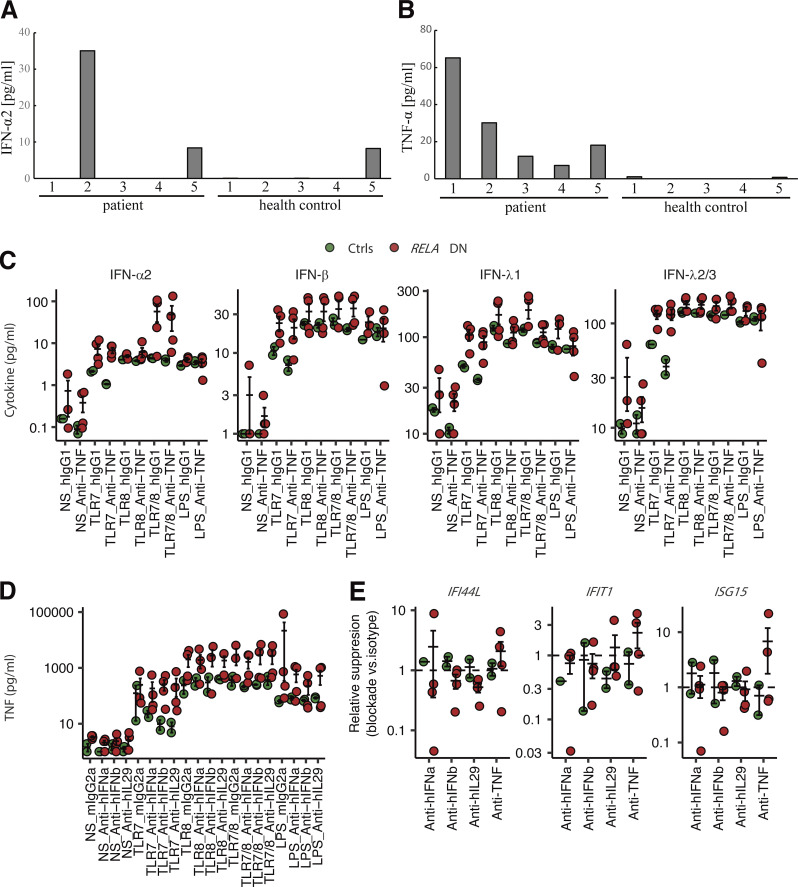
Enhanced expression of type I and III IFNs and ISGs. (A and B) The concentration of (A) IFN-α2 and (B) TNF in the sera of five patients and five healthy controls. With the exception of P1, P5, and healthy control #5, the IFN-α2 concentration was below the limit of quantification (<4 pg/ml). (C–E) PBMC stimulation assay with neutralizing antibodies. PBMCs from P2/3/5 (biological duplicates for P3) and healthy donors were stimulated with TLR7, TLR8, TLR7/8 agonists, or LPS for 24 h, together with monoclonal antibodies neutralizing TNF, IFN-α, IFN-β, or IL-29, or corresponding isotype control antibodies. (C and D) Secreted cytokines were measured by a LEGENDplex assay. (E) ISG mRNA levels were determined by qPCR. GUSB was used as an internal control. Relative suppression was defined as RQT, Neut × RQNS, Isotype/RQNS, Neut × RQT, Isotype, where RQ is a GUSB-normalized relative expression level, T is a TLR7/8 agonist, NS is a nonstimulated condition, Neut denotes a neutralizing antibody, and Isotype a corresponding isotype control antibody. Values below 1 indicate suppression of a given ISG by addition of a given neutralizing antibody. In C–E, bars represent the mean and SEM.
IFN blockade partially inhibits TLR-dependent hyperinduction of ISGs in vitro
To mechanistically characterize relationships between type I/III IFNs, TNF, and ISGs, we next investigated the impact of recombinant neutralizing antibodies against these cytokines in the leukocytes of the patients (P2/3/5; biological duplicates for P3) and healthy controls. We noted that TNF blockade (variable region identical to adalimumab) slightly impaired production of type I/III IFNs triggered by a TLR7 agonist in healthy controls (Fig. S3 C). However, the patients’ cells produced excessive type I/III IFNs regardless of TNF blockade (Fig. S3 C). Similarly, we did not observe any impact of blockade of IFN-α2, IFN-β, and IFN-λ1 on the production of TNF (Fig. S3 D). We next analyzed whether IFN/TNF blockade reduces induction of ISGs. We focused on the TLR7/8 agonist, which induced significantly higher levels of IFI44L, IFIT1, and ISG15 in the patients’ leukocytes (Fig. 4 D). Neutralizing mAbs against IFN-β and IFN-λ1 suppressed induction of IFI44L in the patients compared to the isotype control mAb, but cells from healthy donors were not affected by these mAbs (Fig. S3 E). Moreover, induction of IFIT1 was also inhibited by anti–IFN-β/λ1 mAbs in P3 and P5 but not in P2. Notably, IFIT1 induction was completely abolished by anti–IFN-α2 mAb in P2, indicating a heterogeneous dependence on different IFNs among patients. In contrast, TNF blockade did not inhibit induction of ISGs under the conditions tested. Thus, excessive production of type I/III IFNs in response to TLR agonists is responsible for excessive expression of ISGs in leukocytes of patients with RELA DN mutations.
Normal response to recombinant type I IFNs in T lymphocytes and monocytes in vitro
We next sought to determine whether cells with RELA DN mutations are hypersensitive to IFNs with respect to ISG expression. To this end, we first stimulated T cell blasts of P1/2/3/4/5 and controls with recombinant IFN-α2 or IFN-γ. Contrary to our expectation, however, IFN-α2 stimulation induced normal levels of ISGs, such as IFI6, ISG15, and RSAD2 (Fig. 5 A). IFN-γ did not induce any ISGs tested in T cell blasts (Fig. 5 A). Next, we assessed the responses of sorted monocytes of P1/2/4/5 to recombinant IFN-α2, IFN-γ, or LPS. Again, monocytes with RELA DN mutations showed IFN-γ–driven secretion of IFN-λ1 at a level comparable to that of cells from healthy controls (Fig. 5 B). Notably, monocytes with RELA DN mutations secreted significantly less CXCL10 (also known as IFN-γ–inducible protein 10) in response to IFN-γ than cells from controls, indicating a repressed, rather than augmented, response to type II IFNs (Fig. 5 B). We further dissected the cellular response of monocytes with RELA DN mutations to IFN-α2, IFN-γ, and LPS through RNA sequencing (RNASeq). Principal component analysis did not reveal any global difference in monocytes with or without RELA DN mutations (Fig. 5 C). Differential expression (DE) analysis comparing monocytes from controls and RELA DN patients identified only 121 and 59 genes among 12,275 genes tested that were differentially induced or repressed by IFN-α2 or IFN-γ, respectively (Fig. 5 D), suggesting that RELA DN mutations do not affect >99% of the genes expressed in response to type I or type II IFNs in monocytes. Consistently, gene set enrichment analysis (GSEA) of the fold-change-based ranks of DE genes in RELA DN patients compared to controls in response to IFN-α2, IFN-γ, or LPS did not identify any hallmark gene sets that were significantly enriched or depleted. Thus, RELA DN mutations do not augment cellular responses to type I IFNs in either T lymphocytes or monocytes.
Figure 5.
Cellular responses to type I and II IFNs. (A) T cell blast stimulation assay. T cells were expanded for ∼13 d and restimulated with the indicated reagents for 4 h. Gene expression levels were determined by qPCR. GUSB was used as an internal control. (B–D) Monocyte stimulation assay. CD14+ monocytes were sorted, rested overnight, and then stimulated with the indicated reagents for 4 h. (B) Secreted cytokines as measured by a LEGENDplex assay. Statistical significance of the difference between RELA DN patients and healthy controls was determined by two-tailed Wilcoxon’s rank sum test with FDR adjustment. (C and D) RNA sequencing. (C) Global transcriptional profiles were summarized via uniform manifold approximation and projection (UMAP). (D) Differential expression analysis. Genes with log2 fold-changes below −1 or above 1, together with FDR-adjusted P values below 0.05, are colored. In A and B, bars represent the mean and SEM.
