Abstract
Objective:
Systemic sclerosis is characterized by endothelial dysfunction, autoimmunity abnormalities, and fibrosis of the skin and internal organs. The pathogenetic mechanisms underlying systemic sclerosis vasculopathy are still not clarified. A complex cellular and extracellular network of interactions has been studied, but it is currently unclear what drives the activation of fibroblasts/myofibroblasts and the extracellular matrix deposition.
Methods:
Using RNA sequencing, the aim of the work was to identify potential functional pathways implied in systemic sclerosis pathogenesis and markers of endothelial dysfunction and fibrosis in systemic sclerosis patients. RNA-sequencing analysis was performed on RNA obtained from biopsies from three systemic sclerosis patients and three healthy controls enrolled in our University Hospital. RNA was used to generate sequencing libraries that were sequenced according to proper transcriptomic analyses. Subsequently, we performed gene set enrichment analysis of differentially expressed genes on the entire list of genes that compose the RNA-sequencing expression matrix.
Results:
Gene set enrichment analysis revealed that healthy controls were characterized by gene signatures related to stromal stem cells proliferation, cytokine–cytokine receptor interaction, macrophage-enriched metabolic network, whereas systemic sclerosis tissues were enriched in signatures associated with keratinization, cornification, retinoblastoma 1 and tumor suppressor 53 signaling.
Conclusion:
According to our data, RNA-sequencing and pathway analysis revealed that systemic sclerosis subjects display a discrete pattern of gene expression associated with keratinization, extracellular matrix generation, and negative regulation of angiogenesis and stromal stem cells proliferation. Further analysis on larger numbers of patients is needed; however, our findings provide an interesting framework for the development of biomarkers useful to explore potential future therapeutic approaches.
Keywords: Systemic sclerosis, RNA-sequencing analysis, gene expression, pathogenetic pathways
Introduction
Systemic sclerosis (SSc) is a rare and life-threatening connective tissue disease, characterized by endothelial dysfunction, autoimmunity abnormalities, and aberrant fibrosis of the skin and internal organs.1–3 Disease pathogenesis is characterized by early microvascular changes with endothelial cells (ECs) alteration, followed by dysfunctional mechanisms promoting their transition into myofibroblasts, the cells responsible for fibrosis and collagen deposition in the tissues. 4 It has been observed that microvascular damage might be the first symptom of SSc; based on these factors, myofibroblast generation process may link two pivotal events in SSc: microvascular injury and fibrosis. 5 Production of activating cytokines, disruption of vascular permeability with extravasation of growth factors, and induction of hypoxia possibly contribute to the pool of myofibroblasts through endothelial-to-mesenchymal transition. A complex autoimmune activation, involving innate and adaptive immunity with peculiar autoantibody production, also characterizes the disease.
To resume, a complex network of interactions between ECs, pericytes, myofibroblasts, and the extracellular matrix (ECM), together with growth factors and cytokines, participate in disease diffusion and evolution, but it is currently poorly clear what drives the activation of fibroblasts and the increased ECM deposition responsible for the fibrotic changes well known in SSc vasculopathy.6,7
These modifications drive some of the most noticeable SSc clinical manifestations, such as Raynaud’s phenomenon (RP), digital ulcers (DUs), and pulmonary arterial hypertension (PAH).6,7
Altered gene expression seems to contribute to these aberrant mechanisms. Prior gene expression profiling studies and proteome-side analyses partially elucidated the molecular pathways affected in SSc patients.8,9 However, these studies do not account for cellular heterogeneity and differential cell composition of target tissues, and their results are limited.
Using RNA-sequencing (RNA-seq), our report aims to identify potential functional pathways possibly involved in SSc pathogenesis and markers that could potentially be used to better understand endothelial damage and fibrosis mechanisms in SSc patients.
Methods
Patients and healthy volunteers
Three SSc patients and three age- and sex-matched healthy controls (HCs) were enrolled in our University Hospital between January 2019 and December 2020. Written informed consent was obtained from all of the participants. This study was approved by local ethical committees (protocol no. 275/2016) and performed in accordance with the latest version of the Helsinki declaration.
RNA extraction
Skin biopsies were performed under local anesthetic with a skin biopsy punch (size range, 2–5 mm), in the site-surgery (perioral skin) before autologous fat grafting (lipofilling) and one sample was taken at a time. The biopsy was transferred to a labeled cryovial that was then immediately immersed in liquid nitrogen. All samples were logged in accordance with standard operating procedures and stored in liquid nitrogen until use. Standard precautions to prevent contamination with RNases were employed. The sample was removed from the liquid nitrogen and transferred on dry ice. Samples were immediately placed into a tube containing stainless steel beads and cold lysis buffer (RLT) with beta-mercaptoethanol (RNeasy Plus Mini kit; Qiagen, Hilden, Germany). Samples were homogenized first in the TissueLyser LT (Qiagen) and then in the QIAshredder (Qiagen). Then, RNA was obtained using the RNeasy Plus Mini kit, following manufacturer’s instructions. Eluted RNA was measured using the Nanodrop Microvolume Spectrophotometer and RNA quality was measured using a microfluidic gel electrophoresis chip (Bioanalyzer RNA 6000 Nano Chip, Agilent, UK). RNA integrity numbers were obtained with the software provided (2100 Expert Software) with the Agilent 2100 Bioanalyzer (Agilent, UK). For every sample, RNA integrity number (RIN) was >8. A260/280 and A260/230 ratios were also obtained and were >1.9 for all samples.
Transcriptomic analyses
RNA from each sample was used to generate sequencing libraries that were sequenced using an Illumina Hiseq 2500, giving 30 million paired end reads per sample which were 100 bp in length. FASTQ files were checked for quality using FastQC version 0.11.9 and aligned using the splice aware aligner program STAR to generate alignment files (GENCODE Human Release 37, reference genome sequence GRCh38/hg38). The read counts for each sample file were obtained using the R package Rsubread v2.4.3. Differential gene expression analysis was carried out using edgeR package v3.32.1. Library size normalization by trimmed mean of M (TMM) values was performed using the “calcNormFactors” function embedded in edgeR. Differential gene expression was assessed using “exactTest” function of egdeR, using default parameters. Benjamini–Hochberg correction was applied to estimate the false discovery rate (FDR). Differentially expressed genes (DEGs) were selected using as threshold FDR ⩽ 0.01 and log2FC > 1. Gene ontology enrichment analysis was performed using clusterProfiler v4.2.2.
Gene set enrichment analysis
Gene set enrichment analysis (GSEA) was applied on the entire list of genes that compose the RNA-seq expression matrix. Genes were ranked based on their fold change calculated by pairwise comparison between HC and SSc groups, and analyzed by GSEA in pre-ranked mode. As enrichment statistic, we adopted a more conservative scoring approach by setting enrichment statistic = “classic,” which is the recommended approach for RNA-seq data. The number of permutations has been set to 1000, while Max size and Min size (to exclude larger or smaller sets) have been set to 500 and 15, respectively. To normalize the enrichment scores (ESs) across analyzed gene, we adopted “meandiv” mode. All gene sets of interest were retrieved from the curated signatures collection (c2.all.v7.1 and c5.all.v7.1) of the Molecular Signatures Database.
Digital cytometry
CIBERSORTx is a machine learning method and was used to impute cell fraction without physical cell isolation. 10 Briefly, we built a custom matrix file with a human skin signature using a publicly available single-cell RNA-seq data set (GSE130973). Then, we used this matrix to infer cell fractions from our bulk RNA-seq samples. CIBERSORTx was executed in “absolute mode” to calculate a score that reflects the absolute proportion of each cell type in our bulk RNA-seq mixture using 100 permutations (the quantile normalization was disabled as recommended for RNA-seq data).
Results
RNA-seq analysis was performed on RNA extracted from the biopsies obtained with a skin biopsy punch (size range, 2–5 mm) of three SSc female patients and three female HCs. Biopsies were performed in the site-surgery before autologous fat grafting (lipofilling) procedure for SSc subjects as SSc regenerative treatment for the face (mouth); while HC without comorbidities received various aesthetic face-lifting approaches with concomitant skin biopsies.
The demographic and clinical data of participants are listed in Table 1. The median ages were 51.3 ± 8.1 SD years (range, 42–56) and 50.6 ± 6.6 SD years (range, 43–55) for SSc cases and HC, respectively.
Table 1.
