Summary
The coronavirus (CoV) family includes several viruses infecting humans, highlighting the importance of exploring pan-CoV vaccine strategies to provide broad adaptive immune protection. We analyze T cell reactivity against representative Alpha (NL63) and Beta (OC43) common cold CoVs (CCCs) in pre-pandemic samples. S, N, M, and nsp3 antigens are immunodominant, as shown for severe acute respiratory syndrome 2 (SARS2), while nsp2 and nsp12 are Alpha or Beta specific. We further identify 78 OC43- and 87 NL63-specific epitopes, and, for a subset of those, we assess the T cell capability to cross-recognize sequences from representative viruses belonging to AlphaCoV, sarbecoCoV, and Beta-non-sarbecoCoV groups. We find T cell cross-reactivity within the Alpha and Beta groups, in 89% of the instances associated with sequence conservation >67%. However, despite conservation, limited cross-reactivity is observed for sarbecoCoV, indicating that previous CoV exposure is a contributing factor in determining cross-reactivity. Overall, these results provide critical insights in developing future pan-CoV vaccines.
Keywords: T cells, Coronavirus, OC43, NL63, epitopes, common cold, cross-reactivity, sarbeco, CCC, HuCoV
Graphical abstract
Highlights
-
•
CD4 T cells recognize six immunodominant common cold coronavirus (CoV) antigens
-
•
Identify 165 epitopes and test 18 of these for cross-reactivity across CoVs
-
•
In 89% of instances, cross-reactivity was predicted by sequence conservation >67%
-
•
Previous CoV exposure likely affects T cell cross-reactivity to other CoVs
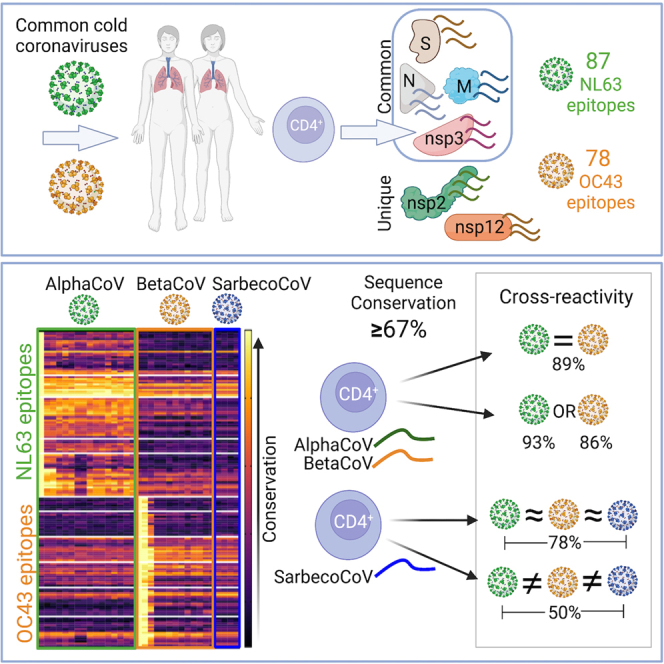
Tarke et al. investigate CD4 T cells in common cold coronaviruses (CCCs) and find S, N, M, and nsp3 antigens are commonly recognized also in SARS2; 165 CCC CD4 T cell epitopes are identified. Cross-reactivity is observed between coronavirus (CoV) sequences with >67% homology in Alpha and Beta CoV groups.
Introduction
Coronaviruses (CoVs) remain a general concern because of their pandemic potential, as illustrated not only by the severe acute respiratory syndrome coronavirus (SARS-CoV)-2 pandemic but also by previous CoV outbreaks, including SARS-CoV and the Middle East respiratory syndrome coronavirus (MERS-CoV). In this context, the development of a pan-CoV vaccine to preemptively provide adaptive immunity against the threat of an upcoming CoV outbreak resulting from zoonotic spillover from an animal reservoir species into humans is of interest.1 Indeed, all CoVs associated with the recent outbreaks had zoonotic origins, infecting bats, pigs, pangolins, and rodents before being transferred to humans. Zoonotic and human coronaviruses, are broadly classified in two main genera: Alpha and Beta. The Beta coronaviruses are subdivided into additional subgenera, with the Beta sarbecoCoV group being of the most pandemic concern.2 Compared with AlphaCoV, the BetaCoV genus has been evolutionary more prolific with multiple subgenera infecting humans with various degrees of phylogenetic relation, including merbecoCoV (MERS-CoV) and sarbecoCoV (SARS and SARS-CoV-2).2,3,4 Within AlphaCoV, two CoVs infect humans seasonally (HCoV-229E and HCoV-NL63), and, within the BetaCoV, HCoV-HKU1 and HCoV-OC43 cause common colds upon infection, with cyclical and alternating patterns of prevalence in different populations and geographical locations.5 These four common cold coronaviruses (CCCs) are prevalent worldwide and usually cause mild illness primarily affecting the upper respiratory tract.5
Despite the seasonality and prevalence of common cold viruses, scarce data were available regarding general immunity, with a prevalent focus on humoral immunity and no information on cellular immunity before the pandemic.6 In contrast, both components of adaptive immunity have been more investigated in the context of the sarbecoCoV, with particular emphasis on SARS-CoV-2.1,7,8
In the context of SARS-CoV-2 and with particular emphasis on cellular immunity, several studies have shown that early broad and polyantigenic T cell responses play a potential part in the resolution of SARS-CoV-2 infection and COVID-19.7,9,10,11 In non-human primates, T cells can also contribute to the reduction of SARS-CoV-2 viral loads.12 When individuals with agammaglobulinemia and B cell-depletion are infected with SARS-CoV-2, there is only a small increase in the risk of hospitalization,13 indicating the T cells could be providing protection against more severe disease. Indeed, individuals with multiple sclerosis (MS) who are without antibody responses while treated with ocrelizumab exhibit mild COVID-19 upon infection, suggesting that COVID-19 can be modulated without antibody responses.14,15 Furthermore, the preservation of T cell reactivity against variants in which binding of neutralizing antibodies is impaired correlates with the preservation of protection against severe disease but decreased protection from infection. Specifically, several studies showed that T cell responses were largely preserved at the population level to SARS-CoV-2 variants, including Omicron.16,17,18,19,20,21,22
The impact of cross-reactive T cells recognizing viral variants has also been reported in the influenza system. It has been shown that pre-existing T cell immunity to influenza correlates with disease protection,23,24 including protection from symptomatic infection.25 Additionally, influenza-specific T cell epitopes have been identified and shown to be recognized by multiple donors and conserved in multiple influenza strains.26 In the absence of cross-reactive neutralizing antibodies, conserved virus-specific T cell responses correlated with cross-protection from symptomatic influenza.27 This evidence points to the potential value of cross-reactive T cells in the context of influenza viral infection and that could be applicable to other viral infections, including coronaviruses.
In this context, several studies have reported pre-existing cross-reactive memory T cells associated with CCC, and pre-existing T cells were shown to associate with milder disease and better vaccination responses.7,28,29,30,31,32 Specifically, T cell responses to SARS-CoV-2 have been identified in unexposed subjects,7,28,33,34,35 which, in some instances, have been shown to map to cross-reactive recognition of the SARS-CoV-2 sequences by T cells induced by endemic CCC.36,37,38,39 SARS-CoV-2-specific T cells were also able to cross-recognize other human CoVs, including SARS-CoV-1 and MERS-CoV,40,41,42,43,44 and other viral species.29,45,46 Based on these data, it has been proposed that immunodominant T cell regions conserved across CoVs may be of interest for inducing a pan-CoV T cell response,1 to be considered not as an alternative but in conjunction with the induction of broadly reactive antibody responses.47,48
However, while over 100 different studies have investigated the T cell epitope repertoire induced after infection with SARS-CoV-2, as reviewed by Grifoni et al.,49 sparse data are available for other human circulating CoVs. We have recently shown that CCC T cell immunity is readily detectable in the general population with unknown CCC exposure, and showed that it is sustained over time.50 This suggested the feasibility of the study of the pattern of protein immunodominance and T cell epitope repertoire recognized by the general population after CCC exposure. Accordingly, in this study, we define CCC CD4+ T cell targets using NL63 and OC43 as prototypes for Alpha and Beta CCC viruses to study which antigens and epitopes are recognized and to what extent broad T cell responses can be identified and predicted on the basis of sequence conservation.
Results
Characteristics of a cohort of healthy blood donors who donated pre-SARS-CoV-2 pandemic samples
We investigated the pattern of immunodominance in T cell responses using peripheral blood mononuclear cells (PBMCs) isolated from 88 healthy adult participants (Table S1; indicated hereafter as “first cohort”), spanning a wide age range (ages from 19 to 84 years; median 46 years), with a balanced gender ratio (48:52, male:female). The ethnic breakdown was reflective of the demographics of the local enrolled population with a prevalence of white, not Hispanic or Latino (60%), and a 22% representation of other ethnicities; 18% of the cohort has not reported information regarding ethnicity. Human leukocyte antigen (HLA) typing of these donors is presented in Table S2. Blood samples were collected from March 2020 to February 2021, 71% of the samples were collected prior to June 2020, while only 29% were collected between November 2020 and February 2021 when SARS-CoV-2 infection rates were minimal in San Diego (Figures S1A and S1B). Accordingly, and based on epidemiological data on CCC seasonality for the 2019–2021 years,5 we selected NL63 and OC43 as representative prototypes for recent exposure to AlphaCoV and BetaCoV CCC, respectively.
All donors in this cohort were seronegative for SARS-CoV-2 Spike receptor-binding domain (RBD) antibodies at the time of sample collection, ensuring that any responses detected would not be related to SARS-CoV-2 exposure or vaccination. Likewise, as expected based on expected previous CCC exposure, these donors were seropositive for antibodies to the RBD domains of the NL63 and OC43 viruses, as measured by immunoglobulin G (IgG) ELISA (Figure S1C).
Immunodominant antigens recognized by CCC-specific T cell responses
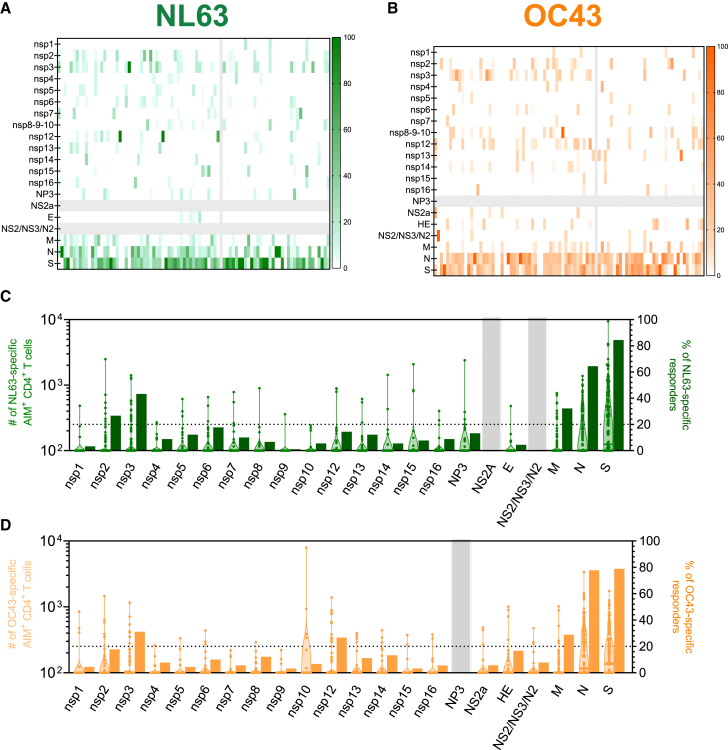
In contrast with the wealth of information available on antigen immunodominance resulting from SARS-CoV-2 infection,49 a systematic analysis of which antigens are dominantly recognized in T cell responses elicited by CCC infection is currently lacking. To define the specific antigens recognized by CD4+ T cells from donors previously exposed to human Alpha and Beta coronaviruses (as determined by NL63 and OC43 seropositivity), we tested PBMCs from the first cohort donors described in Table S1, with sets of overlapping peptides spanning proteins from the entire viral proteome (Figure 1). The same approach was previously used to define the SARS-CoV-2 antigens recognized by T cells33,51 in SARS-CoV-2 convalescent donors. CD4+ T cell responses were measured by the activation-induced marker (AIM) assay utilizing the OX40 and CD137 markers. The gating strategy of the flow cytometry-based AIM assay is detailed in Figure S2A. For each antigen/donor combination, the total magnitude of response is shown as a heatmap to illustrate the donor-to-donor variability (Figures 1A and 1B). Additionally, for each protein, both the total magnitude, calculated by summing all responses observed for a given antigen in the study cohort, and frequency of responses were derived (Figures 1C and 1D).
