Abstract
Single-cell multi-omics technologies and methods characterize cell states and activities by simultaneously integrating various single-modality omics methods that profile the transcriptome, genome, epigenome, epitranscriptome, proteome, metabolome and other (emerging) omics. Collectively, these methods are revolutionizing molecular cell biology research. In this comprehensive Review, we discuss established multi-omics technologies as well as cutting-edge and state-of-the-art methods in the field. We discuss how multi-omics technologies have been adapted and improved over the past decade using a framework characterized by optimization of throughput and resolution, modality integration, uniqueness and accuracy, and we also discuss multi-omics limitations. We highlight the impact that single-cell multi-omics technologies have had in cell lineage tracing, tissue-specific and cell-specific atlas production, tumour immunology and cancer genetics, and in mapping of cellular spatial information in fundamental and translational research. Finally, we discuss bioinformatics tools that have been developed to link different omics modalities and elucidate functionality through the use of better mathematical modelling and computational methods.
Subject terms: Next-generation sequencing, Transcriptomics, Protein-protein interaction networks
Single-cell multi-omics methods are essential for characterizing cell states and types. The past decade has ushered in improvements in spatial resolution and computational data integration and in new omics modalities. Consequently, single-cell multi-omics have advanced fundamental and translational research, including, for example, in production of cell atlases and in tumour immunology therapeutics.
Introduction
Various single-omics methods have generated a plethora of single-modality data aiming to dissect the mechanistic basis of gene regulation or reveal aspects of human diseases. Cell diversity within the human body is complex as cells undergo proliferation, differentiation and death, and this diversity is amplified when considering the tissue in which these cells reside — their local and distant environments. Thus, integrative capture and analyses of multiple cellular processes is desirable to resolve their biological complexity.
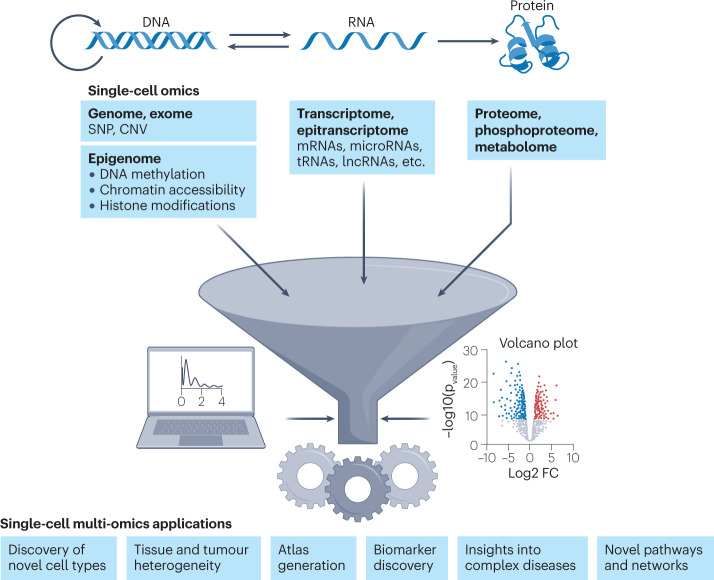
Methodological and technological advances now allow the simultaneous profiling of genome, epigenome, transcriptome, proteome and other (emerging) omics modalities in an effort to better understand biological mechanisms and genotype-to-phenotype relationships. By using laser capture microdissection (LCM), robotic micromanipulation, fluorescence-activated cell sorting or microfluidic platforms1, single cells can be isolated into individual compartments where they undergo barcoding of multiple types of molecules, thereby enabling genome-scale characterization of cell-type-specific genomic information at an unprecedented resolution. Over the past decade, single-cell multi-omics technologies have been developed to directly characterize the phenotypes that are usually inferred only from single-omics methods. These technologies have generated a huge amount of data that enable the discovery of mechanisms of gene regulation, protein expression dynamics, epigenetic variation and cell perturbation models, which have revolutionized how we understand cell heterogeneity in health and disease (Fig. 1). As single-cell technologies have continually advanced, emerging methods capable of spatially mapping gene expression within the tissue drove the conception of ‘spatial multi-omics’.
Fig. 1. From single omics to multi-omics and their broad applications.
Single-omics technologies consist of many modalities, each pertaining to specific parts of the central dogma of DNA–RNA–protein. Single-cell single-modality methods utilize technologies for a variety of applications including but not limited to quantification of genomic, exomic and epigenomic (DNA methylation, chromatin accessibility and histone modifications) information from DNA; transcriptomic and epitranscriptomic information from RNA; and proteomic, phosphoproteomic and metabolomic data from proteins. Integration of single-omics modalities, aided by computational analysis methods, contributes to a broad array of fundamental and clinical research applications ranging from generation of cell and tissue atlases to interrogation of complex disease biology. CNV, copy number variation; FC, fold change; lncRNA, long non-coding RNA; SNP, single-nucleotide polymorphism.
Gaining a clearer understanding of the origin, state and fate of a cell in a physiological and specifically immunological context2 is a pressing need within biological and biomedical research. Continuous advancements in multiplexing, throughput, resolution and accuracy in single-cell multi-omics technologies have aided the comprehensive delineation of the genetic landscape of a cell and revolutionized our capacity to identify cellular heterogeneities and relationships (Table 1). The broad implementation of these technologies has been transformative in enabling applications such as tracing cell lineages3, producing cell-type atlases of various organs4 and dissecting mechanisms in disease settings such as tumour immunology and cancer genetics5. In this Review, we discuss how single-cell multi-omics technologies have been improved and adapted over the past decade, the biological and clinical impact of these technologies, and the bioinformatics tools that have become integral to functional multi-omics.
Table 1.
Single-cell and spatial omics methods by date
| Year | Method | Technology | Data types provided | Resolution | Ref. |
|---|---|---|---|---|---|
| 2014 | Co-detection and sequencing of genes and transcripts | Microchannel-based microfluidics | DNA, mRNA sequence | Single cell | 20 |
| Smart-seq2 | Plate-based sequencing | DNA, mRNA | Single cell | 13 | |
| 2015 | G&T-seq | Plate-based sequencing | DNA, mRNA | Single cell | 21 |
| DR-seq | Mouth-pipetting sequencing | DNA, mRNA | Single cell | 22 | |
| 2016 | scM&T-seq | Bead-based sequencing | DNA methylation, mRNA | Single cell | 28 |
| scTrio-seq | Pipette cell-picking sequencing | DNA, RNA, DNA methylation | Single cell | 33 | |
| PLAYR | Mass cytometry | mRNA, protein | Single cell | 67 | |
| Perturb-seq | Droplet-based microfluidics sequencing | sgRNA, mRNA | Single cell | 82 | |
| Spatial transcriptomics | Microarray-based sequencing | mRNA | Spatial, 100 μm | 116 | |
| 2017 | scNOMe-seq | Plate-based sequencing | DNA methylation, chromatin accessibility | Single cell | 35 |
| CITE-seq | Droplet-based microfluidics sequencing | mRNA, protein | Single cell | 68 | |
| REAP-seq | Droplet-based microfluidics sequencing | mRNA, protein | Single cell | 69 | |
| Geo-seq | Laser capture microdissection and single-cell RNA-seq | Transcriptome | Spatial | 134 | |
| 2018 | SIDR | Microplate-based sequencing | DNA, mRNA | Single cell | 24 |
| Sci-CAR-seq | Plate-based sequencing | Chromatin accessibility, mRNA | Single cell | 56 | |
| SPLiT-seq | Plate-based sequencing | Chromatin accessibility, mRNA | Single cell | 58 | |
| 10x Visium | Microarray-based sequencing | mRNA | Spatial, 55 μm | 117 | |
| 2019 | Target-seq | Plate-based sequencing | Genomic and coding DNA, mRNA | Single cell | 23 |
| scMT-seq | Micro-pipetting sequencing | DNA methylation, mRNA | Single cell | 31 | |
| scChIP–seq | Droplet-based microfluidics sequencing | Histone modifications, mRNA | Single cell | 45 | |
| ECCITE-seq | Droplet-based microfluidics sequencing | mRNA, T cell receptor clonotype, protein, sgRNA | Single cell | 81 | |
| SNARE-seq | Droplet-based microfluidics sequencing | Accessible chromatin, mRNA | Single cell | 57 | |
| RAID | Plate-based sequencing | Intracellular proteins, phosphorylated proteins, mRNA | Single cell | 77 | |
| Slide-seq | Microarray-based sequencing | mRNA | Spatial, 10 μm | 118 | |
| HDST | Microarray-based sequencing | mRNA | Spatial, 2 μm | 120 | |
| 2020 | DBiT-seq | Microchannel-based microfluidics | mRNA, protein | Spatial, 10–50 μm | 123 |
| ASTAR-seq | Microfluidics-based sequencing | Chromatin accessibility, mRNA | Single cell | 54 | |
| 2021 | Paired-Tag | Plate-based sequencing | Histone modifications, mRNA | Single cell | 51 |
| CoTECH | Plate-based sequencing | Chromatin occupancy, mRNA | Single cell | 52 | |
| ASAP-seq and DOGMA-seq | Droplet-based microfluidics sequencing | Accessible chromatin, mRNA, proteins, mitochondrial DNA | Single cell | 78 | |
| TEA-seq | Droplet-based microfluidics sequencing | Accessible chromatin, proteins, mRNA | Single cell | 61 | |
| Slide-seqV2 | Microarray-based sequencing | mRNA | Spatial, 10 μm | 119 | |
| 2022 | scCUT&Tag pro | Droplet-based microfluidics sequencing | Histone modifications, proteins | Single cell | 50 |
| ISSAAC-seq | Droplet-based sequencing | Chromatin accessibility, mRNA | Single cell | 62 | |
| PHAGE-ATAC | Droplet-based microfluidics sequencing | Accessible chromatin, proteins, mitochondrial DNA | Single cell | 74 | |
| NEAT-seq | Droplet-based microfluidics sequencing | Intracellular proteins, accessible chromatin, mRNA | Single cell | 80 | |
| scGET-seq | Droplet-based microfluidics sequencing | Accessible chromatin, heterochromatin, CNVs, SNVs | Single cell | 182 | |
| Slide-DNA-seq | Microarray-based sequencing | CNVs, clonal populations, spatial transcriptome | Spatial, 25 μm | 157 | |
| Spatial ATAC-seq | Microchannel-based microfluidics sequencing | Chromatin accessibility | Spatial, 10–50 μm | 124 | |
| Spatial CUT&Tag | Microchannel-based microfluidics sequencing | Histone modification | Spatial, 10–50 μm | 125 | |
| SPARC-seq | Microchannel-based microfluidics sequencing | Chromatin accessibility, transcriptome | Spatial, 50 μm | 128 | |
| SM-Omics | Microarray-based sequencing | mRNA, protein | Spatial, 100 μm | 129 | |
| Slide-TCR-seq | Microarray-based sequencing | mRNA, T cell receptor clonotype | Spatial, 10 μm | 132 | |
| 2023 | Spatial CITE-seq | Microchannel-based microfluidics sequencing | mRNA, protein | Spatial, 10–50 μm | 126 |
| Spatial ATAC–RNA-seq | Microchannel-based microfluidics sequencing | Chromatin accessibility, transcriptome | Spatial, 10–50 μm | 127 | |
| Spatial CUT&Tag–RNA-seq | Microchannel-based microfluidics sequencing | Histone modification, transcriptome | Spatial, 10–50 μm | 127 | |
| SPOTS | Microarray-based sequencing | mRNA, protein | Spatial, 55 μm | 130 |
ASAP-seq, ATAC with select antigen profiling by sequencing; ASTAR-seq, assay for single-cell transcriptome and accessibility regions; ATAC-seq, assay for transposase-accessible chromatin with sequencing; CITE-seq, cellular indexing of transcriptomes and epitopes by sequencing; CNVs, copy number variants; CoTECH, combinatorial barcoding method allowing high-throughput single-cell joint detection of chromatin occupancy and transcriptome; CRISPR, clustered regularly interspaced short palindromic repeats; CUT&Tag, cleavage under targets and tagmentation; DBiT-seq, deterministic barcoding in tissue for spatial omics sequencing; DOGMA-seq, an adaptation of CITE-seq for measuring gene activity across the central dogma of gene regulation; DR-seq, genomic DNA–mRNA sequencing; ECCITE-seq, expanded CRISPR-compatible CITE-seq; G&T-seq, genome and transcriptome sequencing; Geo-seq, geographical position sequencing; HDST, high-definition spatial transcriptomics; ISSAAC-seq, in situ sequencing hetero RNA–DNA-hybrid after assay for transposase-accessible chromatin sequencing; NEAT-seq, sequencing of nuclear protein epitope abundance, chromatin accessibility and the transcriptome in single cells; Paired-Tag, parallel analysis of individual cells for RNA expression and DNA from targeted tagmentation by sequencing; PLAYR, proximity ligation assay for RNA; RAID, RNA and immunodetection; REAP-seq, RNA expression and protein sequencing assay; scChIP–seq, single-cell chromatin immunoprecipitation followed by sequencing; scCUT&Tag pro, a multimodal assay for profiling protein–DNA interactions coupled with the abundance of surface proteins in single cells; scGET-seq, single-cell genome and epigenome by transposases sequencing; Sci-CAR-seq, combinatorial indexing-based co-assay that jointly profiles chromatin accessibility and mRNA; scM&T-seq, single-cell genome-wide methylome and transcriptome sequencing; scMT-seq, simultaneously profile DNA methylome and transcriptome from one single cell; scNOMe-seq, simultaneously measure chromatin accessibility and endogenous DNA methylation in single cells; scTrio-seq, single-cell triple omics sequencing technique; sgRNA, single guide RNA; SIDR, simultaneous isolation of genomic DNA and total RNA; Smart-seq, switching mechanism at the 5′ end of RNA template; SM-Omics, spatial multi-omics; SNARE-seq, single-nucleus chromatin accessibility and mRNA expression sequencing; SNVs, single-nucleotide variants; SPARC-seq, microfluidic indexing-based spatial ATAC and RNA sequencing; SPLiT-seq, split-pool ligation-based transcriptome sequencing; SPOTS, spatial protein and transcriptome sequencing; TCR, T cell receptor; TEA-seq, a trimodal assay for integrated single-cell measurement of transcription, epitopes and chromatin accessibility.