Enhanced type I IFN signature across lymphoid and myeloid leukocyte subsets
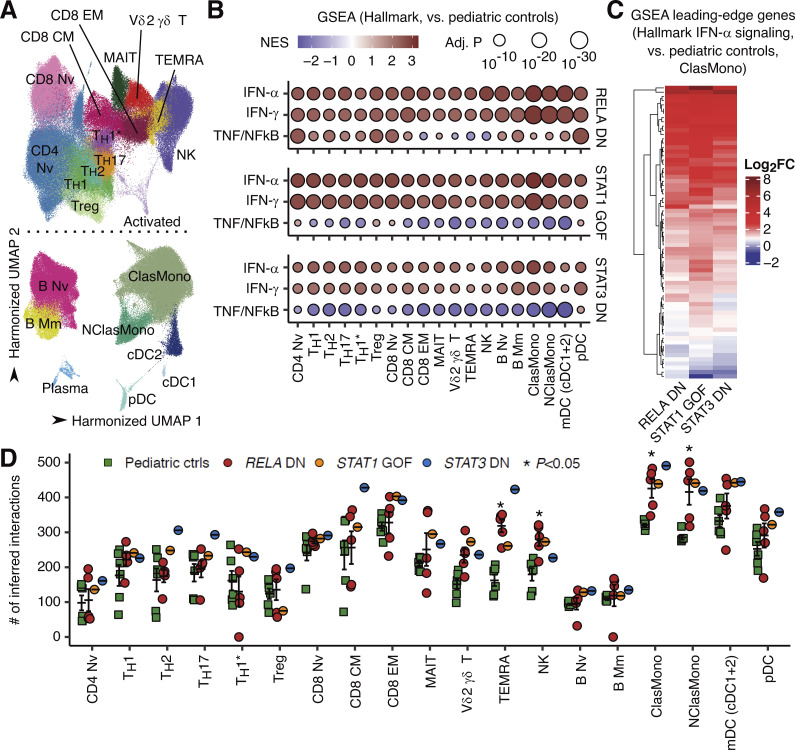
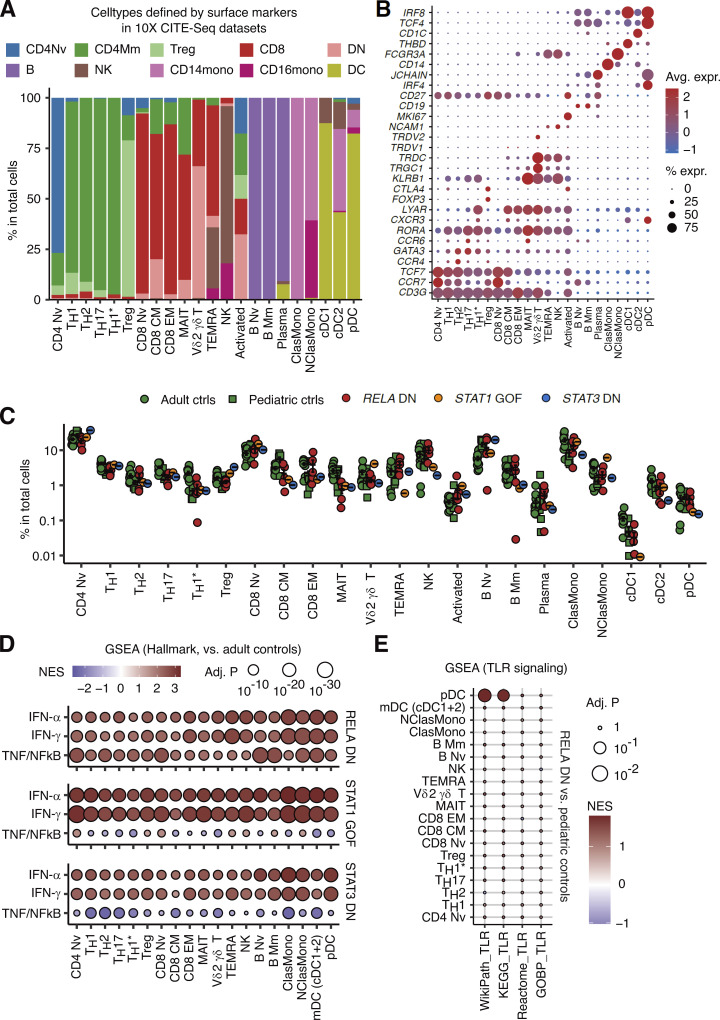
To delineate the cellular and molecular basis of the observed type I IFN signatures and excessive production of type I/III IFNs, we analyzed PBMCs of P1/2/3/4/5, together with patients with STAT1 GOF and STAT3 DN mutations (N = 1 each), through single-cell RNASeq (scRNASeq). Batch-corrected clustering analysis identified 22 distinct leukocyte subsets (Fig. 6 A and Fig. S4, A and B). The development of lymphoid and myeloid leukocyte subsets in the patients was not affected (Fig. S4 C). Pseudobulk DE analysis revealed massive upregulation of ISGs across a broad range of leukocyte subsets in patients with RELA DN, STAT1 GOF, and STAT3 DN mutations compared to healthy controls (both adults and children; Fig. 6, B and C; and Fig. S4 D). Furthermore, intercellular communication analysis using CellChat (Jin et al., 2021) inferred significantly higher numbers of ligand‒receptor interactions in cytotoxic lymphocytes (terminally differentiated effector T and natural killer lymphocytes) and classical and nonclassical monocytes of patients with RELA DN, STAT1 GOF, and STAT3 DN mutations compared to healthy pediatric controls, again implicating physiological similarity among these monogenic conditions (Fig. 6 D). Thus, a broad range of lymphoid and myeloid cells with RELA DN mutations exhibits a type I IFN signature, similar to classical type I interferonopathy triggered by STAT1 GOF or STAT3 DN mutations.
Figure 6.
Type I IFN signatures across lymphoid and myeloid leukocyte subsets. scRNASeq analysis was performed on cryopreserved PBMCs from P1/2/3/4/5, patients with STAT1 GOF (N = 2), or STAT3 DN (N = 1) mutations, and healthy adult and pediatric controls. (A) Unsupervised clustering. Graph-based clustering was performed after batch correction with Harmony, and clusters were manually identified with the aid of the SingleR pipeline informed by the MonacoImmuneDataset. (B) GSEA. Genes were ranked based on the fold-change estimated through pseudobulk differential expression analysis between patients and healthy pediatric controls. Gene ranking was projected against the Hallmark gene sets (http://www.gsea-msigdb.org/gsea/msigdb/genesets.jsp?collection=H). Only gene sets with FDR-adjusted P values below 10−20 in at least one cell type are shown. NES, normalized enrichment score. (C) Heatmap showing the log2 fold-change values between patients and healthy pediatric controls for the GSEA leading-edge genes for the hallmark IFN-α signaling gene set in classical monocytes. (D) Intercellular communication analysis with CellChat. Statistical significance for the difference between RELA DN patients and healthy children was determined through two-tailed Wilcoxon’s rank sum test with FDR adjustment. Bars represent the mean and SEM.
Figure S4.
Single-cell transcriptomic analysis of leukocyte subsets. (A and B) Cluster annotations. (A) Fraction of cell subsets defined based on the combination of canonical surface markers in 10X public CITE-Seq datasets superimposed onto each manually annotated cluster in our in-house dataset. Treg, regulatory T cell; CM, central memory T cell; EM, effector memory T cell; TEMRA, effector memory-expressing CD45RA T cell; MAIT, mucosal-associated invariant T cell, B Nv, naïve B cell; B Mm, memory B cell; ClasMono, classical monocyte; NClasMono, non-classical monocyte; cDC, classical dendritic cell. (B) Expression levels of select marker genes in each manually annotated cluster. (C) Relative abundance of cell types defined through clustering. Bars represent the mean and SEM. (D) GSEA for hallmark gene sets compared to healthy adult controls, as described in Fig. 6 B. (E) GSEA for TLR-related gene sets taken from the MsigDB database. Statistically nonsignificant (FDR-adjusted P values of 0.05 or higher) pathway–cell type pairs are shown by a small dot. NES, normalized enrichment score.