Figure 1.
CCC-specific CD4+ T cell reactivity per protein
(A–D) PBMCs from healthy donors (n = 88) were analyzed for reactivity against NL63 (green; A and C) and OC43 (orange; B and D).
(A and B) T cell reactivity across the CCC proteome is shown as heatmaps as a function of the donor tested. The x axis shows individual donors’ responses for each protein (y axis).
(C and D) Immunodominance at the antigen level and for the frequency of T cell responders. Magnitude data for each single donor/protein combination are shown as truncated violin plots on the left y axis. The frequency of donors responding to the specific protein are shown as bar plots on the right y axis. See also Figures S1 and S2.
The overall pattern of recognition of NL63 and OC43 was similar; specifically, 20% or more of responses were ascribed to the S, N, M, or nsp3 proteins for both viruses, with S and N proteins most dominantly recognized, followed by M and other non-structural proteins (Figures 1C and 1D). Additionally, nsp2 and nsp12 responses were found to be more frequent in NL63 (Figure 1C) or OC43 (Figure 1D), respectively. These six proteins account for 85% and 81% of the overall responses for NL63 and OC43, respectively (data not shown).
The protein antigens found here to be dominant in CCC responses were similar to those previously shown to be dominantly recognized in the context of SARS-CoV-2 responses.51 In the case of CCC responses, the top six antigens accounted for 80% or more of the Alpha and Beta non-SARS-CoV-2 responses, compared with the eight or nine protein antigens required to cover 80% of the SARS-CoV-2-specific response.51
To assess relative dominance hierarchies, we next plotted the total T cell reactivity detected for each OC43 and NL63 antigen in the current study and compared this with the total reactivity to the various SARS-CoV-2 antigens, previously measured in a cohort of mostly mild COVID-19 convalescent donors using the same methodology.51 The majority of the antigens were similarly recognized in the different viruses, with Spearman correlation analysis (Figure S2B) showing significant positive correlations, especially for OC43/SARS-CoV-2 (R = 0.5649 and p = 0.0095) and NL63/SARS-CoV-2 (R = 0.6140 and p = 0.0067). This is consistent with those two viruses belonging to the BetaCoV genus, and therefore being phylogenetically more similar to each other than with the NL63, which belongs to the AlphaCoV genus.2
The total SARS-CoV-2 response previously observed in SARS-CoV-2 convalescents51 was significantly higher than what was observed with the other two CCC (Kruskal-Wallis; p < 0.0001), consistent with the more recent SARS-CoV-2 exposure in the COVID-19 convalescent cohort, which was analyzed 1 month after infection, compared with the unknown timing of exposure to the other CCCs in the donors analyzed in this study (Figure S2C). Overall, these results provide ant unbiased genome-wide analysis of CD4+ T cell reactivity to two ubiquitous CCCs.
Identification of CCC-derived CD4+ T cell epitopes
The analysis of the NL63 and OC43 proteome-wide immunogenicity pinpointed specific donor-antigen combinations associated with good reactivity for CD4+ T cells that were deconvoluted based on the availability of donor cells. To identify specific CD4+ T cell epitopes, we deconvoluted peptide pools corresponding to the six immunodominant antigens (S, M, N, nsp2, nsp3, and nsp12) identified above as accounting for 80% or more of the NL63 and OC43 CD4+ T cell activity (Figure 1). Epitope deconvolution was performed in at least eight independent donors per antigen. CD4+ T cell epitopes were defined using an HLA-unbiased approach. First, overlapping peptides spanning the entire sequence of the antigen in question were pooled in intermediate pools of about 10 peptides each and tested for reactivity in the AIM assay. The intermediate pools found to be positive in the AIM assays were then deconvoluted to identify the specific peptides associated with the positive response in a second round of experiments.51 The positivity threshold was defined as >100 net AIM+ cell counts (background subtracted by the average of triplicate negative controls) and a stimulation index (SI) >2, as previously described.51,52
Table S3 provides a summary of the 165 epitopes identified, fairly evenly distributed between NL63 (n = 87) and OC43 (n = 78). Overall, these results provide an unbiased genome-wide CD4+ T cell epitope identification screen to two CCCs.
Selection of a panel of Alpha, Beta, and sarbeco virus representatives of coronaviruses of human concern
We next addressed to what degree the CCC epitopes identified in this study were conserved within different coronavirus species, with the ultimate goal of identifying CCC-specific epitopes that would be predicted to cross-react with other Alpha and Beta coronaviruses, and potentially broadly reactive with other different HCoVs, including sarbecoCoV of potentially pandemic concern, as well. Several studies have reported pre-existing memory SARS-CoV-2 CD4+ T cell reactivity in unexposed donors and have shown cross-reactivity between SARS-CoV-2 and CCC sequences,33,35,36,39 demonstrating the presence of T cell memory clones able to cross-recognize multiple HCoVs.
We selected a representative set of viruses according to the criteria summarized in Figure S3. The selection included clustering based on genomic sequence identity, sorting clusters based on cluster size, and a phylogeny and metadata-based sampling method to select representatives (blue) in major phylogenetic clusters. A total of 33 sequences were selected, including the prototype sequences used to identity NL63 and OC43 epitopes. Of those, 16 sequences were selected to represent the AlphaCoV genus and 17 to represent BetaCoV genus. The BetaCoV were further divided into a group of four sequences specifically related to the subgenus sarbecoCoV and a group of 13 non-sarbecoCoV (Table 1).
Table 1.
List of the representative CoVs
| Subgenus | GenBank Sequence ID | Source | Isolate | Host | |
|---|---|---|---|---|---|
| AlphaCoV (n = 16) | NC_005831 | coronavirus NL63 | Amsterdam I | human | |
| NC_002645 | coronavirus 229E | 229E | human | ||
| NC_028752 | isolate camel/Riyadh/Ry141/2015 | camel/Riyadh/Ry141/2015 | camel | ||
| NC_009657 | Scotophilus bat coronavirus 512 | BtCoV/512/2005 | bat | ||
| NC_018871 | Rousettus bat coronavirus HKU10 | 183A | bat | ||
| MH687935 | Alphacoronavirus species | VZ_AlphaCoV_16715_24 | bat | ||
| NC_009988 | Rhinolophus bat coronavirus HKU2 | HKU2/GD/430/2006 | bat | ||
| NC_028824 | BtRf-AlphaCoV/YN2012 | BtRf-YN2012 | bat | ||
| NC_010437 | bat coronavirus 1A | AFCD62 | bat | ||
| NC_010438 | Miniopterus bat coronavirus HKU8 | AFCD77 | bat | ||
| KJ473798 | BtMf-AlphaCoV/HuB2013 | BtMf-HuB2013 | bat | ||
| NC_048216 | NL63-related bat coronavirus | BtKYNL63-9b | bat | ||
| NC_022103 | bat coronavirus CDPHE15/USA/2006 | bat/USA/CDPHE15/2006 | bat | ||
| NC_046964 | bat-CoV/P.kuhlii/Italy/3398-19/2015 | bat-CoV/P.kuhlii/Italy/3398-19/2015 | bat | ||
| NC_028814 | BtRf-AlphaCoV/HuB2013 | BtRf-HuB2013 | bat | ||
| MK720945 | bat coronavirus HKU32 | TCL26A | bat | ||
| Beta (n = 17) | non-sarbeco CoV (n = 13) | NC_019843 | Middle East respiratory syndrome-related coronavirus (MERS-CoV) | HCoV-EMC/2012 | human |
| NC_038294 | Betacoronavirus England 1 | H123990006 | human | ||
| NC_003045 | bovine coronavirus | BCoV-ENT | bovine | ||
| NC_006213 | coronavirus OC43 (HCoV-OC43) | OC43 | human | ||
| NC_006577 | coronavirus HKU1 | HKU1 | human | ||
| NC_039207 | Erinaceus/VMC/DEU/2012 | ErinaceusCoV/2012-174/GER/2012 | hedgehog | ||
| NC_009019 | Tylonycteris bat coronavirus HKU4 | HKU4-1 B04f | bat | ||
| NC_009020 | Pipistrellus bat coronavirus HKU5 | HKU5-1 LMH03f | bat | ||
| MT337386 | coronavirus BtRt-BetaCoV/GX2018 | MCL_19_bat_606_2 | bat | ||
| KC869678 | coronavirus Neoromicia/PML-PHE1/RSA/2011 | Neoromicia/PML-PHE1/RSA/2011 | bat | ||
| MG596802 | MERS-CoV | bat-CoV/H.savii/Italy/206645-40/2011 | bat | ||
| HM211100 | bat coronavirus HKU9-10-1 | HKU9-10-1 | bat | ||
| HM211101 | bat coronavirus HKU9-10-2 | HKU9-10-2 | bat | ||
| sarbecoCoV (n = 4) | MK211374 | coronavirus BtRl-BetaCoV/SC2018 | BtRl-BetaCoV/SC2018 | bat | |
| MT121216 | pangolin coronavirus | MP789 | pangolin | ||
| NC_004718 | SARS coronavirus Tor2 | Tor2 | human | ||
| MT952134 | SARS-CoV-2 | SARS-CoV-2/human/USA/WA-CDC-WA1/2020 | human | ||
Sequence conservation of CCC CD4+ T cell epitopes in other CoVs
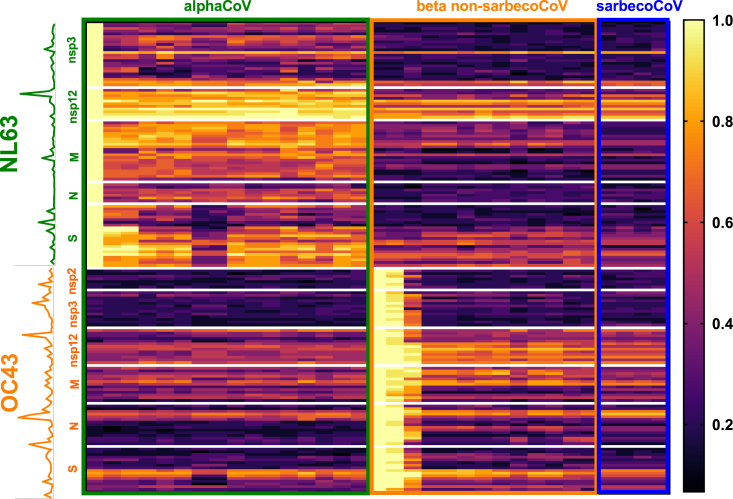
We first calculated the degree of conservation (percentage sequence conservation) of the sequences of each of the NL63 and OC43 T cell epitopes in each of the representative sequences (Figure S4). We next calculated the median overall conservation for each of the epitopes and the median conservation within AlphaCoV, sarbeco, and non-sarbeco BetaCoV groups (Table S3). The results are also graphically summarized in Figure 2. Information regarding the protein location of each epitope and the total magnitude of responses associated with each epitope is shown in the vertical line graph.
Figure 2.
Epitope conservation across coronaviruses
Heatmap of conservation of NL63 (green text) and OC43 (orange text) T cell epitopes in each of the coronavirus representative sequences divided into AlphaCoV (n = 16; green outline), BetaCoV non-sarbecoCoV (n = 13; orange outline), and sarbecoCoV (n = 4; blue outline) groups. Each row represents a different epitope, and each column a different coronavirus representative. The color intensity, following the gradient shown on the right of the heatmap, shows the degree of calculated sequence conservation for each epitope/virus combination. The y axis shows information regarding the protein of each epitope, and a line graph illustrates the total response associated with each epitope. See also Figures S3 and S4.
Overall, NL63 epitopes showed the highest degree of conservation across AlphaCoV representative sequences, with some specific epitopes, especially in the nsp12 protein, associated with broad conservation across all HCoVs. OC43 epitopes show the broadest conservation, particularly for nsp12, followed by specific sub-regions pertaining to M, N, and S proteins. Thus, the nsp12 protein is associated with the highest combined level of conservation and immunogenicity, for both CCC viruses analyzed, making it a potential candidate to stimulate broadly cross-reactive T cell responses. In summary, the combined experiments identify a number of different epitopes and antigen regions, derived from CCC, immunogenic in humans, and with different degrees of conservation in coronaviruses of human concern.