The landscape of single-cell multi-omics
The most mature of the single-cell omics methods, single-cell transcriptomics, is often paired with other omics to study the connection between gene expression and phenotypic heterogeneity in an unbiased manner (Fig. 2). In 2009, the first single-cell genome-wide mRNA sequencing (scRNA-seq) method was reported6, thanks in part to the emergence of next-generation sequencing (NGS). The field has rapidly developed ever since and is divided into two major categories: microfluidics droplet-based transcriptomics7–10 and microwell plate-based11–13 transcriptomics (Box 1). Single-cell sequencing was designated ‘Method of the Year’ in 2013 owing to its availability, versatility and robustness14. Several scRNA-seq methods developed in the recent decade have become essential for capturing intercellular variation and its impact on cell functions at the population scale15–19. Building on scRNA-seq, multi-omics technologies preserve the progress made in characterizing transcriptomic information while simultaneously capturing other modalities and increasing throughput through cell multiplexing, cell tagging, cell hashing and other methods (Fig. 3).
Fig. 2. The landscape of multi-omics sequencing.
Many multi-omics methods function with transcriptome profiling as their ‘anchor’ to facilitate single-cell multi-omics interrogation. Genome and transcriptome sequencing (G&T-seq) is a representative multi-omics technology for profiling both the genome and the transcriptome in single cells using biotinylated oligo(dT) capture primers and streptavidin-coated magnetic beads to target the poly(A) tail of mRNA, thereby separating the mRNA from the genomic material of the cell. Single-cell assay for transposase-accessible chromatin with sequencing (scATAC-seq) profiles the transcriptome together with the epigenome by simultaneously tagging and fragmenting DNA sequences in open chromatin regions using a DNA transposase (Tn5). This method has paved the way for other technologies that profile both the epigenome and transcriptome in single nuclei, such as Sci-CAR-seq, single-nucleus chromatin accessibility and mRNA expression sequencing (SNARE-seq) and 10x Multiome. Cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq) labels single cells with antibody conjugates bound to biotinylated DNA barcodes. By using droplet-based microfluidics, mRNA and cell-surface protein barcodes are converted to cDNA, which can be split downstream into two respective libraries using size selection. V(D)J sequencing can be used in tandem with transcriptome sequencing to analyse in the same cell full-length V(D)J sequences of B cell and T cell receptors using gene 5′-end sequencing. In contrast to conventional 3′-end sequencing that is traditionally used in transcriptome sequencing, barcodes are not adjacent to the poly d(T) primer, but instead are adjacent to a sequence in the 5′-end of the transcript. V(D)J sequencing and transcriptome sequencing can be performed together in the same cell and in tandem with other modalities such as ATAC-seq, CITE-seq and other methods. These representative technologies are just some of the many examples of multi-modality technologies used in single-cell multi-omics research. ADT, antibody-derived tag; RT, reverse transcription; Sci-CAR-seq, combinatorial indexing-based co-assay that jointly profiles chromatin accessibility and mRNA.
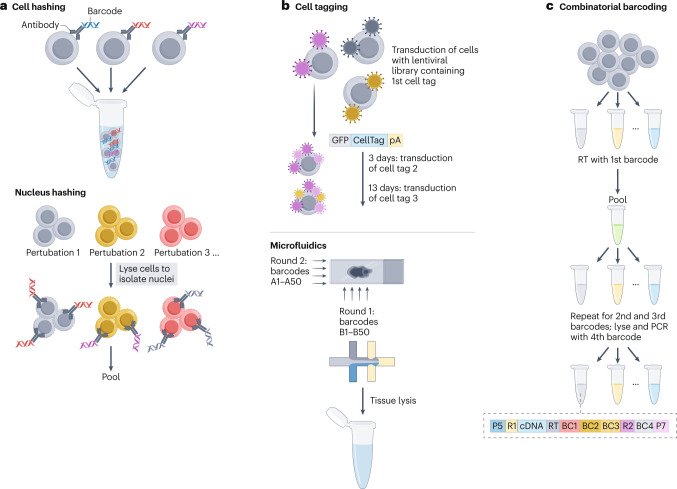
Fig. 3. Towards achieving higher throughput: multi-omics methods that increase throughput in single experiments.
a, In cell hashing, ubiquitously expressed cell-surface proteins (or nucleus-surface proteins, in the case of nuclear hashing) are bound by antibodies that are conjugated to an oligonucleotide or a specific sequence acting as a specific ‘barcode’, thereby enabling the profiling of multiple cells in a single experiment. b, Cell tagging is a combinatorial cell-indexing and high-throughput cell tracking method, in which sequential rounds of cell labelling with unique nucleic acid sequences deliver heritable barcode combinations to single cells. Microfluidic methods, such as used in deterministic barcoding in tissue for spatial omics sequencing (DBiT-seq), deliver barcodes (A1…A50, B1…B50) to cells on a tissue slide through perpendicular microfluidic channels to form a spatial grid (A1,B1…A50,B50) that is sequenced at high throughput to construct spatial omics maps. c, Combinatorial barcoding of single cells involves splitting and pooling either cells or nuclei into wells where barcodes are introduced in situ. Following multiple rounds of barcoding, molecules within the same cell are labelled with a unique barcode combination (BC1–BC4). GFP, green fluorescent protein; P5 and P7, sequencing index primers; pA, poly(A) tail; R1 and R2, read-one and read-two primers; RT, reverse transcription primer.
Box 1 Droplet-based versus plate-based transcriptomics.
Single-cell transcriptomics methods can be broadly distinguished as being either droplet-based or plate-based technologies (see the figure). Both profiling methods are akin to one another in that they involve reverse transcription (RT) of RNA into cDNA followed by PCR amplification to generate enough DNA transcripts for sequencing analysis.
Plate-based methods can generate full-length transcripts that carry ample information to allow detection of genes that are not highly expressed and other rare transcripts. However, these methods are limited by plate size and the quantity of cells available for analysis.
Droplet-based methods can analyse single cells at a much higher throughput (thousands of cells profiled at once) by utilizing microfluidic devices, but only from the 5′ or 3′ end of the transcript, thereby omitting the detection of allele-specific expression and other isoforms. These methods require both high quantity and high quality of cells.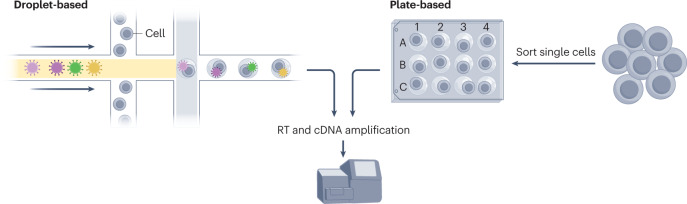
Integrating genome and transcriptome
Parallel profiling of the genome and transcriptome of the same single cell can reflect the transcriptional state of the genome. On its own, scRNA-seq is unable to provide coverage of key mutations and does not provide genome–transcriptome correlations in cells that ultimately lead to the discovery of novel gene-expression programmes in disease. Overcoming these shortcomings, we reported the first amplification and sequencing of a whole genome and whole transcriptome from a single cell using a microfluidics-facilitated technology in 2014, which captured single cells using microvalve-based control channels, selectively lysed and separated cytoplasmic and nuclear contents, and then performed on-chip amplification, thereby enabling multiple genomic measurements20.
The next year, genome and transcriptome sequencing (G&T-seq)21 and gDNA–mRNA sequencing (DR-seq)22 were reported. For G&T-seq, oligo(dT)-coated magnetic beads were used to separate genomic DNA from full-length mRNAs in 220 single cells following a modified Smart-seq213 protocol, in which cells were isolated and lysed to release genomic DNA and mRNA for whole-genome and whole-transcriptome analyses, respectively (Fig. 2). DR-seq is a low-throughput method that primarily deviates from G&T-seq in the amplification step, in which the DNA and RNA amplification occurs before separation, thereby allowing for reduction of contamination and RNA loss. Both methods are similar in that DNA and RNA must be physically separated, which may result in loss of genetic material and sensitivity or in amplification bias, which limits the ability to confidently detect small pathogenic alterations. In 2019, TARGET-seq, a plate-based method, demonstrated increased throughput of ~5,000 single cells profiled per run through the use of barcoding and pooling of libraries in reduced reaction volumes and was set apart from previously mentioned methods by improving mutation coverage using protein digestion, which improves the release of DNA and RNA during cell lysis23. Another plate-based method, simultaneous isolation of genomic DNA and total RNA (SIDR), involves incubation of single cells with antibody-conjugated magnetic beads, which are subsequently sorted into a microplate in which hypotonic selective lysis produces a separated supernatant and pellet solution that distinguishes between DNA and total RNA24.
Integrating transcriptome and epigenome
Our understanding of how phenotypes are derived from single cells is based on information that is more complex than that provided by the coupling of genome and transcriptome profiling, which cannot address the question of how the same sequence of DNA can have varying expression patterns in different cells. Single-cell epigenome analysis in tandem with transcriptomics allows the direct elucidation of DNA epigenetic features such as DNA methylation, DNA accessibility and histone modifications in relation to the transcriptome they produce.
DNA methylation at CpG islands generally regulates gene expression25. Single-cell DNA methylation profiling methods with single-base resolution can be categorized according to two core technologies: reduced-range bisulfite sequencing (RRBS) and whole-genome bisulfite sequencing (WGBS), both using bisulfite conversion followed by NGS. The primary differences between the two categories are coverage and cost: WGBS has higher coverage of CpG islands in the genome whereas RRBS is less expensive. Early methods aimed to characterize the correlation between gene expression and DNA methylation at CpG sites through post-bisulfite adaptor tagging26 used in WGBS methods, or through RRBS measuring high CpG-content regions27. Many groups have built from this foundation to develop technologies that co-profile the DNA methylome and transcriptome28–31. The first method of WGBS multi-omics, single-cell genome-wide methylome and transcriptome sequencing (scM&T-seq)28, borrowed already-established single-omics techniques from G&T-seq21 and scBS-seq26 to measure DNA methylation heterogeneity in single cells, combined with Smart-seq2 to generate transcriptional data concurrently with genome-wide methylation data. Similarly, scMT-seq31 manually separates the cytoplasm and nucleus, with the cytoplasm containing the mRNA for scRNA-seq and the nucleus containing DNA for methylome profiling by scRRBS32.
Subsequently, DNA methylation co-profiling technologies were developed for tri-omics analysis: single-cell triple omics sequencing (scTrio-seq) combined profiling of genome, methylome and transcriptome in 25 single cancer cells by applying scRRBS and whole-genome sequencing33; the WGBS method scTrio-seq2 combines scTrio-seq and scBS-seq to profile methylation with mRNA and copy number variations34; and scNOMe-seq characterizes nucleosome positioning, chromatin accessibility and DNA methylation by uniquely recovering reads regardless of signal and allelic drop-outs, thereby distinguishing them from reads of inaccessible chromatin35. Adding to the comprehensive tri-omics methods and WGBS technologies is single-cell nucleosome occupancy mapping, which is demonstrated in scNMT-seq29 and scNOMeRe-seq36 and by which DNA methylation, chromatin accessibility and transcripts from the same DNA molecule can be profiled.
Chromatin accessibility is integral to detecting genomic activity as it reveals enhancer activity, transcription factor binding sites and other DNA regulatory elements. Chromatin accessibility paired with transcriptional analysis can reveal regulatory elements responsible for driving gene expression. Most recent single-cell multi-omics technologies that profile chromatin accessibility are primarily adapted from assay for transposase-accessible chromatin sequencing (ATAC-seq37), which uses the transposase Tn5 to fragment open chromatin and tag adaptors to the DNA38–42. ATAC-seq allows the identification of regions within the genome that have open chromatin states associated with transcription in low numbers of cells and is an easy and time-friendly method as it does not include extraction or enzymatic-digestion steps. To determine open chromatin regions in heterogeneous cell populations, two methods of single-cell level ATAC-seq were further developed. The first one, scATAC-seq43, used the Fluidigm C1 microfluidics platform to capture single cells, followed by tagmentation and library amplification with cell-identifying barcoded primers (Fig. 2), whereas the other, a plate-based approach, used a combinatorial cell indexing strategy and tagmentation with Tn5 loaded with unique adaptors to measure chromatin accessibility in over 15,000 cells44.
Histone modifications at single-cell resolution are informative in understanding epigenetic programmes and differentiation trajectories of cells, thereby aiding in cell-state prediction. In addition to the well-established chromatin immunoprecipitation followed by sequencing (ChIP–seq)45 and CUT&RUN46 methods, cleavage under targets and tagmentation (CUT&Tag), which was developed in 2019, uses Tn5 transposase to directly tagment the antibody binding site47. This method has been modified further by other groups to profile histone modifications at the single-cell level: methods such as scCUT&Tag48 and scCUT&Tag2for149 use tagmentation directed by antibodies to profile active and silenced regulatory elements genome-wide by targeting domains bound by, respectively, RNA polymerase II and Polycomb repressive complexes. This methodology was further developed into scCUT&TAG-pro, which is a multimodal assay for profiling protein–DNA interactions and the abundance of surface proteins in single cells50. These methods have been extended to characterize RNA and histone modifications using multi-omics techniques such as Paired-Tag51 and CoTECH52, which use combinatorial barcoding to allow high-throughput detection of both transcriptome and chromatin occupancy.