Enhanced expression of molecular components involved in TLR7 signaling
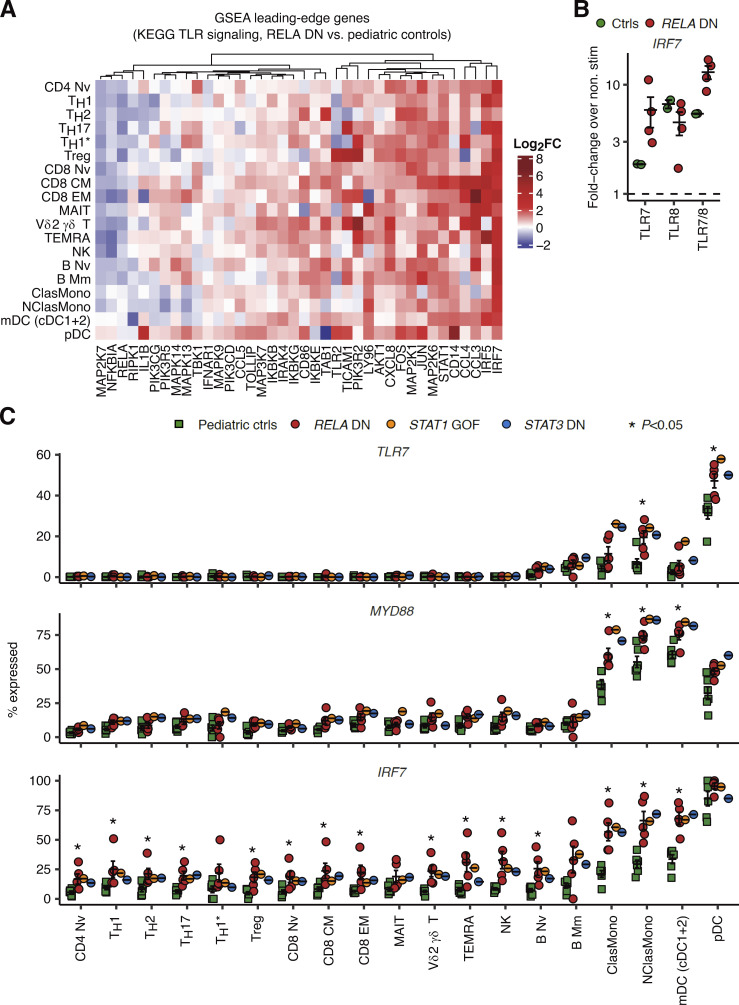
We explored scRNASeq data to investigate the molecular pathogenesis underlying TLR7-dependent hyperproduction of type I/III IFNs by leukocytes with RELA DN mutations. First, GSEA based on pseudobulk DE analysis revealed significant upregulation of genes related to TLR signaling only in plasmacytoid dendritic cells (pDCs), a specialized myeloid cell subset producing a large amount of type I IFNs in response to TLR7 agonists (Asano et al., 2021a; Asano et al., 2021b), in patients with RELA DN mutations compared to healthy children (Fig. S4 E). The leading-edge genes upregulated in the patients include genes encoding TLR7 signal mediators such as IRF7, IRAK4, and IKBKB (Kawai and Akira, 2007). Surprisingly, we found that most of the leading-edge genes for pDCs, most notably IRF7, were actually upregulated in various lymphoid and myeloid leukocyte subsets in RELA DN patients (Fig. 7 A). Consistent with the observed elevated expression of IRF7 mRNA at baseline, we found that TLR7 and TLR7/8 agonists, but not a TLR8 agonist, induced higher expression of IRF7 mRNA in the patients’ leukocytes (Fig. 7 B). IRF7 is critically required for type I IFN production by pDCs in mice (Honda et al., 2005). These observations led us to hypothesize that DN RelA induces transcription of genes encoding components involved in TLR7 signaling. Indeed, reanalysis of the scRNASeq data revealed significantly enhanced expression of TLR7 and MYD88 in classical and nonclassical monocytes, myeloid DCs (mDCs), and pDCs, as well as IRF7 in a broad range of lymphoid and myeloid leukocyte subsets, in RELA DN patients compared to healthy children (Fig. 7 C). Thus, RELA DN mutations underlie type I interferonopathy by unleashing TLR7-dependent induction of type I/III IFNs, probably in both pDCs and non-pDC myeloid cells.
Figure 7.
Enhanced expression of molecular components involved in TLR7 signaling. (A) Heatmap showing log2 fold-change values between RELA DN patients and healthy pediatric controls for pseudobulk GSEA leading-edge genes for the Kyoto Encyclopedia of Genes and Genomes (KEGG) TLR signaling gene set in pDCs. (B) Fold-change induction over nonstimulated conditions of IRF7 mRNA in PBMCs from RELA DN patients (P2/3/5; biological duplicates for P3) and healthy controls stimulated with TLR agonists for 24 h. GUSB was used as an internal control. (C) Fraction of cells expressing mRNA for genes involved in TLR7 signaling. Statistical significance for the difference between RELA DN patients and healthy children was determined through a two-tailed Wilcoxon’s rank sum test with FDR adjustment. Bars represent the mean and SEM.
Discussion
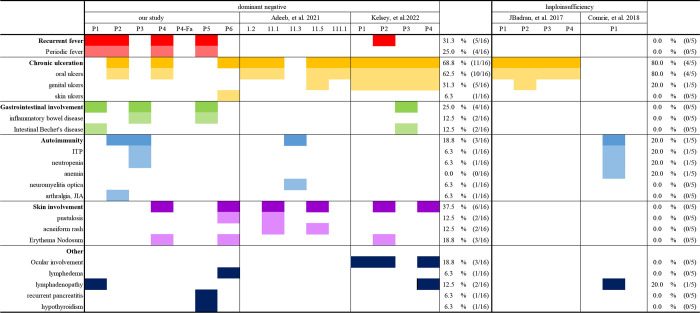
Here, we report a novel form of human AD RELA deficiency, with DN mutations in six patients from five unrelated families. Previously, 14 patients from four families with AD RELA haploinsufficiency have been described (Badran et al., 2017; Comrie et al., 2018; Adeeb et al., 2021; Lecerf et al., 2022). The first study reported four cases from a two-generation family with a heterozygous mutation in RELA (c.599 + 1G>A) that caused RELA haploinsufficiency (Badran et al., 2017). The patients presented chronic mucocutaneous ulcers in their oral and genital regions with onset in their first years of life. One of the patients suffered from relatively severe symptoms associated with recurrent acute ileitis. The second report of RELA haploinsufficiency described a patient with autoimmune hematological disorders (neutropenia and thrombocytopenia), splenomegaly, and mild lymphadenopathy associated with RELA heterozygosity (c.736C>T, p.R246*; Comrie et al., 2018). In contrast, five cases from a three-generation family harbored a monoallelic single nucleotide deletion, c.1459delC (p.H487Tfs*7), and four cases presented with p.Y349Lfs*13 mutation in the 3′ portion of RELA, which was found to be a DN mutation in the present study (Adeeb et al., 2021; Lecerf et al., 2022). Here, we stratified each case into a phenotypic category on the basis of DN or haploinsufficiency (Table 1). The patients with RELA DN mutations shared clinical phenotypes with RELA haploinsufficiency, presenting chronic mucocutaneous ulcerations and autoimmune hematological disorders such as immune thrombocytopenia (ITP) and neutropenia. However, patients with RELA DN mutations additionally presented periodic fever, inflammatory bowel diseases (IBDs), juvenile idiopathic arthritis (JIA), and skin involvement. Complete penetrance was observed for IBD. Moreover, one patient needed hematopoietic stem cell transplantation (HSCT) to treat refractory thrombocytopenia. These clinical observations suggest that patients with RELA DN mutations apparently have severer phenotypes and worse clinical outcomes than those with RELA haploinsufficiency. The comprehensive characterization of the mechanisms of negative dominance in AD IEI conditions improves both our understanding of the pathogenesis and its clinical management (Asano et al., 2021a; Asano et al., 2021b; Béziat et al., 2020).
Table 1.
The comparison of clinical manifestations between RELA DN and RELA haploinsufficiency
The RELA mutations newly identified in the current study accumulated in the 3′ portion of the RELA gene and are expected to escape the RNA decay system by creating a premature stop codon within the last exon or ∼55 bp of the penultimate exon of the RELA gene. As expected, stable expression of mutant allele-derived mRNA and the corresponding truncated protein products were experimentally confirmed in the patient samples. The establishment of RELA KO HEK293 cells enabled us to perform a detailed characterization of the identified variants. The NF-κB reporter assay revealed that four RELA mutations (p.R329*, p.Q389*, p.Y349*, and two mutant isoforms [P5-1 and P5-2] due to c.1034-1G>A), as well as p.R246* and p.Y349Lfs*13 mutations, are complete LOF. On the other hand, p.E473Rfs*18 and p.H487Tfs*7 mutations were hypomorphic, presenting some residual activities. The five RELA mutations identified in the current study, together with the p.Y349Lfs*13 and p.H487Tfs*7 mutations reported in previous studies (Adeeb et al., 2021; Lecerf et al., 2022) exerted a DN effect against the WT protein in a dose-dependent manner, whereas p.R246* RELA, a known LOF mutation that leads to RELA haploinsufficiency, did not affect WT protein-induced NF-κB upregulation. Curiously, hypomorphic RELA mutants (p.E473Rfs*18 and p.H487Tfs*7) also exerted a DN effect, but this effect was subtle compared with those in the other four LOF and DN mutations. This observation may explain the relatively mild symptoms in P4 and the presence of asymptomatic carriers (P4’s father) in Family C.