Demographic and clinical data of SSc patients and HC.
| Patients | ||
|---|---|---|
| SSc3 | HC3 | |
| General features | ||
| Mean age, M ± SD years (range) | 51.3 ± 8.1 (42–56) | 50.6 ± 6.6 (43–55) |
| Females | 3 (100%) | 3 (100%) |
| Mean disease duration, M ± SD years (range) | 11.0 ± 5.6 (6–17) | / |
| SSc subset | ||
| Diffuse cutaneous | 2 (66.7%) | / |
| Limited cutaneous | 1 (33.3%) | / |
| Antibodies | ||
| Scl-70 | 3 (100%) | / |
| Comorbidities | ||
| None | / | 3 (100%) |
| Digital ulcers | 3 (100%) | / |
| Pulmonary hypertension | 1 (33.3%) | / |
| Pulmonary fibrosis | 2 (66.7%) | / |
| Treatments | ||
| Prostanoids | 3 (100%) | / |
| CCB | 3 (100%) | / |
| ERA | 2 (66.7%) | / |
| PDE5Inh | 2 (66.7%) | / |
| DMARDs/bDMARDs | 2 (66.7%) | / |
SSc: systemic sclerosis; HC: healthy control; CCB: calcium channel blockers; ERA: endothelin receptor antagonist; PDE5Inh: phosphodiesterase type 5 inhibitors; DMARDs/bDMARDs: disease-modifying anti-rheumatic drugs traditional/biologics.
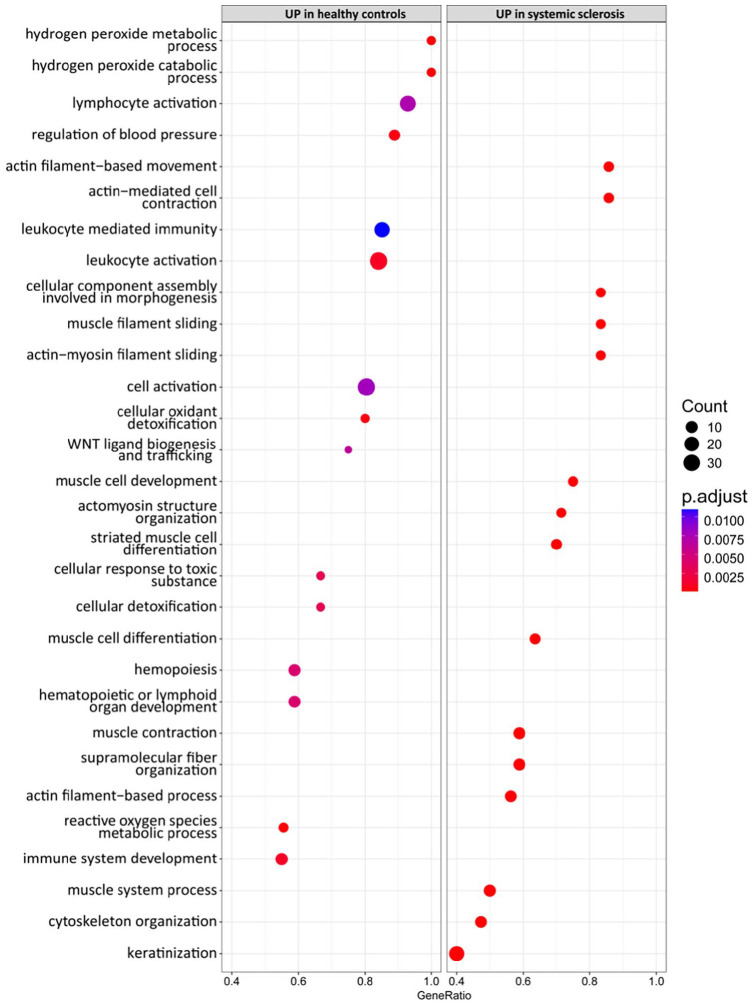
We performed DEGs analysis between SSc and HC, and we identified 305 DEGs that were up- or downregulated at least two-fold (Table 2). In particular, 175 genes were upregulated and 130 genes were downregulated (Table 2). A marked upregulation of genes involved in Wnt signaling, including Wnt family member (WNT) 4, WNT9B, WNT3, WNT16, with 2.5-,11.3- 6.5-, 5.6-folds, respectively, was present in HC if compared with SSc (Table 2 and Figure 1). The upregulation of collagen type VI alpha 5 chain (COL6A5), Fos proto-oncogene, AP-1 transcription factor subunit (FOS), glycoprotein M6A (GPM6A), extracellular matrix protein 2 (ECM2), adhesion G protein-coupled receptor E3 (ADGRE3), vascular endothelial growth factor D (VEGFD), LIF interleukin 6 family cytokine (LIF), among others, was also observed (Table 2 and Figure 1). Conversely, a marked downregulation of late cornified envelope (LCE) 3D, LCE3E, LCE1E, with 22-, 26- and 22-folds, respectively, and of genes encoding for keratins, including keratin (KRT) 35, KRT38, KRT82, was present in HC versus SSc samples (Table 2 and Figure 1). Gene ontology enrichment analysis revealed that DEGs in SSc biopsies were enriched for gene sets involved in actin filament-based movement, actin-mediated cell contraction, actomyosin structure organization, cell response to toxic substance, among others (Figure 2).
Table 2.
Differentially expressed genes between HC and SSc.
| Entrezid | Symbol | Gene name | logFC | P value |
|---|---|---|---|---|
| 101926892 | LOC101926892 | Uncharacterized LOC101926892 | 8.85 | 9.64E–11 |
| 155 | ADRB3 | Adrenoceptor beta 3 | 7.63 | 1.50E–05 |
| 402381 | SOHLH1 | Spermatogenesis and oogenesis-specific basic helix–loop–helix 1 | 7.57 | 5.30E–08 |
| 84658 | ADGRE3 | Adhesion G protein-coupled receptor E3 | 5.56 | 3.24E–07 |
| 23547 | LILRA4 | Leukocyte immunoglobulin-like receptor A4 | 5.48 | 7.33E–09 |
| 400120 | SERTM1 | Serine-rich and transmembrane domain containing 1 | 5.31 | 2.53E–07 |
| 3045 | HBD | Hemoglobin subunit delta | 4.97 | 4.43E–09 |
| 101929777 | LOC101929777 | Uncharacterized LOC101929777 | 4.77 | 1.51E–07 |
| 1360 | CPB1 | Carboxypeptidase B1 | 4.69 | 8.72E–05 |
| 3043 | HBB | Hemoglobin subunit beta | 4.68 | 1.25E–08 |
| 101927350 | LINC01254 | Long intergenic non-protein-coding RNA 1254 | 4.65 | 3.05E–12 |
| 404266 | HOXB-AS3 | HOXB cluster antisense RNA 3 | 4.54 | 8.47E–05 |
| 1178 | CLC | Charcot-Leyden crystal galectin | 4.54 | 1.71E–05 |
| 8785 | MATN4 | Matrilin 4 | 4.50 | 1.51E–07 |
| 3040 | HBA2 | Hemoglobin subunit alpha 2 | 4.31 | 2.15E–08 |
| 340273 | ABCB5 | ATP-binding cassette subfamily B member 5 | 4.30 | 6.29E–09 |
| 3952 | LEP | Leptin | 4.25 | 2.93E–05 |
| 3039 | HBA1 | Hemoglobin subunit alpha 1 | 4.21 | 2.76E–09 |
| 25975 | EGFL6 | EGF-like domain multiple 6 | 4.14 | 8.03E–06 |
| 80763 | SPX | Spexin hormone | 4.07 | 0.00011464 |
| 389903 | CSAG3 | CSAG family member 3 | 4.06 | 1.58E–05 |
| 345275 | HSD17B13 | Hydroxysteroid 17-beta dehydrogenase 13 | 3.81 | 8.78E–12 |
| 931 | MS4A1 | Membrane spanning 4-domains A1 | 3.77 | 2.22E–07 |
| 60675 | PROK2 | Prokineticin 2 | 3.67 | 1.43E–05 |
| 933 | CD22 | CD22 molecule | 3.57 | 3.20E–05 |
| 201516 | ZSCAN4 | Zinc finger and SCAN domain containing 4 | 3.50 | 7.55E–06 |
| 7484 | WNT9B | Wnt family member 9B | 3.47 | 3.95E–12 |
| 60385 | TSKS | Testis-specific serine kinase substrate | 3.47 | 0.000538562 |
| 114043 | TSPEAR-AS2 | TSPEAR antisense RNA 2 | 3.44 | 6.00E–05 |
| 54084 | TSPEAR | Thrombospondin-type laminin G domain and EAR repeats | 3.36 | 2.34E–06 |
| 1046 | CDX4 | Caudal type homeobox 4 | 3.36 | 0.000517385 |
| 8972 | MGAM | Maltase-glucoamylase | 3.35 | 3.80E–07 |
| 10249 | GLYAT | Glycine-N-acyltransferase | 3.34 | 5.20E–05 |
| 8875 | VNN2 | Vanin 2 | 3.33 | 4.32E–06 |
| 64407 | RGS18 | Regulator of G protein signaling 18 | 3.31 | 1.57E–05 |
| 6530 | SLC6A2 | Solute carrier family 6 member 2 | 3.25 | 3.71E–14 |
| 256076 | COL6A5 | Collagen type VI alpha 5 chain | 3.10 | 1.16E–05 |
