Key Points
Question
Among patients in the Surveillance, Epidemiology, and End Results registries diagnosed with cancer between 2013 and 2019, what was the prevalence of germline genetic testing?
Findings
In this observational study that included 1 369 602 patients diagnosed with cancer in California and Georgia, germline genetic testing after cancer diagnosis was low (6.8%; n = 93 052). Testing was highest in males with breast cancer (50%) and in patients with ovarian cancer (38.6%).
Meaning
Few patients diagnosed with cancer between 2013 and 2019 in California and Georgia underwent germline testing.
Abstract
Importance
Germline genetic testing is recommended by practice guidelines for patients diagnosed with cancer to enable genetically targeted treatment and identify relatives who may benefit from personalized cancer screening and prevention.
Objective
To describe the prevalence of germline genetic testing among patients diagnosed with cancer in California and Georgia between 2013 and 2019.
Design, Setting, and Participants
Observational study including patients aged 20 years or older who had been diagnosed with any type of cancer between January 1, 2013, and March 31, 2019, that was reported to statewide Surveillance, Epidemiology, and End Results registries in California and Georgia. These patients were linked to genetic testing results from 4 laboratories that performed most germline testing for California and Georgia.
Main Outcomes and Measures
The primary outcome was germline genetic testing within 2 years of a cancer diagnosis. Testing trends were analyzed with logistic regression modeling. The results of sequencing each gene, including variants associated with increased cancer risk (pathogenic results) and variants whose cancer risk association was unknown (uncertain results), were evaluated. The genes were categorized according to their primary cancer association, including breast or ovarian, gastrointestinal, and other, and whether practice guidelines recommended germline testing.
Results
Among 1 369 602 patients diagnosed with cancer between 2013 and 2019 in California and Georgia, 93 052 (6.8%) underwent germline testing through March 31, 2021. The proportion of patients tested varied by cancer type: male breast (50%), ovarian (38.6%), female breast (26%), multiple (7.5%), endometrial (6.4%), pancreatic (5.6%), colorectal (5.6%), prostate (1.1%), and lung (0.3%). In a logistic regression model, compared with the 31% (95% CI, 30%-31%) of non-Hispanic White patients with male breast cancer, female breast cancer, or ovarian cancer who underwent testing, patients of other races and ethnicities underwent testing less often: 22% (95% CI, 21%-22%) of Asian patients, 25% (95% CI, 24%-25%) of Black patients, and 23% (95% CI, 23%-23%) of Hispanic patients (P < .001 using the χ2 test). Of all pathogenic results, 67.5% to 94.9% of variants were identified in genes for which practice guidelines recommend testing and 68.3% to 83.8% of variants were identified in genes associated with the diagnosed cancer type.
Conclusions and Relevance
Among patients diagnosed with cancer in California and Georgia between 2013 and 2019, only 6.8% underwent germline genetic testing. Compared with non-Hispanic White patients, rates of testing were lower among Asian, Black, and Hispanic patients.
This observational study describes the prevalence of germline genetic testing in California and Georgia between 2013 and 2019 among patients diagnosed with cancer.
Introduction
Genetic testing for inherited cancer risk can improve survival for patients diagnosed with cancer by enabling genetically targeted therapies such as poly(adenosine diphosphate–ribose) polymerase inhibitors.1,2 Genetic testing can also improve outcomes for relatives of patients with cancer by modifying cancer screening and preventive therapies.3,4 Germline testing, in which inherited DNA is sequenced, is recommended by practice guidelines for patients with several cancer types, including breast, ovarian, pancreatic, colorectal, and prostate.5,6,7 With advances in sequencing technology, the number of genes tested has increased and costs have declined.
However, little is known about germline genetic testing or results in patients diagnosed with cancer. The rates and results of germline genetic testing were analyzed in patients diagnosed with cancer in California and Georgia between 2013 and 2019 according to the statewide Surveillance, Epidemiology, and End Results (SEER) registries.
Methods
Patient Cohort
Patients aged 20 years or older who had been diagnosed with any type of cancer from January 1, 2013, through March 31, 2019, and were reported to SEER registries as part of statewide surveillance in California and Georgia were identified. These patients were linked to germline genetic testing results from the 4 laboratories (Ambry Genetics, GeneDx, Invitae, and Myriad Genetics) that performed the majority of testing for patients in these states.8,9 The exclusion criteria were (1) cancer diagnosis only on death certificate or autopsy, (2) younger than 20 years of age, (3) sex coded as other, (4) genetic testing before cancer diagnosis, which could be ascertained beginning January 1, 2012, and (5) incomplete data on genetic testing (eFigure 1 in Supplement 1).
The analytic data set contained linked SEER registry variables and the test results from the laboratories, but omitted all personal identifiable information.10 Participant consent was waived. The institutional review boards overseeing the SEER registries approved the research.
Testing Results
Three of 4 laboratories submitted results for all germline cancer genetic tests performed in the US from January 1, 2013, through March 31, 2021, which helped to identify results for patients who had cancer and then moved out of state. The fourth laboratory, which contributed 1% of tests in this data set, submitted results from tests performed in California and Georgia only. Laboratories submitted gene-level interpretations provided to the ordering clinician according to criteria from the American College of Medical Genetics and Genomics.11
The laboratory results were categorized as pathogenic (defined as variants associated with an increased risk of cancer), benign (defined as variants not associated with an increased risk of cancer), and uncertain (defined as variants for which the associated risk of cancer was unknown). Patients with both pathogenic and uncertain results in different genes were categorized as having pathogenic results, whereas patients with only uncertain results were categorized as having uncertain results. The results from all 4 laboratories were combined for the analysis, comprising 107 tested genes. Additional details appear in the eMethods in Supplement 1.