Aberrantly enhanced type I IFN signaling is a hallmark of type I interferonopathy, which manifests as various forms of autoimmune and autoinflammatory disease (Jeremiah et al., 2014; Kacar et al., 2019; Crow and Stetson, 2022). We detected elevated expression of multiple ISGs, known as type I IFN signature, in patients with RELA DN mutations. Consistently, our patients developed JIA, thyroiditis, and other symptoms that can be associated with type I interferonopathy (Tables S1 and S2). In the current study, we showed that (1) sera from patients with RELA DN mutations contained aberrantly elevated levels of TNF; (2) leukocytes with RELA DN mutations secreted excessive amounts of TNF in response to LPS, TLR8 agonist, and TLR7/8 agonist; (3) myeloid cells (classical and nonclassical monocytes, mDCs, and pDCs) with RELA DN mutations showed enhanced TLR7 and MYD88 mRNA expression; (4) leukocytes with RELA DN mutations secreted excessive amounts of type I/III IFNs and IRF7 mRNA in response to a TLR7 agonist; and (5) blockade of type I/III IFNs partially inhibited TLR7/8-driven upregulation of ISGs. On the other hand, we did not observe any enhanced response to type I or type II IFNs in patient T lymphocytes or monocytes. Although TLR7 is predominantly and constitutively expressed in pDCs, TNF is known to induce transcription of TLR7 mRNA in an NF-κB–dependent manner in cells other than pDCs (Lee et al., 2009). Similarly, transcription of IRF7 mRNA is governed by both IFN-driven ISG factor 3 and TNF-driven RelA-p50 heterodimers in human monocytes (Lu et al., 2002). TLR7 recognizes single-stranded RNA (typically of viral origin) and functions with MyD88 to activate the RelA-p50 heterodimer and induce phosphorylation and homodimerization of IRF7 (Kawai and Akira, 2007). Therefore, we postulate that excessive TNF produced in response to various environmental stimuli causes upregulation of TLR7 and IRF7 in myeloid leukocytes other than pDCs, such as monocytes and mDCs, thus driving excessive TLR7-dependent production of type I/III IFNs and induction of ISGs. Most patients with RELA haploinsufficiency are effectively treated with anti-TNF agents, as was the case for P1 in this report. Consistently, Rela+/− mice develop cutaneous ulceration when exposed to TNF and exhibit severe dextran sodium sulfate–induced colitis ameliorated by TNF inhibition (Badran et al., 2017). Clinical and experimental evidence suggests that excessive TNF-dependent immunopathology is a common pathogenic factor in patients with AD RELA deficiency. On the other hand, the apparent absence of type I interferonopathy-associated clinical phenotypes in patients with RELA haploinsufficiency suggests that the presence of DN RelA mutants is indispensable for induction of type I IFN signatures. One possibility is that prolonged binding of DN RelA mutants prevents condensation of the chromatin region harboring the IRF7 promoter, as topoisomerase inhibitors such as etoposide spontaneously induce transcription of IRF7 in a human B lymphoma cell line (Lu et al., 2002). Given the clinical and cellular features of type I interferonopathy in patients with RELA DN mutations, Janus kinase inhibitors are a promising option for cases refractory to anti-TNF or other therapy (Jamilloux et al., 2019). Overall, this report establishes a novel form of AD IEI caused by RELA DN mutations leading to chronic mucocutaneous ulcerations and other autoinflammatory and autoimmune manifestations. This study also mechanistically connect the hitherto separate groups of IEI of NF-κB pathway and type I interferonopathies.
Materials and methods
Case reports
P1 (Family A) is a Japanese male who suffered from periodic fever, cervical lymphadenopathy, and tonsillitis from the first month after birth (Tables S1 and S2). He began to complain of daily abdominal pain and refractory diarrhea at 6 yr of age. The patient had scoliosis of the thoracic vertebra. Mucosal inflammation in the entire colon and multiple annular ulcers were observed in the ascending colon during endoscopy. Pathological examination showed the infiltration of neutrophils and crypt abscesses. Only the anti-TNF antibody infliximab was effective in controlling his symptoms. His mother (P2) had recurrent aphthous stomatitis without detectable HSV, Candida, or other specific pathogens, as well as periodic fever and arthralgia beginning in childhood, and was diagnosed with JIA. She also exhibited the same heterozygous variant as previously described (Uchida et al., 2020).
P3 (Family B) is a Japanese male who has presented with perianal abscesses since the neonatal period (Tables S1 and S2). He developed ITP at the age of 2 yr and intractable diarrhea and neutropenia at the age of 4 yr (Table S3). Endoscopy showed a longitudinal ulcer and mucosal edema from the ileal valve to the terminal ileum without detectable HSV, Candida, or the other specific pathogens. He was given a diagnosis of chronic ITP with elevated PA-IgG levels, autoimmune neutropenia with positive antineutrophil antibodies, and IBD. Steroid pulse therapy, prednisolone, tacrolimus, rituximab, and other immunosuppressive therapies were administered, but the thrombocytopenia was refractory. At the age of 9 yr, after the genetic diagnosis was confirmed, HSCT with a reduced-intensity regimen (etoposide, 450 mg/m2; fludarabine, 180 mg/m2; melphalan, 140 mg/m2; total-body irradiation, 3 Gy; rabbit anti-thymocyte globulin, 5 mg/kg) was performed. Complete donor chimerism was achieved with mild intestinal graft versus host disease. Immunosuppressant therapy was discontinued, and the patient remained in complete remission for more than 1 yr after HSCT.
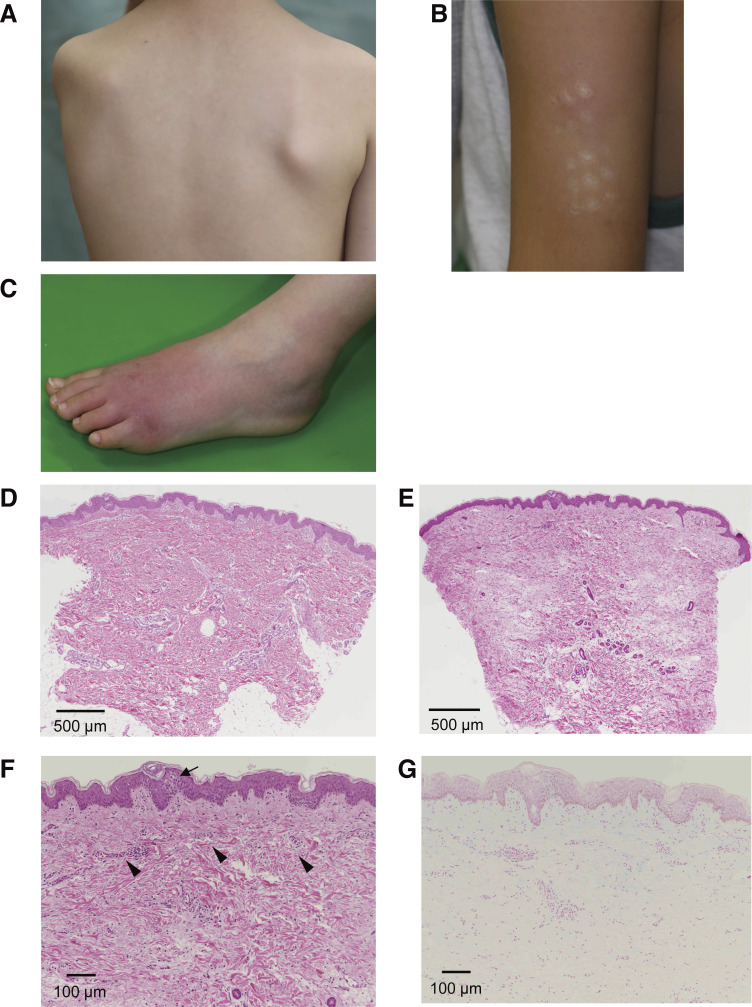
P4 (Family C) is a 10-yr-old Japanese male whose symptoms, such as periodic fever and painful subcutaneous nodules, began at the age of 2 yr (Tables S1 and S2). The patient exhibited no mental retardation, but he had short stature (−2.2 SD) and low body weight (−2.1 SD). His febrile attacks occurred once a month to once a year and lasted 1–3 d. In his early childhood, febrile episodes were always accompanied by painful subcutaneous nodules in his trunk and extremities (Fig. S5, A–C). The subcutaneous nodules were up to several centimeters in size and were accompanied by redness and swelling of the superficial skin. Skin biopsy revealed mild epidermal spongiosis and slight infiltration of mononuclear cells around the small vessels of the upper dermis (Fig. S5, D–G). In his late childhood, skin symptoms became less frequent, while he began complaining of generalized myalgia with a duration ranging from hours to a full day in almost every febrile episode. Stomatitis was occasionally present, but no other gastrointestinal symptoms were noted. Blood tests revealed mild elevation of acute phase reactants, elevated IgG (22.0 g/liter) and IgD (53.8 mg/dl) as well as IL-1β (42 pg/ml) and weakly positive antinuclear antibody. Oral cimetidine did not affect the frequency of febrile attacks.
Figure S5.
Skin manifestation and pathological findings at skin biopsy of P4. (A–C) Subcutaneous nodule on the trunk (A) and rash on the left upper arm (B) and left dorsum of the foot (C). (D–G) H&E staining (D–F) and Alcian blue staining (G) of skin biopsies. The pathological findings showed slight infiltration of mononuclear cells around the small vessels of the upper dermis of the trunk (D) and in the samples from the upper dermis of the left upper arm (E; scale bar is 500 μm; original magnification, 2×); higher magnification (F) clearly showed some spongiosis in the epidermis (arrow) and slight infiltration of mononuclear cells around the small vessels (arrowheads). Sparse spaces between collagen fibers observed in F were not stained with Alcian blue (G; scale bar is 100 μm; original magnification, 40×), which may be an artifact of specimen preparation.
