Abstract
Farnesoid X receptor (FXR) is a ligand-activated transcription factor known as bile acid receptor (BAR). FXR plays critical roles in various biological processes, including metabolism, immune inflammation, liver regeneration and liver carcinogenesis. FXR forms a heterodimer with the retinoid X receptor (RXR) and binds to diverse FXR response elements (FXREs) to exert its various biological functions. However, the mechanism by which the FXR/RXR heterodimer binds the DNA elements remains unclear. In this study, we aimed to use structural, biochemical and bioinformatics analyses to study the mechanism of FXR binding to the typical FXRE, such as the IR1 site, and the heterodimer interactions in the FXR-DBD/RXR-DBD complex. Further biochemical assays showed that RAR, THR and NR4A2 do not form heterodimers with RXR when bound to the IR1 sites, which indicates that IR1 may be a unique binding site for the FXR/RXR heterodimer. Our studies may provide a further understanding of the dimerization specificity of nuclear receptors.
Key words: Farnesoid X receptor, Retinoid X receptor, Heterodimer, Inverted repeat DNA, Crystal structure
Graphical Abstract
1. Introduction
The farnesoid X receptor (FXR, also named NR1H4) is a ligand-activated transcription factor and belongs to the nuclear receptor superfamily [1]. Expressed mainly in the liver, intestine, and kidney, FXR is an endogenous biosensor for bile acids and is known as the bile acid receptor (BAR) [2]. FXR affects numerous signaling pathways by directly regulating the transcription of several genes, including small heterodimer partner (SHP), multidrug resistance protein 3 (MDR3), organic solute transporter alpha (OSTα) and intestinal bile acid binding protein (IBABP) [3]. FXR plays critical roles in metabolism, immune inflammation, liver regeneration and liver carcinogenesis [4], [5]. Transcription termination after Arg176 (R176 *) or inserting a Lys into Tyr139 and Asn140 (Tyr139_Asn140insLys) may cause a low GGT form of severe progressive familial intrahepatic cholestasis (PFIC) [6]. Given its importance, FXR has been identified as an effective drug target to treat related diseases. Obeticholic acid (OCA), an FXR agonist, has been approved as a treatment for primary biliary cholangitis (PBC) since 2016 and is under evaluation in primary sclerosing cholangitis (PSC) and nonalcoholic steatohepatitis (NASH) [7], [8]. The decrease in FXR signaling could downregulate the expression of ACE2, which may protect individuals from SARS-CoV-2 infection in vivo and in vitro [9].
FXR regulates gene expression in different manners. Upon activation, FXR can bind diverse FXR response elements (FXREs) with its heterodimeric partner, retinoid X receptor (RXR), to exert its various biological functions. The most well-known FXRE motif is IR1, which consists of two copies of a six-nucleotide sequence (AGGTCA) arranged as inverted repeats separated by one nucleotide (Fig. 1A) [3]. The FXR/RXR heterodimer also binds other FXREs, such as the IR0 motif, everted repeats with two spacers (ER2) and direct repeats separated by one nucleotide (DR1) [10], [11]. In addition, FXR binds as a monomer or homodimer to the negative FXREs in the promoter region of the ApoA1 gene to repress its expression [12].
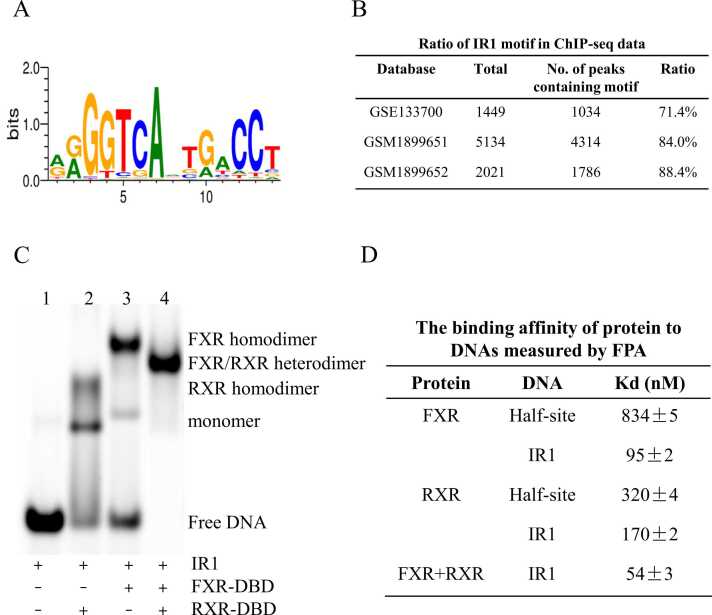
Fig. 1.
The binding characteristic of FXR to the IR1 site. A: DNA matrix of the IR1 element. B: Occurrence of the IR1 binding motif in the FXR-binding sites. C: Binding features of FXR with IR1 in the absence of RXR or in the presence of RXR were determined by EMSA. D: DNA binding affinities of FXR and RXR to IR1 were measured by FPA.
RXR is known as a heterodimer partner with type II nuclear receptors, including retinoic acid receptor (RAR), FXR, thyroid hormone receptor (THR), and NR4A2 [13], [14], [15], [16]. RXR heterodimers promote the transcription of downstream target genes and regulate diverse biological processes in response to endogenous ligands and therapeutic drugs [17]. RXR heterodimers generally bind direct repeat DNA sites separated by 1–5 nucleotides (DR1–5)[13], [18], [19], [20], [21]. A set of crystal structures of RXR-DBD heterodimers bound with direct repeat DNA sites, including RXR/RAR/DR0 (6XWH) [22], RXR/RAR/DR1 (1DSZ) [23], RXR/VDR/DR3 (1YNW) [24], RXR/THR/DR4 (2NLL) [25], RXR/RAR/DR5 (6XWG) [22], and some DBD/DNA structures of multiple-domain nuclear receptors organized on DNA direct repeats such as PPAR/RXR/DR1 (3DZY) [26], RARβ/RXRα/DR1 (5UAN) [27], HNF4α/DR1 (4IQR) [28] and RXR/LXR/DR4 (4NQA) [29], [30] has revealed how RXR and other nuclear receptors bind to their cognate sites selectively in a head-to-tail orientation. The FXR/RXR heterodimer is known to preferentially target IR1 response elements, which is distinct from the DNA binding requirements of the RXR heterodimer described above.