Multi-omics technologies continue to be developed to increase both throughput and the modalities that can be profiled in a single experiment. Medium-throughput methods involving manipulation of intact cells rather than nuclei and profiling both epigenome and transcriptome such as scCAT-seq53 and assay for single-cell transcriptome and accessibility regions (ASTAR-seq)54 have been developed. In the ASTAR-seq protocol, individual cells are isolated at distinct capture sites, to which reagents are added in specialized compartments of a Fluidigm C1 microfluidics chip. Following tagmentation, the cDNA is biotinylated and separated from genomic DNA using streptavidin beads. To achieve higher throughput in single-cell analysis, Sci-CAR-seq combined single-cell combinatorial indexing55 with ATAC-seq to provide deep insights into chromatin states jointly with gene expression in over 11,200 nuclei56. As an alternative, single-nucleus chromatin accessibility and mRNA expression sequencing (SNARE-seq) used droplet-based microfluidics to co-profile over 10,000 mRNA transcripts with chromatin accessibility from the same single cell57. Improvements in single-cell isolation or barcoding methods led to the development of ultra-high-throughput technologies. Split-pool ligation-based transcriptome sequencing (SPLiT-seq), a plate-based transcriptomics method, uniquely labelled over 100,000 nuclei in one experiment using four rounds of combinatorial barcoding58. This strategy was implemented shortly after by Paired-seq, which used three rounds of ligation-based combinatorial indexing to process 1 million nuclei in a single experiment for parallel transcriptome and chromatin accessibility analyses59. Increasing throughput, SHARE-seq was built based on SPLiT-seq and Paired-seq and uses several rounds of hybridization blocking to label mRNA and chromatin fragments in single cells60. Recently, TEA-seq, a method for trimodal single-cell measurement of transcripts, epitopes and chromatin accessibility, used the scATAC-seq workflow and the 10x Genomics Multiome ATAC Plus Gene expression kit to elucidate the modulators of gene regulation in thousands of single cells61. Another recent high-throughput method, ISSAAC-seq (in situ sequencing hetero RNA–DNA-hybrid after assay for transposase-accessible chromatin sequencing), used a two-tagmentation strategy: an initial tagmentation reaction on accessible chromatin followed by reverse transcription and another tagmentation reaction on cDNA–RNA hybrids62. This final reaction was then separated into its DNA and RNA constituents using fluorescence-activated cell sorting (FACS) or microfluidics technology, ready for downstream multi-omics analysis.
Integrating transcriptome and proteome
All cellular processes and functions revolve around proteins, as they contribute to the structure of cells and perform biochemical processes by functioning as enzymes. Thus, it is essential to characterize proteins at the single-cell level through their post-translational modifications and interactions. Many multi-omics studies, in which protein and mRNA are co-profiled, rely on the use of predetermined, curated antibody panels to confer single-cell phenotypes, thereby losing novel and unexpected proteomic information63–66. For example, in 2016, the proximity ligation assay for RNA (PLAYR) was developed to simultaneously quantify more than 40 targeted mRNAs and proteins from 10,000 single cells in several cell types67. PLAYR labels proteins with antibodies conjugated to unique metal isotopes, while RNA transcripts are conjugated to isotope-labelled probes that can be measured by mass cytometry67. Another method used a homogeneous affinity-based proximity extension assay (PEA) to interrogate ~100 protein targets in single-cell lysates by linking matched pairs of oligonucleotide–antibody conjugates to target antigens and simultaneously co-detecting RNA by real-time PCR64. An integrated protein and RNA detection method combining PEA and specific target amplification was also developed in 2016 to detect RNAs and proteins from the same single cell in a single reaction chamber of the Fluidigm C1 platform65. In this method, cells are lysed with buffer containing protein-binding PEA probes, and reverse transcriptase is used on cellular RNA, relying on extension of PEA nucleotides to enable the synthesis of cDNAs with random primers.
A great leap in throughput for co-profiling the transcriptome and proteome came in 2017 with the development of cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq). This method combines highly multiplexed protein-marker detection with unbiased transcriptome profiling for thousands of single cells68. Specifically, cell-surface proteins were detected by antibodies conjugated to oligonucleotides containing a PCR handle that can be captured by oligo(dT) or probe-specific primers compatible with Drop-seq9 and 10x Genomics microfluidic systems, making this method adaptable for integration with most scRNA-seq methods. Once mRNA and oligonucleotide-tagged antibodies are released from lysed single cells, they are bound to magnetic beads, which allow their separation according to size and quantification by NGS (Fig. 2). Similarly, RNA expression and protein sequencing assay (REAP-seq) demonstrated quantification of surface proteins with 82 antibodies and genome-scale mRNAs in a single workflow69. REAP-seq differs from CITE-seq in how the DNA barcode is conjugated to the antibody: in CITE-seq, streptavidin is conjugated to each antibody, and in REAP-seq, small stable covalent bonds are formed between the antibody and the DNA barcode. Later, combinatorial indexing of splint oligonucleotides to barcode antibodies was demonstrated with SCITO-seq, which enables multiplexing of cells and multi-omics profiling of over 150 surface proteins in parallel with mRNA expression in single cells70. The combination of sample multiplexing and droplet-based platforms such as 10x Genomics or microwell-based platforms such as BD Rhapsody71 have increased the throughput of mRNA and protein co-profiling.
Other single-cell proteomics methods that do not rely on antibodies produce a more comprehensive characterization of the cellular proteome, as they are not limited by antigen-specific reagents and can profile thousands of proteins in single cells. SCoPE-MS and SCoPE2 are such methods using mass spectrometry, but they have no available integration method with other omics, leaving room for future development72,73. PHAGE-ATAC74, a recently developed method based on epitope recognition by nanobody-displaying phages, stands in contrast to methods in which fluorescent or oligonucleotide-conjugated antibodies are used. Importantly, the hypervariable complementarity-determining region 3 (CDR3) of the nanobody-encoding phagemid acts as a unique genetic barcode, which is identified by downstream sequencing and acts as a proxy for antigen detection. This multi-omics method allows profiling of thousands of single cells while reliably detecting cell-surface proteins, mitochondrial DNA genotypes and epigenomic modifications.
Intracellular post-translational modifications such as those responsible for regulation of signalling pathways and metabolic activity are essential to single-cell proteomics. Technologies that can profile both intracellular and cell-surface proteins can better elucidate the signalling pathways that required for the function of a single cell75,76. RNA and immunodetection (RAID), a method developed for immunodetection of intracellular proteins or phosphorylated proteins together with mRNA profiling, enables correlation analysis of environmental stimuli with heterogeneous cellular responses77. Recently, ATAC with select antigen profiling by sequencing (ASAP-seq) and DOGMA-seq78 demonstrated simultaneous profiling of chromatin accessibility, gene expression and protein expression by using reagents and protocols similar to those used in mtscATAC-seq79, CITE-seq68 and scATAC-seq43. Notably, ASAP-seq uses molecular bridging to enable the use of existing antibody conjugates, making it a technology that is user-friendly and widely accessible. Incorporating even more modalities is NEAT-seq80, which co-profiles the abundance of nuclear protein epitopes, chromatin accessibility and the transcriptome in single cells. In NEAT-seq, blocking the charge of antibody–oligonucleotide conjugates with Escherichia coli single-strand DNA improves the signal-to-noise ratio. Nuclear proteins are actively involved in gene regulation, and their characterization and linkage to the epigenetic and transcriptional status of the cell are a powerful approach for studying mechanisms of gene regulation. Finally, expanded CRISPR-compatible CITE-seq (ECCITE-seq) expanded the protein multi-omics field to incorporate CRISPR-compatible transcriptome, immunity repertoire and proteome indexing, including both clonotype profiling and CRISPR-directed perturbations at high sensitivity and single-cell resolution81. Thus, multi-omics technologies have the capacity to measure the effects of cell perturbations and signalling pathways on the state of single cells. Profiling cellular or secreted proteins and other modalities have led to commercialization of such technologies by several companies.
Other multi-omics modalities
Understanding cellular heterogeneity and its inherent complexity at the single-cell level involves the ability to simultaneously characterize as many functions of the cell as possible. By capturing multiple layers of cellular information using emerging omics methods, we can gain more-specific data and greater clarity of cell function. For example, scRNA-seq can be used in CRISPR screens to link CRISPR–Cas9-induced genetic perturbations to transcriptomic phenotypes. Perturb-seq is one of these methods, in which thousands of CRISPR-mediated perturbations and mRNA profiles are analysed in a single experiment of 200,000 cells82. CROP-seq, another CRISPR-based genetic screening method combined with single-cell transcriptomics, directly links expression of a single guide RNA (sgRNA) to transcriptome responses in thousands of individual cells83. The sgRNA-expression vector was re-engineered from a common construct (lentiGuide-Puro84) to embed the sgRNA in a polyadenylated mRNA transcript, thereby allowing its easy integration into most scRNA-seq platforms. In 2016, CRISP-seq, a method to capture mRNA and genomic perturbations from CRISPR-pooled screens emerged, elucidating the function of multiple regulatory factors in a single experiment85. Another method, Mosaic-seq, was developed to systematically perturb enhancers and measure their endogenous activities while jointly measuring the transcriptome at single-cell resolution86. CRISPR screens have been coupled with other methods such as scATAC-seq — for example, in developing Spear-ATAC-seq, which characterized over 100,000 single-cell epigenetic states and over 400 CRISPR perturbations87.
Other modalities of multi-omics include the characterization of the complex genomic repertoire of T cell receptors (TCRs) and B cell receptors (BCRs), which are crucial components of adaptive immunity. These data can provide an in-depth understanding of immunity spectra in individuals and suggest how various receptor configurations contribute to adaptive immunity responses, autoimmunity and tumour growth88. Several methods have been published pairing gene expression with TCR sequences in single cells89–92. Integration of specific TCR and BCR (DNA) clonotypes out of a cell population repertoire with scRNA-seq paves the way for a better understanding of the relationships between transcriptome, clone composition and unique sequence features of antigen receptors such as their CDR3 length and presence of specific sequence motifs93. The V(D)J regions of rearranged TCR and BCR mRNAs are located in the first ~500 nucleotides of the 5′ end of the transcripts, and short-read scRNA-seq is not always sufficient to resolve the genetic complexity of these isoforms while preserving accurate gene expression quantification at the 3′ end of the transcripts90. Therefore, most of the technologies that can capture TCR and BCR isoform sequences while maintaining whole-transcriptome information of single cells have implemented 5′-end RNA amplification, which has been made commercially available by 10x Genomics and has also been extended to include proteomics (ECCITE-seq81) and chromatin accessibility (T-ATAC-seq94) (Fig. 2). 10x Genomics and Oxford Nanopore recently announced their partnership to sequence full-length transcripts in single cells to characterize antigen-receptor sequences and transcriptomes at high accuracy and sensitivity95. Immunity repertoire sequencing, together with technological advances and integration with other omics modalities, has the capacity to provide deeper understanding of how T-cell and B cell clonality and specificity influence immune responses.
Multi-omics goes spatial
The organization of cellular compartments and macro-structures and of intercellular interactions is fundamental to the function of multicellular organisms. The aforementioned single-cell multi-omics methods require dissociation of cells from their tissue and thus lose information about physical interactions within and between cells, despite several computational efforts made to infer cell–cell interactions96,97. Emerging spatial multi-omics technologies seek to expand mapping of interactions between single cells within intact tissues at a genome scale (Fig. 4). It is essential to address the importance of this cutting-edge field.
Fig. 4. Towards spatially resolved multi-omics.
a, Examples of spatial technologies used for multi-omics analysis. Imaging-based technologies such as fluorescence in situ hybridization (FISH) and sequential FISH (seqFISH+) use sequential imaging of different fluorescent probes to characterize spatial organization within cells. Microarray-based and microfluidics technologies use barcodes to label the structural organization of single cells in tissues. Methods such as spatial transcriptomics profile tissue by placing it on a glass slide that is coated with an array of spots containing oligonucleotides designed to extract spatial information from a tissue section, whereas methods such as deterministic barcoding in tissue for spatial omics sequencing (DBiT-seq) use microfluidics channels to deliver reagents and barcodes to a tissue on a glass slide, thereby creating a barcoded grid in which each grid space contains a single cell. Marker genes with known function and physical location are used as references. ‘Genes/pixel’ indicates the number of genes detected in each spatially barcoded pixel. Laser capture microdissection (LCM)-based technologies allow full profiling of a gene or protein within a single cell by cutting a tissue section with a laser. b, Spatial-omics microfluidics-based tissue barcoding modalities have expanded in the past decade, enabling integration of complex datasets with spatial context. DBiT-seq has the integrative capacity to profile many modalities including spatial assay for transposase-accessible chromatin with sequencing (ATAC-seq), cleavage under targets and tagmentation (CUT&Tag), cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq) and other modalities. c, Spatial multi-omics data analysis methods generally partition data into clusters to find functional cell subtypes within different microenvironments. These high-dimensional data are typically visualized using dimensionality reduction techniques such as uniform manifold approximation and projection (UMAP) to view multimodal single-cell measurements. Various computational clustering algorithms can be applied to downstream multimodal analysis to automatically estimate cell types and compare clustering attributes of different modalities. Within and between cell clusters, heterogeneous archetypes of signalling pathways inferred from cell–cell interaction analyses can be used to characterize local cellular microenvironments. Geo-seq, geographical position sequencing; PDMS, polydimethylsiloxane; SMI, spatial molecular imager; UMI, unique molecular identifier. Part a adapted with permission from ref. 123, Elsevier.
Imaging-based technologies
Fluorescence in situ hybridization (FISH) is a technique that has existed since the 1980s98. Early attempts at characterizing single cells and their gene expression profiles with spatial resolution utilized multiple short probes targeting different regions of the same transcript through spectral barcoding or sequential imaging with single-molecule FISH (smFISH)99 (Fig. 4a). In 2014, a multiplexed smFISH approach was shown to detect individual transcripts through serial rounds of probe hybridization, imaging and probe removal of 24 probes labelled with unique combinations of fluorophores100. A year later, MERFISH substantially increased the number of RNA molecules that could be simultaneously imaged in single cells by utilizing combinatorial labelling and sequential imaging of multiple readout hybridizations101. In MERFISH, computational error-correction after each readout accounts for imperfect hybridizations. Other FISH technologies developed subsequently mainly aimed to shorten the imaging time, reduce optical crowding and increase multiplexing efficiency. Sequential FISH (seqFISH+) demonstrated the ability to perform discovery-driven studies of the spatial organization of tissues with its capability of imaging mRNAs for 10,000 genes in single cells102. The use of a larger palette of ‘pseudocolours’ allowed seqFISH+ to obtain readouts at the transcriptome-level scale. To assess both RNA and proteins, RNAscope, commercialized in 2011 by Advanced Cell Diagnostics103, hybridizes DNA with probes tagged with metal instead of fluorophores, followed by the addition to the tissue section of metal-conjugated antibodies and mass-cytometry measurements of metal abundance104,105. Several other multiplex FISH methods have been reported to characterize spatial organization of cell types identified from scRNA-seq101,106–111.