P5 (Family D) is a Japanese male who suffered from periodic fever lasting from 1 to 3 d every 8 d since the age of 6 mo (Tables S1 and S2). Under the diagnosis of an autoinflammatory disease of unknown etiology, tocilizumab therapy was initiated, in addition to regular oral nonsteroidal anti-inflammatory drugs. He presented growth failure with no mental retardation. We also measured his height (−3.3 SD) and weight (−2.5 SD). The patient had mild aortic regurgitation, hypothyroidism, and recurrent pancreatitis without pancreatic ductal anomaly. No autoantibodies other than antinuclear antibodies were detected (Table S2). Endoscopy at 11 yr of age showed multiple erosive ulcers in the ascending, transverse, and descending colon and inflammation of the entire colon. He received a diagnosis of IBD and was switched from tocilizumab to anti-TNF antibody (adalimumab).
P6 (Family E) is a 21-yr-old male of Moroccan origin whose symptoms, such as repeated bacterial and mycotic infections, began at the age of 18 mo. There was no family history except his sister who presented atopy. He had a short stature (−3.0 SD) and delayed puberty (Tanner stages: stage 1 of pubic hair scale, stage 2 of male external genitalia scale) without mental retardation. In his early childhood, he presented with bullous lesions in the toes and trunk that then disseminated to the body. He also had conjunctivitis. Dermatological examination showed facial edema with macrochelitis, diffuse xerosis, fissured intertrigo of the neck, subcutaneous nodules of his body, and adherent crusts of the scalp. Skin symptoms became less frequent after topical corticosteroids, antibiotics and antimycotics, but symptoms reappeared each time and were responsible for frequent hospitalizations. The last patient presented with lymphedema of the right leg with irregular erythematous plaque and the beginning of desquamation associated with well-defined ulcerations. Blood tests revealed lymphopenia (0.80 × 109/liter), elevated IgG (22.1 g/liter), elevated IgE (2,950 IU/ml), and elevated inflammatory biomarkers, such as C-reactive protein. The other tests were normal. The clinical diagnosis of hyper IgE syndrome was retained.
Full-length RT-PCR for RELA and qPCR analysis
Total RNA was extracted by an RNeasy mini kit (Qiagen) and used as the template for cDNA synthesis with Super Script II reverse transcriptase (Life Technologies). The cDNA for the full-length transcript was amplified with the following primers: forward primer: 5′-GCGAATGGCTCGTCTGTAGT-3′, reverse primer: 5′-AGAAGCTGGAGGATGGGGAT-3′. RT-qPCR was performed with the Biosystems Assays-on-Demand probe/primer for TaqMan probes for RELA (Hs0104201_m1; Thermo Fisher Scientific). β-actin was used for normalization. The results are expressed according to the 2−ΔΔC(t) method, as described by the manufacturer.
Immunoblotting
Total protein extracts were prepared by mixing cells with lysis buffer (CellLytic M, Sigma-Aldrich) containing protease inhibitor cocktail (Sigma-Aldrich). Then, cytoplasmic and nuclear proteins were extracted with a Nuclear/Cytosolic Fractionation Kit (Cell Biolabs, Inc.) according to the manufacturer’s instructions. To confirm whether the cytoplasmic and nuclear proteins were properly separated, the membranes were probed with anti–α tubulin antibody (B-7; Santa Cruz Biotechnology) and anti–lamin A/C antibody (E-1; Santa Cruz Biotechnology). Equal amounts of protein (according to the Pierce BCA Protein Assay Kit; Thermo Fisher Scientific) were resolved by sodium dodecyl sulfate‒polyacrylamide gel electrophoresis in a Mini-PROTEAN TGX 10% Precast Gel (Bio-Rad) and transferred to a polyvinylidene difluoride membrane. The membrane was probed using an antibody against RelA (F-6; Santa Cruz Biotechnology) or β-actin (017-24551; Wako). The appropriate horseradish peroxidase–conjugated secondary antibodies were incubated with the membrane, and antibody binding was detected using Amersham ECL Prime Western Blotting Detection Reagents (GE Healthcare) and a ChemiDoc MP system (Bio-Rad).
Co-immunoprecipitation
The pCMV4 plasmids with a Myc or Flag tag inserted at the 5′ end of RELA and a Myc tag at the 5′ end of NFKB1 encoding p50 were generated by PCR. RELA KO HEK293 cells were seeded in 6-well plates (7.5 × 105/well) and transfected with 2.5 µg of Myc-RELA or Myc-NFKB1/pCMV4 and 2.5 µg of Flag-RELA/pCMV4 carrying WT or each variant using Lipofectamine LTX (Thermo Fisher Scientific) according to the manufacturer’s protocol. The transfected cells were divided into two fractions, which were used for direct immunoblotting and immunoprecipitation followed by immunoblotting. Co-immunoprecipitation was performed using a Dynabeads Co-Immunoprecipitation Kit (Invitrogen). 10 μg of anti–c-Myc antibody (Sigma-Aldrich) was added to 1.5 mg of Dynabeads and incubated overnight at 37°C for coupling. Cell pellets were lysed and incubated with the antibody-coupled beads on a rotator for 30 min at 4°C, washed four times with Extraction Buffer A containing 100 mM NaCl, and finally washed with washing buffer and then eluted from the beads.
Establishment of RELA KO HEK293 cells
HEK293 cells stably transfected with the human TLR4a, MD2, and CD14 genes were purchased (293-htlr4md2cd14; Invitrogen). The RELA gene was knocked out using the clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 system. A single-guide RNA targeting the RELA gene (target sequence; 5′-TCCTTTCCTACAAGCTCGTG-3′) and Cas9 expression plasmid was constructed using the PX458 vector (48138; Addgene). 10 μg of plasmid was electroporated into 1 × 106 cells with a NEPA21 electroporator (Nepa Gene Co). 2 d after electroporation, single clones obtained by limiting dilution were cultured. After two rounds of electroporation and single clone selection, RELA KO cells were established. Successful RELA KO was confirmed by direct sequencing of genomic DNA and Western blotting of candidate clones.
Transfection of RELA-deficient HEK293 cells and expression of RELA variants
The pCMV4 vector encoding human RELA was purchased from Addgene. The construct carrying the mutant allele was generated by direct mutagenesis with the QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies). Direct mutagenesis was used to generate the RELA mutation in the WT plasmid. Cells were transfected for 24 h using Lipofectamine LTX according to the manufacturer’s protocol, after which RelA protein was detected by immunoblotting with monoclonal anti-RelA antibody (F-6; Santa Cruz Biotechnology) and an antibody against β-actin (loading control).
NF-κB reporter assay
RELA KO HEK293 cells were cotransfected with the pCMV4 expression vector containing WT or mutant RELA, the reporter vector (in combination with 100 ng of IgkCona-Luc and 5 ng of pRL-TK [Promega]) using Lipofectamine LTX. The total amount of WT or mutant RELA vector was kept constant at 50 ng by supplementation with EV. 24 h later, the cells were lysed in passive lysis buffer, and luciferase activities were measured in the Dual-Luciferase Reporter Assay (Promega). The experiments were performed in quadruplicate, and the data are expressed in relative luciferase units. The results shown are representative of three independent experiments.
Type I IFN signature analysis in whole-blood leukocytes
The type I IFN signature was determined as previously described (Oda et al., 2014). Total RNA was extracted from whole blood collected into PAXgene Blood RNA tubes (762165; PreAnalytiX) and reverse-transcribed by a PrimeScript II 1st strand cDNA synthesis kit (6210A; Takara Bio). qPCR was performed with TaqMan Gene Expression Master Mix (4369016; Applied Biosystems) with probes (IFI27, Hs01086370_m1; IFIT1, Hs00356631_g1; RSAD2, Hs01057264_m1; SIGLEC1, Hs00988063_m1; ISG15, Hs00192713_m1; IFI44L, Hs00199115_m1; BACT, Hs01060665_g1; Applied Biosystems) on a StepOnePlus Real-Time PCR system (Thermo Fisher Scientific). The expression levels of each transcript were determined in triplicate and normalized to the level of β-actin. The results are shown relative to a single calibrator. The median relative quantification value of the six ISGs was used to calculate the “IFN score” for each patient. IFN scores greater than +2 SD of the average of 11 healthy controls (5.04) were designated as positive. For each DN RELA patient except P3, multiple blood draws were taken at regular follow-up appointments in the absence of obvious signs of infection or fever. For P3, only one sample, taken just before bone marrow transplant (under conditioning), was available. For disease controls, data from three AGS patients (two genetically diagnosed: IFIH1 [p.R779H/WT] and IFIH1 [p.R720Q/WT], and one clinically diagnosed) were used.
scRNASeq of primary leukocytes at steady state
Cryopreserved PBMCs from P1/2/3/4/5, one patient with a heterozygous STAT1 GOF mutation, and one patient with a heterozygous STAT3 DN mutation were analyzed through scRNASeq. Briefly, thawed cells were washed with medium and filtered with a 70-µm MACS SmartStrainer (Cat: 130-098-462; Miltenyi Biotec) to remove large debris. The cells were then washed three times with PBS plus 0.5% FBS and finally filtered again with a 40-µm Falcon Cell Strainer (Cat: 352340; Corning) before being subjected to single-cell capture via a 10X Genomics Chromium chip. Libraries were prepared with a Chromium Single Cell 3′ Reagent Kit (v3 Chemistry) and sequenced with an Illumina NovaSeq 6000 sequencer. Sequences were preprocessed with CellRanger. Approximately 10,000 cells were sequenced per sample, with ∼30,000 reads per cell on average.