FXR shares a common structural organization with other nuclear receptors, which share highly conserved DNA-binding domains (DBDs) and moderately conserved ligand-binding domains (LBDs). The reported FXR-LBD structure provided key information about how bile acids and the drug currently being developed for NASH bind to this pocket [31]. The crystal structure of the FXR-LBD/RXR-LBD heterodimer showed that dimerization with RXR induced conformational changes in FXR, which improved the recruitment of coactivators and enhanced the transcriptional activity of FXR [32]. However, the mechanism by which the FXR/RXR heterodimer binds DNA elements remains unclear. In this study, we aimed to use structural, biochemical and bioinformatics analyses to study the mechanism of FXR binding to the IR1 site and the heterodimer interactions in the FXR-DBD/RXR-DBD complex.
2. Results
2.1. The IR1 motif is a potent FXR binding site
The matrix of the IR1 site used for subsequent research was obtained in the footprintDB and generated using ESPript 3.0 [33] (Fig. 1A). Our bioinformatic analysis showed that the IR1 motif accounts for 71.4% of the total FXR binding site in the ChIP-seq database GSE133700 [10] (Fig. 1B). To further test the potential biological relevance of this motif, we investigated the occurrences of the IR1 motif in two other FXR ChIP-Seq datasets, GSM1899651 and GSM1899652 [34]. The frequencies of the IR1 motif were 84.0% and 88.4%, respectively (Fig. 1B). IR1 motifs were found in the promoter region of some known FXR target genes, including SHP, MDR3 and TYPM6 [35], [36]. These results demonstrated that the IR1 site is an endogenous and important regulatory element for FXR.
2.2. The binding characteristic of FXR to the IR1 site
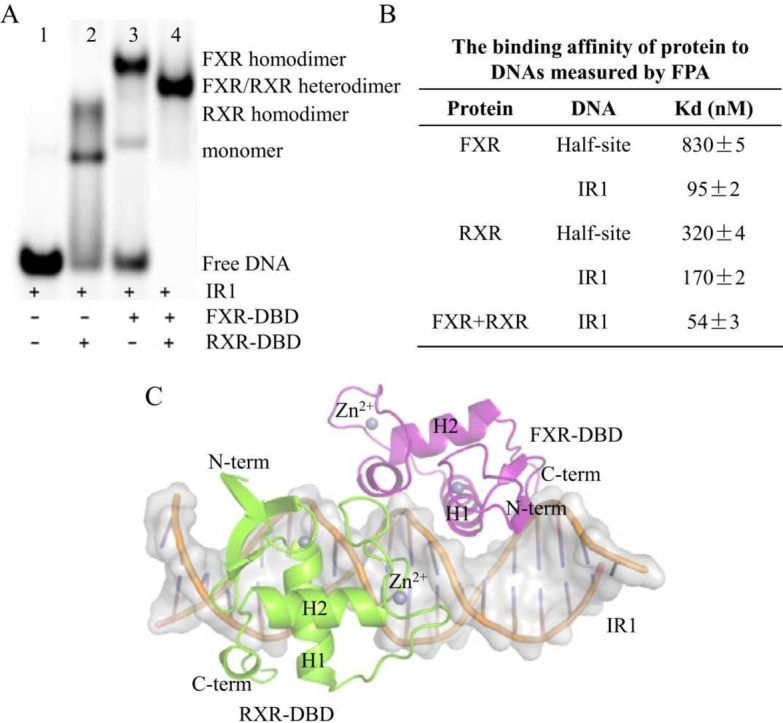
We first performed EMSAs to further characterize the binding of FXR and RXR to the IR1 site (Fig. 1C). In the absence of RXR or FXR, the other protein formed two mobility complex bands when incubated with the IR1 site (lane 2 and lane 3 in Fig. 1C). The migrated lower band indicated the monomer band. The upper, slow-migrating band represented the simultaneous binding of two identical proteins to DNA, which is called the homologous complex. These results indicated that both FXR and RXR could bind as a homodimer or monomer to the IR1 sequences. As shown in lane 4 of Fig. 1C, in the presence of RXR, a mobility complex band was formed upon incubation with FXR and the IR1 element, which migrated in a position between the FXR homodimer band and the RXR homodimer band. This band represents one FXR molecule and one RXR molecule simultaneously bound to IR1 DNA, i.e., an FXR/RXR heterodimer. These results showed that FXR could form a heterodimer with RXR when incubated with the IR1 site at the same time.
We then employed a fluorescence polarization assay (FPA) to measure the binding affinities of protein to DNAs (Fig. 1D and Fig. S1). The binding affinity (Kd) of FXR binding to IR1 is 95 nM, while the Kd value of binding to the half-site of IR1 is 830 nM, indicating that the presence of two half-sites enhanced the binding ability of FXR to DNA. In the presence of RXR, the Kd value of FXR binding to IR1 is approximately 54 nM. The same trend was observed for RXR binding to DNA. These results indicated that heterodimerization with RXR enhanced the binding affinity between FXR and the IR1 site.
2.3. Overall structure of the FXR-DBD/RXR-DBD/IR1 complex
To better characterize the mechanism by which the FXR/RXR heterodimer recognizes the IR1 site, we determined the crystal structure of the FXR-DBD/RXR-DBD/IR1 complex (Fig. 2A). After multiple rounds of screening and optimization, only one crystal was obtained. The complex structure was finally solved at 3.3 Å and crystallized in the P 212121 space group with two complexes per asymmetric unit. Each complex contains one FXR molecule (magentas), one RXR molecule (chartreuse) and a double-stranded IR1 molecule (orange). FXR and RXR occupied the two half-sites of the IR1 site, and two molecules were arranged in a head-to-head orientation. Table S1 summarizes the crystallographic data and refinement statistics for the FXR-DBD/RXR-DBD/IR1 complex.
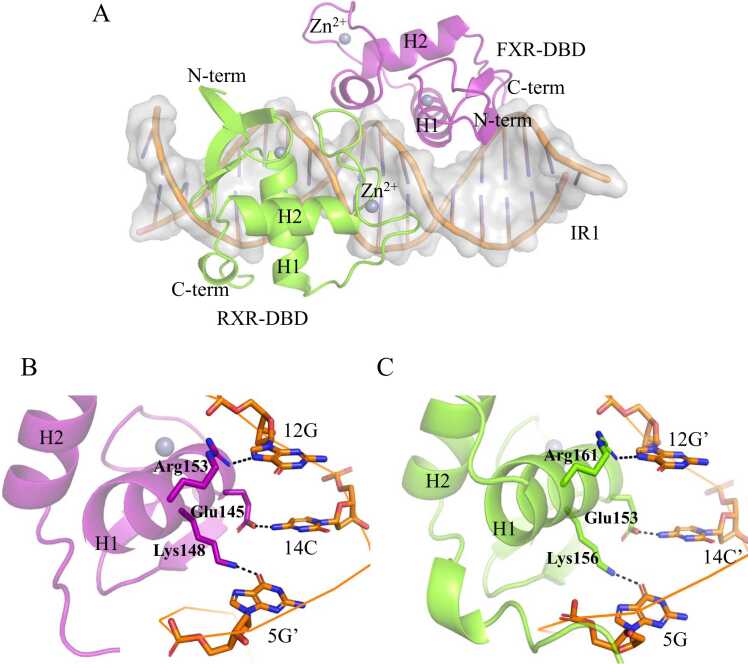
Fig. 2.