Outcome Measures
The primary outcome was germline genetic testing within 2 years of the cancer diagnosis. Tests performed later are less consistently related to the index cancer. The SEER variables included sex (male, female, other, unknown), cancer stage, age at cancer diagnosis, race and ethnicity (patients who are Asian [including Pacific Islanders], Black, Hispanic, non-Hispanic White [hereafter, White], or Other [Native American, unknown, and Other]), poverty assessed at the US Census tract level (<10%, 10%-19%, ≥20%), whether the patient lived in an urban or rural zip code as classified by SEER, and state (California and Georgia).12 Race and ethnicity data were collected because they are social determinants of health and to identify disparities by race and ethnicity in testing. Race and ethnicity were abstracted from medical records by trained staff at the state tumor registries and the categories were based on definitions from the US Census Bureau.
We evaluated testing for all cancer types in the SEER site recode variable. The analyses focused on 8 cancer types, of which 6 had established germline genetic testing indications: these were breast; colorectal; endometrial; epithelial ovarian, fallopian, and peritoneal (hereafter, ovarian); pancreatic; and prostate.5,6,7 Lung cancer was included because of its recently discovered association with pathogenic results in various genes.13 Patients with multiple primary tumor types were included because prior studies showed frequent pathogenic results in patients with multiple cancer types.14 Patients with 1 or more cancer diagnosis (either before their index cancer or ≤2 years afterward) were included in the category of multiple cancer types.
Genes were grouped by associated cancer types or syndromes and by those recommended for testing by practice guidelines. All genes recommended for testing by practice guidelines from the National Comprehensive Cancer Network,5,6,7 the American College of Medical Genetics and Genomics11 (n = 62; eTable 1 in Supplement 1), or both were categorized as follows: breast or ovarian cancer–associated genes including BRCA1 and BRCA2 (BRCA1/2); gastrointestinal cancer–associated genes not previously included in the breast or ovarian category, including Lynch syndrome genes; and other hereditary cancer syndrome genes. The genes not recommended for testing after a cancer diagnosis were categorized as non–guideline-recommended genes.
Statistical Analysis
The individual patient was the unit of analysis. We evaluated the presence and results of genetic testing according to age, race and ethnicity, and cancer type using SEER data. Testing trends were analyzed with logistic regression modeling, controlling for age, cancer type, and diagnosis year. In a separate logistic regression model, we assessed whether testing varied across race and ethnicity among the 3 cancer types (male breast, female breast, and ovarian5,15) with the highest testing rates overall, which were recommended for testing by practice guidelines throughout the study period, and by year, holding age constant and allowing testing trends and racial and ethnic differences to vary by year.
We examined the results by the categories defined above as breast and ovarian cancer–associated genes, gastrointestinal cancer–associated genes, other hereditary cancer syndrome genes, and non–guideline-recommended genes across selected cancer types. Additional details appear in the eMethods in Supplement 1. The statistical analyses were performed using SAS version 9.4 (SAS Institute Inc).
Results
Testing Use
From January 1, 2013, through March 31, 2019, there were 1 369 602 patients diagnosed with cancer who met the inclusion criteria (eFigure 1 in Supplement 1) and 93 052 (6.8%; 95% CI, 6.8%-6.8%) underwent germline genetic testing through March 31, 2021 (Table 1 and Table 2). The proportion of patients tested varied by cancer type: male breast (50%), ovarian (38.6%), female breast (26%), multiple (7.5%), endometrial (6.4%), pancreatic (5.6%), colorectal (5.6%), prostate (1.1%), and lung (0.3%). Genetic testing for all other cancer types appears in eTable 2 in Supplement 1. The rates of testing increased over time, particularly for patients with pancreatic cancer (from 1.2% in 2013 to 18.6% in 2019). Testing remained low in patients with lung cancer (from 0.1% in 2013 to 0.8% in 2019; eTable 3 in Supplement 1).