The spatial proteomics field has also expanded with the development of technologies such as co-detection by indexing (CODEX), which uses a multiplexed cytometric imaging approach to quantitatively characterize protein expression and define tissue architecture112. After multiple cycles of antibody imaging and fluorophore washing, a multiparameter image is constructed, with the capacity to characterize over 100 antibodies in a panel to read out antibody-conjugated barcodes with fluorescent hybridized nucleotides. Very recently, an automated and integrated platform, spatial molecular imager, was developed to resolve multi-omics spatial distribution of 980 RNAs and 108 proteins in formalin-fixed, paraffin-embedded tissues at single-cell and subcellular resolution, using a strategy similar to that of CODEX by performing multiple cycles of nucleic acid hybridization of fluorescent molecular barcodes113. Spatial molecular imager has recently been expanded to 6,000 genes and 100 proteins.
Sequencing-based technologies
Sequencing-based technologies can be performed directly on the biomolecule of interest (DNA, RNA or protein) within an intact tissue (Table 1). The first in situ sequencing study used padlock probes to target known short RNA fragments in tissue sections114 and was later commercialized (now part of the Visium platform of 10x Genomics). Fluorescent in situ RNA sequencing (FISSEQ) was the first untargeted in situ sequencing method to capture the whole transcriptome by sequencing single-stranded 200–400-nm DNA nanoballs through sequencing-by-ligation115.
Sequencing-based spatial technologies moved from in situ to ex situ sequencing by extracting molecules from the tissue. Along with this transition, it became integral to achieve higher resolution, improve RNA capture efficiency and expand spatial transcriptomics to other modalities (Fig. 4b). In 2016, spatial transcriptomics was introduced116, demonstrating a 100-μm spot size on a microarray slide with arrayed oligonucleotides to capture spatial gene expression information. Reverse transcription was performed on the intact tissue, and the resulting cDNA was left coupled to the oligonucleotides on the slide before tissue lysis, ultimately generating NGS libraries. This technology was later adapted by 10x Genomics as ‘10x Visium’, with increased resolution of 55 μm117. Later, micrometre-scale bead arrays were developed to capture spatial transcriptomics data at the cellular level. Slide-seq, and later Slide-seqV2, barcode mRNA in tissues using a method conceptually similar to spatial transcriptomics, but a different strategy: instead of using arrayed barcoded reagents printed on slides, the barcoded reagents are placed directly onto the glass slide in solution, thereby creating a monolayer-packed array on a glass coverslip118,119 (Fig. 4a). In high-definition spatial transcriptomics, subcellular resolution was achieved by using beads with a diameter of 2 μm, which later was reduced even further120. GeoMx (from Nanostring) uses a targeted FISH detection method rather than direct sequencing of transcripts, allowing the detection of up to 10,000 genes in a select microscopic region of tissue through photocleaving and sequencing the DNA probes, which are hybridized to target genes, and the probe panel is now approaching a whole-transcriptome level of coverage121.
All these methods follow the same fundamental principle, namely ‘barcoded solid-phase RNA capture for spatial transcriptomics profiling’122. In 2020, we reported a fundamentally different approach to spatial omics sequencing. Instead of capturing RNA onto a barcoded solid-phase substrate, a microfluidic approach allows the delivering of DNA barcodes into a tissue section in a spatially resolved manner, termed deterministic barcoding in tissue for spatial multi-omics sequencing (DBiT-seq), which was performed by flowing DNA barcodes through two perpendicular microfluidic chips to create a mosaic of 10–50-μm spatially barcoded pixels123 (Fig. 4a). DBiT-seq was the first reported spatial multi-omics technology that demonstrated co-mapping of tens of proteins and with a whole transcriptome, and this method has further expanded to spatial epigenomics profiling of chromatin accessibility124 and histone modification125. Recently, DBiT-seq has further demonstrated its versatility by spatial co-profiling of mRNA transcriptome and hundreds of cellular proteins as part of spatial CITE-seq126 and of transcriptome and epigenome as part of spatial ATAC–RNA-seq127, spatial CUT&Tag–RNA-seq127 and (reported in a preprint) SPARC-seq128. Combined spatial transcriptomics or the Visium platform with CITE-seq chemistry, SM-Omics129 and SPOTS130 also achieved co-analysis of transcriptome and a panel of proteins in tissue sections, although not at single-cell resolution. Expanding to spatial mapping of T-cell clonotype repertoires, Slide-TCR-seq was recently reported to sequence whole transcriptomes and TCRs by integrating Slide-seqV2 with rhTCR-seq131 (a method used to amplify TCR transcripts), revealing the interaction between clonality, neighbouring cell types and gene expression in tissues at single-cell resolution132.
Laser capture microdissection-based technologies
LCM is a decades-old mainstay method for isolating in micrometre-scale tissue or even single cells while retaining spatial information to link histology with molecular measurements. In LCM, a region of interest in the tissue section is isolated through laser cutting. Multiple modalities can be characterized simultaneously based on LCM, as demonstrated in a work that constructed dynamic regulatory networks by co-profiling of mRNAs, microRNAs, DNA methylation and protein expression133. LCM can also be combined with other technologies to elucidate cellular heterogeneity and spatial variance, as demonstrated in Geo-seq134. Recently, LCM coupled with fluorescence imaging enabled deep proteomic characterization of formalin-fixed, paraffin-embedded tissues using mass spectrometry135. LCM-based methods allow full gene or protein profiling at a single-cell level including positional information, but they can assay only a small number of cells. Despite this limitation in throughput and inability to map a whole tissue section pixel-by-pixel at a single-cell level, LCM has the versatility to isolate a spatially defined tissue region for multi-omics analysis.
Research and clinical applications of single-cell multi-omics
Application of single-cell multi-omics to molecular and cell biology is still in early stages, but it promises to be instrumental in detailing complex cellular landscapes. In this section, we discuss how single-cell multi-omics improves our understanding of the molecular mechanisms of tissue biology and the study of diseases, by uncovering cellular heterogeneity and its mechanistic underpinnings and building integral cell lineage and cell atlas resources.
Tracing cell lineages
A principal application of single-cell sequencing is the establishment and building of cellular lineage trees or phylogenies of disease evolution. These lineage trees include discovery of novel cell types, cell lineage segregation, and identification of biomolecular markers through densely characterizing cells at different stages in their development. Here, we describe how the multi-omics technologies discussed in this paper contribute to cell lineage analysis; we refer the reader to more detailed reviews of the definition and single-omics applications of cell lineage tracing136,137.
Recent high-throughput, multiplexed sequencing techniques have paved new directions for lineage tracing. Transcriptomes contain ample information about cell identity such as cell cycle phase, spatially restricted expression of marker genes, expression signatures and metabolic states of cells3. Clonal information can be encoded by DNA oligonucleotide barcodes to record all cell division events within an organism, which is sequenced and combined with other omics methods. Methods discussed above such as scTRIO-seq28 and scNMT-seq26 can be used to simultaneously sample genomic copy number variations, the DNA methylome, nucleosome occupancy and the transcriptome of single cells to reveal new cell types and their roles in a studied lineage. Technologies for profiling the transcriptome in combination with chromatin accessibility, DNA methylation, histone modifications and nucleosome organization provide new insights into cell-type identity and elucidate epigenetic processes involved in lineage priming138. Furthermore, combining the epigenome and transcriptome of single cells can enable inference as to how DNA modifications can facilitate cell differentiation, which can be tricky because epigenomic landscapes vary considerably between cells, but is nonetheless informative in understanding the lineage-specific state of DNA methylation during early differentiation stages.
Single-cell proteomics methods contribute to our understanding of how protein levels change as cells differentiate by analysing lineage-specific transcription factors and their abundance over time, ultimately leading to the maintenance or emergence of a lineage trajectory139. The recently developed iTracer combines reporter barcodes with inducible CRISPR–Cas9 in induced pluripotent stem cell-derived organoids, and is compatible with single-cell and spatial transcriptomics140. In iTracer, clone tracing and lineage recording at distinct time points occurs through CRISPR-mediated gene perturbation incorporated into a lineage recorder system that is based on the Sleeping Beauty transposon system, which can undergo transposition into multiple genomic loci within the cells141. Ultimately, this system can efficiently trace clones from a pool of induced pluripotent stem cells, dynamically trace cell lineage and align it with molecular state and location information using the ‘Spatial iTracer’ software.
Finally, combining genome, transcriptome and lineage reporter methods enables lineage tracing in diseases such as cancer, in which elucidating the persistence of epigenetic states together with genetic mutations can lay a foundation for rational design of therapeutics, as shown by a study elucidating the relevance of single-cell lineages in glioblastoma drug resistance142. Another study combined single-cell transcriptome and DNA methylome data to construct a chronic lymphocytic leukaemia (CLL) lineage tree based on random DNA methylation changes, finding that certain CLL lineages were preferentially affected by drug treatment143. The understanding of these CLL lineages was further enhanced by transcriptomics data, revealing that the cells affected by drug treatment differentially expressed genes involved in certain cell signalling pathways. Tracing cell lineage can also provide valuable insights into how immune cells react to infection, how they differentiate and the mechanisms that underlie their fate, as shown in a recent study combining scRNA-seq and scTCR-seq for tracing clonal expansion and differentiation of CD8+ T cells, ultimately delineating pathways leading to cell exhaustion144,145. Lineage tracing may also be performed with spatial resolution in combination with scRNA-seq and other single-cell modalities to characterize cell organization dynamics and to differentiate between molecular properties and morphology129. Altogether, single-cell multi-omics has had a large impact on cell lineage classification for disease, tumour classification and our understanding of evolution of cell states and types.
Production of cell-type atlases of various organs
Single-cell multi-omics datasets and their inherent complexity increase across the number of samples, conditions and methods used for data acquisition. Current methods of integrating multi-omics data focus on alleviating batch effects and maintaining biological variation. Multiple groups have published atlas resources for community use, but in this Review, we only highlight atlases encompassing multi-omics datasets (Table 2). The Cancer Genome Atlas (TCGA) is a cancer-specific multi-omics data resource characterizing over 20,000 primary cancer and corresponding normal samples from 33 cancer types, representing data from genomic, epigenomic, transcriptomic and proteomic modalities. As the largest repository of cancer multi-omics data, TCGA resources are invaluable for the scientific community. One group combined prostate cancer RNA-seq and SNP data to identify risk-modulating non-coding RNA, demonstrating the application of multi-omics TCGA data integration146,147. The Encyclopedia of DNA Elements (ENCODE) project consortium was piloted in 2003 and has since scaled up to define and annotate functional elements in both human and mouse genomes using multi-omics sequencing148. Recently, part of its bulk epigenomics data was deconvolved into specific cell-type assignments using scRNA-seq149, which further refines the data annotation. Other consortia aim to provide comprehensive multi-omics atlases of the heart, lung, blood, sleep disorders and ageing, all incorporating thousands of datasets to identify cell types associated with disease onset and progression150.
Table 2.
Multi-omics contributions to cell atlases
| Atlas name | Source material | Incorporated data types | Refs. |
|---|---|---|---|
| Human Cell Atlas (HCA) | Human tissues and cells | Genomics, transcriptomics, single-nucleus RNA-seq, epigenetics, proteomics, spatial omics | 151,152 |
| Encyclopedia of DNA Elements (ENCODE) | Human and mouse tissues and cells | Functional genomics, transcriptomics, DNA accessibility, RNA binding, DNase-seq, DNA methylation, 3D chromatin structure | 148 |
| Genotype-Tissue Expression (GTEx) | Human tissues and cells | Transcriptomics, QTLs, histology images | 183 |
| Human Protein Atlas (HPA) | Human tissues and cells | Proteomics, transcriptomics | 184 |
| The Cancer Genome Atlas (TCGA) | Cancer and matched-normal human tissues and cells | Genomics, epigenomics, transcriptomics, proteomics | 147 |
| Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) | Human breast cancer tumours and cell types | Transcriptome, CNVs, SNPs | 185 |
| Omics Discovery Index (OmicsDI) | Humans, model organisms, non-model organisms | Genomics, transcriptomics, proteomics, metabolomics | 186 |
| COvid-19 Multi-omics Blood ATlas (COMBATdb) | Human blood from persons with COVID-19 and healthy individuals | Transcriptomics, epigenomics, proteomics, TCRs and BCRs | 187 |
| Aging Atlas (AA) | Human and mouse tissues and cells | Transcriptomics, epigenomics, proteomics, pharmacogenomics | 188 |
BCRs, B cell receptors; CNVs, copy number variants; DNase-seq, DNase I hypersensitive site sequencing; QTLs, quantitative trait loci; SNPs, single-nucleotide polymorphisms; TCRs, T cell receptors.
The first universal human cell atlas, Human Cell Atlas Project, was launched in 2017, aiming to provide an open-access resource used to integrate all single-cell omics data in one atlas to deepen our understanding of cellular development, physiology and intercellular interactions, and to predict the effects of cellular perturbations or mutations on ultimately every cell type in the human body151,152. The data collection of the project mainly uses single-cell genomics methods including single-cell multi-omics and spatial technologies. Recently, spatial multi-omics data have been used to add tissue context to cell genomic annotations through integration of multiple complementary technologies. This combination adds a valuable parameter to atlas production as spatial distribution of cells can tie multi-omics signatures of specific cell types with their localization within the tissue153.
Tumour immunology and cancer genetics
Single-cell multi-omics technologies have added unprecedented breadth and depth into research of a variety of diseases including viral infections such as SARS-CoV-2 (ref. 154), cardiovascular diseases155, neurological disorders156 and others. However, more comprehensive and transformative insights were generated in immuno-oncology research, including but not limited to defining tumour and immune cell states and revealing the interplay between them in specific disease contexts, inferring the complex nature of antigen–immune receptor dynamics, identifying predictive biomarkers of treatment response and providing directions for the development of therapeutics for multiple cancer types. In this section, we discuss a few recent examples of the impact of single-cell multi-omics on cancer research.