Selection of a panel of CCC T cell epitopes to investigate cross-recognition within other CoVs
We next experimentally investigated whether T cells specific for CCC epitopes could cross-recognize peptides corresponding to the different representative coronaviruses described above. To address this point, we generated specific T cell lines expanded with a single epitope known to be recognized ex vivo by a given donor and asked the question of how broadly a T cell line specific for a given CCC epitope can recognize the homologous sequences encoded in representative CoVs. This approach expands the epitope-specific memory T cells and allows a stringent determination of T cell cross-reactivity based on the specific peptide investigated. In addition to providing direct evidence for cross-reactivity within CoVs, this set of experiments was designed to determine how frequently cross-reactive recognition by human memory T cells of coronavirus sequences could be observed, and which level of homology would correlate with cross-reactivity. Previous studies had indicated a level of conservation of 67% (0.67) as being associated with cross-reactivity of SARS-CoV-2 reactive T cells with CCC sequences.36
To address these points, we calculated the median conservation for each of the three groups analyzed (AlphaCoVs, Beta non-sarbecoCoVs, and sarbecoCoVs) and selected for analysis 18 representative epitopes, defined as the sequence for which reactivity was detected in ex vivo experiments, associated with different degrees of conservation in the different coronavirus groups (Alpha, Beta non-sarbeco, and sarbeco). Table 2 details the epitopes selected, with the first column reporting the virus species from which the epitope was identified and the specific protein and residues. The next columns describe the median conservation in the different viral groups. More specifically, 11 of the 18 representative epitopes were conserved within AlphaCoV, nine epitopes were conserved within Beta non-sarbecoCoV, and eight epitopes were conserved within sarbecoCoV with a sequence identity >0.67 (Table 2). Based on their pattern of conservation, as indicated by the next column in Table 2, five epitopes were classified as common (that is, conserved in all three viral groups), five epitopes were conserved only in AlphaCoV, two conserved only in BetaCoV (not conserved in Alpha, but conserved in all BetaCoV), two peptides were conserved only in BetaCoV-non-sarbeco, and one peptide was conserved only in sarbeco viruses. The last two peptides had sequence identities of <0.67 and were not well conserved in any of these groups.
Table 2.
List of representative epitope candidates and experimental outcome of TCLs
| TCL reactivity |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Conservation (%) |
First cohort |
Validation cohort |
|||||||
| CCC | Epitope | AlphaCoV | Beta non-sarbecoCoV | SarbecoCoV | Conservation category | Alpha/Beta reactivity | Sarbeco reactivity | Alpha/Beta reactivity | Sarbeco reactivity |
| NL63 | nsp124476 | 0.87 | 0.73 | 0.73 | common | TCL-only | – | ND | ND |
| NL63 | nsp124896 | 1 | 0.87 | 0.87 | common | Alpha/Beta | yes | ND | ND |
| OC43 | N121 | 0.67 | 0.8 | 0.87 | common | Alpha/Beta | – | Alpha/Beta | yes |
| NL63 | nsp31286 | 0.87 | 0.73 | 0.8 | common | Alpha/Beta | – | ND | ND |
| OC43 | S911 | 0.67 | 0.73 | 0.73 | common | Alpha/Beta | yes | Alpha/Beta | yes |
| NL63 | S956 | 0.87 | 0.47 | 0.33 | Alpha | Alpha | – | ND | ND |
| NL63 | S951 | 0.87 | 0.27 | 0.13 | Alpha | Alpha | – | Alpha/Beta | – |
| NL63 | nsp124136 | 0.87 | 0.33 | 0.27 | Alpha | Alpha | – | ND | ND |
| NL63 | N121 | 0.73 | 0.53 | 0.6 | Alpha | ND | ND | Alpha/Beta | yes |
| NL63 | M111 | 0.73 | 0.47 | 0.47 | Alpha | ND | ND | Alpha | – |
| NL63 | nsp124731 | 0.8 | 0.33 | 0.4 | Alpha | ND | ND | Alpha | yes |
| OC43 | N116 | 0.47 | 0.73 | 0.73 | Beta | Beta | yes | Beta | yes |
| OC43 | nsp124731 | 0.47 | 0.73 | 0.73 | Beta | Beta | – | – | ND |
| OC43 | M36 | 0.33 | 0.73 | 0.4 | non-sarbeco | Alpha | – | ND | ND |
| OC43 | M46 | 0.47 | 0.73 | 0.53 | non-sarbeco | ND | ND | Beta | – |
| OC43 | N126 | 0.53 | 0.53 | 0.67 | sarbeco | Alpha | – | ND | ND |
| OC43 | S951 | 0.33 | 0.4 | 0.47 | none | ND | ND | Beta | yes |
| OC43 | S956 | 0.33 | 0.6 | 0.6 | none | ND | ND | Beta | yes |
ND, not determined (indicates not tested or lack of response of the TCL). Dash indicates no T cell reactivity.
Assessment of cross-reactivity patterns of CCC CD4+ T cell epitopes within other CoVs
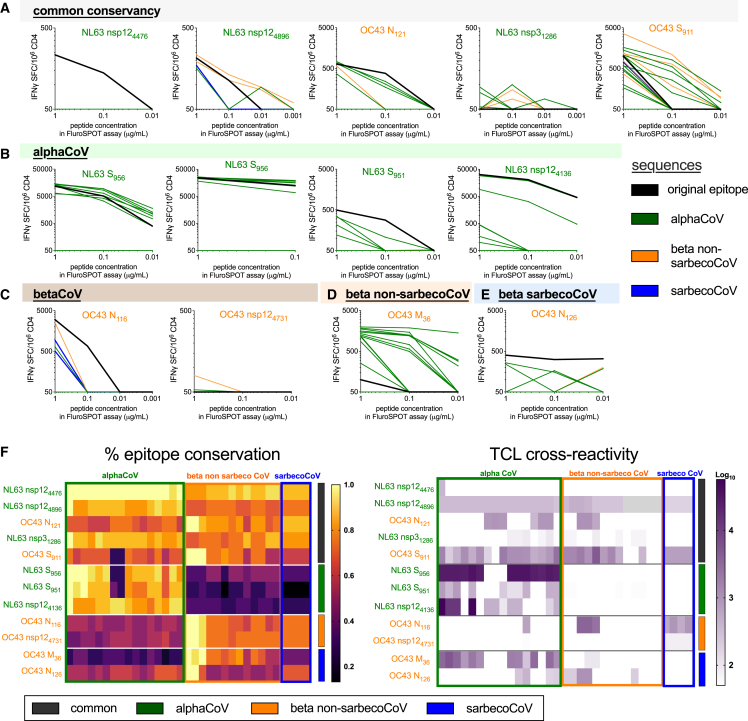
Next, we determined the pattern of cross-reactivity of T cells recognizing the various epitopes by generating epitope-specific short-term T cell lines (TCLs) from PBMCs of a subset of donors from the first cohort. This first round of experiments investigated the reactivity of TCLs specific for 12 different epitopes. These TCLs were tested with a dose range of synthetic peptides corresponding to the sequence of the homolog peptides from each of the virus isolates from Table 1, as previously reported.36 The specific TCL reactivity for each epitope and virus sequence is shown in Figures 3A–3E. Reactivity against AlphaCoV sequences is shown in green, Beta non-sarbecoCoV in orange, and sarbecoCoV in blue. The reactivity of epitopes conserved in all three groups is shown in Figure 3A, Alpha specific (conserved only in Alpha-corona) in Figure 3B, Beta specific (conserved only in Beta-corona) in Figure 3C, Beta non-sarbeco specific in Figure 3D, and sarbeco specific in Figure 3E. The results are also summarized as heatmaps in Figure 3F, depicting the epitope conservation across each viral species and the reactivity of each TCL against each peptide.
Figure 3.
Cross-reactivity of NL63 and OC43 epitopes and homologous CoVs peptides
(A–F) Twelve CCC epitopes were used to stimulate donor PBMCs (n = 12) and generate short-term T cell lines (TCLs) as described in the STAR Methods. These epitopes were selected based on the primary screen with the first cohort, and the TCLs were generated with the same NL63 or OC43 donor-epitope combinations. In these graphs, the TCLs are categorized based on the original epitope selected and predicted on the basis of median sequence conservation >67% to be common (A) or specific for AlphaCoV (B), BetaCoV (C), and further segregating the BetaCoV into non-sarbeco (D) and sarbeco (E) groups. After 14 days of in vitro expansion, each TCL was tested with the CCC epitope used for stimulation (black lines in A, B, and C) and peptides corresponding to analogous sequences from other CoVs (green, AlphaCoV; orange, Beta non-sarbecoCoV; and blue, sarbecoCoV) at three different concentrations (1, 0.1, and 0.01 μg/mL). Interferon gamma (IFNγ) SFCs/106 PBMCs are plotted for TCLs stimulated with each peptide. Sequence identity and overall reactivity are shown as heatmaps in (F). The heatmap for the TCL reactivity represents the log10 scale of the sum reactivity for the three concentrations of peptide tested. The vertical bars in (F) and (G) refer to the sequence conservation (dark gray, common; green, AlphaCoV; orange, Beta non-sarbecoCoV; and blue, sarbecoCoV). See also Figures S3 and S4.
The outcome of these experiments is also summarized in Table 2 under the heading “first cohort.” Overall, seven out of eight epitopes conserved in AlphaCoVs showed T cell reactivity against AlphaCoVs, and six out of eight epitopes conserved in Beta-non-sarbecoCoVs showed T cell reactivity for Beta non-sarbecoCoVs. However, only three out of eight epitopes conserved in Beta-non-sarbecoCoVs showed T cell reactivity for sarbecoCoVs. In conclusion, the results show that cross-reactivity patterns are predicted by sequence conservation for AlphaCoV and Beta non-sarbecoCoVs. Conversely, cross-reactivity within sarbecoCoVs that are phylogenetically more distant from other Beta non-sarbecoCoVs was not well predicted by sequence conservation. Indeed, only SARS-CoV-2 was circulating in humans at the time the samples were obtained, and all donors in this cohort were seronegative for SARS-CoV-2.
Patterns of T cell cross-recognition of other CoVs in an independent cohort
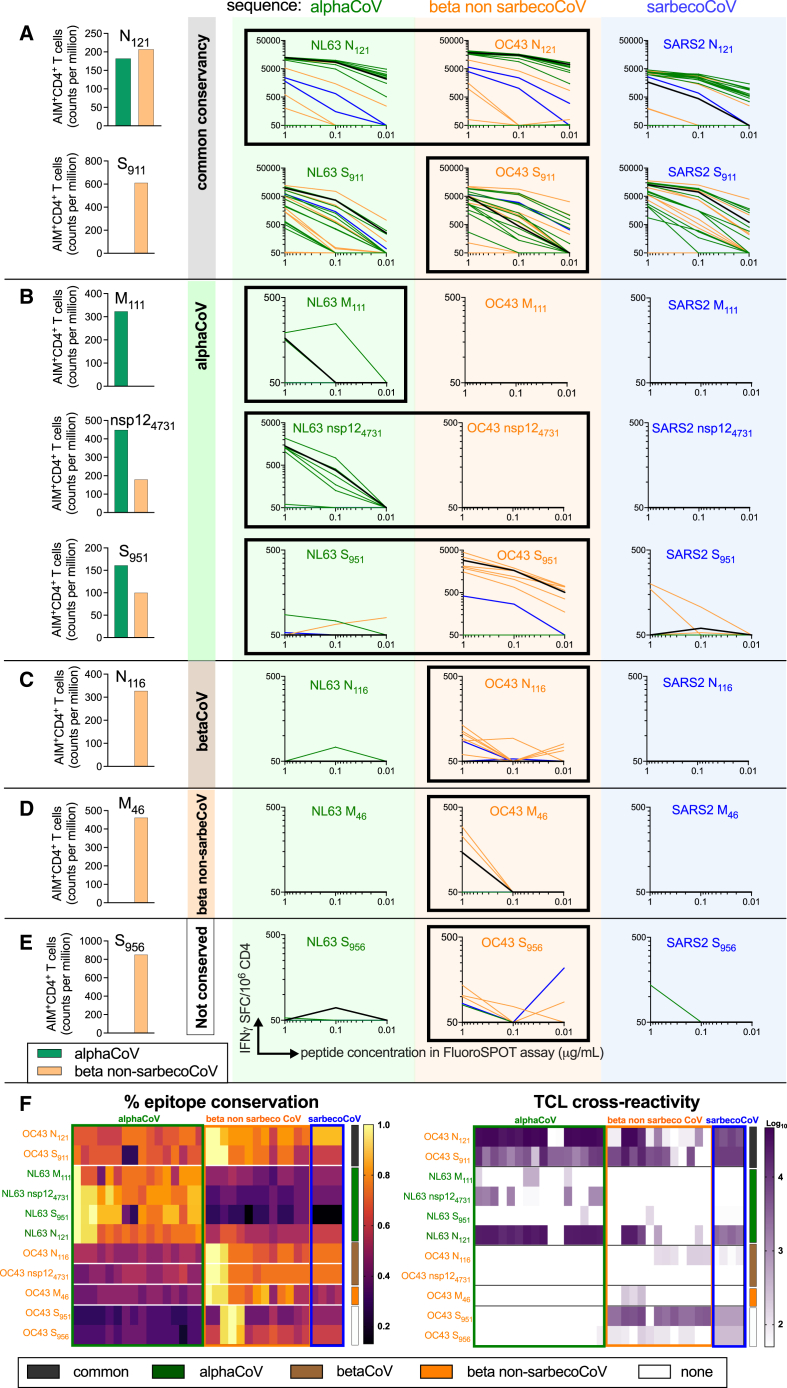
The results presented above show that while cross-reactivity for the AlphaCoV and Beta-non-sarbecoCoV groups could be predicted based on the known sequence conservation, sarbecoCoV reactivity was infrequent and not readily predicted based on sequence conservation. In the next round of experiments, we sought to verify these results in an independent cohort and also investigate whether using AlphaCoV, Beta-non-sarbecoCoV, or sarbecoCoV sequences for the in vitro peptide stimulation might modulate the cross-reactivity patterns. These experiments were performed with a new validation cohort (Table S1) as additional PBMCs from the previous cohort were largely not available. PBMC samples from the validation cohort were collected between October 2018 and August 2019 to ensure that donors had not been exposed to the sarbeco virus SARS-CoV-2 (Table S1). Preliminary experiments determined the ex vivo reactivity in the validation cohort of previously identified epitopes. In these experiments, we tested both peptides corresponding to the OC43 and NL63 sequences and for eight epitopes we detected reactivity to either the AlphaCoV or BetaCoV peptide version, or both (Figures 4A–4D; left-most part of the graphs).