Table 1. Characteristics of Patients Diagnosed With Cancer in California and Georgia and Reported to Surveillance, Epidemiology, and End Results (SEER) Registries From 2013 to 2019 by Germline Genetic Testing Statusa.
| Breast (female) | Breast (male) | Colorectal | Endometrial | Lung | Ovarian | Pancreatic | Prostate | Multiple cancer types | All other cancer types | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | Tested, % | No. | Tested, % | No. | Tested, % | No. | Tested, % | No. | Tested, % | No. | Tested, % | No. | Tested, % | No. | Tested, % | No. | Tested, % | No. | Tested, % | |
| Overall | 203 575 | 26.0 | 1344 | 50.0 | 93 562 | 5.6 | 38 324 | 6.4 | 103 139 | 0.3 | 16 444 | 38.6 | 30 595 | 5.6 | 140 810 | 1.1 | 231 261 | 7.5 | 510 548 | 0.9 |
| State | ||||||||||||||||||||
| California | 159 165 | 24.7 | 984 | 49.1 | 72 281 | 5.6 | 30 728 | 6.1 | 74 026 | 0.3 | 13 347 | 37.5 | 24 047 | 5.9 | 104 471 | 1.2 | 180 456 | 7.4 | 400 579 | 0.9 |
| Georgia | 44 410 | 30.6 | 360 | 52.5 | 21 281 | 5.9 | 7596 | 7.3 | 29 113 | 0.2 | 3097 | 43.1 | 6548 | 4.5 | 36 339 | 0.8 | 50 805 | 7.6 | 109 969 | 0.8 |
| Sexb | ||||||||||||||||||||
| Female | 203 575 | 26.0 | NA | NA | 44 460 | 6.2 | 38 324 | 6.4 | 50 274 | 0.5 | 16 444 | 38.6 | 15 039 | 5.8 | NA | NA | 119 761 | 13.0 | 223 276 | 1.4 |
| Male | NA | NA | 1344 | 50.0 | 49 102 | 5.1 | NA | NA | 52 865 | 0.2 | NA | NA | 15 556 | 5.4 | 140 810 | 1.1 | 111 500 | 1.5 | 287 272 | 0.6 |
| Age group at diagnosis, y | ||||||||||||||||||||
| 20-39 | 10 569 | 69.5 | 36 | 55.6 | 4014 | 28.8 | 1719 | 14.7 | 833 | 2.8 | 946 | 39.4 | 509 | 16.7 | 67 | 6.0 | 2980 | 19.3 | 49 721 | 2.2 |
| 40-49 | 34 645 | 56.7 | 117 | 55.6 | 9160 | 19.4 | 3622 | 10.8 | 2828 | 1.8 | 1871 | 45.9 | 1478 | 12.6 | 3035 | 2.9 | 8527 | 26.8 | 51 496 | 2.0 |
| 50-59 | 50 867 | 25.0 | 284 | 47.5 | 22 419 | 5.1 | 10 462 | 6.8 | 15 329 | 0.5 | 3832 | 44.1 | 5017 | 7.6 | 27 877 | 1.4 | 26 876 | 15.1 | 100 773 | 1.0 |
| 60-69 | 56 394 | 15.7 | 398 | 55.3 | 24 948 | 2.9 | 13 680 | 5.6 | 31 660 | 0.3 | 4564 | 41.6 | 9087 | 6.5 | 60 888 | 1.0 | 59 743 | 9.1 | 134 498 | 0.7 |
| 70-79 | 35 037 | 10.3 | 310 | 49.4 | 18 326 | 2.0 | 6473 | 4.2 | 32 439 | 0.2 | 3370 | 36.1 | 8334 | 4.6 | 37 263 | 0.9 | 73 545 | 5.0 | 103 458 | 0.4 |
| ≥80 | 16 063 | 4.4 | 199 | 39.7 | 14 695 | 0.8 | 2368 | 2.0 | 20 050 | 0 | 1861 | 16.7 | 6170 | 1.5 | 11 680 | 0.6 | 59 590 | 2.0 | 70 602 | 0.1 |
| Race and ethnicityc | ||||||||||||||||||||
| Asian | 26 749 | 23.4 | 117 | 44.4 | 11 828 | 4.6 | 4602 | 6.3 | 11 695 | 0.4 | 2084 | 36.5 | 3476 | 5.9 | 9369 | 1.1 | 15 271 | 10.5 | 44 184 | 0.9 |
| Black | 23 957 | 26.0 | 212 | 44.8 | 11 528 | 4.4 | 4384 | 4.4 | 12 580 | 0.2 | 1390 | 29.4 | 3848 | 3.3 | 24 007 | 0.7 | 20 278 | 7.7 | 41 730 | 0.7 |
| Hispanic | 37 294 | 26.8 | 153 | 43.1 | 18 350 | 6.5 | 8116 | 6.0 | 10 532 | 0.3 | 3396 | 34.3 | 5572 | 4.8 | 21 360 | 0.7 | 24 266 | 9.1 | 91 939 | 1.1 |
| Non-Hispanic White | 112 660 | 26.5 | 844 | 53.7 | 50 356 | 5.9 | 20 705 | 7.0 | 67 657 | 0.3 | 9438 | 41.9 | 17 525 | 6.3 | 78 893 | 1.3 | 169 841 | 6.9 | 313 091 | 0.9 |
| Otherd | 2915 | 20.6 | 18 | 33.3 | 1500 | 4.0 | 517 | 4.4 | 675 | 0.1 | 136 | 40.4 | 174 | 5.2 | 7181 | 0.6 | 1605 | 5.2 | 19 604 | 0.7 |
Abbreviation: NA, not applicable.
Includes genetic tests performed after and within 2 years of diagnosis from January 1, 2013, through March 31, 2021.
Those coded as other (n = 328) were excluded (eFigure 1 in Supplement 1).
Race and ethnicity data were collected by self-report and the categories are mutually exclusive. The categories were based on definitions from the US Census Bureau. Race was derived from the SEER race1 variable (North American Association of Central Cancer Registries [NAACCR] variable 160). Ethnicity (Hispanic ethnicity) was obtained from NAACCR Hispanic identification algorithm–derived Hispanic origin (NAACCR variable 191). All patients identified as Hispanic were coded as Hispanic regardless of race.
Includes patients with SEER race1 variable coded as Native American, unknown, and Other.
Table 2. Additional Characteristics of Patients Diagnosed With Cancer in California and Georgia and Reported to Surveillance, Epidemiology, and End Results (SEER) Registries From 2013 to 2019 by Germline Genetic Testing Statusa.