Structural analysis of the FXR-DBD/RXR-DBD/IR1 complex. A: Overall structure of the FXR-DBD/RXR-DBD/IR1 complex. The FXR-DBD and RXR-DBD molecules are colored magenta and chartreuse, respectively. The DNA is colored orange. B: DNA recognition by the FXR-DBD helix H1. C: DNA recognition by the RXR-DBD helix H1. Black dashed lines are hydrogen bonds.
The global structure of FXR-DBD is similar to those structures of nuclear receptors previously deposited, consisting of an N-terminal b-hairpin, two highly conserved α-helical zinc modules (H1 and H2) and a C-terminal extension (CTE) (Fig. 2A). FXR-CTE includes a T-box and an A-box (Fig. S2A). The RXR-CTE is constituted by nine residues at the C-terminus beyond the GM amino acids, which is named the T-box [37], [38] (Fig. S2B). The structure of RXR-DBD shows an analogous tertiary structure with the structure of FXR-DBD, and the root mean square deviation (RMSD) for the Cα atom superimposition was approximately 0.44 Å. The sequence alignment shows different residues between FXR and RXR (Fig. S3A). Then we calculated the Fo-Fc omit map of these residues to help discriminate the two proteins (Fig. S3B-S3C). Proteins in the one complex has a better-fitted electron density. The electron density of the proteins in the other complexes do not fitting well, while the electron density of DNA matches well. The b factors of chain A (RXR) and chain B (FXR) in the better-fitted complex are 82.21 and 83.61, respectively. The b factors of the proteins in the other complex are 154.09 (RXR) and 135.99 (FXR) (Fig. S3D). Therefore, we chose the better-fitted complex to analyze the DNA-protein and protein-protein interactions.
2.4. Protein–DNA Interactions
In the FXR-DBD/RXR-DBD/IR1 complex, a total of 3464.2 Å2 of solvent-accessible area is buried in DNA binding, 1722.5 Å2 of which is derived from the binding of the RXR subunit, and 1741.7 Å2 from the binding of the FXR subunit. The N-terminal helix H1 of FXR-DBD and RXR-DBD docked into the two major grooves of the IR1 site. Three conserved residues, Glu145, Lys148 and Arg153, in helix H1 of FXR-DBD formed base-specific hydrogen bonds with Cyd14, Gua5′ and Gua12, respectively (Fig. 2B). The corresponding amino acids in RXR-DBD, Glu153, Lys156 and Arg161, also formed base-specific interactions with the other half-site of IR1 (Fig. 2C). In addition to these specific base contacts, the two proteins also interact with the phosphate backbone of DNA through numerous hydrogen bonds and van der Waals interactions.
2.5. The dimer interfaces
A dimer interface between FXR-DBD and RXR-DBD was identified in the FXR-DBD/RXR-DBD/IR1 complex (Fig. 3A). From the analysis of PDBePISA [39], the dimer interface buried a solvent-accessible area of approximately 535 Å2. Two zinc modules in two proteins formed a dimer interface. Residues from the first zinc module of the RXR-DBD are positioned upstream of the interface and interact with residues from the second zinc module of the FXR-DBD. Asp140 from the RXR-DBD made a hydrogen bond contact with Tyr174 from the FXR-DBD (Fig. 3B). The other interface is formed between residues Lys181, Arg182 and Arg186 in the second zinc module of the RXR-DBD and residues Asp132 and Arg133 in the first zinc module of the FXR-DBD. A salt bridge contact is formed between Arg186 of the RXR-DBD and Asp132 of the FXR-DBD (Fig. 3B). Several van der Waals interactions also formed to stabilize the heterodimer interface (Fig. S4).
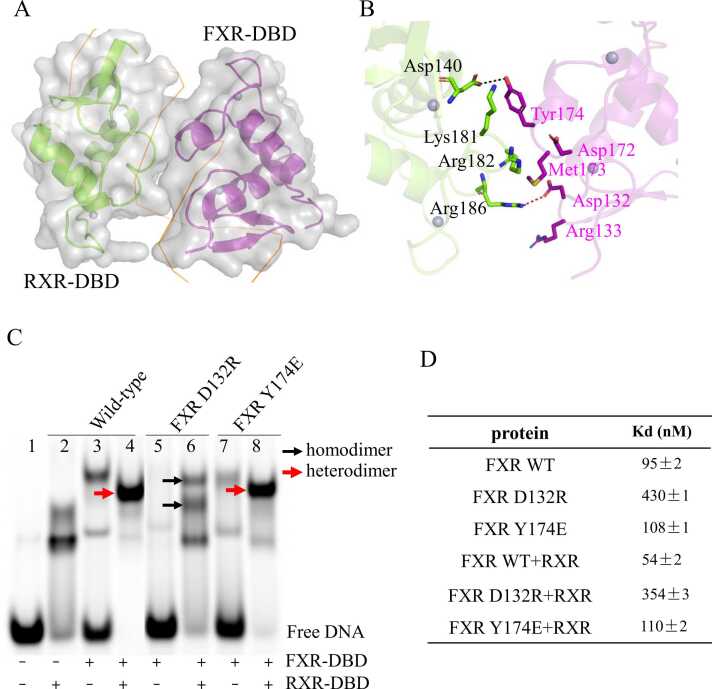
Fig. 3.
Protein—protein interactions in the FXR-DBD/RXR-DBD/IR1 complex. A: Surface representation of the dimerization interface in two molecules. B: Detailed stereo diagram of the residues (shown as sticks) involved in the heterodimer interface. Black dashed lines represent hydrogen bonds, red dashed lines are salt bridges. C: Binding properties of FXR variants to the IR1 site using EMSAs. D: DNA binding affinities of FXR variants to IR1 in the absence of RXR or in the presence of RXR were measured by FPA.