Very recently, a newly developed spatial technology, slide-DNA-seq, was used to preserve local tumour architecture, enabling the discovery of distinct tumour clones and their copy number variations157. The method was applied to a mouse metastasis model and to primary human cancer, revealing spatially distinct clonal populations and uncovering genes associated with clonal variation and the local tumour microenvironment. Another study applied gain and loss of function of CPT1A, which encodes a fatty acids metabolism enzyme, to elucidate genetic and metabolic vulnerabilities in prostate cancer158. Integrated analysis of transcriptomics and metabolomics revealed that CPT1A overexpression is detrimental to individuals with prostate cancer and may support disease progression. Another group performed integrative analysis of metabolomics, transcriptomics and genetic data to confirm metabolic pathways in non-small-cell lung cancer and elucidate metabolites and genes contributing to platelet activation, haemostasis, angiogenesis and cell proliferation159. Furthermore, this study led to the discovery of key factors in the gene–metabolite interaction network, metabolites, cell cycle genes and genes of the p53 signalling pathway, which may have important roles in disease mechanisms. A combination of ChIP–seq and transcriptomics was used to detect enrichment of histone modifications associated with tumour-specific variance in gene expression changes, sites of human papillomavirus integration into the genome and human papillomavirus-associated histone enrichment sites in human cancer cell lines160.
Mediators of cancer progression have also been studied using multi-omics methods. Study of the effects of inactivation of the signal transduction factor SMAD4 on the transcriptome, proteome and secretome led to the identification of three SMAD4-mediated processes that may promote metastasis in individuals with advanced colorectal cancer161. Proteomics, genomics and phosphoproteomics methods were combined to classify hallmarks of lung cancer tumour progression in a large study of non-smoking individuals with early-stage tumours162. Specifically, proteomics clustering distinguished clinical features, biomarkers and druggable targets in early stages of lung adenocarcinoma. Another study applied single-cell multi-omics integration of DNA accessibility, gene expression and protein abundance to mixed-phenotype acute leukaemia models, determining that the gene RUNX1, encoding a transcription factor, is a potential oncogene in mixed-phenotype acute leukaemia associated with poor survival163. Additionally, peak-to-gene link analysis was performed by linking differentially accessible genomic regions, depicted as peaks, to known leukaemia genes, showing that specific peak-to-gene links were enriched in immunity regulation. This study establishes a unique approach to deconstructing cancer-specific features through use of multiple single-cell omics methods, revealing the molecular mechanisms of disease progression.
These methods have also been used to study immune-checkpoint blockade (ICB) therapies for human cancer. One study resolved how tumour-reactive T cells respond to ICB in different tumour types by analysing previously published single-cell transcriptomics, epigenomics and TCR data, finding that the presence CXCL13+CD8+ T cells correlated with favourable response to ICB164. Another study integrated the spatial phenotypes and immunity effectors panel of Akoya Biosciences with multiplexed immunofluorescent imaging, scRNA-seq and TCR repertoire analysis to elucidate T cell evasion pathways in response to ICB in triple-negative breast cancer165. The spatial immunophenotypes were assigned in triple-negative breast cancer through a spatial phenotype classifier, which predicted responses to treatment. Single-cell multi-omics has also been applied to assess chimeric antigen receptor (CAR) T cell therapy in acute lymphoblastic leukaemia, combining transcriptomics and proteomics data to determine the molecular basis of phenotypic differences between complete remission, non-responsive and relapsed individuals166. This analysis led to discovery of TH2 cell pathways and related genes as potential targets for maintaining 5-year remission following CAR T cell immunotherapy. We anticipate that broader applications of spatial and single-cell multi-omics technologies together with computational methods will create an integrative framework with which to understand the complex nature of cancer heterogeneity.
Computational tools used for integration of multi-omics data
Despite the proliferation of and advancements in experimental methods that simultaneously profile more than one omics modality, a major hurdle in the field is the inherent complexity of multi-omics data integration. Different omics layers contain distinct feature spaces that can reveal the underlying mechanisms of the regulation and functionality of diverse cell types, thereby providing a comprehensive understanding of cellular processes (Fig. 4c). Although recent experimental multi-omics strategies can measure different modalities within the same single cell, large amounts of single-cell data consist of unpaired observations on different cells. Current tools used for the joint analysis of multiple single-cell omics modalities can be broadly classified into matched or unmatched pipelines (Table 3). In matched pipelines, various omics layers are measured simultaneously from the same single cell, whereas unmatched pipelines have unaligned omics layers that were produced by different experiments. Integration of unpaired multi-omics data is limited by the inherent information loss that comes from tying together distinct feature spaces of varying multi-omics modalities. A general solution to this obstacle is to project cells into a co-embedded space or nonlinear manifold to find a commonality between all cells. Seurat v3 and linked inference of genomic experimental relationships (LIGER) are examples of unmatched pipelines, which use canonical correlation analysis or non-negative matrix factorization to identify a shared biological space that jointly embeds different omics modalities from different cells by integrating datasets together with pairs of mutual nearest-neighbour cells serving as an ‘anchor’ and representing a similar biological state to guide dataset integration167,168. A recent unmatched method, graph-linked unified embedding (GLUE), achieves triple-omics integration while simultaneously inferring regulatory interactions. In GLUE, a graph variational autoencoder learns feature embeddings from previous biological knowledge, which is then used to reconstruct and link omics data through an inner product with cell embeddings that are aligned further in subsequent steps169. More recently, a suite of methods demonstrated how to integrate unmatched datasets through the use of separately collected multi-omics datasets, which can facilitate accurate integration. These methods include StabMap170, Cobolt171, MultiVI172 and ‘Bridge Integration’ in Seurat v5 (ref. 173). By making use of bona fide multi-omics profiles, these methods can outperform unmatched integration strategies and facilitate an experimental design for cross-modality in which multi-omics datasets are only collected in a subset of experiments.
Table 3.
Timeline and integration capacity of multi-omics computational methods
| Year | Name | Methodology | Integration capacity | Ref. |
|---|---|---|---|---|
| From same single cell (matched) | ||||
| 2019 | SCHEMA | Metric learning-based method | Chromatin accessibility, mRNA, proteins, immunoprofiling, spatial coordinates | 189 |
| 2020 | Seurat v4 | Weighted nearest-neighbour | mRNA, spatial coordinates, protein, accessible chromatin, microRNA | 174 |
| 2021 | DCCA | Variational autoencoders | mRNA, chromatin accessibility | 190 |
| 2021 | DeepMAPS | Autoencoder-like neural networks | mRNA, chromatin accessibility, protein | 175 |
| 2019 | citeFUSE | Network-based method | mRNA, protein | 191 |
| 2020 | MOFA+ | Factor analysis | mRNA, DNA methylation, chromatin accessibility | 192 |
| 2020 | scMVAE | Variational autoencoder | mRNA, chromatin accessibility | 193 |
| 2020 | totalVI | Deep generative | mRNA, protein | 194 |
| 2020 | BREM-SC | Bayesian mixture model | mRNA, protein | 195 |
| 2022 | SCENIC+ | Unsupervised identification model | mRNA, chromatin accessibility | 176 |
| 2022 | FigR | Constrained optimal cell mapping | mRNA, chromatin accessibility | 177 |
| 2021 | MIRA | Probabilistic topic modelling | mRNA, chromatin accessibility | 178 |
| 2023 | CellOracle | Modelling gene regulatory networks | mRNA, CRISPR screening, chromatin accessibility | 179 |
| 2022 | MultiVelo | Probabilistic latent variable model | mRNA, chromatin accessibility | 180 |
| From different single cells (unmatched) | ||||
| 2019 | Spectrum | Weighted nearest-neighbour | microRNA, mRNA, protein | 196 |
| 2020 | BindSC | Canonical correlation | mRNA, chromatin accessibility | 197 |
| 2019 | MMD-MA | Manifold alignment | mRNA, chromatin accessibility, DNA methylation, imaging | 198 |
| 2019 | MuSiC | Unsupervised topic modelling | mRNA, CRISPR screening | 199 |
| 2019 | Seurat v3 | Canonical correlation analysis | mRNA, chromatin accessibility, protein, spatial | 167 |
| 2020 | UnionCom | Manifold alignment | mRNA, DNA methylation, chromatin accessibility | 200 |
| 2019 | CloneAlign | Statistical method | mRNA, DNA | 201 |
| 2021 | Pamona | Manifold alignment | mRNA, chromatin accessibility | 202 |
| 2022 | GLUE | Variational autoencoders | Chromatin accessibility, DNA methylation, mRNA | 169 |
| 2019 | LIGER | Integrative non-negative matrix factorization | mRNA, DNA methylation | 168 |
| 2022 | StabMap | Mosaic data integration | mRNA, chromatin accessibility | 170 |
| 2021 | Cobolt | Multimodal variational autoencoder | mRNA, chromatin accessibility | 171 |
| 2021 | MultiVI | Probabilistic modelling | mRNA, chromatin accessibility | 172 |
| 2022 | Seurat v5 | Bridge integration | mRNA, chromatin accessibility, DNA methylation, protein | 173 |
BREM-SC, Bayesian random effects mixture model; CRISPR, clustered regularly interspaced short palindromic repeats; DCCA, deep cross-omics cycle attention; DeepMAPS, deep learning-based multi-omics analysis platform for single-cell data; FigR, functional inference of gene regulation; GLUE, graph-linked unified embedding; LIGER, linked inference of genomic experimental relationships; MIRA, probabilistic multimodal models for integrated regulatory analysis; MMD-MA, maximal mean discrepancy-manifold alignment; MOFA+, multi-omics factor analysis v2; MultiVI, multi-variational inference; MuSiC, multi-subject single-cell deconvolution; scMVAE, single-cell multimodal variational autoencoder; StabMap, stabilized mapping; totalVI, total variational interference.
Future advancements in computational unmatched data integration are essential because there is a high quantity of single-modality single-cell data generated that can inform new biological insights. By contrast, paired experimental methods (10x Multiome, SHARE-seq60, DBiT-seq123, SNARE-seq57, CITE-seq68 and others) are easier to work with computationally as different modalities can be tethered to a single cell or a single spot; however, the sequencing capacity of each modality causes the overall sample size to decrease. Recently, Seurat v4 used a weighted nearest-neighbour analysis to integrate multiple modalities measured from a single cell and collectively define a cellular state174. This unsupervised strategy learns cell-specific modality ‘weights’ that contain the information from each modality to determine its significance in subsequent analysis steps. Another method, DeepMAPS (deep learning-based multi-omics analysis platform for single-cell data), uses a five-step framework to preprocess data, generate an integrated cell–gene matrix specific to multi-omics data types, use a heterogeneous graph transformer model to generate a score for each cell indicating its gene-to-cell significance, generate clusters and ultimately elucidate diverse biological networks for each cell type175. To maximally utilize all information from multi-omics data sets, it is indispensable to develop integrative methods that include matched and unmatched samples and/or genes from the input data. Despite advancements in the many computational integration tools for multi-omics and multi-layered data, analysis of biological functionality of cell types and gene regulatory networks remains a computational challenge.
Although multi-omics data can improve and refine the definition of cell state, these data also provide a unique resource for identifying regulatory relationships between molecular modalities. These include SCENIC+176, functional inference of gene regulation (FigR)177, and probabilistic multimodal models for integrated regulatory analysis (MIRA)178, which model joint variation across paired transcriptomic and scATAC-seq measurements to link enhancers to genes and to identify transcription factors that drive cellular decisions. Similarly, CellOracle179 leverages multi-omics data, either collected in parallel or integrated computationally, to learn gene regulatory networks and infer the results of in silico perturbations, whereas in silico ChIP–seq predicts transcription factor binding at individual cis-regulatory elements179. These methods can also be extended to model post-transcriptional regulation and predict future cellular states, such as MultiVelo, which integrates chromatin accessibility and gene expression to estimate chromatin accessibility switches and gene splicing states180. These computational approaches will enable multi-omics single-cell analysis not only to improve our understanding of cellular taxonomies, but to further our understanding of fundamental molecular processes such as gene regulation.
Conclusions and future perspective
Single-cell multi-omics have already become integral methods for elucidating the complexity of biological processes, especially when facing rare diseases and cell types. Despite recent advancements, multi-omics techniques need to be improved or explored further to have a more profound and broad impact in molecular cell biology. For example, benchmarking emerging computation tools to aid in multi-omics research has become crucial as most multi-omics methods rely on these computational methods for data integration and analysis. Furthermore, it is difficult to obtain matched or same-experiment datasets characterizing different omics layers along with gold-standard datasets, posing an urgent need for benchmarking multi-omics studies and generating guidelines for data analysis.
Other challenges include high sequencing costs and limited coverage of single cells, for which each omics layer can only be partially profiled, thus resulting in loss of important data. In the effort to capture more informative and complete genomic data, long-read technologies can advance the detection of genetic variation in cells, but still face limitations of low sequencing accuracy and difficulty in obtaining intact DNA and full-length RNA from clinical samples181. The emergence of spatial omics is transforming our understanding of the molecular mechanisms behind disease by providing spatial coordinates of cells, thus tracing back the location of cells (and their associated multi-omics readouts) within tissues. Most of the current spatial technologies are low throughput in the number of tissues they can process simultaneously and therefore cannot fully capture the 3D architecture of a tissue. Furthermore, the relatively small capture area limits their application in the complete profiling of most human tissue sections.
Finally, although many computational methods are offered for pseudotime analysis of scRNA-seq data, these methods are not always accurate in reconstructing the trajectory of cells through their developmental processes, which are complex and dynamic in nature. It would therefore be necessary to apply the dimension of time to single-cell multi-omics to unify the multi-omics layers of different datasets. Improvements and advancements in the single-cell multi-omics field will facilitate design of advanced therapeutic strategies and generate atlases of multiple omics and timescales to aid in our understanding of health and disease.
Acknowledgements
We acknowledge support from a Packard Fellowship for Science and Engineering (to R.F.), a Yale Stem Cell Center Chen Innovation Award (to R.F.) and the US National Institutes of Health (nos. RF1MH128876, U54AG076043, U54AG079759, UG3CA257393, UH3CA257393, R01CA245313 and U54CA274509 to R.F.).