| Breast (female) | Breast (male) | Colorectal | Endometrial | Lung | Ovarian | Pancreatic | Prostate | Multiple cancer types | All other cancer types | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | Tested, % | No. | Tested, % | No. | Tested, % | No. | Tested, % | No. | Tested, % | No. | Tested, % | No. | Tested, % | No. | Tested, % | No. | Tested, % | No. | Tested, % | |
| Overall | 203 575 | 26.0 | 1344 | 50.0 | 93 562 | 5.6 | 38 324 | 6.4 | 103 139 | 0.3 | 16 444 | 38.6 | 30 595 | 5.6 | 140 810 | 1.1 | 231 261 | 7.5 | 510 548 | 0.9 |
| Poverty assessed at the US Census tract level | ||||||||||||||||||||
| Low (<10%) | 106 118 | 27.9 | 710 | 54.1 | 42 010 | 6.5 | 18 226 | 7.0 | 43 766 | 0.4 | 8241 | 43.0 | 14 827 | 7.4 | 72 408 | 1.4 | 125 222 | 8.1 | 255 603 | 1.0 |
| Medium (10%-19%) | 62 000 | 24.4 | 400 | 47.5 | 30 956 | 5.3 | 12 559 | 6.1 | 34 546 | 0.3 | 5149 | 35.9 | 9458 | 4.3 | 42 555 | 0.8 | 68 738 | 6.9 | 158 704 | 0.8 |
| High (≥20%) | 35 457 | 22.9 | 234 | 41.9 | 20 596 | 4.4 | 7539 | 5.2 | 24 827 | 0.2 | 3054 | 31.2 | 6310 | 3.3 | 25 847 | 0.6 | 37 301 | 6.3 | 96 241 | 0.7 |
| Zip code classified by SEER | ||||||||||||||||||||
| Urban | 190 340 | 26.1 | 1245 | 50.1 | 85 984 | 5.7 | 35 673 | 6.4 | 92 393 | 0.3 | 15 351 | 38.6 | 28 264 | 5.7 | 129 858 | 1.1 | 213 129 | 7.6 | 471 730 | 0.9 |
| Rural | 13 235 | 24.2 | 99 | 48.5 | 7578 | 4.9 | 2651 | 5.2 | 10 746 | 0.2 | 1093 | 38.0 | 2331 | 4.4 | 10 952 | 0.9 | 18 132 | 6.3 | 38 818 | 0.7 |
| Cancer stageb | ||||||||||||||||||||
| 0 | 35 224 | 20.3 | 138 | 46.4 | 4640 | 2.5 | 142 | 2.1 | 289 | 0.7 | 38 | 28.9 | 223 | 1.3 | NA | NA | NA | NA | ||
| I | 82 431 | 25.7 | 399 | 54.1 | 19 152 | 4.6 | 26 416 | 6.0 | 18 694 | 0.5 | 3080 | 34.7 | 3503 | 6.0 | 29 557 | 0.5 | NA | NA | NA | NA |
| II | 50 216 | 30.6 | 433 | 54.7 | 19 325 | 6.3 | 1700 | 5.1 | 6953 | 0.2 | 1235 | 44.0 | 6045 | 6.2 | 62 491 | 0.6 | NA | NA | NA | NA |
| III | 17 030 | 32.2 | 180 | 49.4 | 22 690 | 7.4 | 4552 | 8.6 | 18 195 | 0.3 | 5970 | 45.1 | 2892 | 9.8 | 16 696 | 1.8 | NA | NA | NA | NA |
| IV | 9398 | 19.7 | 115 | 40.0 | 19 685 | 6.1 | 3255 | 9.0 | 52 395 | 0.3 | 5156 | 36.1 | 15 285 | 5.0 | 15 620 | 3.7 | NA | NA | NA | NA |
Abbreviation: NA, not applicable.
Includes genetic tests performed after and within 2 years of diagnosis from January 1, 2013, through March 31, 2021.
Stage was obtained from SEER-derived American Joint Committee on Cancer 7 stage group (North American Association of Central Cancer Registries [NAACCR] variable 3430) for cases diagnosed before 2018 and SEER-derived extent of disease 2018 stage group (NAACCR variable 818) for cases diagnosed during or after 2018. Stage was omitted for categories including more than 1 cancer type (eg, multiple cancer types and all other cancer types).
Multivariable Model of Testing
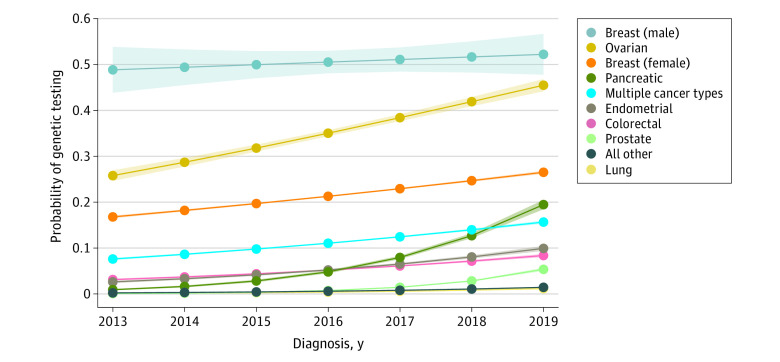
In a logistic regression model, rates of testing were lower in older patients. Eighteen percent were tested at 40 years of age compared with 2% for patients diagnosed at 80 years of age, adjusting for year and type of cancer. Testing probability was highest at 51% (95% CI, 48%-53%) for male breast cancer, 36% (95% CI, 36%-36%) for ovarian cancer, and 22% (95% CI, 22%-22%) for female breast cancer, adjusting for year and age. The modeled probability of testing increased over time for all cancer types, but testing rates exceeded 50% for male breast cancer only (Figure 1 and eTable 4 in Supplement 1).
Figure 1. Modeled Probability of Genetic Testing Over Time in the Most Common Cancer Types Diagnosed From 2013 to 2019.
The probabilities were predicted from a logistic regression model that included variables for age, cancer type, diagnosis year (2013-2019), and a diagnosis year × cancer type interaction, averaging across a constant age distribution for all years and cancer types. The shading around the curves represents 95% CIs.
A second multivariable regression model for testing included race and ethnicity. Controlling for age, cancer type, and year, testing probability differed between racial and ethnic groups overall (χ2 = 2341, P < .001) and was lower for Asian patients (6%; 95% CI, 6%-6%), Black patients (6%; 95% CI, 6%-6%), Hispanic patients (6%; 95% CI, 6%-6%), and Other patients (5%; 95% CI, 5%-5%) compared with White patients (8%; 95% CI, 8%-8%). The racial and ethnic disparities for testing were largest among patients with male breast cancer, female breast cancer, or ovarian cancer (22% [95% CI, 21%-22%] for Asian patients, 25% [95% CI, 24%-25%] for Black patients, 23% [95% CI, 23%-23%] for Hispanic patients, and 31% [95% CI, 30%-31%] for White patients; P < .001 using the χ2 test).