Data generated during this study underwent integrative analysis with historical controls (11 adult and six pediatric controls in nine batches of experiments) and publicly available control PBMC datasets downloaded from the 10X Genomics web portal (https://support.10xgenomics.com/single-cell-gene-expression/datasets). Data were first manually filtered based on common quality-control metrics. The filtered data were then integrated with Harmony (Korsunsky et al., 2019). Two sequential rounds of graph-based clustering were performed. The first round of clustering identified general leukocyte subsets, while the second round of clustering identified memory and effector T lymphocyte subsets and natural killer lymphocytes with a sufficiently high resolution. Clusters were identified based on canonical marker gene expression with the aid of the SingleR pipeline (Aran et al., 2019) guided by MonacoImmuneData (Monaco et al., 2019). The cellular indexing of transcriptomes and epitopes by sequencing (CITE-Seq) datasets obtained from 10X also informed the identity of each cluster. Pseudobulk differential expression analysis was conducted with DESeq2 (Love et al., 2014), excluding all public datasets. GSEA was conducted with the fgsea package by projecting the fold-change ranking onto various MSigDB gene sets (http://www.gsea-msigdb.org/gsea/msigdb/genesets.jsp). Intercellular communication analysis was conducted with CellChat (Jin et al., 2021). All analyses were performed in R v4 (http://www.R-project.org/). The data were uploaded under the SRA ID (PRJNA845112).
PBMC stimulation assay
Cryopreserved PBMCs were thawed, counted, and dispensed into U-bottom 96-well plates at a density of 1 × 105 cells per 100 μl of RPMI plus 10% FBS. The cells were left unstimulated or stimulated with the TLR7 agonist CL264 (Invivogen; 5 μg/ml), the TLR8 agonist TL8-506 (Invivogen; 100 ng/ml), the TLR7/8 agonist R848 (Invivogen; 1 μg/ml), or LPS (Sigma-Aldrich; 10 ng/ml) for 24 h. For studies with neutralizing antibodies, anti–TNF-hIgG1 or anti–β-Gal-hIgG1, anti–IFN-α, anti–IFN-β, anti–IL-29, or mouse IgG2a isotype control (all purchased from Invivogen; 1 μg/ml) was used. Supernatants were harvested and stored at −20°C until analysis. Levels of secreted cytokines were determined with a LEGENDplex assay (BioLegend). Cell pellets were lysed with DNA/RNA Shield (Zymo Research) and stored at −20°C until analysis. Extracted total RNA was used for RT-qPCR analysis. FAM-MGB TaqMan probes (IFI44L, Hs00199115_m1; IFIT1, Hs00356631_g1; ISG15, Hs00192713_m1; IRF7, Hs01014809_g1) were purchased from Thermo Fisher Scientific. The GUSB VIC-TAMRA probe was used as an internal control.
Blood collection
Blood samples were collected from all patients and relatives after written informed consent had been obtained. Blood samples were drawn as part of the diagnostic procedure. The study was approved by the Tohoku University School of Medicine. Heparin-treated blood samples were obtained from healthy relatives of the patient. All these procedures were conducted in accordance with the 1975 Declaration of Helsinki, as revised in 2013.
Whole-exome sequencing
Genomic DNA (3 µg) extracted from the peripheral blood cells of the patient was sheared with a Covaris S2 Ultrasonicator (Covaris). An adaptor-ligated library was prepared with the Paired-End Sample Prep kit V1 (Illumina). Exome capture was performed with the Sure Select Human All Exon kit (Agilent Technologies). Single-end sequencing was performed on an Illumina Genome Analyzer IIx (Illumina), generating 72-base reads.
Sanger sequencing
Genomic DNA was isolated from whole blood cells. RELA genomic DNA was amplified with specific primers (PCR amplification conditions and primer sequences available upon request). PCR products were analyzed by electrophoresis in 1% agarose gels, sequenced with the BigDye Terminator cycle sequencing kit (Applied Biosystems), and analyzed on an ABI Prism 3700 instrument (Applied Biosystems).
Cell isolation and culture
Human PBMCs were isolated by Ficoll–Hypaque density centrifugation (Amersham Pharmacia Biotech) from whole blood and cultured in RPMI medium supplemented with 10% FBS (Gibco BRL, Invitrogen). Immortalized B lymphoblastoid cell lines were induced in-house from PBMCs and cultured in RPMI medium supplemented with 10% FBS. Primary human fibroblasts were obtained from skin biopsy specimens and cultured in DMEM (Gibco BRL, Invitrogen) supplemented with 10% FBS. T cell blasts were induced from PBMCs with ImmunoCult CD3/CD28/CD2 T cell activator and ImmunoCult XF T Cell Expansion Medium (STEMCELL) supplemented with IL-2 (Roche, 10 ng/ml). All cells were cultured at 37°C under an atmosphere containing 5% CO2.
FACS of monocytes
Freshly thawed PBMCs were stained with reagents in FACS buffer (2% FBS and 2 mM EDTA in PBS, filter-sterilized) for 1 h at 4°C in the dark. CD3−CD19−CD14+ monocytes were sorted.
Stimulation assay with IFNs in vitro
Sorted monocytes (1 × 104 cells per well) or expanded T cell blasts (day 13 after induction; 5 × 105 cells per well) were left unstimulated or stimulated with human IFN-α2 (100 IU/ml), IFN-γ (105 IU/ml), PMA and ionomycin (1:1,000; eBioscience), or LPS (10 ng/ml) for 4 h. Secreted cytokine levels were determined by a LEGENDplex assay (BioLegend); mRNA levels were quantified by qPCR with the following TaqMan probes: IFI6, Hs00242571_m1; ISG15, Hs00192713_m1; and RSAD2, Hs00369813_m1. GUSB was used as an internal control.
RNASeq
RNA was extracted from sorted and stimulated monocytes with RNeasy Plus Micro Kit (Qiagen). Full-length cDNA was generated from 1 ng of total RNA with SMART-Seq v4 Ultra Low Input RNA Kit (634888; Clontech), and 1 ng of cDNA was used to prepare libraries with Nextera XT DNA Library Preparation Kit (FC-131-1024; Illumina). Libraries with unique barcodes were pooled at equal molar ratios and sequenced using an Illumina NovaSeq 6000 sequencer with V1.5 reagents and NovaSeq Control Software V1.7.0 to generate 100-bp paired-end reads, according to the manufacturer’s protocol. The FASTQ files generated were first inspected by fastqc to check sequencing quality. Sequences were aligned with the GENCODE human reference genome GRCh37.p13 with STAR aligner v2.6, and alignment quality was evaluated with RSeQC. Gene-level features were quantified with featureCounts v1.6.0 based on the GENCODE GRCh37.p13 gene annotation. Count data were normalized through variance-stabilizing transformation implemented in the DESeq2 package. DE analysis was performed with DESeq2 (Love et al., 2014).
Optimization of co-immunoprecipitation conditions
To verify whether RELA variants can form dimers with WT RELA or WT p50, we confirmed co-immunoprecipitation using Myc-RELA or Myc-p50 as bait. Myc-mock or Myc-STAT1 was used as bait controls. RELA KO HEK293 cells were transfected with 2.5 µg of Myc-RELA (pCMV4), Myc-NFKB1 (pCMV4), Myc-STAT1 (pcDNA3), or Myc-EV (pcDNA3) and 2.5 µg of Flag-RELA (pCMV4) carrying WT or each variant. The transfected cells were divided into two fractions used for direct immunoblotting and immunoprecipitation followed by immunoblotting.
Healthy controls
The healthy controls were volunteer blood donors from Japan and the United States.
Ethics statement
This study was conducted in accordance with the Helsinki Declaration and approved by the Ethics Committee of Hiroshima University (No. hi-126-5), Tohoku University School of Medicine (2019-1-561), Kyoto University Hospital (No. G1118, G1233, and G0729), Kyushu University (#531-03), and the Rockefeller University (institutional review board protocol no. JCA-0708).
Online supplemental material
Fig. S1 shows analysis of the RELA variants found in the patients. Fig. S2 presents functional characterization of the RelA mutants in an overexpression system. Fig. S3 provides additional information about enhanced expression of type I and III IFNs and ISGs. Fig. S4 presents additional evidence of single-cell transcriptomic analysis of leukocyte subsets. Fig. S5 shows skin manifestations and pathological findings at skin biopsy of P4. Table S1, Table S2, and Table S3 provide the clinical spectrum of patients and clinical findings associated with type I interferonopathies, and immunological features of the patients. Table S4, Table S5, and Table S6 provide targeted gene panels for early-onset refractory diarrhea and list of frequencies and combined annotation-dependent depletion (CADD) scores of patient mutations, and the list of candidate variants for P3–P6.