To identify the importance of these residues involved in the heterodimer interface, we mutated residues Asp132 and Tyr174 in the FXR-DBD. We first performed EMSA to assess the DNA binding ability of these structure-guided mutants to IR1. As shown in Fig. 3C, compared to FXR WT (lane 3), the homodimer band formed by FXR mutants was decreased when they were incubated with IR1 alone (lane 5 and 7). The heterodimer band formed by FXR D132R was disappeared when incubated with RXR/IR1. The two bands in lane 6 indicated by the black arrow represent two homodimers: the upper band is the FXR D132R homodimer, and the lower band is the RXR homodimer. The other mutant Y174E still forms heterodimers with RXR bound to IR1. However, compared to the FXR WT (lane 4), the unbound free DNA in lane 8 in the EMSA gel also supports that Y174E decreased the interaction between RXR/FXR and IR1. We also measured the binding affinities of FXR variants to IR1 in the absence or presence of RXR using FPA (Fig. 3D and Fig. S5). FXR mutants exhibited a reduced binding affinity for the IR1 site in the absence of RXR, as well as in the presence of RXR, especially FXR D132R. Then, we used circular dichroism (CD) to test whether these mutations affect the secondary structure of FXR-DBD (Fig. S6A). The overall CD spectra of FXR WT and two FXR mutants indicate that the Y174E mutant does not cause a significant change in the secondary structure of FXR-DBD, while the FXR D132R mutant causes a significant change in the secondary structure of FXR-DBD.
To further identify the heterodimer interface, we constructed two RXR mutants: D140R and R186E. First, we used CD to test whether these mutations affect the secondary structure of RXR-DBD (Fig. S6B). The overall CD spectra of wild-type and two RXR-DBD mutants suggest that those mutations do not affect the secondary structure of RXR-DBD. Then, we performed EMSA to assess the DNA binding ability of these RXR mutants to IR1. Two RXR mutants exhibit decreased heterodimeric binding to IR1 with FXR. The two bands in lane 6 and lane 8 indicated by the black arrow represent two RXR homodimers (Fig. S7). Taken together, these structure-guided mutagenesis assays supported the interfaces defined by the FXR-DBD/RXR-DBD/IR1 complex structure.
2.6. The binding affinity of FXR to other FXRE sites
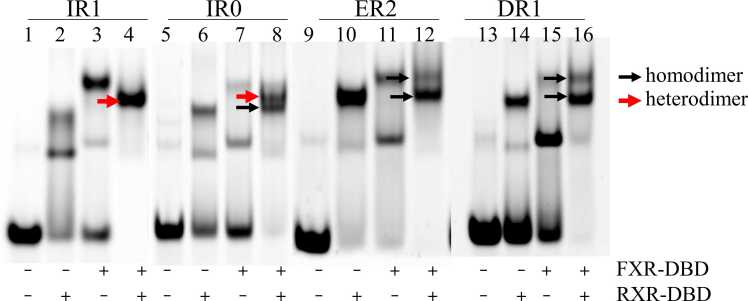
Recent studies have shown that FXR also binds to other FXRE sites, including IR0, DR1 and ER2 [10], [11]. To understand the binding affinity of FXR to these FXRE sites, we first carried out EMSA to analyze the heterodimeric abilities of FXR-DBD/RXR-DBD bound to these elements. As shown in Fig. 4, FXR also formed a heterodimer with RXR when recognizing the IR0 site, while FXR did not form a heterodimeric band with RXR when recognizing the DR1 or ER2 site in the EMSA gels. The upper band represents the FXR homodimer, and the lower band indicates the RXR homodimer. These results suggest that in addition to IR1, FXR-DBD and RXR-DBD also bind heterodimers to the IR0 site.
Fig. 4.
The binding characteristic of FXR to other FXRE sites. The binding properties of FXR to other FXRE sites were measured by EMSA. The first panel is same as Fig. 1C.
Then, we measured the binding abilities of FXR-DBD/RXR-DBD to IR0, DR1 and ER2 by FPA (Fig. S8A). In the absence of RXR, the Kd value of FXR to IR0 is 170 nM, and in the presence of RXR, the Kd value shows a 2-fold (80 nM) increase. The binding affinity of FXR to ER2 is similar in the absence (140 nM) or presence (136 nM) of RXR. The binding affinity of FXR/RXR to the DR1 element is similar to that of FXR to DR1. Then we investigated the occurrence of the IR0 and DR1 motifs in the FXR ChIP-seq dataset (GSE133700). The IR0 and DR1 sites accounted for 9.7% and 8.6% of the total FXR binding sites, respectively (Fig. S8B). These results suggest that FXR binds to IR0, DR1 and ER2 sites and the FXR/RXR heterodimer prefers to IR1 or IR0 sites. However, the proteins used in these experiments are DBDs, so we are unbale to exclude the full-length proteins of FXR and RXR binding as heterodimers to DR1 and ER2 sites in vivo.
2.7. The binding characteristic of other nuclear receptors to the IR1 site
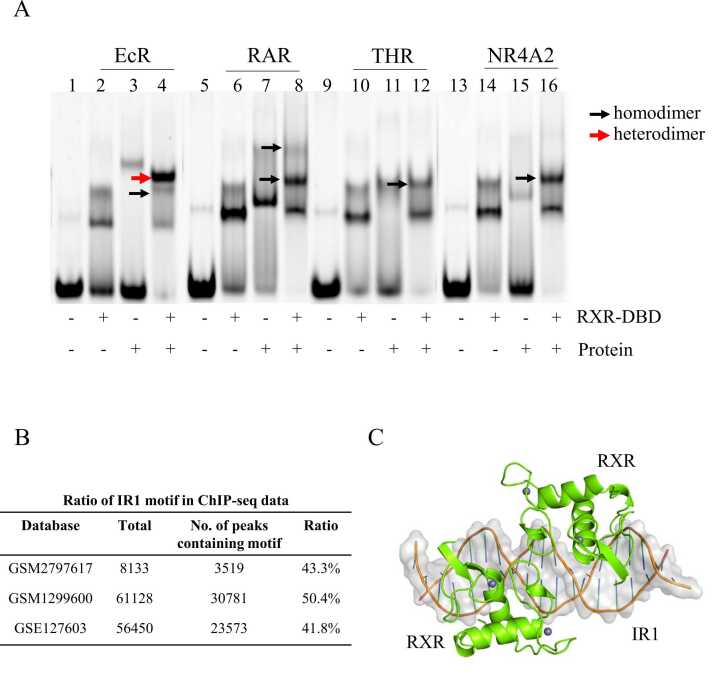
To test whether other nuclear receptors could form a heterodimer with RXR binding to the IR1 site, we studied the IR1 binding features of four other nuclear receptor proteins. Three other type II nuclear receptors, RAR, THR, and NR4A2, did not seem to form a heterodimeric band with RXR bound to IR1 (Fig. 5A), which suggested that IR1 might be a unique binding site for FXR. The Drosophila ecdysteroid hormone receptor (EcR) binds to the IR1 element as a heterodimer with RXR (Fig. 5A). We first analyzed the sequences of FXR and EcR, which showed more than 80% similarity (Fig. S9A). The structural comparison with the previously published EcR-DBD/RXR-DBD/IR1 structure (PDB: 1R0N) [40] with an RMSD of 0.38 showed that EcR formed a similar dimer interface with RXR through two zinc modules on the IR1 element. To further compare the dimer interfaces between the two complexes, we analyzed the details of the interactions in the EcR-DBD/RXR-DBD/IR1 structure (Fig. S9B-S9C). The hydrogen bonds and salt bridge contacts are also existed in the EcR-DBD/RXR-DBD/IR1 structure. Several van der Waals interactions existed in our structure are not found in the interface of the EcR-DBD/RXR-DBD/IR1 structure (Fig. S9D-S9E).