The modeled probability of genetic testing by year and race and ethnicity among patients with male breast, female breast, and ovarian cancer types vs other cancer types appears in Figure 2. A race and ethnicity × diagnosis year interaction term showed no improvement in racial and ethnic differences over time. Compared with White patients, the odds of testing by year decreased for Hispanic patients (odds ratio [OR], 0.98 [95% CI, 0.97-0.99]) and were unchanged for Asian patients (OR, 1.00 [95% CI, 0.98-1.01]), Black patients (OR, 0.99 [95% CI, 0.98-1.01]), and Other patients (OR, 0.99 [95% CI, 0.95-1.03]); the comparisons yielded statistically significant results (likelihood ratio χ24 = 9.76, P = .045; eTable 5 in Supplement 1).
Figure 2. Modeled Probability of Genetic Testing Over Time Across Racial and Ethnic Groups.
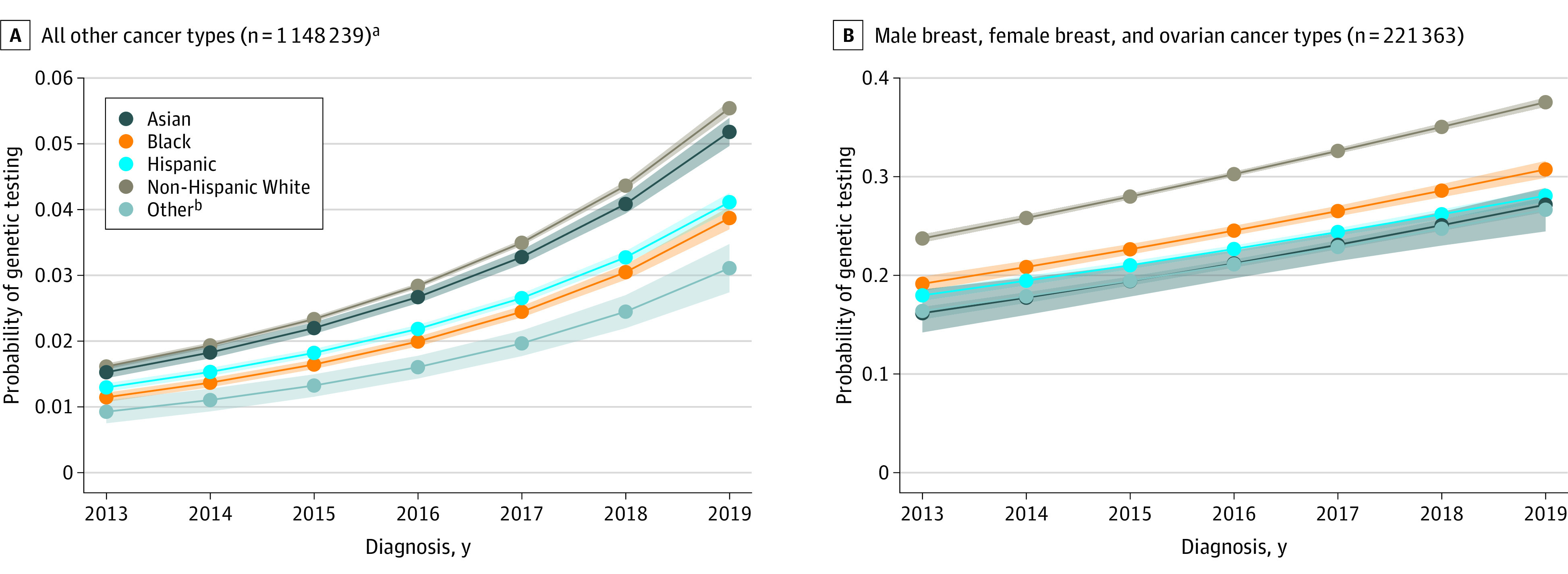
The probabilities were predicted from a logistic regression model that included age, cancer type, diagnosis year (2013-2019), an indicator for the 3 cancer types (male breast, female breast, and ovarian) with the highest testing rates that had testing guidelines throughout the study period, and 3 interactions: (1) cancer type and age, (2) racial and ethnic group and year, and (3) the 3 cancer types (male breast, female breast, and ovarian) with the highest testing rates and racial and ethnic group. The estimates were averaged across constant age and cancer type distributions within each of (1) other cancer types (excluded male breast, female breast, and ovarian) and (2) only male breast, female breast, and ovarian cancer types. The racial and ethnic categories were mutually exclusive. The shading around the curves represents 95% CIs.
aIncludes all cancer types not included in panel B.
bIncludes patients with Surveillance, Epidemiology, and End Results race1 variable coded as Native American, unknown, and Other.
Pathogenic and Uncertain Results by Race, Ethnicity, and Cancer Type
The median number of genes tested increased by year from 2 in 2013 to 34 in 2019. The frequency of pathogenic results was similar across cancer types (10%-30%) and stable over time (eTable 6 in Supplement 1). Uncertain results increased at a greater rate in races and ethnicities other than White (40.0% in Asian patients in 2019 vs 12.2% in 2013, 39.0% in Black patients in 2019 vs 7.5% in 2013, 29.3% in Hispanic patients in 2019 vs 8.6% in 2013, 24.9% in White patients in 2019 vs 6.3% in 2013, and 34.6% in Other patients in 2019 vs 3.8% in 2013; χ2 = 1808, P < .001; eTable 7 in Supplement 1). The ratio of uncertain to pathogenic results varied by race and ethnicity: White patients diagnosed in 2019 were 1.73 (95% CI, 1.56-1.92) times more likely to receive uncertain results (24.9%) than pathogenic results (14.4%), whereas Asian and Black patients diagnosed in 2019 were 3.74 (95% CI, 3.13-4.46) times more likely to receive uncertain results (40.0% for Asian patients and 39.0% for Black patients) than pathogenic results (10.2% for Asian patients and 11.0% for Black patients) (eTable 7 in Supplement 1).