| 79865 | TREML2 | Triggering receptor expressed on myeloid cells like 2 | 3.09 | 7.98E–05 |
| 100294720 | NHEG1 | Neuroblastoma highly expressed 1 | 3.05 | 1.59E–05 |
| 1002 | CDH4 | Cadherin 4 | 3.03 | 2.50E–05 |
| 64167 | ERAP2 | Endoplasmic reticulum aminopeptidase 2 | 2.98 | 7.49E–12 |
| 1441 | CSF3R | Colony stimulating factor 3 receptor | 2.91 | 1.64E–06 |
| 26166 | RGS22 | Regulator of G protein signaling 22 | 2.90 | 5.99E–05 |
| 30009 | TBX21 | T-box 21 | 2.84 | 6.12E–05 |
| 94031 | HTRA3 | HtrA serine peptidase 3 | 2.83 | 0.000107899 |
| 10578 | GNLY | Granulysin | 2.81 | 0.000267073 |
| 114780 | PKD1L2 | Polycystin 1 like 2 (gene/pseudogene) | 2.78 | 0.000744359 |
| 23743 | BHMT2 | Betaine–homocysteine S-methyltransferase 2 | 2.77 | 0.000709063 |
| 7473 | WNT3 | Wnt family member 3 | 2.77 | 1.39E–11 |
| 9597 | SMAD5-AS1 | SMAD5 antisense RNA 1 | 2.72 | 3.10E–10 |
| 26577 | PCOLCE2 | Procollagen C-endopeptidase enhancer 2 | 2.71 | 0.000187824 |
| 3202 | HOXA5 | Homeobox A5 | 2.69 | 0.000581136 |
| 2353 | FOS | Fos proto-oncogene, AP-1 transcription factor subunit | 2.66 | 0.00025139 |
| 2219 | FCN1 | Ficolin 1 | 2.64 | 8.76E–05 |
| 5593 | PRKG2 | Protein kinase cGMP-dependent 2 | 2.63 | 2.73E–07 |
| 969 | CD69 | CD69 molecule | 2.62 | 1.23E–06 |
| 2999 | GZMH | Granzyme H | 2.62 | 0.000205026 |
| 10631 | POSTN | Periostin | 2.61 | 0.000446456 |
| 2668 | GDNF | Glial cell-derived neurotrophic factor | 2.61 | 3.05E–05 |
| 146556 | C16orf89 | Chromosome 16 open reading frame 89 | 2.60 | 0.00037576 |
| 221476 | PI16 | Peptidase inhibitor 16 | 2.60 | 0.000200419 |
| 6402 | SELL | Selectin L | 2.60 | 2.58E–07 |
| 1805 | DPT | Dermatopontin | 2.60 | 9.67E–05 |
| 4969 | OGN | Osteoglycin | 2.59 | 1.43E–05 |
| 93035 | PKHD1L1 | PKHD1 like 1 | 2.58 | 1.93E–05 |
| 3953 | LEPR | Leptin receptor | 2.55 | 3.48E–09 |
| 11027 | LILRA2 | Leukocyte immunoglobulin like receptor A2 | 2.54 | 5.36E–05 |
| 3976 | LIF | LIF interleukin 6 family cytokine | 2.54 | 1.38E–07 |
| 29909 | GPR171 | G protein-coupled receptor 171 | 2.51 | 3.73E–05 |
| 100130231 | LINC00861 | Long intergenic non-protein coding RNA 861 | 2.51 | 0.000196239 |
| 51554 | ACKR4 | Atypical chemokine receptor 4 | 2.50 | 5.31E–05 |
| 9796 | PHYHIP | Phytanoyl-CoA 2-hydroxylase interacting protein | 2.50 | 1.75E–06 |
| 51384 | WNT16 | Wnt family member 16 | 2.49 | 4.62E–06 |
| 91851 | CHRDL1 | Chordin like 1 | 2.47 | 0.000492589 |
| 206338 | LVRN | Laeverin | 2.46 | 0.00082358 |
| 1066 | CES1 | Carboxylesterase 1 | 2.39 | 0.000609521 |
| 2823 | GPM6A | Glycoprotein M6A | 2.35 | 0.000234398 |
| 3575 | IL7R | Interleukin 7 receptor | 2.35 | 0.000342746 |
| 54857 | GDPD2 | Glycerophosphodiester phosphodiesterase domain containing 2 | 2.35 | 8.63E–06 |
| 53829 | P2RY13 | Purinergic receptor P2Y13 | 2.30 | 4.83E–05 |
| 3119 | HLA-DQB1 | Major histocompatibility complex, class II, DQ beta 1 | 2.30 | 1.30E–07 |
| 4069 | LYZ | Lysozyme | 2.28 | 1.50E–07 |
| 161753 | ODF3L1 | Outer dense fiber of sperm tails 3 like 1 | 2.28 | 0.000810101 |
| 5540 | NPY4R | Neuropeptide Y receptor Y4 | 2.26 | 4.25E–05 |
| 440738 | MAP1LC3C | Microtubule-associated protein 1 light chain 3 gamma | 2.25 | 0.000553398 |
| 10800 | CYSLTR1 | Cysteinyl leukotriene receptor 1 | 2.20 | 4.50E–06 |
| 286530 | P2RY8 | P2Y receptor family member 8 | 2.19 | 0.000221151 |
| 1842 | ECM2 | Extracellular matrix protein 2 | 2.19 | 0.000420098 |
| 5551 | PRF1 | Perforin 1 | 2.18 | 0.000531069 |
| 151887 | CCDC80 | Coiled-coil domain containing 80 | 2.15 | 0.000242366 |
| 399823 | FOXI2 | Forkhead box I2 | 2.13 | 0.000413967 |
| 54518 | APBB1IP | Amyloid beta precursor protein binding family B member 1 interacting protein | 2.11 | 0.00044138 |
| 118738 | ZNF488 | Zinc finger protein 488 | 2.11 | 0.000419495 |
| 3561 | IL2RG | Interleukin 2 receptor subunit gamma | 2.10 | 3.58E–05 |
| 10686 | CLDN16 | Claudin 16 | 2.09 | 4.44E–05 |
| 3683 | ITGAL | Integrin subunit alpha L | 2.08 | 0.000509098 |
| 3687 | ITGAX | Integrin subunit alpha X | 2.07 | 0.000174999 |
| 94234 | FOXQ1 | Forkhead box Q1 | 2.06 | 5.87E–10 |
| 117289 | TAGAP | T-cell activation Rho GTPase activating protein | 2.04 | 6.58E–05 |
| 8434 | RECK | Reversion inducing cysteine rich protein with kazal motifs | 2.04 | 2.48E–05 |
| 6352 | CCL5 | C–C motif chemokine ligand 5 | 2.00 | 0.000219667 |
| 90865 | IL33 | Interleukin 33 | 2.00 | 3.95E–08 |
| 5794 | PTPRH | Protein tyrosine phosphatase receptor type H | 1.97 | 0.000693056 |
| 2326 | FMO1 | Flavin containing monooxygenase 1 | 1.97 | 2.38E–08 |
| 6366 | CCL21 | C–C motif chemokine ligand 21 | 1.96 | 1.24E–06 |
| 2124 | EVI2B | Ecotropic viral integration site 2B | 1.95 | 6.78E–05 |
| 64333 | ARHGAP9 | Rho GTPase activating protein 9 | 1.94 | 8.27E–05 |
| 100379345 | MIR181A2HG | MIR181A2 host gene | 1.92 | 0.000817622 |
| 440584 | SLC2A1-AS1 | SLC2A1 antisense RNA 1 | 1.91 | 0.000137611 |
| 1043 | CD52 | CD52 molecule | 1.90 | 0.000310066 |
| 57007 | ACKR3 | Atypical chemokine receptor 3 | 1.90 | 1.74E–05 |