Figure 4.
Cross-reactivity as a function of the initial peptide used for TCL generation
(A–F) Shown here are eight epitopes that had an ex vivo response in the validation cohort of healthy donors (n = 7). (A–D) The bar graphs show the T cell responses ex vivo to the NL63 (green) and OC43 (orange) epitopes for each donor as quantified by AIM assay. Subsequently, we generated three TCLs for each donor based on NL63 (green), OC43 (orange), and SARS-CoV-2 (blue) prototype peptide sequences. The epitopes that originally yielded responses by AIM assay for these donors are indicated by the black highlighted margin around the TCL graphs. After 14 days of in vitro expansion, each TCL was tested with the CCC epitope used for stimulation and peptides corresponding to analogous sequences from other CoVs at three different concentrations (1, 0.1, and 0.01 μg/mL). IFNγ SFCs/106 PBMCs are plotted for TCLs stimulated with each peptide. Common (A), AlphaCoV-specific (B), BetaCoV-specific (C), Beta-non-sarbecoCoV-specific (D), and non-conserved (E) categories were selected based on predicted sequence identity. Sequence identity and overall reactivity are shown as heatmaps in (F). The heatmap for the TCL reactivity represents the log10 scale of the sum reactivity for the three concentrations of peptide tested. The vertical bars in (F)–(G) refer to the sequence conservation (dark gray, common; green, AlphaCoV; orange, Beta non-sarbecoCoV; and blue, sarbecoCoV). See also Figures S3 and S4.
We then generated for each donor-peptide combination separate TCLs using the prototype NL63 or OC43 peptides, and we then tested for cross-reactivity with the other AlphaCoV and BetaCoV sequences. We also used SARS-CoV-2 peptide sequences to generate TCL, and the results are described in the following section. The specific TCL reactivity for each epitope and virus sequence is shown in Figures 4A–4D. Reactivity against AlphaCoV sequences is shown in green, Beta non-sarbecoCoV in orange, and sarbecoCoV in blue. The reactivity of epitopes conserved in all three groups is shown in Figure 4A, Alpha specific (conserved only in AlphaCoV) in Figure 4B, Beta specific (conserved only in BetaCoV) in Figure 4C, Beta non-sarbeco specific in Figure 4D, or not conserved in Figure 4E. The results are also summarized as heatmaps in Figure 4F, depicting the epitope conservation across each viral species and the reactivity of each TCL against each peptide. The outcome of these experiments is also summarized in Table 2 under the heading “validation cohort.”
We first analyzed the reactivity of TCLs generated by in vitro stimulation with the same peptide epitopes for which ex vivo reactivity was detected in that specific donor; these instances are highlighted by a black highlighted margin around the graphs in Figure 4. In those instances, in the case of epitopes that were predicted to be conserved across CoVs, two out of two instances showed cross-reactivity for AlphaCoV, Beta non-sarbecoCoV, and sarbecoCoV when the homologous epitope was used for the TCL generation (Figure 4A). Three epitopes with sequence conservation within AlphaCoV had ex vivo reactivity with the AlphaCoV NL63 sequence and also showed cross-reactivity to other AlphaCoV sequences after TCL expansion (Figure 4B). The NL63 M111 epitope is Alpha specific at the level of conservation and the TCL was associated with a predominant Alpha-specific reactivity (Figure 4B).
In the case of nsp124731 and S951, ex vivo reactivity was detected for both the NL63 and OC43 epitopes, potentially reflective of either multiple exposures and/or cross-reactivity. For nsp124731, the NL63 sequence is conserved within Alpha but not Beta (AlphaCoV = 0.8; Beta non-sarbecoCoV = 0.33), and conversely the OC43 sequence is conserved within Beta but not Alpha (AlphaCoV = 0.47; BetaCoV = 0.73) (Table 2). The TCL obtained by NL63 in vitro stimulation was Alpha specific, while no in vitro expansion was noted in the case of the TCL stimulated with the OC43 sequence (Figure 4B). In the case of S951, the TCL expanded with the OC43 epitope was mostly Beta reactive, and, conversely, the TCL expanded with the NL63 epitope was Alpha-reactive (Figure 4B). These results are consistent with the fact that the NL63 S951 epitope is well conserved within Alpha but not Beta (AlphaCoV = 0.87; Beta non-sarbecoCoV = 0.27) (Table 2). In contrast, the OC43 S951 epitope is not well conserved within Alpha or Beta (AlphaCoV = 0.33; Beta non-sarbecoCoV = 0.4) (Table 2); however, the Beta-specific cross-reactivity could be attributed to the subset of BetaCoV sequences that are conserved for this epitope, as seen in Figure 4F.
The BetaCoV-specific OC43 N116 epitope induced Beta-specific cross-reactivity as expected, based on sequence conservation and ex vivo reactivity (Beta-non-sarbecoCoV = 0.73; sarbecoCoV = 0.73; Figure 4C; Table 2). The Beta-non-sarbeco conserved epitope OC43 M46 was associated with OC43 reactivity ex vivo, and the associated TCL displayed predominant Beta-non-sarbeco reactivity (Figure 4D and Table 2). The S956 epitope was associated with OC43 Beta reactivity in the specific donor tested. Consistent with this observation, the TCL maintained this pattern of prevalent Beta reactivity (Figure 4E). This result is consistent with the fact that the OC43 S956 epitope is well conserved within some Beta but not Alpha (AlphaCoV = 0.4; Beta non-sarbecoCoV = 0.5) (Table 2).
Overall efficacy of sequence conservation and pre-existing reactivity as a predictor of coronavirus cross-reactivity
The overall data from the first and validation cohorts combined were evaluated for parameters that might guide selection of epitopes linked with cross-reactive T cell responses. Table 3 details how frequently sequence conservation of 67%, or above, was associated with experimentally verified cross-reactivity in each of the taxonomic coronavirus groups. When the three groups are considered together, the 67% median sequence conservation threshold predicts T cell cross-reactivity in 78% of the cases (considering 13 + 12 + 6 = 31 instances of cross-reactivity/15 + 13 + 12 = 40 tested). When only AlphaCoV and Beta non-sarbecoCoV are considered, cross-reactivity is correctly predicted in 89% of cases, while in only BetaCoV (Beta non-sarbecoCoV and sarbecoCoV) the prediction accuracy is 69%. When we look within each group separately, the predictive capacity is 93% and 86% if only Alpha or Beta non-sarbecoCoV groups are considered. This is in contrast with the fact that experimental T cell cross-reactivity was observed only in 50% of the cases of sequences conserved within sarbecoCoV (Table 3).
Table 3.
Summary of sequence conservation and TCL cross-reactivity as predictor of CoV cross-reactivity
| Conserved (>67%) | Cross-reactive | % correct | |
|---|---|---|---|
| Alpha | 14 | 13 | 93 |
| Beta non-sarbeco | 14 | 12 | 86 |
| Sarbeco | 12 | 6 | 50 |
| All | 40 | 29 | 73 |
| Alpha + Beta non-sarbeco | 28 | 25 | 89 |
| Beta non-sarbeco + sarbeco | 26 | 18 | 69 |
In conclusion, the results show that cross-reactivity patterns are predicted by sequence conservation for AlphaCoV and Beta non-sarbecoCoVs. Conversely, cross-reactivity with sarbecoCoVs (which were not circulating in humans at the time the samples were obtained) was not well predicted by sequence conservation. Relevant to the hypothesis that pre-exposure might influence the capacity of experimentally observing T cell cross-reactivity, we examined the association between ex vivo reactivity and measured cross-reactivity in the expanded TCLs. Indeed, when we consider the OC43 and NL63 epitopes in Figure 4, for which ex vivo reactivity was experimentally determined, ex vivo reactivity was detected in 11 instances, of which nine showed cross-reactivity in the expanded TCLs. Of the five epitopes for which ex vivo reactivity was not detected prior to in vitro expansion to derive specific TCLs, only one yielded cross-reactive TCLs (p = 0.0357 by the Fisher exact test). This observation demonstrates a correlation of ex vivo reactivity with cross-reactivity after TCL expansion.
As stated above, in parallel experiments we also used SARS-CoV-2 peptide sequences for re-stimulation to examine whether this approach could increase the frequency and extent of CCC cross-reactivity within sarbecoCoV sequences. The results are shown in Figure 4 and summarized in Table 2.
Overall, when either NL63 or OC43 sequences were used for re-stimulation, sarbecoCoV cross-reactivity was noted in three out of 12 instances for the first cohort (25%), and in seven out of 16 (44%) for the validation cohort, for an overall frequency of 10 out of 28 (36%). When the SARS-CoV-2 sequences were utilized in the TCL generation, sarbeco cross-reactivity was noted in two out of eight instances (25%). Thus, the use of SARS-CoV-2 sequences to expand TCLs was not associated with an increase in cross-reactivity and was actually associated with a trend toward lower reactivity. This might reflect the possibility that stimulation with CCC epitopes might be most related to the original in vivo immunogen in this pre-pandemic cohort studied, and thereby also a more effective in vitro stimulus for TCL expansion.
Discussion
The data from the present study addresses three main issues: the pattern of antigens recognized by human CD4+ memory T cells recognizing CCC, the relation of the associated epitope repertoire with the SARS-CoV-2-associated epitope repertoire, and to what extent cross-reactive T cell responses between different taxonomic CoV groups can be observed and predicted. In addition, the studies revealed a prominent role of pre-existing immunity as a driver of development of cross-reactive T cell responses.
In terms of the antigens recognized as immunodominant by T cell responses, we were able to systematically evaluate which antigens are recognized by human memory CD4+ T cell responses in Alpha and Beta CCCs. NL63 and OC43 immunodominant proteins included the structural proteins S, M, and N, and the non-structural protein nsp3. This pattern of immunodominance is similar to what was previously observed in the context of SARS-CoV-2, suggesting these antigens are common targets across multiple CoVs.39,49,51 This is in line with the work of others who described immunodominant CD4+ T cell responses to CCC-conserved epitopes,29,35,36,37,38,45,53,54,55,56,57,58 which have been previously reviewed.30,59 Thus, the dominance of these antigens in coronavirus recognition might reflect conserved and common mechanisms, such as high levels of expression.33
Our study also revealed interesting features of immunodominance specific to CCCs. NL63-specific T cell responses prominently recognized nsp2, and OC43-specific T cells recognized nsp12, previously described in the SARS-CoV-2 context49,51 as recognized by cross-reactive T cells in the context of asymptomatic SARS-CoV-2 infection in healthcare workers.60 Differences in the profile of antigens recognized by different coronaviruses is also to be expected given that some antigens are specifically encoded in some but not other coronavirus genomes, such as the NL63 NP3 protein, which is not found in OC43 or SARS-CoV-2, and the OC43 NS2a protein, which is not encoded in NL63 or SARS-CoV-2 (see STAR Methods for virus sequence information). While these antigens could perhaps have some diagnostic value, our data suggest that they are relatively minor targets for human T cells. Conversely, the fact that certain antigens are broadly conserved targets for human T cell recognition supports the notion that these antigens could be utilized to elicit broadly reactive responses.
