Abstract
Version 2.1 of ASDB (Alternative Splicing Data Base) contains 1922 protein and 2486 DNA sequences. The protein entries from SWISS-PROT are joined into clusters corresponding to alternatively spliced variants of one gene. The DNA division consists of complete genes with alternative splicing mentioned or annotated in GenBank. The search engine allows one to search over SWISS-PROT and GenBank fields and then follow the links to all variants. The database can be assessed at the URL http://cbcg.nersc.gov/asdb
DATABASE DESCRIPTION
Version 2.1 of ASDB consists of two divisions, ASDB(proteins), which contains amino acid sequences, and ASDB(nucleotides) with genomic sequences. ASDB(nucleotides) was developed in 1999, while ASDB(proteins) was updated with the latest data from SWISS-PROT and improved clustering procedures described below.
SWISS-PROT uses two formats for description of alternative splicing. Thus the protein sequences were selected from SWISS-PROT (1) using full text search for the words ‘alternative splicing’ (usually in the CC lines) and ‘varsplic’ (in the FT lines). This search generated 1922 initial entries. Some entries describe just one alternatively spliced variant, some (those that include the ‘varsplic’ field) indicate several.
In order to group proteins that could arise by alternative splicing of the same gene, we developed the clustering procedure. Two proteins were linked if they had a common fragment of at least 20 amino acids, and clusters were initially defined as maximum connected groups of linked proteins (2). Each cluster was represented by multiple alignment of its members constructed using CLUSTALW (3).
It turned out that some clusters were chimeric, in the sense that they contained members of multigene families, but not alternatively spliced variants of one gene. Therefore the multiple alignments were subject to additional analysis aimed at detection of chimeric clusters. This post-processing was based on the following assumption: bona fide alternative variants have few relatively long mismatching regions, as opposed to homologous proteins whose alignment contains multiple short mismatches. However, random matching residues within alternative fragments complicate implementation of this criterion. Thus, as the first step, short runs of matches between mismatches (up to three matching amino acids) were marked as mismatches. During the second step the mismatched fragments were counted. If there were less than five such fragments, the proteins were assumed to be alternatively spliced variants. If there were more than 15, the proteins were assumed to be homologues. The intermediate cases were subject to case by case manual analysis. Through the use of this procedure all chimeric clusters were removed. The distribution of cluster size, representation of species and other relevant statistics can be retrieved from the ASDB Web site.
This processing covers the cases when alternatively spliced variants are described in separate SWISS-PROT entries. The other kind of ASDB records, originating from the SWISS-PROT entries with the ‘varsplic’ field in the feature table, usually describe the proteins that are not part of any cluster. In these cases, information on the variable fragments of the several proteins which result from the alternative splicing of a single gene is contained in the entry itself. ASDB(proteins) entries are marked with different symbols to allow for easy differentiation among the three types: those proteins which are part of the ASDB clusters and the corresponding multi-alignments, those which have information on different variants in the associated SWISS-PROT entries, and those for which information on the variants is not available at the present time. ASDB contains internal links between entries and/or clusters, as well as external links to MEDLINE, GenBank and SWISS-PROT entries.
The ASDB(nucleotides) division was generated by collecting all GenBank (4) entries containing the words ‘alternative splicing’ and further selection of those entries that contain complete gene sequences (all CDS fields are complete, i.e., they do not have continuation signs). The distribution of the number of entries over the most represented species is given in Figure 1.
Figure 1.
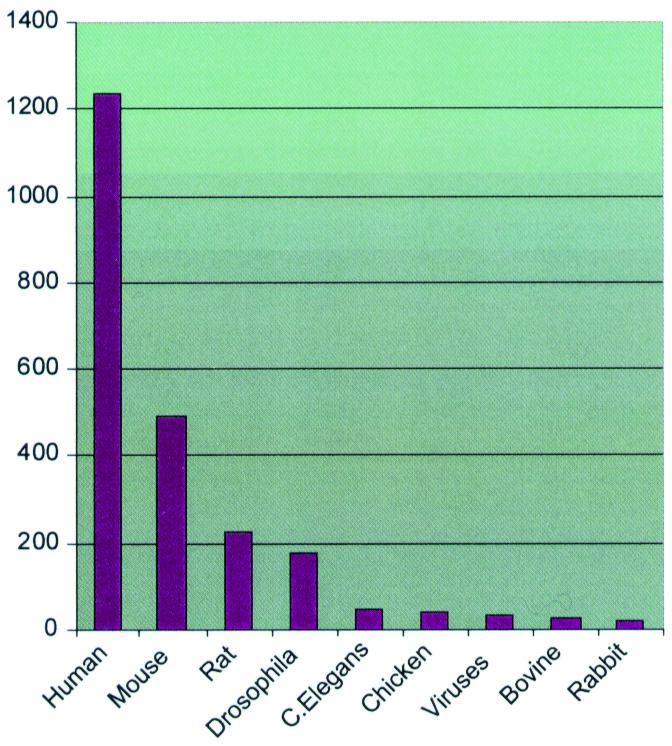
The number of ASDB(nucleotide) entries for the most represented species
The database can be searched using MEDLINE, SWISS-PROT (1) and GenBank (4) identifiers and accession numbers. Standard context search can be performed over SWISS-PROT and GenBank keywords, description, taxonomy, comment fields and feature tables. Standard boolean logic is supported to allow for a wider range of queries.
SUPPLEMENTARY MATERIAL
Instructions on using ASDB and explanation of its various features are available via NAR Online.
AVAILABILITY
ASDB is available at the URL http://cbcg.nersc.gov/asdb . The administrator of the database can be contacted by Email at asdb@lbl.gov
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Sylvia Spengler, Andrey Mironov and Pavel Pevzner for useful discussions. This work was supported by the Director, Office of Energy Research, Office of Biological and Environmental Research, of the US Department of Energy under Contract No. DE-ACO3-76SF00098. M.G. was partially supported by grants from the Russian Fund of Basic Research (99-04-48347), the Russian State Scientific Program ‘Human Genome’ (65/99), and the Merck Genome Research Institute (244).
REFERENCES
- 1.Bairoch A. and Apweiler,R. (1999) Nucleic Acids Res., 27, 49–54. Updated article in this issue: Nucleic Acids Res. (2000), 28, 45–48.9847139 [Google Scholar]
- 2.Gelfand M.S., Dubchak,I., Dralyuk,I. and Zorn,M. (1999) Nucleic Acids Res., 27, 301–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thompson J.D., Gibson,T.J., Plewniak,F., Jeanmougin,F. and Higgins,D.G. (1997) Nucleic Acids Res., 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benson D.A., Boguski,M.S., Lipman,D.J., Ostell,J., Ouellette,B.F.F., Rapp,B.A. and Wheeler,D.L. (1999) Nucleic Acids Res., 27, 12–17. Updated article in this issue: Nucleic Acids Res. (2000), 28, 15–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