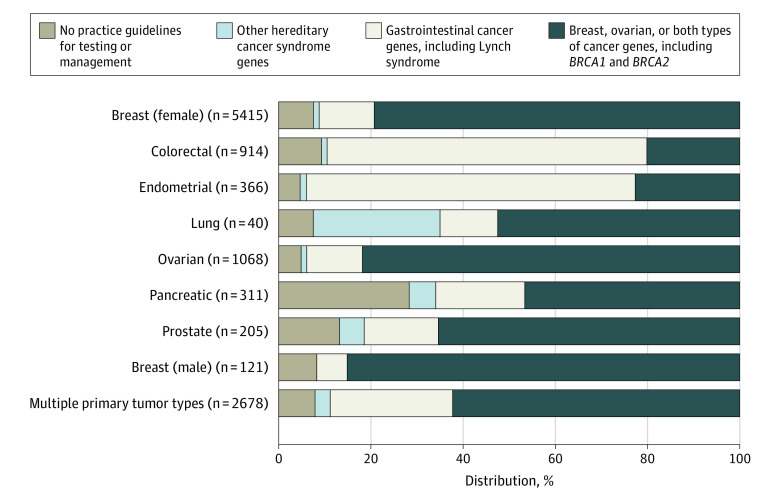
Genes With Pathogenic Results by Cancer Type
Of all pathogenic results, 67.5% to 94.9% of variants were identified in genes for which practice guidelines recommend testing and 68.3% to 83.8% of variants were identified in genes associated with the diagnosed cancer type. The distribution of genes for individuals with pathogenic results (by cancer-associated gene categories) appear in Figure 3. Gastrointestinal cancer–associated genes represented 68.3% of pathogenic results in colorectal cancer and 71.8% of pathogenic results in endometrial cancer. Breast and ovarian cancer–associated genes represented 79.5% of pathogenic results in female breast cancer, 83.8% in male breast cancer, and 82.0% in ovarian cancer. Non–guideline-recommended genes represented between 5.1% of pathogenic results in endometrial cancer to 28.1% of pathogenic results in pancreatic cancer. In a sensitivity analysis, MUTYH was recoded as a non–guideline-recommended gene, and the non–guideline-recommended genes increased to 10.4% of pathogenic results in endometrial cancer and 32.5% in pancreatic cancer (eFigure 2 in Supplement 1).
Figure 3. Distribution of Genes for Patients With Pathogenic Results.
The results are grouped by associated cancer types or hereditary syndromes and by practice guideline indications for testing across selected cancer types (additional details appear in eTable 1 in Supplement 1).
Discussion
In this population-based study conducted in California and Georgia between 2013 and 2021, use of germline genetic testing was only 6.8% after a cancer diagnosis. Genetic testing rates increased over time, but even in 2021 were far lower than 100% for specific cancer types, such as ovarian, male breast, and pancreatic, recommended by practice guidelines. Because clinical trials have demonstrated that germline-directed cancer screening, preventive surgery, and targeted therapies can improve survival,1,2,3,4 low rates of germline genetic testing may contribute to higher rates of cancer mortality.
The racial and ethnic disparities of lower rates of testing in Asian, Black, and Hispanic patients compared with White patients were largest for the 3 cancer types that had the highest testing rates overall and that had established recommendations from practice guidelines throughout the study period5,15: male breast, female breast, and ovarian. This finding is consistent with prior studies.9,16,17 Racial and ethnic differences in testing persisted through 2021. Although there are many possible explanations (including individual preferences and insurance coverage), strategies such as education of clinicians, incorporating genetic counselors into oncology practices, telemedicine, and electronic health record reminders warrant study to address testing gaps affecting patients who are Asian, Black, or Hispanic.18,19
Testing rates were heterogeneous between syndromes with higher rates of testing in primarily BRCA1/2-associated (26.0% for breast cancer and 38.6% for ovarian cancer) than Lynch syndrome–associated cancer types (5.6% for colorectal cancer and 6.4% for endometrial cancer). Persistent undertesting of Lynch syndrome–associated cancer types represents a target for improvement because population frequencies of pathogenic results in Lynch syndrome genes and BRCA1/2 are similar.20,21 Germline genetic testing rates did not differ by cancer stage; this appears inconsistent with previous reports that sequencing advanced cancer types often identifies results warranting confirmatory germline testing.22 The lack of association between cancer stage and germline genetic testing may reflect insufficient confirmatory testing of patients with advanced cancer and failure to offer testing to patients’ relatives.23,24
Although testing failed to meet practice guidelines, it increased substantially over time. This might be explained by the growing evidence demonstrating benefits of treatment with poly(ADP-ribose) polymerase inhibitors. These drugs were approved for BRCA1/2-associated ovarian cancer in 2014, breast cancer in 2018, pancreatic cancer in 2019, and prostate cancer in 2020.2,25,26,27
Pathogenic results were most common in genes with management guidelines, such as prophylactic salpingo-oophorectomy or frequent screening colonoscopy.5,6 This suggests that most pathogenic results may facilitate personalized, risk-adapted care. Pathogenic results often occurred in diagnosis-concordant genes (eg, breast cancer– and ovarian cancer–associated genes in patients with breast cancer),5,6 but were also observed in diagnosis-discordant categories. These results offer support for panel testing of multiple genes across cancer types with appropriate counseling.
By contrast, higher rates of uncertain results, particularly in Asian and Black patients, have the potential to result in suboptimal care because some studies reported mismanagement of uncertain results with preventive surgeries.28,29 Prior studies have shown that uncertain results are more frequent among patients from racial and ethnic groups that have had less access to genetic testing.18,30 Even though germline genetic testing costs have declined and insurance coverage has increased, out-of-pocket costs in the range of $100 to $250 may present a barrier to testing. Increased access to both clinical testing and genetics research is needed for underrepresented groups.