Fig. 5.
The binding characteristic of other nuclear receptors to the IR1 site. A: Binding properties of four nuclear receptors to the IR1 site using EMSAs. B: Occurrence of the IR1 binding motif in the RXR-binding sites. C: Structural model of the RXR/RXR homodimer on the IR1 site.
As RXR also shows a strong tendency to form homodimers or heterodimers on IR1, we investigated the occurrence of the IR1 motif in several RXR ChIP-seq datasets (GSM2797617, GSM1299600 and GSE127603) [41]. The IR1 site accounted for more than 40% of the total RXR binding sites in these three datasets (Fig. 5B). Superimposition RXR on the structure of FXR/RXR/IR1 shows the potential for RXR binding to IR1 (Fig. 5C). These results suggest that the different binding characteristics of other nuclear receptors to the IR1 site and IR1 are also important binding sites for RXR.
2.8. Comparison with other RXR heterodimer complexes
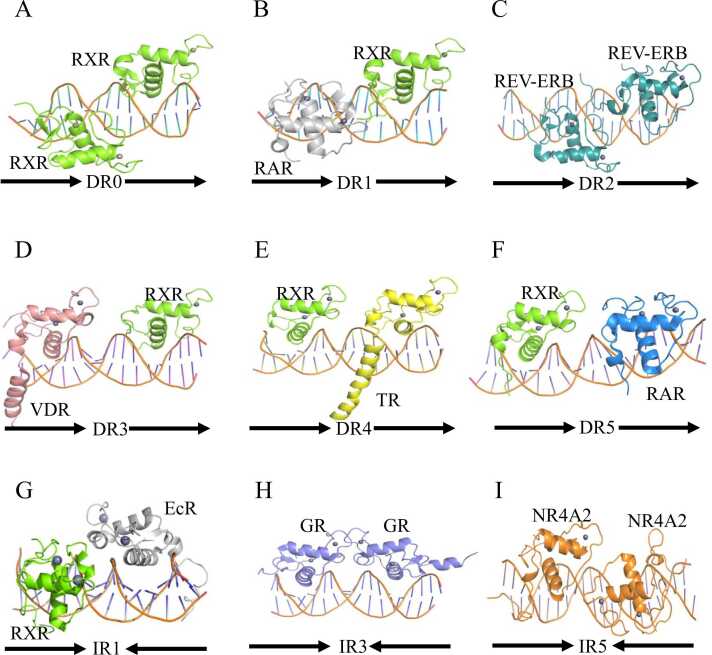
The nuclear receptors DBD recognized the consensus half-sites in an identical manner, forming unique protein-protein dimerization contacts by binding the bipartite response element. The protein-DNA interactions were reinforced by these dimerization interactions. We analyzed the recently reported structures of RXR dimers (Fig. 6). No dimer interactions were observed in the structures of RXR/RXR/DR0 [22] and VDR/RXR/DR3 [24] (Figs. 6A and 6D). The CTE of RXR formed a dimer interaction with the second zinc modules of the RAR molecule in the structure of the RAR/RXR/DR1 [23] complex (Fig. 6B), similar to the REV-ERB/REV-ERB/DR2 complex [38] (Fig. 6C). The second zinc modules of the RXR molecule form a dimer interface with the other molecules in the THR/RXR/DR4 [25] complex and RAR/RXR/DR5 [22] complex (Figs. 6E and 6F). In our structure of the FXR/RXR/IR1 complex and EcR/RXR/IR1 (Fig. 6G), the two zinc modules were involved in the heterodimer interface, which buried a larger surface than previously studied RXR-DBD homo and heterodimers. Two GR molecules formed a dimer interface on the IR3 element through the two second zinc modules [42] (Fig. 6H). Two NR4A2 molecules formed a dimer interface through the reverse arrayed loop on the IR5 element [43] (Fig. 6I). These analyses suggested that the adaptable surfaces of nuclear receptor DBDs are susceptible to structural rearrangements that help to accommodate their association with dimeric partners and differently oriented and spaced response elements [18], [44].
Fig. 6.
Comparison with other RXR heterodimer complexes. A: RXR/RXR/DR0 (PDB code: 6XWH). B: RAR/RXR/DR1 (PDB code: 1DSZ). C: REV-ERB/REV-ERB/DR2 (PDB code: 1GA5). D: VDR/RXR/DR3 (PDB code: 1YNW). E: THR/RXR/DR4 (PDB code: 2NLL). F: RAR/RXR/DR5 (PDB code: 6XWG). G: EcR/RXR/IR1 (PDB code: 1R0N). H: GR/GR/IR3 (PDB code: 3G6P). I: NR4A2/NR4A2/IR5 (PDB code: 6L6L).
3. Discussion
FXR is highly expressed in the liver and intestine and plays important roles in bile acid homeostasis and the development of hepatitis, hepatic fibrosis, and even hepatocellular carcinoma [4], [5]. As a transcription factor, FXR binds to various FXREs to regulate the expression of multiple genes involved in bile acids, lipid, and glucose metabolism. The preferred DNA-binding site for FXR within its target promoters is typically the IR1 motif. ChIP-Seq analysis showed that IR1 accounts for more than seventy percent of these FXRE sites (Fig. 1). Our results also suggest that FXR and RXR bind to the IR1 sites with higher affinity when they are part of a heterodimeric complex than when they are bound individually (Fig. 1). Therefore, elucidating the structural mechanism of FXR/RXR/IR1 heterodimerization is important for understanding the functional regulation of FXR in various biological processes. Here, we present the first crystal structure of the FXR-DBD/RXR-DBD/IR1 complex, and we comprehensively analyze how FXR recognizes DNA and the dimer interface between FXR-DBD and RXR-DBD (Fig. 2, Fig. 3). Given the importance of FXR in the metabolism of bile acids and many other materials in vivo, it is important to understand these interactions and to know how the structure of FXR affects its function.
Our EMSAs suggested that in addition to IR1, FXR-DBD also forms a heterodimer with RXR-DBD on the IR0 response element (Fig. 4). The IR0 site was found to be existed in the FXR binding sites (Fig. S8). However, we were unable to exclude the possibility that FXR and RXR form heterodimers in vivo to recognize these DNAs. Although the structure of full-length FXR receptors has not been determined, the previously published crystal structures of the FXR-LBD/RXR-LBD heterodimer reveal that both heterodimerization and ligand binding induce conformational changes in FXR-LBD, which affect the stability of FXR and its binding to coactivators [30], [45]. In addition, the FXR-LBD, together with the DBDs, might help to stabilize the complex on the DNA and fine-tune gene regulation [46].