Glossary
- Barcoding
Labelling of individual cells with unique nucleic acid sequences (barcodes).
- Cell multiplexing
The labelling of a sample of cells or nuclei with molecular tags or oligonucleotides, followed by mixing of this sample with other tagged or labelled samples.
- Chimeric antigen receptor (CAR) T cell therapy
A form of immunotherapy that harnesses the power of the immune system to attack cancer cells by weaponizing the T cells of the patient with a chimeric antigen receptor.
- Copy number variations
Variations in the number of copies of a specific DNA segment in the genomes of different individuals owing to duplications, deletions and insertions.
- CpG islands
Regions in gene promoters with over 50% CpG dinucleotide content, which are often methylated and function in gene regulation.
- Feature spaces
The n dimensions in which your variables live.
- Immune-checkpoint blockade
(ICB) A type of immunotherapy that blocks immune checkpoint proteins from binding their ligands.
- Molecular bridging
A phenomenon in which two molecules are linked or bridged together by another molecule, often an antibody or a DNA molecule.
- Multi-omics
Two or more omics datasets originating from different cellular features.
- Multiplexing
Consolidation of multiple data sets.
- Nonlinear manifold
The recovery of the true dimensionality of data.
- Omics
Technologies characterizing biomolecules that constitute and determine the structure, function and dynamics of an organism.
- Padlock probes
Oligonucleotides whose ends are complementary to adjacent target sequences.
- Phagemid
A type of DNA-based cloning vector.
- Proximity extension assay
(PEA) An immunohistochemistry tool used for the detection of low-abundance proteins in the blood.
- Pseudotime
Latent dimension measuring the progression of a cell along its differentiation trajectory.
- Sequencing-by-ligation
DNA sequencing using multiple primers offset by a single base in the 3′ end of the adapter, using the mismatch sensitivity of DNA ligase to determine a given set of nucleotides that make up a DNA sequence.
- Signal and allelic drop-outs
The loss of an allele during DNA amplification through PCR.
- Splint oligonucleotides
Short, synthetic single-stranded DNA molecules that are used to help circularize and amplify a target DNA sequence.
- Spot size
In spatial profiling, the size of a region that has a unique barcode, which distinguishes it from other regions.
- Tagmentation
Fragmentation and tagging of DNA molecules before library preparation and sequencing.
- V(D)J regions
The variable (V), diversity (D) and joining (J) gene segments of the immunoglobulin (Ig) and T cell receptor (TCR) genes.
Author contributions
A.B. and Z.B. researched data for the article. All authors contributed substantially to discussion of the content, wrote the article and reviewed and/or edited the manuscript before submission.
Peer review
Peer review information
Nature Reviews Molecular Cell Biology thanks Leng Han, Rafael Kramann and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Competing interests
R.F. is scientific founder and adviser for IsoPlexis, Singleron Biotechnologies and AtlasXomics. The interests of R.F. were reviewed and managed by Yale University Provost’s Office in accordance with the University’s conflict of interest policies. In the past three years, R.S. has worked as a consultant for Bristol-Myers Squibb, Regeneron and Kallyope and served as an Scientific Advisory Board member for ImmunAI, Resolve Biosciences, Nanostring and the NYC Pandemic Response Lab. A.B. and Z.B. declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
The Cancer Genome Atlas: https://www.cancer.gov/ccg/research/genome-sequencing/tcga
These authors contributed equally: Alev Baysoy, Zhiliang Bai.
References
- 1.Lee J, Hyeon DY, Hwang D. Single-cell multiomics: technologies and data analysis methods. Exp. Mol. Med. 2020;52:1428–1442. doi: 10.1038/s12276-020-0420-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davis MM, Tato CM, Furman D. Systems immunology: just getting started. Nat. Immunol. 2017;18:725–732. doi: 10.1038/ni.3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wagner DE, Klein AM. Lineage tracing meets single-cell omics: opportunities and challenges. Nat. Rev. Genet. 2020;21:410–427. doi: 10.1038/s41576-020-0223-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elmentaite R, Domínguez Conde C, Yang L, Teichmann SA. Single-cell atlases: shared and tissue-specific cell types across human organs. Nat. Rev. Genet. 2022;23:395–410. doi: 10.1038/s41576-022-00449-w. [DOI] [PubMed] [Google Scholar]
- 5.Yofe I, Dahan R, Amit I. Single-cell genomic approaches for developing the next generation of immunotherapies. Nat. Med. 2020;26:171–177. doi: 10.1038/s41591-019-0736-4. [DOI] [PubMed] [Google Scholar]
- 6.Tang F, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 7.Gierahn TM, et al. Seq-Well: portable, low-cost RNA sequencing of single cells at high throughput. Nat. Methods. 2017;14:395–398. doi: 10.1038/nmeth.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klein AM, et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161:1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Macosko EZ, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng GX, et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hagemann-Jensen M, et al. Single-cell RNA counting at allele and isoform resolution using Smart-seq3. Nat. Biotechnol. 2020;38:708–714. doi: 10.1038/s41587-020-0497-0. [DOI] [PubMed] [Google Scholar]
- 12.Natarajan KN. Single-cell tagged reverse transcription (STRT-seq) Methods Mol. Biol. 2019;1979:133–153. doi: 10.1007/978-1-4939-9240-9_9. [DOI] [PubMed] [Google Scholar]
- 13.Picelli S, et al. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 14.No Authors Listed. Method of the year 2013. Nat. Methods. 2014;11:1. doi: 10.1038/nmeth.2801. [DOI] [PubMed] [Google Scholar]
- 15.Hashimshony T, Wagner F, Sher N, Yanai I. CEL-seq: single-cell RNA-seq by multiplexed linear amplification. Cell Rep. 2012;2:666–673. doi: 10.1016/j.celrep.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 16.Islam S, et al. Highly multiplexed and strand-specific single-cell RNA 5′ end sequencing. Nat. Protoc. 2012;7:813–828. doi: 10.1038/nprot.2012.022. [DOI] [PubMed] [Google Scholar]
- 17.Jaitin DA, et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 2014;343:776–779. doi: 10.1126/science.1247651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramskold D, et al. Full-length mRNA-seq from single-cell levels of RNA and individual circulating tumor cells. Nat. Biotechnol. 2012;30:777–782. doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sasagawa Y, et al. Quartz-Seq: a highly reproducible and sensitive single-cell RNA sequencing method, reveals non-genetic gene-expression heterogeneity. Genome Biol. 2013;14:R31. doi: 10.1186/gb-2013-14-4-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han L, et al. Co-detection and sequencing of genes and transcripts from the same single cells facilitated by a microfluidics platform. Sci. Rep. 2014;4:6485. doi: 10.1038/srep06485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Macaulay IC, et al. G&T-seq: parallel sequencing of single-cell genomes and transcriptomes. Nat. Methods. 2015;12:519–522. doi: 10.1038/nmeth.3370. [DOI] [PubMed] [Google Scholar]
- 22.Dey SS, Kester L, Spanjaard B, Bienko M, van Oudenaarden A. Integrated genome and transcriptome sequencing of the same cell. Nat. Biotechnol. 2015;33:285–289. doi: 10.1038/nbt.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodriguez-Meira A, et al. Unravelling intratumoral heterogeneity through high-sensitivity single-cell mutational analysis and parallel RNA sequencing. Mol. Cell. 2019;73:1292. doi: 10.1016/j.molcel.2019.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han KY, et al. SIDR: simultaneous isolation and parallel sequencing of genomic DNA and total RNA from single cells. Genome Res. 2018;28:75–87. doi: 10.1101/gr.223263.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moore D. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38:23–38. doi: 10.1038/npp.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smallwood SA, et al. Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat. Methods. 2014;11:817–820. doi: 10.1038/nmeth.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ogbeide S, Giannese F, Mincarelli L, Macaulay IC. Into the multiverse: advances in single-cell multiomic profiling. Trends Genet. 2022;38:831–843. doi: 10.1016/j.tig.2022.03.015. [DOI] [PubMed] [Google Scholar]
- 28.Angermueller C, et al. Parallel single-cell sequencing links transcriptional and epigenetic heterogeneity. Nat. Methods. 2016;13:229–232. doi: 10.1038/nmeth.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clark SJ, et al. scNMT-seq enables joint profiling of chromatin accessibility DNA methylation and transcription in single cells. Nat. Commun. 2018;9:781. doi: 10.1038/s41467-018-03149-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo F, et al. Single-cell multi-omics sequencing of mouse early embryos and embryonic stem cells. Cell Res. 2017;27:967–988. doi: 10.1038/cr.2017.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu Y, et al. Simultaneous profiling of mRNA transcriptome and DNA methylome from a single cell. Methods Mol. Biol. 2019;1979:363–377. doi: 10.1007/978-1-4939-9240-9_21. [DOI] [PubMed] [Google Scholar]
- 32.Guo HS, et al. Profiling DNA methylome landscapes of mammalian cells with single-cell reduced-representation bisulfite sequencing. Nat. Protoc. 2015;10:645–659. doi: 10.1038/nprot.2015.039. [DOI] [PubMed] [Google Scholar]
- 33.Hou Y, et al. Single-cell triple omics sequencing reveals genetic, epigenetic, and transcriptomic heterogeneity in hepatocellular carcinomas. Cell Res. 2016;26:304–319. doi: 10.1038/cr.2016.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bian S, et al. Single-cell multiomics sequencing and analyses of human colorectal cancer. Science. 2018;362:1060–1063. doi: 10.1126/science.aao3791. [DOI] [PubMed] [Google Scholar]
- 35.Pott S. Simultaneous measurement of chromatin accessibility, DNA methylation, and nucleosome phasing in single cells. eLife. 2017 doi: 10.7554/eLife.23203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y, et al. Single-cell multiomics sequencing reveals the functional regulatory landscape of early embryos. Nat. Commun. 2021;12:1247. doi: 10.1038/s41467-021-21409-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen X, et al. Joint single-cell DNA accessibility and protein epitope profiling reveals environmental regulation of epigenomic heterogeneity. Nat. Commun. 2018;9:4590. doi: 10.1038/s41467-018-07115-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen X, Miragaia RJ, Natarajan KN, Teichmann SA. A rapid and robust method for single cell chromatin accessibility profiling. Nat. Commun. 2018;9:5345. doi: 10.1038/s41467-018-07771-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mezger A, et al. High-throughput chromatin accessibility profiling at single-cell resolution. Nat. Commun. 2018;9:3647. doi: 10.1038/s41467-018-05887-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Satpathy AT, et al. Massively parallel single-cell chromatin landscapes of human immune cell development and intratumoral T cell exhaustion. Nat. Biotechnol. 2019;37:925–936. doi: 10.1038/s41587-019-0206-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu W, et al. A plate-based single-cell ATAC-seq workflow for fast and robust profiling of chromatin accessibility. Nat. Protoc. 2021;16:4084–4107. doi: 10.1038/s41596-021-00583-5. [DOI] [PubMed] [Google Scholar]
- 43.Buenrostro JD, et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. 2015;523:486–490. doi: 10.1038/nature14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cusanovich DA, et al. Multiplex single cell profiling of chromatin accessibility by combinatorial cellular indexing. Science. 2015;348:910–914. doi: 10.1126/science.aab1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grosselin K, et al. High-throughput single-cell ChIP-seq identifies heterogeneity of chromatin states in breast cancer. Nat. Genet. 2019;51:1060–1066. doi: 10.1038/s41588-019-0424-9. [DOI] [PubMed] [Google Scholar]
- 46.Skene PJ, Henikoff S. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. eLife. 2017 doi: 10.7554/eLife.21856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kaya-Okur HS, et al. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun. 2019;10:1930. doi: 10.1038/s41467-019-09982-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bartosovic M, Kabbe M, Castelo-Branco G. Single-cell CUT&Tag profiles histone modifications and transcription factors in complex tissues. Nat. Biotechnol. 2021;39:825–835. doi: 10.1038/s41587-021-00869-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Janssens DH, et al. CUT&Tag2for1: a modified method for simultaneous profiling of the accessible and silenced regulome in single cells. Genome Biol. 2022;23:81. doi: 10.1186/s13059-022-02642-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang B, et al. Characterizing cellular heterogeneity in chromatin state with scCUT&Tag-pro. Nat. Biotechnol. 2022;40:1220–1230. doi: 10.1038/s41587-022-01250-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu C, et al. Joint profiling of histone modifications and transcriptome in single cells from mouse brain. Nat. Methods. 2021;18:283–292. doi: 10.1038/s41592-021-01060-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xiong H, Luo Y, Wang Q, Yu X, He A. Single-cell joint detection of chromatin occupancy and transcriptome enables higher-dimensional epigenomic reconstructions. Nat. Methods. 2021;18:652–660. doi: 10.1038/s41592-021-01129-z. [DOI] [PubMed] [Google Scholar]