| 7940 | LST1 | Leukocyte-specific transcript 1 | 1.89 | 5.80E–05 |
| 2192 | FBLN1 | Fibulin 1 | 1.89 | 0.00038501 |
| 55713 | ZNF334 | Zinc finger protein 334 | 1.88 | 2.44E–05 |
| 11118 | BTN3A2 | Butyrophilin subfamily 3 member A2 | 1.88 | 2.48E–08 |
| 403323 | LOC403323 | Uncharacterized LOC403323 | 1.87 | 0.000255561 |
| 10090 | UST | Uronyl 2-sulfotransferase | 1.87 | 3.44E–09 |
| 643650 | LINC00842 | Long intergenic non-protein coding RNA 842 | 1.86 | 0.000183802 |
| 728643 | HNRNPA1P33 | Heterogeneous nuclear ribonucleoprotein A1 pseudogene 33 | 1.86 | 1.36E–07 |
| 23531 | MMD | Monocyte to macrophage differentiation associated | 1.82 | 0.000727749 |
| 129049 | SGSM1 | Small G protein signaling modulator 1 | 1.81 | 2.12E–05 |
| 5788 | PTPRC | Protein tyrosine phosphatase receptor type C | 1.80 | 0.000102226 |
| 55026 | TMEM255A | Transmembrane protein 255A | 1.79 | 0.000135186 |
| 123591 | TMEM266 | Transmembrane protein 266 | 1.77 | 0.000298357 |
| 120425 | JAML | Junction adhesion molecule like | 1.76 | 4.07E–05 |
| 79626 | TNFAIP8L2 | TNF-alpha-induced protein 8 like 2 | 1.76 | 0.000298517 |
| 10320 | IKZF1 | IKAROS family zinc finger 1 | 1.72 | 0.000830127 |
| 9098 | USP6 | Ubiquitin-specific peptidase 6 | 1.71 | 0.00013812 |
| 6252 | RTN1 | Reticulon 1 | 1.70 | 0.000642901 |
| 11119 | BTN3A1 | Butyrophilin subfamily 3 member A1 | 1.68 | 5.19E–06 |
| 1524 | CX3CR1 | C–X3C motif chemokine receptor 1 | 1.67 | 0.000852827 |
| 26157 | GIMAP2 | GTPase, IMAP family member 2 | 1.66 | 0.000169944 |
| 79891 | ZNF671 | Zinc finger protein 671 | 1.66 | 0.000260642 |
| 3117 | HLA-DQA1 | Major histocompatibility complex, class II, DQ alpha 1 | 1.63 | 4.20E–05 |
| 388011 | LINC01550 | Long intergenic non-protein coding RNA 1550 | 1.61 | 0.000368953 |
| 8935 | SKAP2 | Src kinase-associated phosphoprotein 2 | 1.60 | 1.85E–05 |
| 10384 | BTN3A3 | Butyrophilin subfamily 3 member A3 | 1.59 | 5.40E–05 |
| 27306 | HPGDS | Hematopoietic prostaglandin D synthase | 1.59 | 0.000213676 |
| 55244 | SLC47A1 | Solute carrier family 47 member 1 | 1.58 | 0.000876973 |
| 101927164 | LOC101927164 | Uncharacterized LOC101927164 | 1.58 | 6.33E–05 |
| 7058 | THBS2 | Thrombospondin 2 | 1.53 | 1.82E–05 |
| 57758 | SCUBE2 | Signal peptide, CUB domain, and EGF-like domain containing 2 | 1.53 | 3.22E–05 |
| 963 | CD53 | CD53 molecule | 1.50 | 0.000642744 |
| 54504 | CPVL | Carboxypeptidase vitellogenic like | 1.49 | 3.32E–05 |
| 2530 | FUT8 | Fucosyltransferase 8 | 1.48 | 1.93E–05 |
| 2625 | GATA3 | GATA-binding protein 3 | 1.47 | 5.22E–05 |
| 6453 | ITSN1 | Intersectin 1 | 1.45 | 0.000168102 |
| 5997 | RGS2 | Regulator of G protein signaling 2 | 1.44 | 3.56E–05 |
| 79901 | CYBRD1 | Cytochrome b reductase 1 | 1.43 | 0.000662612 |
| 53833 | IL20RB | Interleukin 20 receptor subunit beta | 1.42 | 0.000208175 |
| 1520 | CTSS | Cathepsin S | 1.40 | 0.000105691 |
| 10866 | HCP5 | HLA complex P5 | 1.38 | 1.56E–05 |
| 5328 | PLAU | Plasminogen activator, urokinase | 1.38 | 0.000448423 |
| 28971 | AAMDC | Adipogenesis-associated Mth938 domain containing | 1.36 | 2.19E–05 |
| 6571 | SLC18A2 | Solute carrier family 18 member A2 | 1.34 | 4.92E–05 |
| 51302 | CYP39A1 | Cytochrome P450 family 39 subfamily A member 1 | 1.30 | 0.000753341 |
| 51097 | SCCPDH | Saccharopine dehydrogenase (putative) | 1.30 | 0.000160085 |
| 54361 | WNT4 | Wnt family member 4 | 1.30 | 0.000167086 |
| 7805 | LAPTM5 | Lysosomal protein transmembrane 5 | 1.28 | 0.000372444 |
| 4688 | NCF2 | Neutrophil cytosolic factor 2 | 1.25 | 5.32E–05 |
| 90634 | N4BP2L1 | NEDD4-binding protein 2 like 1 | 1.24 | 0.000709105 |
| 3176 | HNMT | Histamine N-methyltransferase | 1.22 | 0.000848657 |
| 79734 | KCTD17 | Potassium channel tetramerization domain containing 17 | 1.22 | 0.000714539 |
| 493812 | HCG11 | HLA complex group 11 | 1.22 | 0.000212103 |
| 131616 | TMEM42 | Transmembrane protein 42 | 1.22 | 0.000859229 |
| 7128 | TNFAIP3 | TNF-alpha-induced protein 3 | 1.19 | 0.000609645 |
| 9536 | PTGES | Prostaglandin E synthase | 1.19 | 0.000670154 |
| 79690 | GAL3ST4 | Galactose-3-O-sulfotransferase 4 | 1.19 | 0.000663895 |
| 9891 | NUAK1 | NUAK family kinase 1 | 1.18 | 0.000205103 |
| 6304 | SATB1 | SATB homeobox 1 | 1.17 | 4.63E–05 |
| 4088 | SMAD3 | SMAD family member 3 | 1.14 | 5.50E–05 |
| 4792 | NFKBIA | NFKB inhibitor alpha | 1.08 | 0.000807994 |
| 2619 | GAS1 | Growth arrest-specific 1 | 1.08 | 0.000508176 |
| 9805 | SCRN1 | Secernin 1 | 1.04 | 0.000197048 |
| 153020 | RASGEF1B | RasGEF domain family member 1B | 1.01 | 0.000580739 |
| 57475 | PLEKHH1 | Pleckstrin homology, MyTH4 and FERM domain containing H1 | −1.08 | 0.000666038 |
| 23223 | RRP12 | Ribosomal RNA processing 12 homolog | −1.14 | 0.000181227 |
| 50487 | PLA2G3 | Phospholipase A2 group III | −1.19 | 0.000544955 |
| 5507 | PPP1R3C | Protein phosphatase 1 regulatory subunit 3C | −1.20 | 0.000443646 |
| 54751 | FBLIM1 | Filamin-binding LIM protein 1 | −1.27 | 0.000580775 |
| 5187 | PER1 | Period circadian regulator 1 | −1.29 | 6.01E–05 |