Type II nuclear receptors form heterodimers with RXR to regulate gene transcription. Recent studies have shown that the relative orientations and spacing between the two half-sites play an important role in the DNA binding specificity of nuclear receptors [43]. Nuclear receptor/RXR heterodimers usually have their own preferences when recognizing DNA sites. For example, RAR/RXR dimers prefer binding to DR elements, whereas THR/RXR dimers prefer DR4 elements [22], [25]. FXR/RXR dimers prefer IR1 elements. We compared the dimer interfaces of the published crystal structures of nuclear receptor dimers with our structure (Fig. 6). These studies support the opinion that extensible and adjustable dimerization surfaces in DBDs are responsible for their cooperation on various response elements [38], [47]. In addition, EcR also forms heterodimers with RXR and binds to IR1 sites [40], [47]. The EMSA results suggest that FXR and EcR recognize the IR1 motif with RXR in a similar manner (Fig. 5). Sequence alignment and structural comparison showed that EcR formed a similar dimer interface with RXR through two zinc modules on the IR1 element (Fig. S9).
In summary, we determined a crystal structure of FXR-DBD/RXR-DBD/IR1 complexes and provided the molecular basis for DNA recognition by the FXR/RXR heterodimer. Our structural and biochemical studies provide evidence that FXR and RXR bind the IR1 site as a heterodimer and that heterodimerization enhances the binding affinity for DNA. Our structural, biochemical, and bioinformatics analyses not only provide the molecular basis of DNA recognition by the FXR/RXR heterodimer but also provide a better understanding of the dimerization specificity of nuclear receptors.
4. Materials and methods
4.1. Plasmid construction
The gene fragments encoding human FXR-DBD (residues 117–217) and human RXR-DBD (residues 130–212) were cloned into the expression vector pGEX6P1 using homologous recombination. Site-directed mutagenesis of FXR-DBD and RXR-DBD was performed according to the manufacturer’s instructions for the ClonExpress II One Step Cloning Kit (Vazyme) using the recombinant plasmids above as the template. All these plasmids were validated by DNA sequencing at Tsingke Changsha, China.
4.2. Protein expression and purification
The recombinant proteins were purified as previously described [40]. Briefly, the vectors were transformed into Escherichia coli Rosetta BL21 (DE3) cells. After induced with 0.5 mM isopropyl-D-1-thiogalactopyranoside for 12 h at 25 ℃, the cells were harvested by centrifugation. After homogenization (PhD Technology LLC), the supernatant was loaded onto a GST affinity column. After PreScission protease digestion, the cleaved proteins were further purified by cation exchange column and size exclusion chromatography Superdex75. The proteins were identified by SDS—PAGE and concentrated to 15 mg/ml in storage buffer containing 20 mM HEPES pH 7.5, 200 mM NaCl and 1 mM TCEP. The mutants were purified as wild-type proteins.
4.3. Crystallization
The IR1 sequences for crystallization were 5-CCGAGGTCAATGACCTCG-3 and 5-CCGAGGTCATTGACCTCG-3, which were synthesized by GenScript (Suzhou, China) at an annealed concentration of 1.35 mM. The FXR-DBD/RXR-DBD/IR1 complex was prepared by mixing purified FXR-DBD, RXR-DBD and IR1 at a 1:1:1.2 molar ratio. Crystals of the FXR-DBD/RXR-DBD/IR1 complex were grown under the conditions of 100 mM BTP pH 6.7, 10 mM MgCl2, 50 mM MgSO4, 50 mM Li2SO4, 5 mM DTT, and 16% PEG3350. Crystals were harvested with cryoprotectant containing the corresponding crystallization reservoir plus 10% glycerol, flash frozen and stored in liquid nitrogen for further data collection.
4.4. Data collection and structure determination
The best data were collected in the Beamline BL17U1 of Shanghai Synchrotron Radiation Facility (SSRF) and reduced by AutoPX [48]. The structure of the FXR-DBD/RXR-DBD/IR1 complex was solved by molecular replacement (MR) using phaser from the PHENIX package using the ecdysone receptor (EcR)/ultraspiracle (USP)/IR1 (PDB ID: 1R0N) as a search model [49]. The structures were then refined with XYZ positional refinement, NCS refinement and TLS refinement in PHENIX [50]. Zinc ions were added to the model. The statistics of the final refined structure are presented in Table S1. Figures were generated using PyMol [51].
4.5. Electrophoretic mobility shift assay (EMSA)
EMSAs were performed as previously described [52]. The binding reactions were performed in a total mixture volume of 6–8 μL and incubated at room temperature for 10–20 min. The EMSA buffer used here consisted of 20 mM HEPES pH 7.0, 200 mM NaCl, and 10 mM MgCl2. The proteins and DNAs were all diluted to an initial concentration of 45 μM for use. The mixtures were then loaded onto 8% (w/v) native polyacrylamide gels to separate free DNA and protein/DNA complexes.
4.6. Fluorescence polarization assay (FPA)
FPA was performed as previously described [53]. 5′− 5FAM- CCGAGGTCAATGACCTCG and the unlabeled reverse complementary DNA strand were purchased from Tsingke (Changsha, China) and annealed to duplex DNA. Proteins at diluted concentrations (ranging from 10 μM to 1 nM) in 0.5 × PBS were incubated with 5FAM-labeled DNAs (20 nM) in 96-well plates at room temperature for 10–15 min. The fluorescence was measured on an Envision multilabel reader (Perkin Elmer) with ex = 480 nm and em= 535 nm. All assays were performed in triplicate, and Kd values were determined from a nonlinear dose—response curve in GraphPad Prism 7.0.
4.7. Circular dichroism (CD)
CD was performed as previously described[54]. Briefly, CD spectra in the far UV region (190–260 nm) were performed on a Jasco J-815CD spectrometers using a 0.1 cm cell at room temperature. The proteins (wild-type and six mutants) were prepared in 20 mM sodium phosphate (pH7.5) at the concentration of 0.1 mg/ml. Each spectrum was obtained by averaging 3 scans at 1-nm increments.
4.8. Bioinformatics analysis
The bed files of the ChIP-seq data for FXR and RXR were downloaded from the GEO datasets. The peaks were annotated, and sequences were fetched using the R package ChIPpeakAnno [55]. The matrix of the IR1 sequence was obtained from the footprintDB database [56]. The matrix was searched against the ChIP-seq peak sequences using the R package TFBSTools [57].