The present study also provides the epitope repertoire associated with human CCC-specific memory CD4+ T cells. We found an average breadth of seven epitopes (range 1–24) per donor being recognized. This is 2- to 3-fold fewer than what we previously observed in the case of SARS-CoV-2, where an average of 19 CD4 epitopes/donor (range 1–63) were identified using a similar experimental strategy.51 The lower number of epitopes detected for the CCC is likely a reflection of the fact that the SARS-CoV-2 epitope identification studies were performed 2 months after SARS-CoV-2 infection,51 while the present CCC studies were performed in subjects of unknown and presumably less recent exposure. The identified epitopes were associated with predicted promiscuous binding capacity to a panel of frequent HLA alleles, confirming data obtained in several other systems61,62,63,64 and utilized to develop algorithms to predict dominant CD4+ T cell epitopes.65,66,67
The epitope repertoire identified by ex vivo reactivity in NL63 and OC43 encompassed a total of 165 epitopes. These epitopes were largely undescribed, with only 10 epitopes overlapping with epitopes previously described in the Immune Epitope Database (IEDB),68 even by allowing a rather loose criterion of 67% sequence homology. The overlap between the repertoire of epitopes recognized in NL63 and OC43 in the current study, as also defined by a 67% sequence homology or more, was limited to two epitopes. Furthermore, of relevance to the issue of T cell cross-reactivity across different coronaviruses, discussed below, the NL63 and OC43 epitope repertoire was largely non-overlapping (six epitopes; 4% overlap) with the repertoire of SARS-CoV-2-specific CD4+ T cell responses previously described.51 This is consistent with previous observations that described pre-existing cross-reactive memory SARS-CoV-2 responses from unexposed individuals that share sequence homology with CCC,33,36,39 but also that the T cell repertoire that develops upon SARS-CoV-2 infection is largely non-overlapping with the repertoire recognized by pre-existing cross-reactive memory SARS-CoV-2 responses from unexposed individuals.51As discussed above in the introduction, several lines of evidence suggest that T cells play a role in limiting disease severity and terminating infection, in the context of SARS-CoV-2 infection.7,9,10,11 The preservation of T cell reactivity against variants and the impairment of neutralizing antibodies correlates with preservation of protection against severe disease and decreased protection from infection.16,18,19,69 Directly applicable to the present study are the observations, also summarized in the introduction, that pre-existing cross-reactive immunity associated with CCC are beneficial in the context of SARS-CoV-2 infection and vaccination.31,32,60,70 These observations are consistent with studies71 that point out that Hu CoV infections are seasonal and common but associated with limited disease severity, again consistent with a role of T cells in modulating disease rather than preventing infection. Based on this rationale, it has been proposed that expanding T cell-specific for regions conserved across CoVs may be of interest in the context of inducing a pan-CoV T cell response aimed at reducing or preventing severe disease. Based on this rationale, it has been proposed that immunodominant T cell regions conserved across CoVs may be of interest in the context of inducing a pan-CoV T cell response.1
What strategies can be utilized to predict and detect such cross-reactive responses? Bioinformatic analysis of sequence conservation in panels of different viral species was shown to be effective in informing selection of potential cross-reactive T cell epitopes.33,36,67 Cross-reactive epitopes were associated with overall 67% or greater sequence conservation, in agreement with previous studies in the context of SARS-CoV-2 and other viral infections such as Zika virus (ZIKV) and varicella zoster virus (VZV).36,56,72,73 Our results provide the largest dataset available to address this issue, with cross-reactivity data involving 18 different epitopes and 37 TCLs, testing a total of 594 different viral variant epitope sequences. Within the Alpha and Beta-non-sarbeco groups, the degree of sequence conservation was frequently reflected in CD4+ T cell cross-reactivity. Across Alpha and BetaCoV groups, we correctly predicted cross-reactivity in 29 out of 40 CoV-conserved epitopes considered (73% of the cases). This result validates the use of the 67% sequence identity value to predict T cell cross-reactivity. Previous studies on different viral species reported conserved T cell epitopes were also observed. In the influenza system, CD4+ and CD8+ T cell epitopes conservation in multiple influenza strains were reported.26,74 In the context of flaviviruses, despite an overall low degree of cross-reactivity between different flaviviruses,75 such as dengue, yellow fever, and Zika, it has been shown that T cell epitopes conserved among these viruses can protect against disease in animal models.76,77 Overall, cross-reactivity between viruses is more often observed across viruses with closer phylogenetic relations and greater sequence homology.40,44
A remarkably different situation was observed when the cross-reactivity with sarbecoCoV was considered. In this case, there were few instances of T cell cross-reactivity, even in cases with higher sequence conservation, and a total of six out 12 instances of cross-reactivity was observed, corresponding to 50% of cases. The most straightforward explanation of this result in pre-pandemic or SARS-CoV-2-seronegative samples is that previous exposure to Alpha and Beta-non-sarbeco viruses is an important determinant of cross-reactivity alongside the degree of conservation. Indeed, sarbecoCoV are more phylogenetically distant and more different from other BetaCoVs, including OC43. Accordingly, the lack of previous exposure to sarbecoCoVs in these samples, being collected pre-pandemic, was associated with lower degree of cross-reactivity among different representatives of sarbecoCoV sequences, including SARS-CoV-2, and even if those sequences were relatively conserved. Hence, the lower cross-reactivity observed in this study fits with the lack of previous exposure to sarbecoCoVs and suggests this is an important determinant of cross-reactivity alongside the degree of conservation. We then asked if this bias in cross-reactivity using the in vitro expansion was due to the fact that we used only OC43 or NL63 epitopes, and accordingly we simultaneously stimulated the same donor with the three peptide variants and we found that the cross-reactivity was not increased by using SARS-CoV-2 sequences. Indeed, our experiments identify peptides that are associated with cross-reactive capacity. Whether these T cells would naturally expand in individuals infected by different CoVs is a related question. The available data suggest that this is not likely to be the case, and support the notion that immunization with coronavirus cross-reactive regions, such as the ones identified by the approach described herein, might be necessary to avoid the concomitant expansion of non-cross-reactive ones. This is based on the observation that the data in the present study show that cross-reactive T cells are a minor fraction of the overall T cell repertoire, and the NL63 and OC43 epitope repertoire was largely non-overlapping (six epitopes; 4% overlap) with the repertoire of SARS-CoV-2 specific CD4+ T cell responses previously described. These observations are consistent with a previous report51that the minority of epitope repertoire of cross-reactive T cells in pre-pandemic samples36 overlapped with the repertoire of T cells in SARS-CoV-2 convalescent samples.51 These results are consistent with the notion that, although a cross-reactive repertoire is present in unexposed donors, SARS-CoV-2 infection elicits a vast repertoire of additional T cell specificities, and cross-reactive T cells might be diluted post SARS-CoV-2 infection.
Our overall goal is to be able to generate and amplify CD4+ T cells broadly cross-reactive with sarbecoCoVs of zoonotic origin and of potential pandemic concern. Our data suggest that exposure is a contributing factor; therefore, future studies should focus on immunity following SARS-CoV-2 exposure. The large numbers of individuals that have been exposed to SARS-CoV-2 worldwide suggests that this should be a feasible goal. Selecting SARS-CoV-2 epitopes and antigenic regions associated with high immunodominance, indicating their recognition in the general human population, and broad conservation among other sarbecoCoVs and possibly other Beta and Alpha coronaviruses, will give the best opportunity of expanding and eliciting cross-CD4+ T cells broadly reactive for a number of different CoV species.
Limitations of the study
One limitation of this study is that the time of the most recent CoV exposure of the donors tested is unknown. We hypothesize that some of the reactivity we observed was affected by the most recent CoV exposure. In this study, our cohort was collected early in the pandemic and only 29% were collected between November 2020 and February 2021. While we used a negative SARS-CoV-2 serology as an additional criterion of selection, we cannot exclude that those samples may have been exposed to SARS-CoV-2 infection.78,79 Additionally, our study did not evaluate cross-reactivity at the level of CD8+ T cells and in SARS-CoV-2-exposed or -vaccinated subjects. Previous studies have shown that the presence of CD4+ pre-existing T cells against SARS-CoV-2 is more readily detected than CD8+ T cells in the context of samples collected pre-pandemic.33 Future studies should investigate further CD8+ T cell epitope repertoires and assess cross-reactivity with other CoVs. Another limitation of this study is that we have not addressed the HLA restrictions of the epitopes identified, which in this specific study are to be defined as immunogenic peptides. Future studies should investigate further the HLA restriction of the currently defined epitopes. An additional limitation is that we did not perform serology tests to check for exposure to other zoonotic CoV species that may contribute to T cell cross-reactivity; however, exposure to other zoonotic CoVs has not been reported in the area of sample collection, and it is reasonable to assume no or minor impact on the study’s results.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-human CD8 BUV496 (clone RPA-T8) | BD Biosciences | Cat# 612942; RRID:AB_2870223 |
| Mouse anti-human CD3 AF700 (clone UCHT1) | BD Biosciences | Cat# 56-0038-42; RRID: AB_10597906 |
| Mouse anti-human CD14 V500 (clone M5E2) | BD Biosciences | Cat# 561391; RRID:AB_10611856 |
| Mouse anti-human CD19 V500 (clone HIB19) | BD Biosciences | Cat# 561121; RRID:AB_10562391 |
| Mouse anti-human CD4 BV605 (clone RPA-T4) | BD Biosciences | Cat# 562658; RRID:AB_2744420 |
| Mouse anti-human CD69 PE (clone FN50) | BD Biosciences | Cat# 555531; RRID:AB_395916 |
| Mouse anti-human CD134 (OX40) PE-Cy7 (clone Ber-ACT35) | BioLegend | Cat# 350012; RRID:AB_10901161 |
| Mouse anti-human CD137 APC (clone 4B4-1) | BioLegend | Cat# 309810; RRID:AB_830672 |
| Biological samples | ||
| Healthy donor PBMCs samples | LJI Clinical Core | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Brilliant Staining Buffer Plus | BD Biosciences | Cat# 566385 |
| Live/Dead Viability Dye eFluor506 | Invitrogen (Thermo Fisher Scientific) | Cat# 65-0866-14 |
| Synthetic peptides | TC Peptide Lab | https://www.tcpeptide.com |
| Software and algorithms | ||
| GraphPad Prism 9 | GraphPad | https://www.graphpad.com/; RRID:SCR_002798 |
| FlowJo 10.8.1 | FlowJo | https://www.flowjo.com/; RRID:SCR_008520 |
| IEDB | Immune Epitope DataBase | https://www.iedb.org; RRID:SCR_006604 |
| ViPR | Virus Pathogen Resource | http://www.viprbrc.org; RID:SCR_010685 |
| Code to identify conserved T cell epitopes | Public Github repository | https://github.com/yzhang2168/t-cell-epitopes |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to the lead contact, Dr. Alba Grifoni, agrifoni@lji.org (A.G.).
Materials availability
Epitope pools used in this study will be made available to the scientific community upon request, and following execution of a material transfer agreement, by contacting Dr. Alba Grifoni (agrifoni@lji.org).
Experimental model and subject details
Human subjects
Blood samples from healthy adult donors were obtained from the San Diego Blood Bank (SDBB) or by donation collected at the La Jolla institute Clinical Core. The study protocol was approved by the local ethic committee under the La Jolla Institute for Immunology Institutional Review Board (IRB#VD-214 and VD-184). All enrolled patients have provided informed consent. Subjects were considered eligible for this study if they fulfilled the SDBB criteria to donate blood and if they were tested and found negative for SARS-CoV-2 RBD IgG serology. Overview of the cohort analyzed is summarized in Table S1. Whole blood was collected from all donors in heparin coated blood bags and processed as previously described (Tarke et al., 2021). Briefly, peripheral blood mononuclear cells (PBMC) were isolated by density-gradient sedimentation using Ficoll-Paque (Lymphoprep, and 90% heat-inactivated fetal bovine serum (FBS; Hyclone Laboratories, Logan, UT) and stored in liquid nitrogen until used in the assays. Each sample was HLA typed by Murdoch University in Western Australia, an ASHI-accredited laboratory. Typing was performed for the class II DRB1, DQB1, and DPB1 loci.
Method details
Peptide pools
Preparation of 15-mer peptides and subsequent megapools and mesopools
To identify CCC-specific T cell epitopes, we synthesized 15-mer peptides overlapping by 10 amino acids and spanning the entire NL63 and OC43 proteomes. All peptides were synthesized as crude material (TC Lab, San Diego, CA) and individually resuspended in dimethyl sulfoxide (DMSO) at a concentration of 20 mg/mL. Aliquots of peptides were either pooled by antigen (megapools; MP) or by ten peptides each (mesopools). The MP required an additional step of sequential lyophilization as previously reported (Carrasco Pro et al., 2015). MPs were resuspended at 1 mg/mL in DMSO, while mesopools were resuspended at 2 mg/mL.