| 11254 | SLC6A14 | Solute carrier family 6 member 14 | −1.36 | 0.000117055 |
| 3371 | TNC | Tenascin C | −1.42 | 0.000164153 |
| 8497 | PPFIA4 | PTPRF-interacting protein alpha 4 | −1.42 | 0.000453085 |
| 151354 | LRATD1 | LRAT domain containing 1 | −1.43 | 1.01E–05 |
| 3768 | KCNJ12 | Potassium voltage-gated channel subfamily J member 12 | −1.43 | 0.000693952 |
| 84254 | CAMKK1 | Calcium/calmodulin dependent protein kinase 1 | −1.44 | 4.13E–05 |
| 10804 | GJB6 | Gap junction protein beta 6 | −1.50 | 1.83E–06 |
| 5208 | PFKFB2 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 | −1.53 | 0.000797632 |
| 79017 | GGCT | Gamma-glutamylcyclotransferase | −1.58 | 0.000306746 |
| 8863 | PER3 | Period circadian regulator 3 | −1.61 | 6.62E–07 |
| 158158 | RASEF | RAS and EF-hand domain containing | −1.61 | 0.000109187 |
| 9687 | GREB1 | Growth regulating estrogen receptor binding 1 | −1.62 | 0.000385599 |
| 8153 | RND2 | Rho family GTPase 2 | −1.63 | 0.000167306 |
| 402778 | IFITM10 | Interferon-induced transmembrane protein 10 | −1.73 | 0.000119745 |
| 7804 | LRP8 | LDL receptor-related protein 8 | −1.77 | 0.000438195 |
| 374383 | NCR3LG1 | Natural killer cell cytotoxicity receptor 3 ligand 1 | −1.85 | 0.000645021 |
| 22979 | EFR3B | EFR3 homolog B | −1.86 | 0.000757246 |
| 118430 | MUCL1 | Mucin like 1 | −1.87 | 0.000104517 |
| 11226 | GALNT6 | Polypeptide N-acetylgalactosaminyltransferase 6 | −1.96 | 4.03E–08 |
| 192683 | SCAMP5 | Secretory carrier membrane protein 5 | −1.97 | 0.000664492 |
| 7227 | TRPS1 | Transcriptional repressor GATA binding 1 | −1.99 | 5.48E–07 |
| 29842 | TFCP2L1 | Transcription factor CP2 like 1 | −2.03 | 0.000123612 |
| 1285 | COL4A3 | Collagen type IV alpha 3 chain | −2.06 | 0.000635889 |
| 6706 | SPRR2G | Small proline rich protein 2G | −2.06 | 2.71E–08 |
| 84940 | CORO6 | Coronin 6 | −2.10 | 3.02E–05 |
| 765 | CA6 | Carbonic anhydrase 6 | −2.12 | 0.000432192 |
| 83694 | RPS6KL1 | Ribosomal protein S6 kinase like 1 | −2.13 | 0.000374012 |
| 84676 | TRIM63 | Tripartite motif containing 63 | −2.13 | 0.000360823 |
| 2171 | FABP5 | Fatty acid-binding protein 5 | −2.15 | 3.69E–08 |
| 3485 | IGFBP2 | Insulin-like growth factor binding protein 2 | −2.16 | 0.000844872 |
| 65009 | NDRG4 | NDRG family member 4 | −2.17 | 1.40E–05 |
| 79801 | SHCBP1 | SHC-binding and spindle-associated 1 | −2.22 | 1.06E–06 |
| 148523 | CIART | Circadian-associated repressor of transcription | −2.23 | 2.24E–06 |
| 23657 | SLC7A11 | Solute carrier family 7 member 11 | −2.30 | 0.00084804 |
| 23553 | HYAL4 | Hyaluronidase 4 | −2.35 | 0.0003367 |
| 153478 | PLEKHG4B | Pleckstrin homology and RhoGEF domain containing G4B | −2.45 | 8.71E–05 |
| 285489 | DOK7 | Docking protein 7 | −2.48 | 0.000470274 |
| 1734 | DIO2 | Iodothyronine deiodinase 2 | −2.49 | 1.55E–09 |
| 128488 | WFDC12 | WAP four-disulfide core domain 12 | −2.51 | 6.12E–06 |
| 383 | ARG1 | Arginase 1 | −2.52 | 0.000109898 |
| 5820 | PVT1 | Pvt1 oncogene | −2.53 | 1.18E–07 |
| 1745 | DLX1 | Distal-less homeobox 1 | −2.53 | 2.58E–05 |
| 4824 | NKX3-1 | NK3 homeobox 1 | −2.53 | 3.60E–06 |
| 5789 | PTPRD | Protein tyrosine phosphatase receptor type D | −2.54 | 0.000681882 |
| 202299 | LINC01554 | Long intergenic non-protein coding RNA 1554 | −2.56 | 6.65E–08 |
| 1769 | DNAH8 | Dynein axonemal heavy chain 8 | −2.57 | 0.000468749 |
| 1594 | CYP27B1 | Cytochrome P450 family 27 subfamily B member 1 | −2.57 | 0.000213961 |
| 144406 | WDR66 | WD repeat domain 66 | −2.57 | 2.70E–05 |
| 56475 | RPRM | Reprimo, TP53 dependent G2 arrest mediator homolog | −2.58 | 0.000851815 |
| 1768 | DNAH6 | Dynein axonemal heavy chain 6 | −2.62 | 0.000295338 |
| 10218 | ANGPTL7 | Angiopoietin like 7 | −2.71 | 0.000517742 |
| 338667 | VSIG10L2 | V-set and immunoglobulin domain containing 10 like 2 | −2.73 | 0.000311392 |
| 9615 | GDA | Guanine deaminase | −2.77 | 3.70E–05 |
| 4440 | MSI1 | Musashi RNA binding protein 1 | −2.77 | 0.000188836 |
| 140807 | KRT72 | Keratin 72 | −2.79 | 0.000245768 |
| 3755 | KCNG1 | Potassium voltage-gated channel modifier subfamily G member 1 | −2.82 | 0.000372192 |
| 27132 | CPNE7 | Copine 7 | −2.86 | 1.29E–05 |
| 387700 | SLC16A12 | Solute carrier family 16 member 12 | −2.87 | 0.000433402 |
| 387695 | C10orf99 | Chromosome 10 open reading frame 99 | −2.88 | 0.000337971 |
| 6861 | SYT5 | Synaptotagmin 5 | −2.89 | 0.000463435 |
| 148281 | SYT6 | Synaptotagmin 6 | −2.92 | 0.000302434 |
| 319101 | KRT73 | Keratin 73 | −3.00 | 2.30E–05 |
| 105377774 | LOC105377774 | Uncharacterized LOC105377774 | −3.04 | 0.000113072 |
| 387911 | C1QTNF9B | C1q and TNF-related 9B | −3.04 | 0.000748625 |
| 9720 | CCDC144A | Coiled-coil domain containing 144A | −3.06 | 0.000102105 |
| 4753 | NELL2 | Neural EGFL-like 2 | −3.07 | 1.69E–05 |
| 1301 | COL11A1 | Collagen type XI alpha 1 chain | −3.09 | 4.71E–08 |
| 105373551 | LOC105373551 | Uncharacterized LOC105373551 | −3.12 | 1.12E–06 |
| 339768 | ESPNL | Espin like | −3.14 | 0.000376495 |
| 339535 | LINC01139 | Long intergenic non-protein coding RNA 1139 | −3.22 | 2.36E–07 |