Data availability
The FXR-DBD/RXR-DBD/IR1 structure has been deposited with the Protein Data Bank under accession code 8HBM.
Funding
This work was supported by the National Natural Science Foundation of China (grants 81974074 and 82172654 to Y.C.; grant 31900880 to H.W.), Science and Technology Planning Project of Hunan Province (grants 2018TP1017 and 2021RC4012 to Y.C.), and the National Science Foundation of Hunan Province (grant 2022JJ40763 to L. J.).
CRediT authorship contribution statement
Conceptualization, L.J. and X.L.; methodology, L.J., and X.L.; investigation and. data analysis, X.L, S.D., H.W., M.G. Z.C., and D.X.; original draft and visualization, L.J. and X.L.; manuscript preparation, L.J. and X.L. and Y.C.; funding acquisition, Y.C., L.J. and H.W.
Acknowledgments
We thank the staff from the Shanghai Synchrotron Radiation facility (SSRF) beamline BL17U/BL19U/BL02U/BL10U/BL17B for assistance during diffraction data collection.
Declaration of interest
The authors declare no competing interests.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2023.05.026.
Contributor Information
Desheng Xiao, Email: xdsh96@csu.edu.cn.
Yongheng Chen, Email: yonghenc@163.com.
Appendix A. Supplementary material
Supplementary material
.
References
- Goodwin B., Kliewer S.A. Nuclear receptors. I. Nuclear receptors and bile acid homeostasis. Am J Physiol Gastrointest Liver Physiol. 2002;282(6):G926–G931. doi: 10.1152/ajpgi.00044.2002. [DOI] [PubMed] [Google Scholar]
- 2.Chiang J.Y.L., Ferrell J.M. Discovery of farnesoid X receptor and its role in bile acid metabolism. Mol Cell Endocrinol. 2022;548 doi: 10.1016/j.mce.2022.111618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang L., et al. Farnesoid X receptor (FXR): structures and ligands. Comput Struct Biotechnol J. 2021;19:2148–2159. doi: 10.1016/j.csbj.2021.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y.D., et al. FXR: a metabolic regulator and cell protector. Cell Res. 2008;18(11):1087–1095. doi: 10.1038/cr.2008.289. [DOI] [PubMed] [Google Scholar]
- 5.Huang X.F., Zhao W.Y., Huang W.D. FXR and liver carcinogenesis. Acta Pharm Sin. 2015;36(1):37–43. doi: 10.1038/aps.2014.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gomez-Ospina N., et al. Mutations in the nuclear bile acid receptor FXR cause progressive familial intrahepatic cholestasis. Nat Commun. 2016;7:10713. doi: 10.1038/ncomms10713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shah R.A., Kowdley K.V. Obeticholic acid for the treatment of nonalcoholic steatohepatitis. Expert Rev Gastroenterol Hepatol. 2020;14(5):311–321. doi: 10.1080/17474124.2020.1748498. [DOI] [PubMed] [Google Scholar]
- 8.Markham A., Keam S.J. Obeticholic acid: first global approval. Drugs. 2016;76(12):1221–1226. doi: 10.1007/s40265-016-0616-x. [DOI] [PubMed] [Google Scholar]
- 9.Brevini T., et al. FXR inhibition may protect from SARS-CoV-2 infection by reducing ACE2. Nature. 2022 doi: 10.1038/s41586-022-05594-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramos Pittol J.M., et al. FXR isoforms control different metabolic functions in liver cells via binding to specific DNA motifs. Gastroenterology. 2020;159(5):1853–1865. doi: 10.1053/j.gastro.2020.07.036. e1810. [DOI] [PubMed] [Google Scholar]
- 11.Thomas A.M., et al. Genome-wide tissue-specific farnesoid X receptor binding in mouse liver and intestine. Hepatology. 2010;51(4):1410–1419. doi: 10.1002/hep.23450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Claudel T., et al. Bile acid-activated nuclear receptor FXR suppresses apolipoprotein A-I transcription via a negative FXR response element. J Clin Invest. 2002;109(7):961–971. doi: 10.1172/JCI14505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Penvose A., et al. Comprehensive study of nuclear receptor DNA binding provides a revised framework for understanding receptor specificity. Nat Commun. 2019;10(1):2514. doi: 10.1038/s41467-019-10264-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evans R.M., Mangelsdorf D.J. Nuclear receptors, RXR, and the big bang. Cell. 2014;157(1):255–266. doi: 10.1016/j.cell.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khorasanizadeh S., Rastinejad F. Visualizing the architectures and interactions of nuclear receptors. Endocrinology. 2016;157(11):4212–4221. doi: 10.1210/en.2016-1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang P., Chandra V., Rastinejad F. Retinoic acid actions through mammalian nuclear receptors. Chem Rev. 2014;114(1):233–254. doi: 10.1021/cr400161b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rastinejad F. Retinoic acid receptor structures: the journey from single domains to full-length complex. J Mol Endocrinol. 2022;69(4):T25–t36. doi: 10.1530/JME-22-0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khorasanizadeh S., Rastinejad F. Nuclear-receptor interactions on DNA-response elements. Trends Biochem Sci. 2001;26(6):384–390. doi: 10.1016/s0968-0004(01)01800-x. [DOI] [PubMed] [Google Scholar]
- 19.Zhao Q., et al. Structural basis of RXR-DNA interactions. J Mol Biol. 2000;296(2):509–520. doi: 10.1006/jmbi.1999.3457. [DOI] [PubMed] [Google Scholar]
- 20.Rastinejad F. Retinoid X receptor and its partners in the nuclear receptor family. Curr Opin Struct Biol. 2001;11(1):33–38. doi: 10.1016/s0959-440x(00)00165-2. [DOI] [PubMed] [Google Scholar]
- 21.Rastinejad F., et al. Understanding nuclear receptor form and function using structural biology. J Mol Endocrinol. 2013;51(3):T1–t21. doi: 10.1530/JME-13-0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Osz J., et al. Structural basis for DNA recognition and allosteric control of the retinoic acid receptors RAR-RXR. Nucleic Acids Res. 2020;48(17):9969–9985. doi: 10.1093/nar/gkaa697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rastinejad F., et al. Structure of the RXR-RAR DNA-binding complex on the retinoic acid response element DR1. Embo J. 2000;19(5):1045–1054. doi: 10.1093/emboj/19.5.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shaffer P.L., Gewirth D.T. Structural analysis of RXR-VDR interactions on DR3 DNA. J Steroid Biochem Mol Biol. 2004;89–90(1–5):215–219. doi: 10.1016/j.jsbmb.2004.03.084. [DOI] [PubMed] [Google Scholar]
- 25.Rastinejad F., et al. Structural determinants of nuclear receptor assembly on DNA direct repeats. Nature. 1995;375(6528):203–211. doi: 10.1038/375203a0. [DOI] [PubMed] [Google Scholar]