SARS-CoV-2 ELISA
The SARS-CoV-2 RBD ELISA has been described in detail elsewhere (Dan et al., 2021; Grifoni et al., 2020a). Briefly, 96-well half-area plates (ThermoFisher 3690) were coated with 1 μg/mL SARS-CoV-2 Spike (S) Receptor Binding Domain (RBD) and incubated at 4°C overnight. On the following day plates were blocked at room temperature for 2 h with 3% milk in phosphate buffered saline (PBS) containing 0.05% Tween 20. Then, heat-inactivated plasma was added to the plates for another 90-min incubation at room temperature followed by incubation with conjugated secondary antibody, detection, and subsequent data analysis by reading the plates on Spectramax Plate Reader at 450 nm using SoftMax Pro.
CCC ELISA
OC43 and NL63 RBD ELISA were carried out as previously described.80,81 Briefly, coating was performed by Streptavidin (Invitrogen) at 4 μg/mL in Tris-Buffered Saline (TBS) pH 7.4 for 1 h at 37°C followed by blocking with Non-Animal Protein-BLOCKER (GBiosciences). Then biotinylated spike RBD antigens for OC43 and NL63 were added at 1 μg/mL at 37°C for 1 h. All plasma samples were heat-inactivated before usage to minimize risk of residual virus in serum and then incubated at serial dilution followed by multiple washes and incubation with horseradish peroxidase-conjugated secondary Goat Anti-Human secondary IgG (Cat No: 109-035-008, Jackson ImmunoResearch) at 1:40,000 dilution in 3% milk at 37°C for 1 h. The resulting plate was washed and 3,3′,5,5’ -Tetramethylbenzidine (TMB) Liquid Substrate (Sigma-Aldrich) was added for optical density (OD) measurement at 405 nm after stopping the reaction with 50 μL of 1 N HCl.
Flow cytometry
Activation induced cell marker (AIM) assay
The AIM assay for epitope identification was performed mirroring the previously described protocol (Tarke et al., 2021). Cryopreserved PBMCs were thawed in RPMI 1640 media supplemented with 5% human AB serum (Gemini Bioproducts) in the presence of benzonase [20 μL/10 mL]. Cells were stimulated for 24 h in the presence of CCC specific MPs or mesopools at 1 μg/mL and then deconvoluted with 15-mer peptides [10 μg/mL] to reach the epitope level. Stimulation was carried out in 96-wells U bottom plates with 1 × 106 PBMC per well. An equimolar amount of DMSO was used as negative control in triplicates, while stimulation with phytohemagglutinin (PHA, Roche, 1 μg/mL) was included as the positive control. The cells were stained with CD3 AF700 (2:100; Life Technologies Cat# 56-0038-42), CD4 BV605 (1:100; BD Biosciences Cat# 562658), CD8 BUV496 (2:100; Biolegend Cat#612942), CD14 V500 (2:100; BD Biosciences Cat# 561391), CD19 V500 (2:100; BD Biosciences Cat#561121), and Live/Dead eFluor 506 (25:1000; eBioscience Cat# 65-0866-18). Activation was measured by the following markers: CD137 APC (4:100; Biolegend Cat# 309810) and OX40 PE-Cy7 (2:100; Biolegend Cat#350012). All samples were acquired on ZE5 cell analyzer (Bio-rad laboratories) and analyzed with FlowJo software (Tree Star).
In vitro expansion of OC43 and NL63 specific T cells lines (TCLs) and cross-reactivity assessment by FluoroSPOT assays
In vitro expansion of OC43 and NL63 specific T cells was carried out for 14 days to generate epitope-specific T Cell Lines (TCLs). The TCLs were set up using donors selected from the NL63 and OC43 epitope identification screening. The PBMCs were expanded using specific epitope/donor [1 μg/mL] combinations chosen on the basis of the NL63 and OC43 CD4+ T cell epitope screening. IL-2 was added on day 3, 7, and 11. On the 14th day, the cells were harvested and triplicates of 5 × 104 PBMCs were incubated in the presence of the epitope used for expansion and subsequent homologous CoV peptides based on representatives’ sequence selection. Each peptide was tested at 2 to 5 different serial concentrations depending on cells availability after the 14 days of culture (1 μg/mL, 0.1 μg/mL, 0.01 μg/mL, 0.001 μg/mL, and 0.0001 μg/mL) and measured by IFNγ FluoroSPOT assay as previously reported.36 Briefly, cells were incubated in presence of peptide stimulation for 20 h at 37C, 5% CO2 at a concentration of 1 × 105 cells/mL. Cells were then incubated with IFNγ mAb (7-B6-1-BAM Mabtech, Stockholm, Sweden) for 2 h and developed.
Quantification and statistical analysis
Sequence download and quality control
The genome and protein sequences used in this study were downloaded from the Virus Pathogen Resource (ViPR; https://www.viprbrc.org/) and Bacterial and Viral Bioinformatics Resource Center (BV-BRC; https://www.bv-brc.org)82 websites on June 17, 2021. Given the unprecedented number of SARS-CoV-2 genomes, to make the data size more manageable, the SARS-CoV-2 reference dataset (1438 strains as of June 17, 2021) computed by the ViPR team was used. For alphacoronavirus and non-SARS-CoV-2 betacoronavirus, all available sequences in ViPR were used. Potential laboratory strains and low-quality sequences were filtered out using custom scripts.
Representative virus selection
Representative virus selection is summarized in Figure S3. To select viruses that are representative of each taxon group (alphacoronavirus, non-sarbeco betacoronavirus, and sarbecovirus), a targeted sampling approach was used which leverages sequence identity, sequence and annotation quality, host, isolation date and region, RefSeq designation, and phylogenetic structures. First, all genome sequences were clustered based on sequence identity using cd-hit with the non-greedy option that assigns shorter sequences to the closest cluster. To get the desired number of representative viruses, a 0.80 identity threshold was used for the alpha and non-sarbeco beta groups, while a 0.999 threshold was used for the sarbeco group as sarbeco strains have very high similarity in sequence identity. Second, the taxa and metadata (host, isolation country, and isolation year) associated with the sequences in each cluster were extracted using custom scripts. Third, target clusters were selected based on the cluster size (at least 2 for alpha and non-sarbeco beta groups, and at least 10 for the sarbeco group) and host (human, bat, camel, hedgehog, or pangolin). Fourth, representative viruses were selected for each target cluster. Specifically, if a cluster contains NCBI RefSeq sequences (1 or 2 sequences), the RefSeqs were selected as the representatives. Otherwise, a virus with complete metadata, good quality protein annotations and from a recent subcluster was selected as the representative. Finally, since the sarbeco group has a much narrower taxonomic scope, an extra iteration of phylogeny-based sampling was performed to ensure the best diversity representation. Using the genome sequences of the selected sarbeco candidates, a phylogenetic tree was built and visualized on the ViPR website. Representatives were selected to cover the major phylogenetic clusters. This targeted sampling process resulted in the selection of 16 alpha, 13 non-sarbeco beta and 4 sarbecoviruses as representatives.
T cell epitope homolog identification
To find the epitope homologs in the representative viruses, the epitope homologous region in each representative was identified and then the optimal k-mer was found in this region (Figure S4). Specifically, each epitope was mapped to the virus taxon’s RefSeq protein. Then the RefSeq protein sequence harboring the mapped epitope was aligned with each taxon group’s protein sequences using the mafft program einsi mode. In the resulting alignment, the epitope mapped peptide in each virus was defined as the seed. Using the seed, the search space for finding the optimal k-mer, where k is the length of the epitope, was defined. If the seed was k or longer, the search space was the seed itself. Otherwise, the search space was expanded to include (k – seed_length) additional residues on both sides of the seed, unless the boundary of the protein was reached:
search_space = min(max(0, k – seed_length), num_of_upstream_residues) upstream_residues + seed + min(max(0, k – seed_length), num_of_downstream_residues) downstream_residues.
To find the optimal k-mer, each k-mer in the search space was calculated with an identity score, and the k-mer with the maximum score was selected. In case of ties, the leftmost k-mer with the maximum score was selected:
optimal_k-mer = argmax(identity_score(k-mer))
The k-mer identity score was defined as the maximum number of matched residues in all possible non-gap alignments divided by the epitope length k:
identity score = max(num_of_matched_residues_in_alignment)/k.
A non-gap alignment was defined as a pairwise alignment of the k-mer and the input epitope that only allowed shifting and substitutions, but no internal indels. Each k-mer had 2 ∗k - 1 such alignments.
Acknowledgments
This study has been funded by the NIH/NIAD P01 AI168347 to A.G., contract no. 75N93019C00065 to A.S. and D.W., and HHS75N93019C00076 to R.H.S. A.T. was supported by a PhD student fellowship through the Clinical and Experimental Immunology Course at the University of Genoa, Italy. We thank Gina Levi and the LJI clinical core for assistance in sample coordination and blood processing. The graphical abstract accompanying this manuscript was created with BioRender.com, accessed 11 May 2023.
Author contributions
Conceptualization, A.G., R.H.S., and A.S.; data curation and bioinformatic analysis, Y.Z.; formal analysis, A.T., N.M., and T.M.N.; funding acquisition, A.S. and R.H.S.; investigation, A.T., Y.Z., N.M., T.M.N., R.H.S., A.S., and A.G.; project administration, A.F.; resources, D.W., S.M., E.P., J.D., and A.S.; supervision, G.F., L.P., R.H.S., A.S., and A.G.; writing, A.T., Y.Z., R.H.S., A.S., and A.G.
Declaration of interests
A.S. is a consultant for Gritstone Bio, Flow Pharma, Moderna, AstraZeneca, Qiagen, Fortress, Gilead, Sanofi, Merck, RiverVest, MedaCorp, Turnstone, NA Vaccine Institute, Emervax, Gerson Lehrman Group, and Guggenheim. L.J.I. has filed for patent protection for various aspects of T cell epitope and vaccine design work.
Inclusion and diversity
We worked to ensure gender balance in the recruitment of human subjects. We worked to ensure ethnic or other types of diversity in the recruitment of human subjects. We worked to ensure that the study questionnaires were prepared in an inclusive way.
Published: May 29, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2023.101088.
Contributor Information
Richard H. Scheuermann, Email: rscheuermann@jcvi.org.
Alessandro Sette, Email: alex@lji.org.
Alba Grifoni, Email: agrifoni@lji.org.