| 121506 | ERP27 | Endoplasmic reticulum protein 27 | −3.23 | 6.16E–08 |
| 2569 | GABRR1 | Gamma-aminobutyric acid type A receptor rho1 subunit | −3.29 | 0.0003984 |
| 100506217 | NA | NA | −3.30 | 0.000674811 |
| 57016 | AKR1B10 | Aldo-keto reductase family 1 member B10 | −3.30 | 8.00E–07 |
| 7545 | ZIC1 | Zic family member 1 | −3.31 | 0.000392413 |
| 255324 | EPGN | Epithelial mitogen | −3.32 | 0.000478875 |
| 163351 | GBP6 | Guanylate-binding protein family member 6 | −3.36 | 4.40E–08 |
| 55584 | CHRNA9 | Cholinergic receptor nicotinic alpha 9 subunit | −3.37 | 0.000303345 |
| 64208 | POPDC3 | Popeye domain containing 3 | −3.44 | 0.00021796 |
| 176 | ACAN | Aggrecan | −3.47 | 0.000526066 |
| 401074 | LINC00960 | Long intergenic non-protein coding RNA 960 | −3.47 | 5.96E–05 |
| 80309 | SPHKAP | SPHK1 interactor, AKAP domain containing | −3.48 | 1.57E–06 |
| 84107 | ZIC4 | Zic family member 4 | −3.51 | 0.000138626 |
| 729522 | AACSP1 | Acetoacetyl-CoA synthetase pseudogene 1 | −3.57 | 5.13E–06 |
| 92736 | OTOP2 | Otopetrin 2 | −3.66 | 5.50E–07 |
| 1143 | CHRNB4 | Cholinergic receptor nicotinic beta 4 subunit | −3.72 | 0.000636152 |
| 160762 | CCDC63 | Coiled-coil domain containing 63 | −3.79 | 0.000458776 |
| 353134 | LCE1D | Late cornified envelope 1D | −3.80 | 3.35E–08 |
| 121391 | KRT74 | Keratin 74 | −3.92 | 0.000638024 |
| 162632 | USP32P1 | Ubiquitin-specific peptidase 32 pseudogene 1 | −4.01 | 3.80E–09 |
| 4703 | NEB | Nebulin | −4.03 | 0.000467365 |
| 54207 | KCNK10 | Potassium two pore domain channel subfamily K member 10 | −4.07 | 0.000717882 |
| 353135 | LCE1E | Late cornified envelope 1E | −4.20 | 5.96E–09 |
| 84648 | LCE3D | Late cornified envelope 3D | −4.25 | 2.21E–26 |
| 191585 | PLAC4 | Placenta enriched 4 | −4.37 | 1.86E–05 |
| 57795 | BRINP2 | BMP/retinoic acid inducible neural-specific 2 | −4.40 | 0.000184955 |
| 146802 | SLC47A2 | Solute carrier family 47 member 2 | −4.57 | 1.75E–10 |
| 102724541 | NA | NA | −4.61 | 1.55E–08 |
| 4747 | NEFL | Neurofilament light | −4.64 | 0.000177839 |
| 84221 | SPATC1L | Spermatogenesis and centriole-associated 1 like | −4.73 | 4.27E–08 |
| 353145 | LCE3E | Late cornified envelope 3E | −4.78 | 4.73E–22 |
| 84560 | MT4 | Metallothionein 4 | −4.80 | 0.000461366 |
| 440482 | ANKRD20A5P | Ankyrin repeat domain 20 family member A5, pseudogene | −4.94 | 0.00019383 |
| 347741 | OTOP3 | Otopetrin 3 | −4.96 | 2.07E–14 |
| 57586 | SYT13 | Synaptotagmin 13 | −5.15 | 3.77E–05 |
| 93273 | LEMD1 | LEM domain containing 1 | −5.30 | 0.000528385 |
| 389668 | XKR9 | XK-related 9 | −5.31 | 4.64E–10 |
| 3359 | HTR3A | 5-hydroxytryptamine receptor 3A | −5.44 | 1.19E–06 |
| 1311 | COMP | Cartilage oligomeric matrix protein | −5.54 | 3.22E–15 |
| 7273 | TTN | Titin | −5.79 | 2.27E–05 |
| 58503 | OPRPN | Opiorphin prepropeptide | −5.81 | 0.000471129 |
| 4151 | MB | Myoglobin | −6.16 | 0.000584199 |
| 487 | ATP2A1 | ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 | −6.27 | 0.000323427 |
| 643224 | TUBBP5 | Tubulin beta pseudogene 5 | −6.35 | 3.12E–13 |
| 779 | CACNA1S | Calcium voltage-gated channel subunit alpha1S | −6.65 | 0.000296762 |
| 4604 | MYBPC1 | Myosin-binding protein C, slow type | −6.78 | 0.000336416 |
| 116 | ADCYAP1 | Adenylate cyclase activating polypeptide 1 | −6.78 | 0.000849582 |
| 200407 | CREG2 | Cellular repressor of E1A-stimulated genes 2 | −7.12 | 0.000146125 |
| 146481 | FRG2DP | FSHD region gene 2 family member D, pseudogene | −7.16 | 5.70E–09 |
| 8557 | TCAP | Titin-cap | −8.00 | 0.000875387 |
| 284233 | CYP4F35P | Cytochrome P450 family 4 subfamily F member 35, pseudogene | −8.47 | 2.28E–06 |
| 442721 | LMOD2 | Leiomodin 2 | −9.63 | 0.000789905 |
| 58 | ACTA1 | Actin alpha 1, skeletal muscle | −10.85 | 0.000190824 |
| 4620 | MYH2 | Myosin heavy chain 2 | −11.41 | 0.000552856 |
Figure 1.
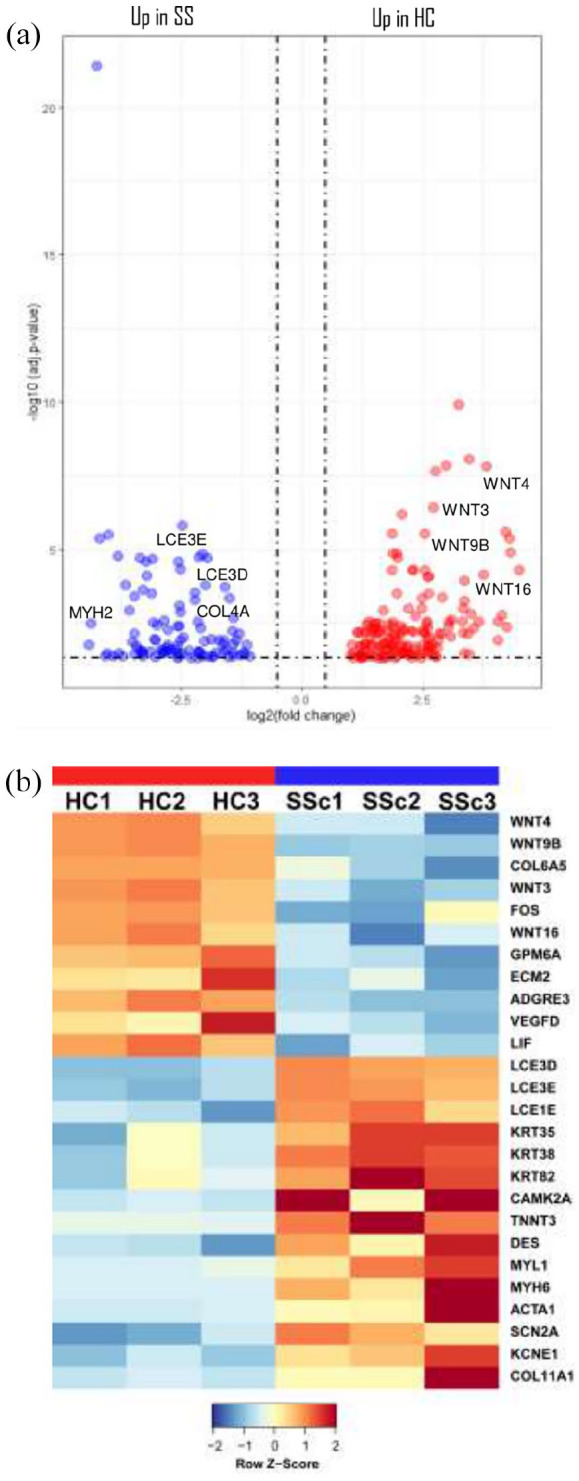
mRNA expression profiling of skin biopsies from HC and SSc patients: (a) volcano plot showing differentially expressed genes in healthy controls (HCs) and systemic sclerosis (SSc) patients and (b) gene expression heatmap of differentially expressed mRNAs in HC versus SSc tissues.
Columns show each patient/individuals (red indicates healthy control (HC); blue indicates systemic sclerosis (SSc) patients). Rows show individual genes. The color of het varies from blue (i.e. downregulated expression) to red (i.e. upregulated expression).
Figure 2.
Dot plot of enriched pathways in HC versus SSc tissues.
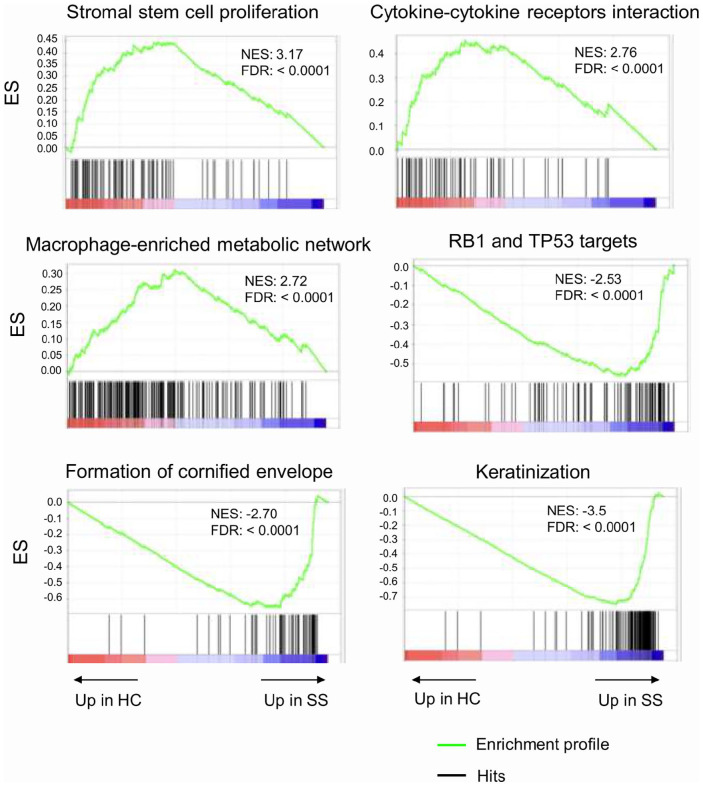
To identify potential functional pathways that could be involved in SSc pathogenesis, we performed GSEA of DEGs. GSEA revealed that HCs were characterized, among others, by gene signatures related to stromal stem cells proliferation, cytokine–cytokine receptor interaction, macrophage-enriched metabolic network, whereas SSc samples were enriched in signatures related to keratinization, cornification, retinoblastoma (RB) 1 and tumor suppressor (TP) 53 signaling (Figure 3).