- 53.Liu L, et al. Deconvolution of single-cell multi-omics layers reveals regulatory heterogeneity. Nat. Commun. 2019;10:470. doi: 10.1038/s41467-018-08205-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xing QR, et al. Parallel bimodal single-cell sequencing of transcriptome and chromatin accessibility. Genome Res. 2020;30:1027–1039. doi: 10.1101/gr.257840.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vitak SA, et al. Sequencing thousands of single-cell genomes with combinatorial indexing. Nat. Methods. 2017;14:302–308. doi: 10.1038/nmeth.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cao J, et al. Joint profiling of chromatin accessibility and gene expression in thousands of single cells. Science. 2018;361:1380–1385. doi: 10.1126/science.aau0730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen S, Lake BB, Zhang K. High-throughput sequencing of the transcriptome and chromatin accessibility in the same cell. Nat. Biotechnol. 2019;37:1452–1457. doi: 10.1038/s41587-019-0290-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rosenberg AB, et al. Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science. 2018;360:176–182. doi: 10.1126/science.aam8999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu C, et al. An ultra high-throughput method for single-cell joint analysis of open chromatin and transcriptome. Nat. Struct. Mol. Biol. 2019;26:1063–1070. doi: 10.1038/s41594-019-0323-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ma S, et al. Chromatin potential identified by shared single-cell profiling of RNA and chromatin. Cell. 2020;183:1103. doi: 10.1016/j.cell.2020.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Swanson E, et al. Simultaneous trimodal single-cell measurement of transcripts, epitopes, and chromatin accessibility using TEA-seq. eLife. 2021 doi: 10.7554/eLife.63632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu W, et al. ISSAAC-seq enables sensitive and flexible multimodal profiling of chromatin accessibility and gene expression in single cells. Nat. Methods. 2022;19:1243–1249. doi: 10.1038/s41592-022-01601-4. [DOI] [PubMed] [Google Scholar]
- 63.Albayrak C, et al. Digital quantification of proteins and mRNA in single mammalian cells. Mol. Cell. 2016;61:914–924. doi: 10.1016/j.molcel.2016.02.030. [DOI] [PubMed] [Google Scholar]
- 64.Darmanis S, et al. Simultaneous multiplexed measurement of RNA and proteins in single cells. Cell Rep. 2016;14:380–389. doi: 10.1016/j.celrep.2015.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Genshaft AS, et al. Multiplexed, targeted profiling of single-cell proteomes and transcriptomes in a single reaction. Genome Biol. 2016;17:188. doi: 10.1186/s13059-016-1045-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stahlberg A, Thomsen C, Ruff D, Aman P. Quantitative PCR analysis of DNA, RNAs, and proteins in the same single cell. Clin. Chem. 2012;58:1682–1691. doi: 10.1373/clinchem.2012.191445. [DOI] [PubMed] [Google Scholar]
- 67.Frei AP, et al. Highly multiplexed simultaneous detection of RNAs and proteins in single cells. Nat. Methods. 2016;13:269–275. doi: 10.1038/nmeth.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stoeckius M, et al. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods. 2017;14:865–868. doi: 10.1038/nmeth.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Peterson VM, et al. Multiplexed quantification of proteins and transcripts in single cells. Nat. Biotechnol. 2017;35:936–939. doi: 10.1038/nbt.3973. [DOI] [PubMed] [Google Scholar]
- 70.Hwang B, et al. SCITO-seq: single-cell combinatorial indexed cytometry sequencing. Nat. Methods. 2021;18:903–911. doi: 10.1038/s41592-021-01222-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shum EY, Walczak EM, Chang C, Christina Fan H. Quantitation of mRNA transcripts and proteins using the BD RhapsodyTM single-cell analysis system. Adv. Exp. Med. Biol. 2019;1129:63–79. doi: 10.1007/978-981-13-6037-4_5. [DOI] [PubMed] [Google Scholar]
- 72.Budnik B, Levy E, Harmange G, Slavov N. SCoPE-MS: mass spectrometry of single mammalian cells quantifies proteome heterogeneity during cell differentiation. Genome Biol. 2018;19:161. doi: 10.1186/s13059-018-1547-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Specht H, et al. Single-cell proteomic and transcriptomic analysis of macrophage heterogeneity using SCoPE2. Genome Biol. 2021;22:50. doi: 10.1186/s13059-021-02267-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fiskin E, et al. Single-cell profiling of proteins and chromatin accessibility using PHAGE-ATAC. Nat. Biotechnol. 2022;40:374–381. doi: 10.1038/s41587-021-01065-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chung H, et al. Joint single-cell measurements of nuclear proteins and RNA in vivo. Nat. Methods. 2021;18:1204–1212. doi: 10.1038/s41592-021-01278-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Katzenelenbogen Y, et al. Coupled scRNA-seq and intracellular protein activity reveal an immunosuppressive role of TREM2 in cancer. Cell. 2020;182:872–885.e19. doi: 10.1016/j.cell.2020.06.032. [DOI] [PubMed] [Google Scholar]
- 77.Gerlach JP, et al. Combined quantification of intracellular (phospho-)proteins and transcriptomics from fixed single cells. Sci. Rep. 2019;9:1469. doi: 10.1038/s41598-018-37977-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mimitou EP, et al. Scalable, multimodal profiling of chromatin accessibility, gene expression and protein levels in single cells. Nat. Biotechnol. 2021;39:1246–1258. doi: 10.1038/s41587-021-00927-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lareau CA, et al. Massively parallel single-cell mitochondrial DNA genotyping and chromatin profiling. Nat. Biotechnol. 2021;39:451–461. doi: 10.1038/s41587-020-0645-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen AF, et al. NEAT-seq: simultaneous profiling of intra-nuclear proteins, chromatin accessibility and gene expression in single cells. Nat. Methods. 2022;19:547–553. doi: 10.1038/s41592-022-01461-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mimitou EP, et al. Multiplexed detection of proteins, transcriptomes, clonotypes and CRISPR perturbations in single cells. Nat. Methods. 2019;16:409–412. doi: 10.1038/s41592-019-0392-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dixit A, et al. Perturb-Seq: dissecting molecular circuits with scalable single-cell RNA profiling of pooled genetic screens. Cell. 2016;167:1853–1866.e17. doi: 10.1016/j.cell.2016.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Datlinger P, et al. Pooled CRISPR screening with single-cell transcriptome readout. Nat. Methods. 2017;14:297–301. doi: 10.1038/nmeth.4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jaitin DA, et al. Dissecting immune circuits by linking CRISPR-pooled screens with single-cell RNA-seq. Cell. 2016;167:1883–1896.e15. doi: 10.1016/j.cell.2016.11.039. [DOI] [PubMed] [Google Scholar]
- 86.Xie S, Duan J, Li B, Zhou P, Hon GC. Multiplexed engineering and analysis of combinatorial enhancer activity in single cells. Mol. Cell. 2017;66:285–299.e5. doi: 10.1016/j.molcel.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 87.Pierce SE, Granja JM, Greenleaf WJ. High-throughput single-cell chromatin accessibility CRISPR screens enable unbiased identification of regulatory networks in cancer. Nat. Commun. 2021;12:2969. doi: 10.1038/s41467-021-23213-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pai JA, Satpathy AT. High-throughput and single-cell T cell receptor sequencing technologies. Nat. Methods. 2021;18:881–892. doi: 10.1038/s41592-021-01201-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Neal JT, et al. Organoid modeling of the tumor immune microenvironment. Cell. 2018;175:1972–1988.e16. doi: 10.1016/j.cell.2018.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Singh M, et al. High-throughput targeted long-read single cell sequencing reveals the clonal and transcriptional landscape of lymphocytes. Nat. Commun. 2019;10:3120. doi: 10.1038/s41467-019-11049-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tu AA, et al. TCR sequencing paired with massively parallel 3′ RNA-seq reveals clonotypic T cell signatures. Nat. Immunol. 2019;20:1692–1699. doi: 10.1038/s41590-019-0544-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zemmour D, et al. Publisher correction: single-cell gene expression reveals a landscape of regulatory T cell phenotypes shaped by the TCR. Nat. Immunol. 2018;19:645. doi: 10.1038/s41590-018-0104-4. [DOI] [PubMed] [Google Scholar]
- 93.Minervina A, Pogorelyy M, Mamedov I. T-cell receptor and B-cell receptor repertoire profiling in adaptive immunity. Transpl. Int. 2019;32:1111–1123. doi: 10.1111/tri.13475. [DOI] [PubMed] [Google Scholar]
- 94.Satpathy AT, et al. Transcript-indexed ATAC-seq for precision immune profiling. Nat. Med. 2018;24:580–590. doi: 10.1038/s41591-018-0008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Oxford Nanopore Technologies. Oxford Nanopore Technologies announces new collaboration with 10x Genomics to make single-cell and spatial full-length isoform transcript sequencing accessible to any laboratory, https://nanoporetech.com/about-us/news/oxford-nanopore-technologies-announces-new-collaboration-10x-genomics-make-single (27 October 2022).
- 96.Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R. CellPhoneDB: inferring cell–cell communication from combined expression of multi-subunit ligand–receptor complexes. Nat. Protoc. 2020;15:1484–1506. doi: 10.1038/s41596-020-0292-x. [DOI] [PubMed] [Google Scholar]
- 97.Browaeys R, Saelens W, Saeys Y. NicheNet: modeling intercellular communication by linking ligands to target genes. Nat. Methods. 2020;17:159–162. doi: 10.1038/s41592-019-0667-5. [DOI] [PubMed] [Google Scholar]
- 98.Langer-Safer PR, Levine M, Ward DC. Immunological method for mapping genes on Drosophila polytene chromosomes. Proc. Natl Acad. Sci. USA. 1982;79:4381–4385. doi: 10.1073/pnas.79.14.4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Femino AM, Fay FS, Fogarty K, Singer RH. Visualization of single RNA transcripts in situ. Science. 1998;280:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- 100.Lubeck E, Coskun AF, Zhiyentayev T, Ahmad M, Cai L. Single-cell in situ RNA profiling by sequential hybridization. Nat. Methods. 2014;11:360–361. doi: 10.1038/nmeth.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X. Spatially resolved, highly multiplexed RNA profiling in single cells. Science. 2015;348:aaa6090. doi: 10.1126/science.aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Eng CL, et al. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH. Nature. 2019;568:235–239. doi: 10.1038/s41586-019-1049-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bio-Techne. Bio-Techne announces commercial release of RNAscope® HiPlex Assay: a multiplex in situ hybridization assay for tissues, https://investors.bio-techne.com/press-releases/detail/148/bio-techne-announces-commercial-release-of-rnascope (2016).
- 104.Schulz D, et al. Simultaneous multiplexed imaging of mRNA and proteins with subcellular resolution in breast cancer tissue samples by mass cytometry. Cell Syst. 2018;6:531. doi: 10.1016/j.cels.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang F, et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J. Mol. Diagn. 2012;14:22–29. doi: 10.1016/j.jmoldx.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Borm LE, et al. Scalable in situ single-cell profiling by electrophoretic capture of mRNA using EEL FISH. Nat. Biotechnol. 2022 doi: 10.1038/s41587-022-01455-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chen F, et al. Nanoscale imaging of RNA with expansion microscopy. Nat. Methods. 2016;13:679–684. doi: 10.1038/nmeth.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Codeluppi S, et al. Spatial organization of the somatosensory cortex revealed by osmFISH. Nat. Methods. 2018;15:932–935. doi: 10.1038/s41592-018-0175-z. [DOI] [PubMed] [Google Scholar]
- 109.Moffitt JR, et al. Molecular, spatial, and functional single-cell profiling of the hypothalamic preoptic region. Science. 2018 doi: 10.1126/science.aau5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Qian X, et al. Probabilistic cell typing enables fine mapping of closely related cell types in situ. Nat. Methods. 2020;17:101–106. doi: 10.1038/s41592-019-0631-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wang X, et al. Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science. 2018 doi: 10.1126/science.aat5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Goltsev Y, et al. Deep profiling of mouse splenic architecture with CODEX multiplexed imaging. Cell. 2018;174:968–981.e15. doi: 10.1016/j.cell.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.He S, et al. High-plex imaging of RNA and proteins at subcellular resolution in fixed tissue by spatial molecular imaging. Nat. Biotechnol. 2022 doi: 10.1038/s41587-022-01483-z. [DOI] [PubMed] [Google Scholar]
- 114.Ke R, et al. In situ sequencing for RNA analysis in preserved tissue and cells. Nat. Methods. 2013;10:857–860. doi: 10.1038/nmeth.2563. [DOI] [PubMed] [Google Scholar]
- 115.Lee JH, et al. Highly multiplexed subcellular RNA sequencing in situ. Science. 2014;343:1360–1363. doi: 10.1126/science.1250212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ståhl PL, et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. 2016;353:78–82. doi: 10.1126/science.aaf2403. [DOI] [PubMed] [Google Scholar]
- 117.10x Genomics. 10x Genomics acquires Spatial Transcriptomics, https://www.genomeweb.com/sequencing/10x-genomics-acquires-spatial-transcriptomics (2018).