Supplemental information
NL63 and OC43 epitopes information pertaining protein composition and location are included together with the sequence identity values related to each of the representative sequence for Sarbecoviruses (n = 10), Betacoronaviruses excluding Sarbecoviruses (n = 15), and Alphacoronaviruses (n = 15)
Data and code availability
The published article includes all data generated or analyzed during this study, and summarized in the accompanying tables, figures and supplemental materials. The code to identify conserved T cell epitopes has been deposited in the Public Github repository and can be accessed with the following link: https://github.com/yzhang2168/t-cell-epitopes. Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
References
- 1.Sette A., Saphire E.O. Inducing broad-based immunity against viruses with pandemic potential. Immunity. 2022;55:738–748. doi: 10.1016/j.immuni.2022.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hu B., Guo H., Zhou P., Shi Z.-L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021;19:141–154. doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Masters P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006;66:193–292. doi: 10.1016/s0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shah M.M., Winn A., Dahl R.M., Kniss K.L., Silk B.J., Killerby M.E. Seasonality of common human coronaviruses, United States, 2014-2021(1) Emerg. Infect. Dis. 2022;28:1970–1976. doi: 10.3201/eid2810.220396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sariol A., Perlman S. Lessons for COVID-19 immunity from other coronavirus infections. Immunity. 2020;53:248–263. doi: 10.1016/j.immuni.2020.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sette A., Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021;184:861–880. doi: 10.1016/j.cell.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dan J., da Silva Antunes R., Grifoni A., Weiskopf D., Crotty S., Sette A. Observations and perspectives on adaptive immunity to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Clin. Infect. Dis. 2022;75:24–29. doi: 10.1093/cid/ciac310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rydyznski Moderbacher C., Ramirez S.I., Dan J.M., Grifoni A., Hastie K.M., Weiskopf D., Belanger S., Abbott R.K., Kim C., Choi J., et al. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell. 2020;183:996–1012.e19. doi: 10.1016/j.cell.2020.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan A.T., Linster M., Tan C.W., Le Bert N., Chia W.N., Kunasegaran K., Zhuang Y., Tham C.Y.L., Chia A., Smith G.J.D., et al. Early induction of functional SARS-CoV-2-specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients. Cell Rep. 2021;34:108728. doi: 10.1016/j.celrep.2021.108728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tarke A., Potesta M., Varchetta S., Fenoglio D., Iannetta M., Sarmati L., Mele D., Dentone C., Bassetti M., Montesano C., et al. Early and polyantigenic CD4 T cell responses correlate with mild disease in acute COVID-19 donors. Int. J. Mol. Sci. 2022;23:7155. doi: 10.3390/ijms23137155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muñoz-Fontela C., Dowling W.E., Funnell S.G.P., Gsell P.S., Riveros-Balta A.X., Albrecht R.A., Andersen H., Baric R.S., Carroll M.W., Cavaleri M., et al. Animal models for COVID-19. Nature. 2020;586:509–515. doi: 10.1038/s41586-020-2787-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soresina A., Moratto D., Chiarini M., Paolillo C., Baresi G., Focà E., Bezzi M., Baronio B., Giacomelli M., Badolato R. Two X-linked agammaglobulinemia patients develop pneumonia as COVID-19 manifestation but recover. Pediatr. Allergy Immunol. 2020;31:565–569. doi: 10.1111/pai.13263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Apostolidis S.A., Kakara M., Painter M.M., Goel R.R., Mathew D., Lenzi K., Rezk A., Patterson K.R., Espinoza D.A., Kadri J.C., et al. Cellular and humoral immune responses following SARS-CoV-2 mRNA vaccination in patients with multiple sclerosis on anti-CD20 therapy. Nat. Med. 2021;27:1990–2001. doi: 10.1038/s41591-021-01507-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sabatino J.J., Jr., Mittl K., Rowles W.M., McPolin K., Rajan J.V., Laurie M.T., Zamecnik C.R., Dandekar R., Alvarenga B.D., Loudermilk R.P., et al. Multiple sclerosis therapies differentially affect SARS-CoV-2 vaccine-induced antibody and T cell immunity and function. JCI Insight. 2022;7:e156978. doi: 10.1172/jci.insight.156978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tarke A., Coelho C.H., Zhang Z., Dan J.M., Yu E.D., Methot N., Bloom N.I., Goodwin B., Phillips E., Mallal S., et al. SARS-CoV-2 vaccination induces immunological T cell memory able to cross-recognize variants from Alpha to Omicron. Cell. 2022;185:847–859.e11. doi: 10.1016/j.cell.2022.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tarke A., Sidney J., Methot N., Yu E.D., Zhang Y., Dan J.M., Goodwin B., Rubiro P., Sutherland A., Wang E., et al. Impact of SARS-CoV-2 variants on the total CD4(+) and CD8(+) T cell reactivity in infected or vaccinated individuals. Cell Rep. Med. 2021;2:100355. doi: 10.1016/j.xcrm.2021.100355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geers D., Shamier M.C., Bogers S., den Hartog G., Gommers L., Nieuwkoop N.N., Schmitz K.S., Rijsbergen L.C., van Osch J.A.T., Dijkhuizen E., et al. SARS-CoV-2 variants of concern partially escape humoral but not T cell responses in COVID-19 convalescent donors and vaccine recipients. Sci. Immunol. 2021;6:eabj1750. doi: 10.1126/sciimmunol.abj1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.GeurtsvanKessel C.H., Geers D., Schmitz K.S., Mykytyn A.Z., Lamers M.M., Bogers S., Scherbeijn S., Gommers L., Sablerolles R.S.G., Nieuwkoop N.N., et al. Divergent SARS-CoV-2 Omicron-reactive T and B cell responses in COVID-19 vaccine recipients. Sci. Immunol. 2022;7:eabo2202. doi: 10.1126/sciimmunol.abo2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Melo-González F., Soto J.A., González L.A., Fernández J., Duarte L.F., Schultz B.M., Gálvez N.M.S., Pacheco G.A., Ríos M., Vázquez Y., et al. Recognition of variants of concern by antibodies and T cells induced by a SARS-CoV-2 inactivated vaccine. Front. Immunol. 2021;12:747830. doi: 10.3389/fimmu.2021.747830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keeton R., Richardson S.I., Moyo-Gwete T., Hermanus T., Tincho M.B., Benede N., Manamela N.P., Baguma R., Makhado Z., Ngomti A., et al. Prior infection with SARS-CoV-2 boosts and broadens Ad26.COV2.S immunogenicity in a variant-dependent manner. Cell Host Microbe. 2021;29:1611–1619.e5. doi: 10.1016/j.chom.2021.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao Y., Cai C., Grifoni A., Müller T.R., Niessl J., Olofsson A., Humbert M., Hansson L., Österborg A., Bergman P., et al. Ancestral SARS-CoV-2-specific T cells cross-recognize the Omicron variant. Nat. Med. 2022;28:472–476. doi: 10.1038/s41591-022-01700-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilkinson T.M., Li C.K.F., Chui C.S.C., Huang A.K.Y., Perkins M., Liebner J.C., Lambkin-Williams R., Gilbert A., Oxford J., Nicholas B., et al. Preexisting influenza-specific CD4+ T cells correlate with disease protection against influenza challenge in humans. Nat. Med. 2012;18:274–280. doi: 10.1038/nm.2612. [DOI] [PubMed] [Google Scholar]
- 24.Wild K., Smits M., Killmer S., Strohmeier S., Neumann-Haefelin C., Bengsch B., Krammer F., Schwemmle M., Hofmann M., Thimme R., et al. Pre-existing immunity and vaccine history determine hemagglutinin-specific CD4 T cell and IgG response following seasonal influenza vaccination. Nat. Commun. 2021;12:6720. doi: 10.1038/s41467-021-27064-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayward A.C., Wang L., Goonetilleke N., Fragaszy E.B., Bermingham A., Copas A., Dukes O., Millett E.R.C., Nazareth I., Nguyen-Van-Tam J.S., et al. Natural T cell-mediated protection against seasonal and pandemic influenza. Results of the flu watch cohort study. Am. J. Respir. Crit. Care Med. 2015;191:1422–1431. doi: 10.1164/rccm.201411-1988OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Assarsson E., Bui H.H., Sidney J., Zhang Q., Glenn J., Oseroff C., Mbawuike I.N., Alexander J., Newman M.J., Grey H., Sette A. Immunomic analysis of the repertoire of T-cell specificities for influenza A virus in humans. J. Virol. 2008;82:12241–12251. doi: 10.1128/jvi.01563-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sridhar S., Begom S., Bermingham A., Hoschler K., Adamson W., Carman W., Bean T., Barclay W., Deeks J.J., Lalvani A. Cellular immune correlates of protection against symptomatic pandemic influenza. Nat. Med. 2013;19:1305–1312. doi: 10.1038/nm.3350. [DOI] [PubMed] [Google Scholar]
- 28.Sette A., Crotty S. Pre-existing immunity to SARS-CoV-2: the knowns and unknowns. Nat. Rev. Immunol. 2020;20:457–458. doi: 10.1038/s41577-020-0389-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bacher P., Rosati E., Esser D., Martini G.R., Saggau C., Schiminsky E., Dargvainiene J., Schröder I., Wieters I., Khodamoradi Y., et al. Low-avidity CD4(+) T cell responses to SARS-CoV-2 in unexposed individuals and humans with severe COVID-19. Immunity. 2020;53:1258–1271.e5. doi: 10.1016/j.immuni.2020.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lipsitch M., Grad Y.H., Sette A., Crotty S. Cross-reactive memory T cells and herd immunity to SARS-CoV-2. Nat. Rev. Immunol. 2020;20:709–713. doi: 10.1038/s41577-020-00460-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sagar M., Reifler K., Rossi M., Miller N.S., Sinha P., White L.F., Mizgerd J.P. Recent endemic coronavirus infection is associated with less-severe COVID-19. J. Clin. Invest. 2021;131:e143380. doi: 10.1172/jci143380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mateus J., Dan J.M., Zhang Z., Rydyznski Moderbacher C., Lammers M., Goodwin B., Sette A., Crotty S., Weiskopf D. Low-dose mRNA-1273 COVID-19 vaccine generates durable memory enhanced by cross-reactive T cells. Science. 2021;374:eabj9853. doi: 10.1126/science.abj9853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., et al. Targets of T Cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weiskopf D., Schmitz K.S., Raadsen M.P., Grifoni A., Okba N.M.A., Endeman H., van den Akker J.P.C., Molenkamp R., Koopmans M.P.G., van Gorp E.C.M., et al. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci. Immunol. 2020;5:eabd2071. doi: 10.1126/sciimmunol.abd2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Braun J., Loyal L., Frentsch M., Wendisch D., Georg P., Kurth F., Hippenstiel S., Dingeldey M., Kruse B., Fauchere F., et al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020;587:270–274. doi: 10.1038/s41586-020-2598-9. [DOI] [PubMed] [Google Scholar]
- 36.Mateus J., Grifoni A., Tarke A., Sidney J., Ramirez S.I., Dan J.M., Burger Z.C., Rawlings S.A., Smith D.M., Phillips E., et al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. 2020;370:89–94. doi: 10.1126/science.abd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loyal L., Braun J., Henze L., Kruse B., Dingeldey M., Reimer U., Kern F., Schwarz T., Mangold M., Unger C., et al. Cross-reactive CD4+ T cells enhance SARS-CoV-2 immune responses upon infection and vaccination. Science. 2021;374:eabh1823. doi: 10.1126/science.abh1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nelde A., Bilich T., Heitmann J.S., Maringer Y., Salih H.R., Roerden M., Lübke M., Bauer J., Rieth J., Wacker M., et al. SARS-CoV-2-derived peptides define heterologous and COVID-19-induced T cell recognition. Nat. Immunol. 2021;22:74–85. doi: 10.1038/s41590-020-00808-x. [DOI] [PubMed] [Google Scholar]
- 39.Le Bert N., Tan A.T., Kunasegaran K., Tham C.Y.L., Hafezi M., Chia A., Chng M.H.Y., Lin M., Tan N., Linster M., et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 40.Habel J.R., Nguyen T.H.O., van de Sandt C.E., Juno J.A., Chaurasia P., Wragg K., Koutsakos M., Hensen L., Jia X., Chua B., et al. Suboptimal SARS-CoV-2-specific CD8(+) T cell response associated with the prominent HLA-A∗02:01 phenotype. Proc. Natl. Acad. Sci. USA. 2020;117:24384–24391. doi: 10.1073/pnas.2015486117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rha B., Lively J.Y., Englund J.A., Staat M.A., Weinberg G.A., Selvarangan R., Halasa N.B., Williams J.V., Boom J.A., Sahni L.C., et al. Severe acute respiratory syndrome coronavirus 2 infections in children: multicenter surveillance, United States, january-march 2020. J. Pediatric Infect. Dis. Soc. 2020;9:609–612. doi: 10.1093/jpids/piaa075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gangaev A., Ketelaars S.L.C., Patiwael S., Dopler A., Isaeva O.I., Hoefakker K., Biasi S.D., Mussini C., Guaraldi G., Girardis M., et al. Profound CD8 T cell responses towards the SARS-CoV-2 ORF1ab in COVID-19 patients. Res. Sq. 2021 doi: 10.21203/rs.3.rs-33197/v1. [DOI] [Google Scholar]