Figure 3.
Gene set enrichment analysis (GSEA) in HC versus SSc tissues.
Enrichment of gene signature was analyzed in transcriptomic data from HC and SSc samples. ES: enrichment score; NES: normalized enrichment score; FDR: false discovery rate.
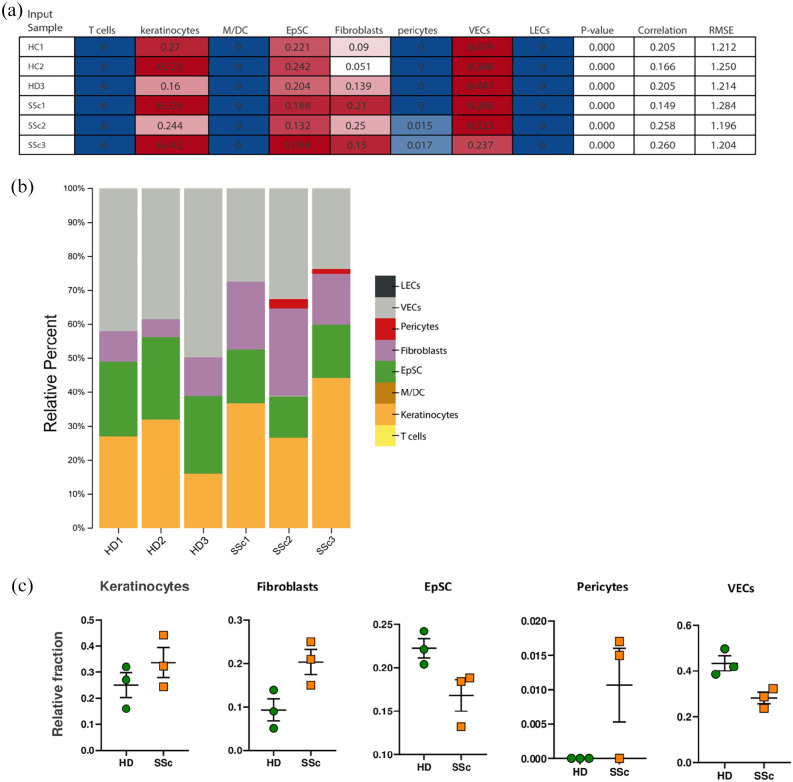
Cell subsets in which differential gene expression occurs were identified and quantified according to the CIBERSORTx algorithm (Figure 4). We found that DEGs were expressed in keratinocytes, epithelial stem cells (EpSC), fibroblasts, pericytes, and vascular endothelial cells (VECs), and that keratinocytes and fibroblasts in SSc, whereas EpSC and VECs were decreased (Figure 4).
Figure 4.
Cell subsets according to the CIBERSORTx algorithm: (a) cell types expressing DEGs obtained by RNA-seq. Columns represent the cell types from the signature genes file and rows represent deconvolution results for each mixture sample. All results are reported as relative fractions normalized to 1 across all cell subsets. M/DC, macrophages/dendritic cells; EpSC, epithelial stem cells; VECs, vascular endothelial cells; LECs, lymphatic endothelial cells; p value, statistical significance of the deconvolution result across all cell subsets; correlation, Pearson’s correlation coefficient (R), generated from comparing the original mixture with the estimated mixture; RMSE, root mean squared error between the original mixture and the imputed mixture. (b) Bar chart showing the relative percentage (relative fractions × 100) of each cell type computed by CIBERSORTx. (c) Dotplots show the computed relative cellular fractions reported in A.
Discussion
As introduced, altered gene expression seems to contribute to the aberrant mechanisms that propagate SSc vasculopathy.8,9 Recent advances in cell transcriptome technology, including RNA-seq analysis, seem to provide an unprecedented point of view into SSc pathogenesis and offer important implications for personalized disease management. 11
Here, we provided a comprehensive analysis of RNA-seq data derived from SSc and HCs skin tissues. According to our data, RNA-seq, differential gene expression and pathway analysis revealed that SSc subjects display a discrete pattern of gene expression associated with keratinization and ECM generation. In detail, according to GSEA, we demonstrated that HC were characterized by gene signatures related to stromal stem cells proliferation, cytokine–cytokine receptor interaction macrophage-enriched metabolic network, among others. On the other hand, SSc tissues were added in signatures related to keratinization, cornification, RB 1 and TP 53 signaling, to the detriment of regulation of angiogenesis and stromal stem cells proliferation.
Various conditions characterized by aberrant fibrosis and vascular dysfunction, such as keloids, were also studied using RNA-seq.12,13 Results from these reports highlighted the roles of tumor growth factor beta (TGF-β) and Eph–ephrin signaling pathways in keloids processes; critical regulators probably involved, such as TWIST1, FOXO3, and SMAD3, were also identified. In addition, tumor-related pathways were activated and dysregulated in keloid fibroblasts and ECs, which could explain malignant features of keloids. These findings will help the clinicians to better recognize fibrotic skin pathogenesis and provide possible targets for fibrotic disease therapies.
Other connective tissue diseases, such as Sjögren’s syndrome, were recently studied through transcriptomic, genomic, epigenetic, cytokine expression and flow cytometry data, to identify groups of patients with distinct patterns of immune dysregulation, in combination with clinical parameters. RNA-Seq was also used and the identified biomarkers were functional to evaluate response to treatments and to develop future target therapies. 14
Identifying the specific immune mechanisms underlying SSc pathogenesis could similarly result in innovative therapies production that selectively target the aberrant immune response, with better efficacy and less toxicity. A comprehensive analysis of T-cell mediated immune responses in the affected skin of SSc patients was performed by means of single-cell RNA-seq with interesting results about distinct signaling activated pathways. 15
Clinical implications are therefore mandatory. In 2019, some authors exploited single-cell RNA-seq by performing pathway analysis with GSEA and ingenuity pathway analysis (IPA) in SSc skin. 12 They finally demonstrated that the SSc EC expression profile is enriched in processes related to ECM generation and negative regulation of angiogenesis and epithelial-to-mesenchymal transition. Two of the top DEGs, HSPG2 and APLNR, were independently verified as potential markers of EC injury. These genes have been associated with vascular dysfunction and fibrosis in different settings, including SSc with its harmful complications, such as lung fibrosis and PAH.16–19
As partially known, myofibroblasts are key effector cells in the remodeling process of interstitial lung disease (ILD) associated with SSc. Transcriptomic analysis using single-cell RNA-seq was performed by some authors 20 to define the transcriptomes of myofibroblasts and other mesenchymal cells in SSc to clarify how alterations in fibroblast phenotypes lead to SSc-ILD fibrosis. Results from this study established a great previously unrecognized fibroblast heterogeneity in SSc-ILD and described multimodal transcriptome-phenotypes associated with these cells. These data considerably highlighted that myofibroblast differentiation and proliferation are crucial pathological mechanisms driving fibrosis in SSc-ILD with interesting new comprehensions into their functional role. 20 Similar findings were also observed in idiopathic pulmonary fibrosis (IPF). 21
EC dysfunction efforts the initiation and contributes to the propagation of PAH too. Integrated analyses, including RNA-seq, aimed to provide a comprehensive atlas of EC in the health lung and PAH condition. These analyses revealed in detail that PAH-induced EC transcriptomic changes could provide novel targets for therapeutic development.22,23
SSc is a rare detrimental disease which offers a challenging study model to speculate into pathologic angiogenesis and fibrogenesis processes. 24 SSc-related complications, such as skin ulcers, ILD, and PAH, still represent frequent causes of morbidity and mortality.3,25 In our experience, we analyzed scleroderma spectrum with special focus on the above conditions and their proper approaches and innovative treatment proposals.26–30 A critical and detailed analysis of such complications is necessary to pursue a personalized therapeutic strategy. Rapid progress in sequencing technologies in recent years provided valuable insights into complex biological systems with interesting potential medical applications and treatment implications. 31
Our SSc subjects underwent autologous fat grafting procedure as SSc regenerative medicine approach to treat their cutaneous scleroderma-related manifestations. According to our preliminary findings, RNA-seq and pathway analysis particularly revealed that SSc subjects displayed a pattern of gene expression associated with keratinization and ECM generation. GSEA also established that SSc tissues were enriched in signatures related to keratinization and cornification, at the expense of regulation of angiogenesis and stromal stem cells proliferation.