Supplementary Material
shows the clinical spectrum of patients with AD RELA deficiency.
shows clinical findings associated with type I interferonopathies.
shows immunological features of the patients with AD RELA.
shows targeted gene panels for early-onset refractory diarrhea.
lists frequencies and CADD scores of patient mutations.
lists candidate variants for P3–P6.
is the source file for Fig. 2.
is the source file for Fig. S1.
is the source file for Fig. S2.
Acknowledgments
We thank the patients and their parents for participating in this study. In addition, the authors thank Miharu Izumikawa and all physicians, nurses, and support personnel for their dedicated care of patients in this study.
K. Moriya (grant number 19K23819 and 21K07791), Y. Honda (grant number 20K16924), and S. Okada (grant number 18KK0228, 19H03620, and 22H03041) were supported by grant from the Japanese Ministry of Health, Labour and Welfare of Japan. The Laboratory of Human Genetics of Infectious Diseases is supported by the Howard Hughes Medical Institute, the Rockefeller University, the St. Giles Foundation, the National Institutes of Health (R01AI127564 and P01AI061093), the National Center for Advancing Translational Sciences, National Institutes of Health Clinical and Translational Science Award program (UL1TR001866), the French National Research Agency (ANR) under the “Investments for the Future” program (ANR-10-IAHU-01), the Integrative Biology of Emerging Infectious Diseases Laboratory of Excellence (ANR-10-LABX-62-IBEID), the ANR “LTh-MSMD-CMCD” project (ANR-18-CE93-0008-01), the ANR “KREM-AIF” project (ANR-21-CE17-0014-03), the French Foundation for Medical Research (EQU201903007798), the Square Foundation, Grandir—Fonds de solidarité pour l’enfance, Institut National de la Santé et de la Recherche Médicale, and the Paris Cité University. M. Ogishi was supported by the David Rockefeller Graduate Program, the Funai Foundation for Information Technology, the Honjo International Scholarship Foundation, the New York Hideyo Noguchi Memorial Society, and the National Cancer Institute F99 Award (F99CA274708). This research was supported by the following grants: Health Labor Sciences Research Grants for Research on Intractable Diseases from the Ministry of Health, Labor and Welfare of Japan (H29-Nanchi-Ippan-020 and JPMH20317089 to K. Izawa, T. Yasumi, and R. Nishikomori); the Practical Research Project for Rare/Intractable Diseases from the Japan Agency for Medical Research and Development (JP20ek0109477, JP21ek0109477 to K. Izawa and R. Nishikomori; JP20ek0109480 to S. Okada).
Author contributions: K. Moriya and S. Okada are the principal investigators, and K. Moriya conceived and designed the study. K. Moriya, T. Nakano, Y. Honda, M. Ogishi, M. Sonoda, M. Tsumura, M. Yoko, T. Uchida, D. Rinchai, and P. Zhang performed the experiments and analyzed the data. M. Ishimura, K. Izawa, F. Kakuta, D. Abukawa, M. Hbibi, A. Puel, and A. Bousfiha were the treating physicians responsible for diagnosis, sample collection, and clinical supervision. All authors revised the manuscript and approved the final manuscript as submitted. K. Moriya, T. Nakano, Y. Honda, M. Ogishi, M. Sonoda, M. Tsumura, B. Boisson, J.-L. Casanova. and S. Okada analyzed the results and wrote the manuscript.
Data availability
The data underlying Figs. 1, 2, 3, 4, 5, 6, and 7 are available in the published article and its online supplemental material. scRNASeq data are uploaded under the SRA ID PRJNA845112.
References
- Adeeb, F., Dorris E.R., Morgan N.E., Lawless D., Maqsood A., Ng W.L., Killeen O., Cummins E.P., Taylor C.T., Savic S., et al. 2021. A novel RELA truncating mutation in a familial behçet’s disease-like mucocutaneous ulcerative condition. Arthritis Rheumatol. 73:490–497. 10.1002/art.41531 [DOI] [PubMed] [Google Scholar]
- Aran, D., Looney A.P., Liu L., Wu E., Fong V., Hsu A., Chak S., Naikawadi R.P., Wolters P.J., Abate A.R., et al. 2019. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 20:163–172. 10.1038/s41590-018-0276-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano, T., Khourieh J., Zhang P., Rapaport F., Spaan A.N., Li J., Lei W.T., Pelham S.J., Hum D., Chrabieh M., et al. 2021a. Human STAT3 variants underlie autosomal dominant hyper-IgE syndrome by negative dominance. J. Exp. Med. 218:e20202592. 10.1084/jem.20202592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano, T., Boisson B., Onodi F., Matuozzo D., Moncada-Velez M., Maglorius Renkilaraj M.R.L., Zhang P., Meertens L., Bolze A., Materna M., et al. 2021b. X-linked recessive TLR7 deficiency in ∼1% of men under 60 years old with life-threatening COVID-19. Sci. Immunol. 6:1–31. 10.1126/sciimmunol.abl4348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badran, Y.R., Dedeoglu F., Leyva Castillo J.M., Bainter W., Ohsumi T.K., Bousvaros A., Goldsmith J.D., Geha R.S., and Chou J.. 2017. Human RELA haploinsufficiency results in autosomal-dominant chronic mucocutaneous ulceration. J. Exp. Med. 214:1937–1947. 10.1084/jem.20160724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béziat, V., Tavernier S.J., Chen Y.H., Ma C.S., Materna M., Laurence A., Staal J., Aschenbrenner D., Roels L., Worley L., et al. 2020. Dominant-negative mutations in human IL6ST underlie hyper-IgE syndrome. J. Exp. Med. 217:e20191804. 10.1084/jem.20191804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisson, B., Puel A., Picard C., and Casanova J.L.. 2017. Human IκBα gain of function: A severe and syndromic immunodeficiency. J. Clin. Immunol. 37:397–412. 10.1007/s10875-017-0400-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinez, C., Miraghazadeh B., Tanita K., da Silva E., Hoshino A., Okada S., Chand R., Asano T., Tsumura M., Yoshida K., et al. 2018. Gain-of-function IKBKB mutation causes human combined immune deficiency. J. Exp. Med. 215:2715–2724. 10.1084/jem.20180639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova, J.-L., Abel L., and Quintana-Murci L.. 2011. Human TLRs and IL-1rs in host defense: Natural insights from evolutionary, epidemiological, and clinical genetics. Annu. Rev. Immunol. 29:447–491. 10.1146/annurev-immunol-030409-101335 [DOI] [PubMed] [Google Scholar]
- Comrie, W.A., Faruqi A.J., Price S., Zhang Y., Rao V.K., Su H.C., and Lenardo M.J.. 2018. RELA haploinsufficiency in CD4 lymphoproliferative disease with autoimmune cytopenias. J. Allergy Clin. Immunol. 141:1507–1510.e8. 10.1016/j.jaci.2017.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtois, G., Smahi A., Reichenbach J., Döffinger R., Cancrini C., Bonnet M., Puel A., Chable-bessia C., Yamaoka S., Feinberg J., et al. 2003. A hypermorphic IκBα mutation is associated with autosomal dominant anhidrotic ectodermal dysplasia and T cell immunodeficiency. J. Clin. Invest. 112:1108–1115. 10.1172/JCI200318714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow, Y.J., and Stetson D.B.. 2022. The type I interferonopathies: 10 years on. Nat. Rev. Immunol. 22:471–483. 10.1038/s41577-021-00633-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliegauf, M., Bryant V.L., Frede N., Slade C., Woon S.T., Lehnert K., Winzer S., Bulashevska A., Scerri T., Leung E., et al. 2015. Haploinsufficiency of the NF-κB1 subunit p50 in common variable immunodeficiency. Am. J. Hum. Genet. 97:389–403. 10.1016/j.ajhg.2015.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel, R.R., Nakabo S., Dizon B.L.P., Urban A., Waldman M., Howard L., Darnell D., Buhaya M., Carmona-Rivera C., Hasni S., et al. 2021. Lupus-like autoimmunity and increased interferon response in patients with STAT3-deficient hyper-IgE syndrome. J. Allergy Clin. Immunol. 147:746–749.e9. 10.1016/j.jaci.2020.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda, K., Ohba Y., Yanai H., Negishi H., Mizutani T., Takaoka A., Taya C., and Taniguchi T.. 2005. Spatiotemporal regulation of MyD88-IRF-7 signalling for robust type-I interferon induction. Nature. 434:1035–1040. 10.1038/nature03547 [DOI] [PubMed] [Google Scholar]
- Jain, A., Ma C.A., Liu S., Brown M., Cohen J., and Strober W.. 2001. Specific missense mutations in NEMO result in hyper-IgM syndrome with hypohydrotic ectodermal dysplasia. Nat. Immunol. 2:223–228. 10.1038/85277 [DOI] [PubMed] [Google Scholar]