- 43.Grifoni A., Sidney J., Zhang Y., Scheuermann R.H., Peters B., Sette A. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host Microbe. 2020;27:671–680.e2. doi: 10.1016/j.chom.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prakash S., Srivastava R., Coulon P.G., Dhanushkodi N.R., Chentoufi A.A., Tifrea D.F., Edwards R.A., Figueroa C.J., Schubl S.D., Hsieh L., et al. Genome-wide B cell, CD4(+), and CD8(+) T cell epitopes that are highly conserved between human and animal coronaviruses, identified from SARS-CoV-2 as targets for preemptive pan-coronavirus vaccines. J. Immunol. 2021;206:2566–2582. doi: 10.4049/jimmunol.2001438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Le Bert N., Clapham H.E., Tan A.T., Chia W.N., Tham C.Y.L., Lim J.M., Kunasegaran K., Tan L.W.L., Dutertre C.A., Shankar N., et al. Highly functional virus-specific cellular immune response in asymptomatic SARS-CoV-2 infection. J. Exp. Med. 2021;218:e20202617. doi: 10.1084/jem.20202617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ahmadi E., Zabihi M.R., Hosseinzadeh R., Mohamed Khosroshahi L., Noorbakhsh F. SARS-CoV-2 spike protein displays sequence similarities with paramyxovirus surface proteins; a bioinformatics study. PLoS One. 2021;16:e0260360. doi: 10.1371/journal.pone.0260360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fan C., Cohen A.A., Park M., Hung A.F.H., Keeffe J.R., Gnanapragasam P.N.P., Lee Y.E., Gao H., Kakutani L.M., Wu Z., et al. Neutralizing monoclonal antibodies elicited by mosaic RBD nanoparticles bind conserved sarbecovirus epitopes. Immunity. 2022;55:2419–2435.e10. doi: 10.1016/j.immuni.2022.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cohen A.A., van Doremalen N., Greaney A.J., Andersen H., Sharma A., Starr T.N., Keeffe J.R., Fan C., Schulz J.E., Gnanapragasam P.N.P., et al. Mosaic RBD nanoparticles protect against challenge by diverse sarbecoviruses in animal models. Science. 2022;377:eabq0839. doi: 10.1126/science.abq0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grifoni A., Sidney J., Vita R., Peters B., Crotty S., Weiskopf D., Sette A. SARS-CoV-2 human T cell epitopes: adaptive immune response against COVID-19. Cell Host Microbe. 2021;29:1076–1092. doi: 10.1016/j.chom.2021.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu E.D., Narowski T.M., Wang E., Garrigan E., Mateus J., Frazier A., Weiskopf D., Grifoni A., Premkumar L., da Silva Antunes R., Sette A. Immunological memory to common cold coronaviruses assessed longitudinally over a three-year period pre-COVID19 pandemic. Cell Host Microbe. 2022;30:1269–1278.e4. doi: 10.1016/j.chom.2022.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tarke A., Sidney J., Kidd C.K., Dan J.M., Ramirez S.I., Yu E.D., Mateus J., da Silva Antunes R., Moore E., Rubiro P., et al. Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases. Cell Rep. Med. 2021;2:100204. doi: 10.1016/j.xcrm.2021.100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.da Silva Antunes R., Quiambao L.G., Sutherland A., Soldevila F., Dhanda S.K., Armstrong S.K., Brickman T.J., Merkel T., Peters B., Sette A. Development and validation of a bordetella pertussis whole-genome screening strategy. J. Immunol. Res. 2020;2020:8202067. doi: 10.1155/2020/8202067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lineburg K.E., Grant E.J., Swaminathan S., Chatzileontiadou D.S.M., Szeto C., Sloane H., Panikkar A., Raju J., Crooks P., Rehan S., et al. CD8(+) T cells specific for an immunodominant SARS-CoV-2 nucleocapsid epitope cross-react with selective seasonal coronaviruses. Immunity. 2021;54:1055–1065.e5. doi: 10.1016/j.immuni.2021.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wirsching S., Harder L., Heymanns M., Gröndahl B., Hilbert K., Kowalzik F., Meyer C., Gehring S. Long-term, CD4(+) memory T cell response to SARS-CoV-2. Front. Immunol. 2022;13:800070. doi: 10.3389/fimmu.2022.800070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dykema A.G., Zhang B., Woldemeskel B.A., Garliss C.C., Cheung L.S., Choudhury D., Zhang J., Aparicio L., Bom S., Rashid R., et al. Functional characterization of CD4+ T cell receptors crossreactive for SARS-CoV-2 and endemic coronaviruses. J. Clin. Invest. 2021;131:e146922. doi: 10.1172/jci146922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johansson A.M., Malhotra U., Kim Y.G., Gomez R., Krist M.P., Wald A., Koelle D.M., Kwok W.W. Cross-reactive and mono-reactive SARS-CoV-2 CD4+ T cells in prepandemic and COVID-19 convalescent individuals. PLoS Pathog. 2021;17:e1010203. doi: 10.1371/journal.ppat.1010203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bartolo L., Afroz S., Pan Y.G., Xu R., Williams L., Lin C.F., Tanes C., Bittinger K., Friedman E.S., Gimotty P.A., et al. SARS-CoV-2-specific T cells in unexposed adults display broad trafficking potential and cross-react with commensal antigens. Sci. Immunol. 2022;7:eabn3127. doi: 10.1126/sciimmunol.abn3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sekine T., Perez-Potti A., Rivera-Ballesteros O., Strålin K., Gorin J.B., Olsson A., Llewellyn-Lacey S., Kamal H., Bogdanovic G., Muschiol S., et al. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell. 2020;183:158–168.e14. doi: 10.1016/j.cell.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Niessl J., Sekine T., Buggert M. T cell immunity to SARS-CoV-2. Semin. Immunol. 2021;55:101505. doi: 10.1016/j.smim.2021.101505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Swadling L., Diniz M.O., Schmidt N.M., Amin O.E., Chandran A., Shaw E., Pade C., Gibbons J.M., Le Bert N., Tan A.T., et al. Pre-existing polymerase-specific T cells expand in abortive seronegative SARS-CoV-2. Nature. 2022;601:110–117. doi: 10.1038/s41586-021-04186-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nascimento E.J.M., Mailliard R.B., Khan A.M., Sidney J., Sette A., Guzman N., Paulaitis M., de Melo A.B., Cordeiro M.T., Gil L.V.G., et al. Identification of conserved and HLA promiscuous DENV3 T-cell epitopes. PLoS Negl. Trop. Dis. 2013;7:e2497. doi: 10.1371/journal.pntd.0002497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ribeiro S.P., De Moura Mattaraia V.G., Almeida R.R., Valentine E.J.G., Sales N.S., Ferreira L.C.S., Sa-Rocha L.C., Jacintho L.C., Santana V.C., Sidney J., et al. A promiscuous T cell epitope-based HIV vaccine providing redundant population coverage of the HLA class II elicits broad, polyfunctional T cell responses in nonhuman primates. Vaccine. 2022;40:239–246. doi: 10.1016/j.vaccine.2021.11.076. [DOI] [PubMed] [Google Scholar]
- 63.Doolan D.L., Southwood S., Chesnut R., Appella E., Gomez E., Richards A., Higashimoto Y.I., Maewal A., Sidney J., Gramzinski R.A., et al. HLA-DR-promiscuous T cell epitopes from Plasmodium falciparum pre-erythrocytic-stage antigens restricted by multiple HLA class II alleles. J. Immunol. 2000;165:1123–1137. doi: 10.4049/jimmunol.165.2.1123. [DOI] [PubMed] [Google Scholar]
- 64.Wilson C.C., Palmer B., Southwood S., Sidney J., Higashimoto Y., Appella E., Chesnut R., Sette A., Livingston B.D. Identification and antigenicity of broadly cross-reactive and conserved human immunodeficiency virus type 1-derived helper T-lymphocyte epitopes. J. Virol. 2001;75:4195–4207. doi: 10.1128/jvi.75.9.4195-4207.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schipper R.F., van Els C.A., D'Amaro J., Oudshoorn M. Minimal phenotype panels. A method for achieving maximum population coverage with a minimum of HLA antigens. Hum. Immunol. 1996;51:95–98. doi: 10.1016/s0198-8859(96)00138-3. [DOI] [PubMed] [Google Scholar]
- 66.Bui H.H., Sidney J., Dinh K., Southwood S., Newman M.J., Sette A. Predicting population coverage of T-cell epitope-based diagnostics and vaccines. BMC Bioinformatics. 2006;7:153. doi: 10.1186/1471-2105-7-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sidney J., Peters B., Sette A. Epitope prediction and identification- adaptive T cell responses in humans. Semin. Immunol. 2020;50:101418. doi: 10.1016/j.smim.2020.101418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vita R., Mahajan S., Overton J.A., Dhanda S.K., Martini S., Cantrell J.R., Wheeler D.K., Sette A., Peters B. The immune epitope database (IEDB): 2018 update. Nucleic Acids Res. 2019;47:339–343. doi: 10.1093/nar/gky1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Alter G., Yu J., Liu J., Chandrashekar A., Borducchi E.N., Tostanoski L.H., McMahan K., Jacob-Dolan C., Martinez D.R., Chang A., et al. Immunogenicity of Ad26.COV2.S vaccine against SARS-CoV-2 variants in humans. Nature. 2021;596:268–272. doi: 10.1038/s41586-021-03681-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.da Silva Antunes R., Pallikkuth S., Williams E., Dawen Yu E., Mateus J., Quiambao L., Wang E., Rawlings S.A., Stadlbauer D., Jiang K., et al. Differential T-cell reactivity to endemic coronaviruses and SARS-CoV-2 in community and health care workers. J. Infect. Dis. 2021;224:70–80. doi: 10.1093/infdis/jiab176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Edridge A.W.D., Kaczorowska J., Hoste A.C.R., Bakker M., Klein M., Loens K., Jebbink M.F., Matser A., Kinsella C.M., Rueda P., et al. Seasonal coronavirus protective immunity is short-lasting. Nat. Med. 2020;26:1691–1693. doi: 10.1038/s41591-020-1083-1. [DOI] [PubMed] [Google Scholar]
- 72.Schouest B., Grifoni A., Pham J., Mateus J., Sydney J., Brien J.D., De Silva A.D., Balmaseda A., Harris E., Sette A., Weiskopf D. Pre-existing T cell memory against Zika virus. J. Virol. 2021;95:001322-21. doi: 10.1128/jvi.00132-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Voic H., de Vries R.D., Sidney J., Rubiro P., Moore E., Phillips E., Mallal S., Schwan B., Weiskopf D., Sette A., Grifoni A. Identification and characterization of CD4(+) T cell epitopes after shingrix vaccination. J. Virol. 2020;94:016411-20. doi: 10.1128/jvi.01641-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Alexander J., Bilsel P., del Guercio M.F., Marinkovic-Petrovic A., Southwood S., Stewart S., Ishioka G., Kotturi M.F., Botten J., Sidney J., et al. Identification of broad binding class I HLA supertype epitopes to provide universal coverage of influenza A virus. Hum. Immunol. 2010;71:468–474. doi: 10.1016/j.humimm.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Grifoni A., Voic H., Dhanda S.K., Kidd C.K., Brien J.D., Buus S., Stryhn A., Durbin A.P., Whitehead S., Diehl S.A., et al. T cell responses induced by attenuated flavivirus vaccination are specific and show limited cross-reactivity with other flavivirus species. J. Virol. 2020;94:000899-20. doi: 10.1128/jvi.00089-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Regla-Nava J.A., Elong Ngono A., Viramontes K.M., Huynh A.T., Wang Y.T., Nguyen A.V.T., Salgado R., Mamidi A., Kim K., Diamond M.S., Shresta S. Cross-reactive Dengue virus-specific CD8(+) T cells protect against Zika virus during pregnancy. Nat. Commun. 2018;9:3042. doi: 10.1038/s41467-018-05458-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wen J., Wang Y.T., Valentine K.M., Dos Santos Alves R.P., Xu Z., Regla-Nava J.A., Ngono A.E., Young M.P., Ferreira L.C.S., Shresta S. CD4(+) T cells cross-reactive with Dengue and Zika viruses protect against Zika virus infection. Cell Rep. 2020;31:107566. doi: 10.1016/j.celrep.2020.107566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Reynolds C.J., Swadling L., Gibbons J.M., Pade C., Jensen M.P., Diniz M.O., Schmidt N.M., Butler D.K., Amin O.E., Bailey S.N.L., et al. Discordant neutralizing antibody and T cell responses in asymptomatic and mild SARS-CoV-2 infection. Sci. Immunol. 2020;5:eabf3698. doi: 10.1126/sciimmunol.abf3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yu E.D., Wang E., Garrigan E., Goodwin B., Sutherland A., Tarke A., Chang J., Gálvez R.I., Mateus J., Ramirez S.I., et al. Development of a T cell-based immunodiagnostic system to effectively distinguish SARS-CoV-2 infection and COVID-19 vaccination status. Cell Host Microbe. 2022;30:388–399.e3. doi: 10.1016/j.chom.2022.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Premkumar L., Segovia-Chumbez B., Jadi R., Martinez D.R., Raut R., Markmann A., Cornaby C., Bartelt L., Weiss S., Park Y., et al. The receptor binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci. Immunol. 2020;5:eabc8413. doi: 10.1126/sciimmunol.abc8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yu E.D., Narowski T.M., Wang E., Garrigan E., Mateus J., Frazier A., Weiskopf D., Grifoni A., Premkumar L., da Silva Antunes R., Sette A. Immunological memory to Common Cold Coronaviruses assessed longitudinally over a three-year period. bioRxiv. 2022 doi: 10.1101/2022.03.01.482548. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Olson R.D., Assaf R., Brettin T., Conrad N., Cucinell C., Davis J.J., Dempsey D.M., Dickerman A., Dietrich E.M., Kenyon R.W., et al. Introducing the bacterial and viral bioinformatics resource center (BV-BRC): a resource combining PATRIC, IRD and ViPR. Nucleic Acids Res. 2023;51:D678–D689. doi: 10.1093/nar/gkac1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
NL63 and OC43 epitopes information pertaining protein composition and location are included together with the sequence identity values related to each of the representative sequence for Sarbecoviruses (n = 10), Betacoronaviruses excluding Sarbecoviruses (n = 15), and Alphacoronaviruses (n = 15)
Data Availability Statement
The published article includes all data generated or analyzed during this study, and summarized in the accompanying tables, figures and supplemental materials. The code to identify conserved T cell epitopes has been deposited in the Public Github repository and can be accessed with the following link: https://github.com/yzhang2168/t-cell-epitopes. Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.