These findings are in line with the proposed pathogenetic mechanisms underlying SSc, especially highlighting the importance of extracellular microenvironment imbalance and cells interaction, which leads to impaired angiogenesis, endothelial and epithelial to mesenchymal transition, fibroblast activation and ECM deposition, finally resulting in fibrosis.
The present study has several limitations. The first limitation is the low number of patients with heterogeneous disease duration used for the analysis, who were chosen on the basis of similar clinical features. Second, functional analysis is needed to clarify the roles of the identified possible pathogenetic mechanisms in SSc. However, our results provide an interesting framework for the identification of valuable biomarkers representing vascular damage and fibrotic alterations in SSc to explore future perspectives and therapeutic targets.
Footnotes
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
ORCID iDs: Marco de Pinto  https://orcid.org/0000-0002-0947-8663
https://orcid.org/0000-0002-0947-8663
Melba Lattanzi  https://orcid.org/0000-0003-1230-7539
https://orcid.org/0000-0003-1230-7539
Dilia Giuggioli  https://orcid.org/0000-0002-0041-3695
https://orcid.org/0000-0002-0041-3695
References
- 1.Hachulla E, Launay D. Diagnosis and classification of systemic sclerosis. Clin Rev Allergy Immunol 2011; 40: 78–83. [DOI] [PubMed] [Google Scholar]
- 2.Van den Hoogen F, Khanna D, Fransen J, et al. 2013 Classification criteria for systemic sclerosis: an American College of Rheumatology/European league against rheumatism collaborative initiative. Arthritis Rheum 2013; 65: 2737–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferri C, Sebastiani M, Lo Monaco A, et al. Systemic sclerosis evolution of disease pathomorphosis and survival. Our experience on Italian patients’ population and review of the literature. Autoimmun Rev 2014; 13(10): 1026–1034. [DOI] [PubMed] [Google Scholar]
- 4.Cutolo M, Soldano S, Smith V. Pathophysiology of systemic sclerosis: current understanding and new insights. Expert Rev Clin Immunol 2019; 15(7): 753–764. [DOI] [PubMed] [Google Scholar]
- 5.Di Benedetto P, Ruscitti P, Liakouli V, et al. The vessels contribute to fibrosis in systemic sclerosis. Isr Med Assoc J 2019; 21(7): 471–474. [PubMed] [Google Scholar]
- 6.Matucci-Cerinic M, Kahaleh B, Wigley FM. Review: evidence that systemic sclerosis is a vascular disease. Arthritis Rheum 2013; 65(8): 1953–1962. [DOI] [PubMed] [Google Scholar]
- 7.Altorok N, Wang Y, Kahaleh B. Endothelial dysfunction in systemic sclerosis. Curr Opin Rheumatol 2014; 26: 615–620. [DOI] [PubMed] [Google Scholar]
- 8.van Bon L, Affandi AJ, Broen J, et al. Proteome-wide analysis and CXCL4 as a biomarker in systemic sclerosis. N Engl J Med 2014; 370: 433–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manetti M, Guiducci S, Romano E, et al. Overexpression of VEGF165b, an inhibitory splice variant of vascular endothelial growth factor, leads to insufficient angiogenesis in patients with systemic sclerosis. Circ Res 2011; 109: e14–e26. [DOI] [PubMed] [Google Scholar]
- 10.Newman AM, Steen CB, Liu CL, et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol 2019; 37(7): 773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Apostolidis SA, Stifano G, Tabib T, et al. Single cell RNA sequencing identifies HSPG2 and APLNR as markers of endothelial cell injury in systemic sclerosis skin. Front Immunol 2018; 9: 2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng CC, Hu YF, Zhu DH, et al. Single-cell RNA-seq reveals fibroblast heterogeneity and increased mesenchymal fibroblasts in human fibrotic skin diseases. Nat Commun 2021; 12(1): 3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu X, Chen W, Zeng Q, et al. Single-cell RNA-sequencing reveals lineage-specific regulatory changes of fibroblasts and vascular endothelial cells in keloids. J Invest Dermatol 2022; 142(1): 124–135. [DOI] [PubMed] [Google Scholar]
- 14.Soret P, Le Dantec C, Desvaux E, et al. A new molecular classification to drive precision treatment strategies in primary Sjögren’s syndrome. Nat Commun 2021; 12(1): 3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaydosik AM, Tabib T, Domsic R, et al. Single-cell transcriptome analysis identifies skin-specific T-cell responses in systemic sclerosis. Ann Rheum Dis 2021; 80(11): 1453–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eyries M, Siegfried G, Ciumas M, et al. Hypoxia-induced apelin expression regulates endothelial cell proliferation and regenerative angiogenesis. Circ Res 2008; 103: 432–440. [DOI] [PubMed] [Google Scholar]
- 17.Kang Y, Kim J, Anderson JP, et al. Apelin-APJ signaling is a critical regulator of endothelial MEF2 activation in cardiovascular development. Circ Res 2013; 113: 22–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baiocchini A, Montaldo C, Conigliaro A, et al. Extracellular matrix molecular remodeling in human liver fibrosis evolution. PLoS ONE 2016; 11(3): e0151736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laplante P, Raymond MA, Gagnon G, et al. Novel fibrogenic pathways are activated in response to endothelial apoptosis: implications in the pathophysiology of systemic sclerosis. J Immunol 2005; 174: 5740–5749. [DOI] [PubMed] [Google Scholar]
- 20.Valenzi E, Bulik M, Tabib T, et al. Single-cell analysis reveals fibroblast heterogeneity and myofibroblasts in systemic sclerosis-associated interstitial lung disease. Ann Rheum Dis 2019; 78(10): 1379–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adams TS, Schupp JC, Poli S, et al. Single-cell RNA-seq reveals ectopic and aberrant lung-resident cell populations in idiopathic pulmonary fibrosis. Sci Adv 2020; 6(28): eaba1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schupp JC, Adams TS, Cosme C, et al. Integrated single-cell atlas of endothelial cells of the human lung. Circulation 2021; 144(4): 286–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodor J, Chen SH, Scanlon JP, et al. Single-cell RNA-seq profiling of mouse endothelial cells in response to pulmonary arterial hypertension. Cardiovasc Res 2021; 118: 2519–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferri C, Arcangeletti MC, Caselli E, et al. Insights into the knowledge of complex diseases: environmental infectious/toxic agents as potential etiopathogenetic factors of systemic sclerosis. J Autoimmun 2021; 124: 102727. [DOI] [PubMed] [Google Scholar]
- 25.Ferri C, Giuggioli D, Guiducci S, et al. Systemic sclerosis Progression INvestiGation (SPRING) Italian registry: demographic and clinico-serological features of the scleroderma spectrum. Clin Exp Rheumatol 2020; 38 (3 Suppl. 125): 40–47. [PubMed] [Google Scholar]
- 26.Giuggioli D, Manfredi A, Lumetti F, et al. Scleroderma skin ulcers definition, classification and treatment strategies our experience and review of the literature. Autoimmun Rev 2018; 17(2): 155–164. [DOI] [PubMed] [Google Scholar]
- 27.Pignatti M, Spinella A, Cocchiara E, et al. Autologous fat grafting for the oral and digital complications of systemic sclerosis: results of a prospective study. Aesthetic Plast Surg 2020; 44(5): 1820–1832. [DOI] [PubMed] [Google Scholar]
- 28.Starnoni M, Pappalardo M, Spinella A, et al. Systemic sclerosis cutaneous expression: management of skin fibrosis and digital ulcers. Ann Med Surg 2021; 71: 102984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Occhipinti M, Bruni C, Camiciottoli G, et al. Quantitative analysis of pulmonary vasculature in systemic sclerosis at spirometry-gated chest CT. Ann Rheum Dis 2020; 79(9): 1210–1217. [DOI] [PubMed] [Google Scholar]
- 30.Giuggioli D, Bruni C, Cacciapaglia F, et al. Pulmonary arterial hypertension: guidelines and unmet clinical needs. Reumatismo 2021; 72(4): 228–246. [DOI] [PubMed] [Google Scholar]
- 31.Lo Tartaro D, De Biasi S, Forcato M, et al. Gene expression analysis of T-Cells by single-cell RNA-Seq. Methods Mol Biol 2021; 2285: 277–296. [DOI] [PubMed] [Google Scholar]