- Jamilloux, Y., El Jammal T., Vuitton L., Gerfaud-Valentin M., Kerever S., and Sève P.. 2019. JAK inhibitors for the treatment of autoimmune and inflammatory diseases. Autoimmun. Rev. 18:102390. 10.1016/j.autrev.2019.102390 [DOI] [PubMed] [Google Scholar]
- Jeremiah, N., Neven B., Gentili M., Callebaut I., Maschalidi S., Stolzenberg M.C., Goudin N., Frémond M.L., Nitschke P., Molina T.J., et al. 2014. Inherited STING-activating mutation underlies a familial inflammatory syndrome with lupus-like manifestations. J. Clin. Invest. 124:5516–5520. 10.1172/JCI79100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, S., Guerrero-Juarez C.F., Zhang L., Chang I., Ramos R., Kuan C.H., Myung P., Plikus M.V., and Nie Q.. 2021. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 12:1088. 10.1038/s41467-021-21246-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kacar, M., Pathak S., and Savic S.. 2019. Hereditary systemic autoinflammatory diseases and Schnitzler’s syndrome. Rheumatology. 58:vi31–vi43. 10.1093/rheumatology/kez448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaleviste, E., Saare M., Leahy T.R., Bondet V., Duffy D., Mogensen T.H., Jørgensen S.E., Nurm H., Ip W., Davies E.G., et al. 2019. Interferon signature in patients with STAT1 gain-of-function mutation is epigenetically determined. Eur. J. Immunol. 49:790–800. 10.1002/eji.201847955 [DOI] [PubMed] [Google Scholar]
- Kaustio, M., Haapaniemi E., Göös H., Hautala T., Park G., Syrjänen J., Einarsdottir E., Sahu B., Kilpinen S., Rounioja S., et al. 2017. Damaging heterozygous mutations in NFKB1 lead to diverse immunologic phenotypes. J. Allergy Clin. Immunol. 140:782–796. 10.1016/j.jaci.2016.10.054 [DOI] [PubMed] [Google Scholar]
- Kawai, T., and Akira S.. 2007. Signaling to NF-kappaB by toll-like receptors. Trends Mol. Med. 13:460–469. 10.1016/j.molmed.2007.09.002 [DOI] [PubMed] [Google Scholar]
- Korsunsky, I., Millard N., Fan J., Slowikowski K., Zhang F., Wei K., Baglaenko Y., Brenner M., Loh P.R., and Raychaudhuri S.. 2019. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods. 16:1289–1296. 10.1038/s41592-019-0619-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecerf, K., Koboldt D.C., Kuehn H.S., Jayaraman V., Lee K., Mihalic Mosher T., Yonkof J.R., Mori M., Hickey S.E., Franklin S., et al. 2022. Case report and review of the literature: immune dysregulation in a large familial cohort due to a novel pathogenic RELA variant. Rheumatology (Oxford). 62:347–359. 10.1093/rheumatology/keac227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J., Hayashi M., Lo J.F., Fearns C., Chu W.M., Luo Y., Xiang R., and Chuang T.H.. 2009. Nuclear factor kappaB (NF-kappaB) activation primes cells to a pro-inflammatory polarized response to a Toll-like receptor 7 (TLR7) agonist. Biochem. J. 421:301–310. 10.1042/BJ20090013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lévy, R., Langlais D., Béziat V., Rapaport F., Rao G., Lazarov T., Bourgey M., Zhou Y.J., Briand C., Moriya K., et al. 2021. Inherited human c-Rel deficiency disrupts myeloid and lymphoid immunity to multiple infectious agents. J. Clin. Invest. 131:e150143. 10.1172/JCI150143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., Lei W.T., Zhang P., Rapaport F., Seeleuthner Y., Lyu B., Asano T., Rosain J., Hammadi B., Zhang Y., et al. 2021. Biochemically deleterious human NFKB1 variants underlie an autosomal dominant form of common variable immunodeficiency. J. Exp. Med. 218:e20210566. 10.1084/jem.20210566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, R., Moore P.A., and Pitha P.M.. 2002. Stimulation of IRF-7 gene expression by tumor necrosis factor α: Requirement for NFkappa B transcription factor and gene accessibility. J. Biol. Chem. 277:16592–16598. 10.1074/jbc.M111440200 [DOI] [PubMed] [Google Scholar]
- Monaco, G., Lee B., Xu W., Mustafah S., Hwang Y.Y., Carré C., Burdin N., Visan L., Ceccarelli M., Poidinger M., et al. 2019. RNA-seq signatures normalized by mRNA abundance allow absolute deconvolution of human immune cell types. Cell Rep. 26:1627–1640.e7. 10.1016/j.celrep.2019.01.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya, K., Sasahara Y., Ohnishi H., Kawai T., and Kanegane H.. 2018. IKBA S32 mutations underlie ectodermal dysplasia with immunodeficiency and severe noninfectious systemic inflammation. J. Clin. Immunol. 38:543–545. 10.1007/s10875-018-0522-y [DOI] [PubMed] [Google Scholar]
- Mousallem, T., Yang J., Urban T.J., Wang H., Adeli M., Parrott R.E., Roberts J.L., Goldstein D.B., Buckley R.H., and Zhong X.P.. 2014. A nonsense mutation in IKBKB causes combined immunodeficiency. Blood. 124:2046–2050. 10.1182/blood-2014-04-571265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, C., Jakobsen M.A., Larsen M.J., Müller A.C., Hansen S., Lillevang S.T., Fisker N., and Barington T.. 2014. Immunodeficiency associated with a nonsense mutation of IKBKB. J. Clin. Immunol. 34:916–921. 10.1007/s10875-014-0097-1 [DOI] [PubMed] [Google Scholar]
- Oda, H., Nakagawa K., Abe J., Awaya T., Funabiki M., Hijikata A., Nishikomori R., Funatsuka M., Ohshima Y., Sugawara Y., et al. 2014. Aicardi-Goutières syndrome is caused by IFIH1 mutations. Am. J. Hum. Genet. 95:121–125. 10.1016/j.ajhg.2014.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannicke, U., Baumann B., Fuchs S., Henneke P., Rensing-Ehl A., Rizzi M., Janda A., Hese K., Schlesier M., Holzmann K., et al. 2013. Deficiency of innate and acquired immunity caused by an IKBKB mutation. N. Engl. J. Med. 369:2504–2514. 10.1056/NEJMoa1309199 [DOI] [PubMed] [Google Scholar]
- Scott, O., Lindsay K., Erwood S., Mollica A., Roifman C.M., Cohn R.D., and Ivakine E.A.. 2021. STAT1 gain-of-function heterozygous cell models reveal diverse interferon-signature gene transcriptional responses. NPJ Genom. Med. 6:34. 10.1038/s41525-021-00196-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tangye, S.G., Al-Herz W., Bousfiha A., Cunningham-Rundles C., Franco J.L., Holland S.M., Klein C., Morio T., Oksenhendler E., Picard C., et al. 2021. The Ever-Increasing Array of Novel Inborn Errors of Immunity: an Interim Update by the IUIS Committee. J. Clin. Immunol. 41:666–679. 10.1007/s10875-021-00980-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tangye, S.G., Al-Herz W., Bousfiha A., Cunningham-Rundles C., Franco J.L., Holland S.M., Klein C., Morio T., Oksenhendler E., Picard C., et al. 2022. Human Inborn Errors of Immunity: 2022 Update on the Classification from the International Union of Immunological Societies Expert Committee. J. Clin. Immunol. 42:1473–1507. 10.1007/s10875-022-01289-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida, T., Suzuki T., Kikuchi A., Kakuta F., Ishige T., Nakayama Y., Kanegane H., Etani Y., Mizuochi T., Fujiwara S.I., et al. 2020. Comprehensive targeted sequencing identifies monogenic disorders in patients with early-onset refractory diarrhea. J. Pediatr. Gastroenterol. Nutr. 71:333–339. 10.1097/MPG.0000000000002796 [DOI] [PubMed] [Google Scholar]
- Zhang, Q., Lenardo M.J., and Baltimore D.. 2017. 30 Years of NF-κB: A blossoming of relevance to human pathobiology. Cell. 168:37–57. 10.1016/j.cell.2016.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, P., Bigio B., Rapaport F., Zhang S.Y., Casanova J.L., Abel L., Boisson B., and Itan Y.. 2018. PopViz: a webserver for visualizing minor allele frequencies and damage prediction scores of human genetic variations. Bioinformatics. 34:4307–4309. 10.1093/bioinformatics/bty536 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows the clinical spectrum of patients with AD RELA deficiency.
shows clinical findings associated with type I interferonopathies.
shows immunological features of the patients with AD RELA.
shows targeted gene panels for early-onset refractory diarrhea.
lists frequencies and CADD scores of patient mutations.
lists candidate variants for P3–P6.
is the source file for Fig. 2.
is the source file for Fig. S1.
is the source file for Fig. S2.
Data Availability Statement
The data underlying Figs. 1, 2, 3, 4, 5, 6, and 7 are available in the published article and its online supplemental material. scRNASeq data are uploaded under the SRA ID PRJNA845112.