Limitations
This study has several limitations. First, germline genetic testing from laboratories other than the 4 laboratories selected for this study, or from direct-to-consumer laboratories, was not ascertained; however, evidence suggested that these 4 participating laboratories were the primary genetic testing laboratories.8,9
Second, SEER lacks data about family history or tumor sequencing. Third, insurance information was incomplete. Fourth, data were not available about why germline genetic testing did not occur, such as when testing was declined by patients. Fifth, data were limited to California and Georgia and may not apply to other states in the US.
Conclusions
Among patients diagnosed with cancer in California and Georgia between 2013 and 2019, only 6.8% underwent germline genetic testing. Compared with non-Hispanic White patients, rates of testing were lower among Asian, Black, and Hispanic patients.
eFigure 1. Flow and decay diagram
eFigure 2. Genes with pathogenic results, grouped by associated cancers and/or hereditary syndromes and by practice guideline indications for clinical testing, with MUTYH re-categorized as a gene with no practice guidelines for testing
eMethods
eTable 1. All tested genes, grouped by cancer type associations and testing guideline recommendations
eTable 2. Genetic testing utilization among individuals with all other cancer types, diagnosed 2013-2019
eTable 3. Genetic testing time trends among individuals diagnosed with cancer in Georgia and California and reported to SEER registries from 2013-2019
eTable 4. Logistic regression model output for individuals diagnosed with cancer in Georgia and California and reported to SEER registries from 2013-2019, controlling for age and cancer type and allowing for a year by cancer type interaction
eTable 5. Three logistic regression model outputs regressing genetic testing on SEER racial and ethnic categories controlling for year, age and a variable that indicates the three cancers with the highest testing rates that had testing guidelines throughout the study period (male breast, female breast, and ovarian cancer) for cancers diagnosed in Georgia and California and reported to SEER registries from 2013-2019
eTable 6. Uncertain and pathogenic results and ratio in individuals with selected cancers diagnosed and reported to Georgia and California SEER registries, 2013-2019
eTable 7. Uncertain and pathogenic results and ratio in individuals with cancers diagnosed and reported to Georgia and California SEER Registries, 2013-2019, by race and ethnicity
Data sharing statement
References
- 1.Geyer CE Jr, Garber JE, Gelber RD, et al. ; OlympiA Clinical Trial Steering Committee and Investigators . Overall survival in the OlympiA phase III trial of adjuvant olaparib in patients with germline pathogenic variants in BRCA1/2 and high-risk, early breast cancer. Ann Oncol. 2022;33(12):1250-1268. doi: 10.1016/j.annonc.2022.09.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hussain M, Mateo J, Fizazi K, et al. ; PROfound Trial Investigators . Survival with olaparib in metastatic castration-resistant prostate cancer. N Engl J Med. 2020;383(24):2345-2357. doi: 10.1056/NEJMoa2022485 [DOI] [PubMed] [Google Scholar]
- 3.Domchek SM, Friebel TM, Singer CF, et al. Association of risk-reducing surgery in BRCA1 or BRCA2 mutation carriers with cancer risk and mortality. JAMA. 2010;304(9):967-975. doi: 10.1001/jama.2010.1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ladabaum U, Wang G, Terdiman J, et al. Strategies to identify the Lynch syndrome among patients with colorectal cancer: a cost-effectiveness analysis. Ann Intern Med. 2011;155(2):69-79. doi: 10.7326/0003-4819-155-2-201107190-00002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.National Comprehensive Cancer Network . Genetic/familial high risk assessment: breast, ovarian and pancreatic: version 3. Published February 13, 2023. Accessed March 19, 2023. https://www.nccn.org
- 6.National Comprehensive Cancer Network . Genetic/familial high-risk assessment: colorectal: version 2. Published December 7, 2022. Accessed March 19, 2023. https://www.nccn.org
- 7.National Comprehensive Cancer Network . Prostate cancer early detection: version 1. Published January 9, 2023. Accessed March 19, 2023. https://www.nccn.org
- 8.Kurian AW, Ward KC, Howlader N, et al. Genetic testing and results in a population-based cohort of breast cancer patients and ovarian cancer patients. J Clin Oncol. 2019;37(15):1305-1315. doi: 10.1200/JCO.18.01854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kurian AW, Griffith KA, Hamilton AS, et al. Genetic testing and counseling among patients with newly diagnosed breast cancer. JAMA. 2017;317(5):531-534. doi: 10.1001/jama.2016.16918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.US Department of Health and Human Services . Guidance regarding methods for de-identification of protected health information in accordance with the Health Insurance Portability and Accountability Act (HIPAA) privacy rule. Accessed March 19, 2023. https://www.hhs.gov/hipaa/for-professionals/privacy/special-topics/de-identification/index.html#safeharborguidance
- 11.Kalia SS, Adelman K, Bale SJ, et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet Med. 2017;19(2):249-255. doi: 10.1038/gim.2016.190 [DOI] [PubMed] [Google Scholar]
- 12.North American Association of Central Cancer Registries . Data dictionary. Accessed March 19, 2023. http://datadictionary.naaccr.org/default.aspx?c=10&Version=23#220
- 13.Sorscher S, LoPiccolo J, Chen E, et al. Landscape of pathogenic germline variants in patients with lung cancer. J Clin Oncol. 2022;40(suppl 36):388570. doi: 10.1200/JCO.2022.40.36_suppl.388570 [DOI] [Google Scholar]