- 118.Rodriques SG, et al. Slide-seq: a scalable technology for measuring genome-wide expression at high spatial resolution. Science. 2019;363:1463–1467. doi: 10.1126/science.aaw1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Stickels RR, et al. Highly sensitive spatial transcriptomics at near-cellular resolution with Slide-seqV2. Nat. Biotechnol. 2021;39:313–319. doi: 10.1038/s41587-020-0739-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Vickovic S, et al. High-definition spatial transcriptomics for in situ tissue profiling. Nat. Methods. 2019;16:987–990. doi: 10.1038/s41592-019-0548-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Merritt CR, et al. Multiplex digital spatial profiling of proteins and RNA in fixed tissue. Nat. Biotechnol. 2020;38:586–599. doi: 10.1038/s41587-020-0472-9. [DOI] [PubMed] [Google Scholar]
- 122.Salmen F. Barcoded solid-phase RNA capture for spatial transcriptomics profiling in mammalian tissue sections. Nature. 2018;13:2501–2534. doi: 10.1038/s41596-018-0045-2. [DOI] [PubMed] [Google Scholar]
- 123.Liu Y, et al. High-spatial-resolution multi-omics sequencing via deterministic barcoding in tissue. Cell. 2020;183:1665–1681.e18. doi: 10.1016/j.cell.2020.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Deng Y, et al. Spatial profiling of chromatin accessibility in mouse and human tissues. Nature. 2022;609:375–383. doi: 10.1038/s41586-022-05094-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Deng Y, et al. Spatial-CUT&Tag: Spatially resolved chromatin modification profiling at the cellular level. Science. 2022;375:681–686. doi: 10.1126/science.abg7216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Liu Y, et al. High-plex protein and whole transcriptome co-mapping at cellular resolution with spatial CITE-seq. Nat. Biotechnol. 2023 doi: 10.1038/s41587-023-01676-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhang D, et al. Spatial epigenome–transcriptome co-profiling of mammalian tissues. Nature. 2023;616:113–122. doi: 10.1038/s41586-023-05795-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jiang F, et al. Simultaneously spatiotemporal gene expression and chromatin accessibility for mouse brain development. bioRxiv. 2022 doi: 10.1101/2022.03.22.485333. [DOI] [Google Scholar]
- 129.Vickovic S, et al. SM-Omics is an automated platform for high-throughput spatial multi-omics. Nat. Commun. 2022;13:795. doi: 10.1038/s41467-022-28445-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ben-Chetrit N, et al. Integration of whole transcriptome spatial profiling with protein markers. Nat. Biotechnol. 2023 doi: 10.1038/s41587-022-01536-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Li S, et al. RNase H-dependent PCR-enabled T-cell receptor sequencing for highly specific and efficient targeted sequencing of T-cell receptor mRNA for single-cell and repertoire analysis. Nat. Protoc. 2019;14:2571–2594. doi: 10.1038/s41596-019-0195-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Liu S, et al. Spatial maps of T cell receptors and transcriptomes reveal distinct immune niches and interactions in the adaptive immune response. Immunity. 2022;55:1940–1952.e5. doi: 10.1016/j.immuni.2022.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ding J, et al. Integrating multiomics longitudinal data to reconstruct networks underlying lung development. Am. J. Physiol. Lung Cell Mol. Physiol. 2019;317:L556–L568. doi: 10.1152/ajplung.00554.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Chen J, et al. Spatial transcriptomic analysis of cryosectioned tissue samples with Geo-seq. Nat. Protoc. 2017;12:566–580. doi: 10.1038/nprot.2017.003. [DOI] [PubMed] [Google Scholar]
- 135.Mund A. Deep visual proteomics defines single-cell identity and heterogeneity. Nat. Biotechnol. 2022;40:1231–1240. doi: 10.1038/s41587-022-01302-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kester L, van Oudenaarden A. Single-cell transcriptomics meets lineage tracing. Cell Stem Cell. 2018;23:166–179. doi: 10.1016/j.stem.2018.04.014. [DOI] [PubMed] [Google Scholar]
- 137.VanHorn S, Morris SA. Next-generation lineage tracing and fate mapping to interrogate development. Dev. Cell. 2021;56:7–21. doi: 10.1016/j.devcel.2020.10.021. [DOI] [PubMed] [Google Scholar]
- 138.Woodworth MB, Girskis KM, Walsh CA. Building a lineage from single cells: genetic techniques for cell lineage tracking. Nat. Rev. Genet. 2017;18:230–244. doi: 10.1038/nrg.2016.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Palii CG, et al. Single-cell proteomics reveal that quantitative changes in co-expressed lineage-specific transcription factors determine cell fate. Cell Stem Cell. 2019;24:812–820.e5. doi: 10.1016/j.stem.2019.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.He Z. Lineage recording in human cerebral organoids. Nat. Methods. 2022;19:90–99. doi: 10.1038/s41592-021-01344-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kowarz E. Optimized Sleeping Beauty transposons rapidly generate stable transgenic cell lines. Biotechnol. J. 2015;10:647–653. doi: 10.1002/biot.201400821. [DOI] [PubMed] [Google Scholar]
- 142.Eyler CE, et al. Single-cell lineage analysis reveals genetic and epigenetic interplay in glioblastoma drug resistance. Genome Biol. 2020;21:174. doi: 10.1186/s13059-020-02085-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Gaiti F. Epigenetic evolution and lineage histories of chronic lymphocytic leukaemia. Nature. 2019;569:576–580. doi: 10.1038/s41586-019-1198-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kasmani MY, et al. Clonal lineage tracing reveals mechanisms skewing CD8+ T cell fate decisions in chronic infection. J. Exp. Med. 2023 doi: 10.1084/jem.20220679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Peng G, Cui G, Ke J, Jing N. Using single-cell and spatial transcriptomes to understand stem cell lineage specification during early embryo development. Annu. Rev. Genomics Hum. Genet. 2020;21:163–181. doi: 10.1146/annurev-genom-120219-083220. [DOI] [PubMed] [Google Scholar]
- 146.Guo H. Modulation of long noncoding RNAs by risk SNPs underlying genetic predispositions to prostate cancer. Nat. Genet. 2016;48:1142–1150. doi: 10.1038/ng.3637. [DOI] [PubMed] [Google Scholar]
- 147.Cancer Genome Atlas Research Network et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Dunham I, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Abascal F, et al. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature. 2020;583:699–710. doi: 10.1038/s41586-020-2493-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Taliun D, et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature. 2021;590:290–299. doi: 10.1038/s41586-021-03205-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Regev A, et al. The Human Cell Atlas. eLife. 2017;6:e27041. doi: 10.7554/eLife.27041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lindeboom RGH, Regev A, Teichmann SA. Towards a human cell atlas: taking notes from the past. Trends Genet. 2021;37:625–630. doi: 10.1016/j.tig.2021.03.007. [DOI] [PubMed] [Google Scholar]
- 153.Madissoon E, et al. A spatially resolved atlas of the human lung characterizes a gland-associated immune niche. Nat. Genet. 2023;55:66–77. doi: 10.1038/s41588-022-01243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Stephenson E, et al. Single-cell multi-omics analysis of the immune response in COVID-19. Nat. Med. 2021;27:904–916. doi: 10.1038/s41591-021-01329-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Miranda AMA, et al. Single-cell transcriptomics for the assessment of cardiac disease. Nat. Rev. Cardiol. 2022 doi: 10.1038/s41569-022-00805-7. [DOI] [PubMed] [Google Scholar]
- 156.Dong X, Liu C, Dozmorov M. Review of multi-omics data resources and integrative analysis for human brain disorders. Brief. Funct. Genomics. 2021;20:223–234. doi: 10.1093/bfgp/elab024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zhao T, et al. Spatial genomics enables multi-modal study of clonal heterogeneity in tissues. Nature. 2022;601:85–91. doi: 10.1038/s41586-021-04217-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Joshi M. CPT1A over-expression increases reactive oxygen species in the mitochondria and promotes antioxidant defenses in prostate cancer. Cancers. 2020;12:3431. doi: 10.3390/cancers12113431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Hoang LT, et al. Metabolomic, transcriptomic and genetic integrative analysis reveals important roles of adenosine diphosphate in haemostasis and platelet activation in non-small-cell lung cancer. Mol. Oncol. 2019;13:2406–2421. doi: 10.1002/1878-0261.12568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Dan K. Integrated analysis of whole-genome ChIP-seq and RNA-seq data of primary head and neck tumor samples associates HPV integration sites with open chromatin marks. Cancer Res. 2017;77:6538–6550. doi: 10.1158/0008-5472.CAN-17-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Dijkstra JJ. Multiomics of colorectal cancer organoids reveals putative mediators of cancer progression resulting from SMAD4 inactivation. J. Proteome Res. 2022;22:138–151. doi: 10.1021/acs.jproteome.2c00551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Chen YJ, et al. Proteogenomics of non-smoking lung cancer in East Asia delineates molecular signatures of pathogenesis and progression. Cell. 2020;182:226–244.e17. doi: 10.1016/j.cell.2020.06.012. [DOI] [PubMed] [Google Scholar]
- 163.Granja JM, et al. Single-cell multiomic analysis identifies regulatory programs in mixed-phenotype acute leukemia. Nat. Biotechnol. 2019;37:1458–1465. doi: 10.1038/s41587-019-0332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Liu B, Zhang Y, Wang D, Hu X, Zhang Z. Single-cell meta-analyses reveal responses of tumor-reactive CXCL13+ T cells to immune-checkpoint blockade. Nat. Cancer. 2022;3:1123–1136. doi: 10.1038/s43018-022-00433-7. [DOI] [PubMed] [Google Scholar]
- 165.Hammerl D, et al. Spatial immunophenotypes predict response to anti-PD1 treatment and capture distinct paths of T cell evasion in triple negative breast cancer. Nat. Commun. 2021;12:5668. doi: 10.1038/s41467-021-25962-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Bai Z, et al. Single-cell antigen-specific landscape of CAR T infusion product identifies determinants of CD19-positive relapse in patients with ALL. Sci. Adv. 2022;8:eabj2820. doi: 10.1126/sciadv.abj2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Stuart T, et al. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Welch J. Single-cell multi-omic integration compares and contrasts features of brain cell identity. Cell. 2019;177:1873–1887. doi: 10.1016/j.cell.2019.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Cao ZJ, Gao G. Multi-omics single-cell data integration and regulatory inference with graph-linked embedding. Nat. Biotechnol. 2022;40:1458–1466. doi: 10.1038/s41587-022-01284-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ghazanfar S. StabMap: Mosaic single cell data integration using non-overlapping features. bioRxiv. 2022 doi: 10.1101/2022.02.24.481823. [DOI] [Google Scholar]
- 171.Gong B. Cobolt: integrative analysis of multimodal single-cell sequencing data. Genome Biol. 2021;22:351. doi: 10.1186/s13059-021-02556-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Ashuach T. MultiVI: deep generative model for the integration of multi-modal data. bioRxiv. 2021 doi: 10.1101/2021.08.20.457057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Hao Y. Dictionary learning for integrative, multimodal, and scalable single-cell analysis. bioRxiv. 2022 doi: 10.1101/2022.02.24.481684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Hao Y, et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–3587.e29. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Ma A, et al. Single-cell biological network inference using a heterogeneous graph transformer. Nat. Commun. 2023;14:964. doi: 10.1038/s41467-023-36559-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Gonzalez-Blas C. SCENIC+: single-cell multiomic inference of enhancers and gene regulatory networks. bioRxiv. 2022 doi: 10.1101/2022.08.19.504505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Kartha V. Functional inference of gene regulation using single-cell multi-omics. Cell Genom. 2022;2:100166. doi: 10.1016/j.xgen.2022.100166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Lynch A. MIRA: joint regulatory modeling of multimodal expression and chromatin accessibility in single cells. Nat. Methods. 2021;19:1097–1108. doi: 10.1038/s41592-022-01595-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Kamimoto K. Dissecting cell identity via network inference and in silico gene perturbation. Nature. 2023;614:742–751. doi: 10.1038/s41586-022-05688-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Li C. Multi-omic single-cell velocity models epigenome–transcriptome interactions and improves cell fate prediction. Nat. Biotechnol. 2023;41:387–398. doi: 10.1038/s41587-022-01476-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Sakamoto Y, Sereewattanawoot S, Suzuki A. A new era of long-read sequencing for cancer genomics. J. Hum. Genet. 2020;65:3–10. doi: 10.1038/s10038-019-0658-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Tedesco M, et al. Chromatin Velocity reveals epigenetic dynamics by single-cell profiling of heterochromatin and euchromatin. Nat. Biotechnol. 2022;40:235–244. doi: 10.1038/s41587-021-01031-1. [DOI] [PubMed] [Google Scholar]
- 183.GTEx Consortium. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Thul PJ, Lindskog C. The human protein atlas: a spatial map of the human proteome. Protein Sci. 2018;27:233–244. doi: 10.1002/pro.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Curtis C, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Perez-Riverol Y, et al. Discovering and linking public omics data sets using the Omics Discovery Index. Nat. Biotechnol. 2017;35:406–409. doi: 10.1038/nbt.3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.COvid-19 Multi-omics Blood ATlas (COMBAT) Consortium. A blood atlas of COVID-19 defines hallmarks of disease severity and specificity. Cell. 2022;185:916–938. doi: 10.1016/j.cell.2022.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Aging Atlas Consortium. Aging Atlas: a multi-omics database for aging biology. Nucleic Acids Res. 2020;49:D825–D830. doi: 10.1093/nar/gkaa894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Singh R, Hie BL, Narayan A, Berger B. Schema: metric learning enables interpretable synthesis of heterogeneous single-cell modalities. Genome Biol. 2021;22:131. doi: 10.1186/s13059-021-02313-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Zuo C, Dai H, Chen L. Deep cross-omics cycle attention model for joint analysis of single-cell multi-omics data. Bioinformatics. 2021 doi: 10.1093/bioinformatics/btab403. [DOI] [PubMed] [Google Scholar]
- 191.Kim HJ, Lin Y, Geddes TA, Yang JYH, Yang P. CiteFuse enables multi-modal analysis of CITE-seq data. Bioinformatics. 2020;36:4137–4143. doi: 10.1093/bioinformatics/btaa282. [DOI] [PubMed] [Google Scholar]
- 192.Argelaguet R, et al. MOFA+: a statistical framework for comprehensive integration of multi-modal single-cell data. Genome Biol. 2020;21:111. doi: 10.1186/s13059-020-02015-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Zuo C, Chen L. Deep-joint-learning analysis model of single cell transcriptome and open chromatin accessibility data. Brief. Bioinform. 2021 doi: 10.1093/bib/bbaa287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Ma A, McDermaid A, Xu J, Chang Y, Ma Q. Integrative methods and practical challenges for single-cell multi-omics. Trends Biotechnol. 2020;38:1007–1022. doi: 10.1016/j.tibtech.2020.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Wang X, et al. BREM-SC: a Bayesian random effects mixture model for joint clustering single cell multi-omics data. Nucleic Acids Res. 2020;48:5814–5824. doi: 10.1093/nar/gkaa314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.John CR, Watson D, Barnes MR, Pitzalis C, Lewis MJ. Spectrum: fast density-aware spectral clustering for single and multi-omic data. Bioinformatics. 2020;36:1159–1166. doi: 10.1093/bioinformatics/btz704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Dou J. Unbiased integration of single cell multi-omics data. bioRxiv. 2020 doi: 10.1101/2020.12.11.422014. [DOI] [Google Scholar]
- 198.Liu J, Huang Y, Singh R, Vert JP, Noble WS. Jointly embedding multiple single-cell omics measurements. Algorithms Bioinform. 2019 doi: 10.4230/LIPIcs.WABI.2019.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Duan B, et al. Model-based understanding of single-cell CRISPR screening. Nat. Commun. 2019;10:2233. doi: 10.1038/s41467-019-10216-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Cao K, Bai X, Hong Y, Wan L. Unsupervised topological alignment for single-cell multi-omics integration. Bioinformatics. 2020;36:i48–i56. doi: 10.1093/bioinformatics/btaa443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Campbell KR, et al. clonealign: Statistical integration of independent single-cell RNA and DNA sequencing data from human cancers. Genome Biol. 2019;20:54. doi: 10.1186/s13059-019-1645-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Cao K, Hong Y, Wan L. Manifold alignment for heterogeneous single-cell multi-omics data integration using Pamona. Bioinformatics. 2021;38:211–219. doi: 10.1093/bioinformatics/btab594. [DOI] [PMC free article] [PubMed] [Google Scholar]