- 26.Chandra V., et al. Structure of the intact PPAR-gamma-RXR- nuclear receptor complex on DNA. Nature. 2008;456(7220):350–356. doi: 10.1038/nature07413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chandra V., et al. The quaternary architecture of RARβ-RXRα heterodimer facilitates domain-domain signal transmission. Nat Commun. 2017;8(1):868. doi: 10.1038/s41467-017-00981-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chandra V., et al. Multidomain integration in the structure of the HNF-4α nuclear receptor complex. Nature. 2013;495(7441):394–398. doi: 10.1038/nature11966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lou X., et al. Structure of the retinoid X receptor α-liver X receptor β (RXRα-LXRβ) heterodimer on DNA. Nat Struct Mol Biol. 2014;21(3):277–281. doi: 10.1038/nsmb.2778. [DOI] [PubMed] [Google Scholar]
- 30.Rastinejad F., Ollendorff V., Polikarpov I. Nuclear receptor full-length architectures: confronting myth and illusion with high resolution. Trends Biochem Sci. 2015;40(1):16–24. doi: 10.1016/j.tibs.2014.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Costantino G., et al. Molecular dynamics simulation of the ligand binding domain of farnesoid X receptor. Insights into helix-12 stability and coactivator peptide stabilization in response to agonist binding. J Med Chem. 2005;48(9):3251–3259. doi: 10.1021/jm049182o. [DOI] [PubMed] [Google Scholar]
- 32.Zheng W., et al. Structural insights into the heterodimeric complex of the nuclear receptors FXR and RXR. J Biol Chem. 2018;293(32):12535–12541. doi: 10.1074/jbc.RA118.004188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robert X., Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42(Web Server issue):W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ijssennagger N., et al. Gene expression profiling in human precision cut liver slices in response to the FXR agonist obeticholic acid. J Hepatol. 2016;64(5):1158–1166. doi: 10.1016/j.jhep.2016.01.016. [DOI] [PubMed] [Google Scholar]
- 35.Clifford B.L., et al. FXR activation protects against NAFLD via bile-acid-dependent reductions in lipid absorption. Cell Metab. 2021;33(8):1671–1684. doi: 10.1016/j.cmet.2021.06.012. e1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ding L., et al. Bile acid nuclear receptor FXR and digestive system diseases. Acta Pharm Sin B. 2015;5(2):135–144. doi: 10.1016/j.apsb.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao Q., et al. Structural elements of an orphan nuclear receptor-DNA complex. Mol Cell. 1998;1(6):849–861. doi: 10.1016/s1097-2765(00)80084-2. [DOI] [PubMed] [Google Scholar]
- 38.Sierk M.L., Zhao Q., Rastinejad F. DNA deformability as a recognition feature in the reverb response element. Biochemistry. 2001;40(43):12833–12843. doi: 10.1021/bi011086r. [DOI] [PubMed] [Google Scholar]
- 39.Krissinel E., Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372(3):774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 40.Devarakonda S., et al. Structure of the heterodimeric ecdysone receptor DNA-binding complex. Embo J. 2003;22(21):5827–5840. doi: 10.1093/emboj/cdg569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He Y., et al. The role of retinoic acid in hepatic lipid homeostasis defined by genomic binding and transcriptome profiling. BMC Genom. 2013;14:575. doi: 10.1186/1471-2164-14-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luisi B.F., et al. Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA. Nature. 1991;352(6335):497–505. doi: 10.1038/352497a0. [DOI] [PubMed] [Google Scholar]
- 43.Jiang L., et al. Structural basis of binding of homodimers of the nuclear receptor NR4A2 to selective Nur-responsive DNA elements. J Biol Chem. 2019;294(51):19795–19803. doi: 10.1074/jbc.RA119.010730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Helsen C., et al. Structural basis for nuclear hormone receptor DNA binding. Mol Cell Endocrinol. 2012;348(2):411–417. doi: 10.1016/j.mce.2011.07.025. [DOI] [PubMed] [Google Scholar]
- 45.Wang N., et al. Ligand binding and heterodimerization with retinoid X receptor α (RXRα) induce farnesoid X receptor (FXR) conformational changes affecting coactivator binding. J Biol Chem. 2018;293(47):18180–18191. doi: 10.1074/jbc.RA118.004652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maletta M., et al. The palindromic DNA-bound USP/EcR nuclear receptor adopts an asymmetric organization with allosteric domain positioning. Nat Commun. 2014;5:4139. doi: 10.1038/ncomms5139. [DOI] [PubMed] [Google Scholar]
- 47.Orlowski M., et al. Plasticity of the ecdysone receptor DNA binding domain. Mol Endocrinol. 2004;18(9):2166–2184. doi: 10.1210/me.2004-0154. [DOI] [PubMed] [Google Scholar]
- 48.Wang L., et al. AutoPX: a new software package to process X-ray diffraction data from biomacromolecular crystals. Acta Crystallogr D Struct Biol. 2022;78(Pt 7):890–902. doi: 10.1107/S2059798322005745. [DOI] [PubMed] [Google Scholar]
- 49.McCoy A.J., et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonine P.V., et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68(Pt 4):352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bramucci E., et al. PyMod: sequence similarity searches, multiple sequence-structure alignments, and homology modeling within PyMOL. BMC Bioinforma. 2012;13 Suppl 4(Suppl 4):S2. doi: 10.1186/1471-2105-13-S4-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen X., et al. Structural basis for DNA recognition by FOXC2. Nucleic Acids Res. 2019;47(7):3752–3764. doi: 10.1093/nar/gkz077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li J., et al. Mechanism of forkhead transcription factors binding to a novel palindromic DNA site. Nucleic Acids Res. 2021;49(6):3573–3583. doi: 10.1093/nar/gkab086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pace C.N., et al. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4(11):2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu L.J., et al. ChIPpeakAnno: a bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinforma. 2010;11:237. doi: 10.1186/1471-2105-11-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Contreras-Moreira B., Sebastian A. FootprintDB: analysis of plant cis-regulatory elements, transcription factors, and binding interfaces. Methods Mol Biol. 2016;1482:259–277. doi: 10.1007/978-1-4939-6396-6_17. [DOI] [PubMed] [Google Scholar]
- 57.Tan G., Lenhard B. TFBSTools: an R/bioconductor package for transcription factor binding site analysis. Bioinformatics. 2016;32(10):1555–1556. doi: 10.1093/bioinformatics/btw024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
The FXR-DBD/RXR-DBD/IR1 structure has been deposited with the Protein Data Bank under accession code 8HBM.