- 14.Maxwell KN, Wenz BM, Kulkarni A, et al. Mutation rates in cancer susceptibility genes in patients with breast cancer with multiple primary cancers. JCO Precision Oncol. 2020;4:916-925. doi: 10.1200/PO.19.00301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daly MB, Pilarski R, Axilbund JE, et al. ; National Comprehensive Cancer Network . Genetic/familial high-risk assessment: breast and ovarian, version 1.2014. J Natl Compr Canc Netw. 2014;12(9):1326-1338. doi: 10.6004/jnccn.2014.0127 [DOI] [PubMed] [Google Scholar]
- 16.McCarthy AM, Bristol M, Domchek SM, et al. Health care segregation, physician recommendation, and racial disparities in BRCA1/2 testing among women with breast cancer. J Clin Oncol. 2016;34(22):2610-2618. doi: 10.1200/JCO.2015.66.0019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dharwadkar P, Greenan G, Stoffel EM, et al. Racial and ethnic disparities in germline genetic testing of patients with young-onset colorectal cancer. Clin Gastroenterol Hepatol. 2022;20(2):353-361.e3. doi: 10.1016/j.cgh.2020.12.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reid S, Spalluto LB, Pal T. Strategies to enhance identification of hereditary breast cancer gene carriers. Expert Rev Mol Diagn. 2020;20(9):861-865. doi: 10.1080/14737159.2020.1816829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin J, Sharaf RN, Saganty R, et al. Achieving universal genetic assessment for women with ovarian cancer: are we there yet? a systematic review and meta-analysis. Gynecol Oncol. 2021;162(2):506-516. doi: 10.1016/j.ygyno.2021.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buchanan AH, Lester Kirchner H, Schwartz MLB, et al. Clinical outcomes of a genomic screening program for actionable genetic conditions. Genet Med. 2020;22(11):1874-1882. doi: 10.1038/s41436-020-0876-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Risch HA, McLaughlin JR, Cole DE, et al. Population BRCA1 and BRCA2 mutation frequencies and cancer penetrances: a kin-cohort study in Ontario, Canada. J Natl Cancer Inst. 2006;98(23):1694-1706. [DOI] [PubMed] [Google Scholar]
- 22.Stadler ZK, Maio A, Chakravarty D, et al. Therapeutic implications of germline testing in patients with advanced cancers. J Clin Oncol. 2021;39(24):2698-2709. doi: 10.1200/JCO.20.03661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Offit K, Tkachuk KA, Stadler ZK, et al. Cascading after peridiagnostic cancer genetic testing: an alternative to population-based screening. J Clin Oncol. 2020;38(13):1398-1408. doi: 10.1200/JCO.19.02010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurian AW, Katz SJ. Emerging opportunity of cascade genetic testing for population-wide cancer prevention and control. J Clin Oncol. 2020;38(13):1371-1374. doi: 10.1200/JCO.20.00140 [DOI] [PubMed] [Google Scholar]
- 25.Kim G, Ison G, McKee AE, et al. FDA approval summary: olaparib monotherapy in patients with deleterious germline BRCA-mutated advanced ovarian cancer treated with three or more lines of chemotherapy. Clin Cancer Res. 2015;21(19):4257-4261. doi: 10.1158/1078-0432.CCR-15-0887 [DOI] [PubMed] [Google Scholar]
- 26.Robson M, Im SA, Senkus E, et al. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N Engl J Med. 2017;377(6):523-533. doi: 10.1056/NEJMoa1706450 [DOI] [PubMed] [Google Scholar]
- 27.Golan T, Hammel P, Reni M, et al. Maintenance olaparib for germline BRCA-mutated metastatic pancreatic cancer. N Engl J Med. 2019;381(4):317-327. doi: 10.1056/NEJMoa1903387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Domchek S, Brower J, Symecko H, et al. Uptake of oophorectomy in women with findings on multigene panel testing: results from the Prospective Registry of Multiplex Testing (PROMPT). J Clin Oncol. 2020;38(1508). doi: 10.1200/JCO.2020.38.15_suppl.1508 [DOI] [Google Scholar]
- 29.Kurian AW, Li Y, Hamilton AS, et al. Gaps in incorporating germline genetic testing into treatment decision-making for early-stage breast cancer. J Clin Oncol. 2017;35(20):2232-2239. doi: 10.1200/JCO.2016.71.6480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manrai AK, Funke BH, Rehm HL, et al. Genetic misdiagnoses and the potential for health disparities. N Engl J Med. 2016;375(7):655-665. doi: 10.1056/NEJMsa1507092 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eFigure 1. Flow and decay diagram
eFigure 2. Genes with pathogenic results, grouped by associated cancers and/or hereditary syndromes and by practice guideline indications for clinical testing, with MUTYH re-categorized as a gene with no practice guidelines for testing
eMethods
eTable 1. All tested genes, grouped by cancer type associations and testing guideline recommendations
eTable 2. Genetic testing utilization among individuals with all other cancer types, diagnosed 2013-2019
eTable 3. Genetic testing time trends among individuals diagnosed with cancer in Georgia and California and reported to SEER registries from 2013-2019
eTable 4. Logistic regression model output for individuals diagnosed with cancer in Georgia and California and reported to SEER registries from 2013-2019, controlling for age and cancer type and allowing for a year by cancer type interaction
eTable 5. Three logistic regression model outputs regressing genetic testing on SEER racial and ethnic categories controlling for year, age and a variable that indicates the three cancers with the highest testing rates that had testing guidelines throughout the study period (male breast, female breast, and ovarian cancer) for cancers diagnosed in Georgia and California and reported to SEER registries from 2013-2019
eTable 6. Uncertain and pathogenic results and ratio in individuals with selected cancers diagnosed and reported to Georgia and California SEER registries, 2013-2019
eTable 7. Uncertain and pathogenic results and ratio in individuals with cancers diagnosed and reported to Georgia and California SEER Registries, 2013-2019, by race and ethnicity
Data sharing statement