Abstract
The recently emerged Omicron subvariants XBB and BQ.1.1 have presented striking immune evasion against most monoclonal neutralizing antibodies and convalescent plasma. Therefore, it is essential to develop broad-spectrum COVID-19 vaccines to combat current and future emerging variants. Here, we found that the human IgG Fc-conjugated RBD of the original SARS-CoV-2 strain (WA1) plus a novel STING agonist-based adjuvant CF501 (CF501/RBD-Fc) could induce highly potent and durable broad-neutralizing antibody (bnAb) responses against Omicron subvariants, including BQ.1.1 and XBB in rhesus macaques with NT50s ranging from 2,118 to 61,742 after three doses. A decline of 0.9- to 4.7-fold was observed in the neutralization activity of sera in the CF501/RBD-Fc group against BA.2.2, BA.2.9, BA.5, BA.2.75, and BF.7 relative to D614G after three doses, while a significant decline of NT50 against BQ.1.1 (26.9-fold) and XBB (22.5-fold) relative to D614G. However, the bnAbs were still effective in neutralizing BQ.1.1 and XBB infection. These results suggest that the conservative but nondominant epitopes in RBD could be stimulated by CF501 to generate bnAbs, providing a proof-of-concept for using “nonchangeable against changeables” strategy to develop pan-sarbecovirus vaccines against sarbecoviruses, including SARS-CoV-2 and its variants.
Keywords: pan-sarbecovirus vaccine, SARS-CoV-2, Omicron subvariants, Vaccines, Adjuvant
With the evolution of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron, various Omicron subvariants have been continuously identified, posing a significant challenge to current COVID-19 vaccines owing to their strong evasion of humoral immunity (1, 2). Even three or four doses of parental messenger RNA (mRNA) vaccine cannot induce strong neutralization against BA.5 (3). Results from the pseudovirus neutralization assay showed that XBB is about 66- to 155-fold more resistant to neutralizing antibodies (nAbs) in the sera from vaccinees and infected individuals than the ancestral strain D614G (4). Moreover, even a booster of the bivalent vaccine containing both BA.5 and ancestral spike proteins has not generated strong enough nAbs against the newly emerged XBB (5). This calls for an urgent development of a pan-sarbecovirus vaccine to combat all SARS-CoV-2 variants and subvariants.
Many mutations were observed in the receptor-binding domain (RBD) of Omicron subvariants (Fig. 1A), and most RBD-specific antibodies showing neutralizing activities against the original SARS-CoV-2 were shown to be inactive against Omicron subvariants (6). This has raised doubts about whether the RBD still contains highly conserved epitopes, and thus, whether it could be used as a proper immunogen to develop COVID-19 vaccines. Even if confirmed, how to stimulate these conserved epitopes to generate broadly neutralizing antibodies (bnAbs) remains to be elucidated.
Fig. 1.
Cross-binding antibodies induced by pan-sarbecovirus vaccine in rhesus macaques. (A) Amino acid mutations in spike protein for the indicated Omicron subvariants compared with WA1. (B) Vaccination procedure. Rhesus macaques were vaccinated three times at days 0, 21, and 115, respectively. Antisera in CF501/RBD-Fc (n = 3) and Alum/RBD-Fc (n = 3) groups were collected on the indicated days. (C–J) Omicron subvariants BA.2.2 (C), BA.2.9 (D), BA.2.12.1 (E), BA.5 (F), BA.2.75 (G), BF.7 (H), BQ.1.1 (I), and XBB (J) RBD-specific binding IgG endpoint titer during days 28 to 191. Data shown are means ± SEM. Statistical analyses were performed using two-way ANOVA. *P < 0.05, **P < 0.001, ***P < 0.0001.
We have previously developed a pan-sarbecovirus vaccine that contains human Fc-conjugated RBD of ancestral SARS-CoV-2 strain as the immunogen and a novel STING agonist as the adjuvant (CF501/RBD-Fc) (7, 8). However, the antibody evasion capability of some Omicron subvariants especially XBB has been demonstrated to reach or even exceed SARS-CoV (1). Here, we investigate whether this STING-adjuvanted pan-sarbecovirus vaccine could also elicit potent bnAbs against infection caused by Omicron subvariants, especially BQ.1.1 and XBB.
Results
Rhesus macaques received three-dose vaccination of the original RBD-Fc adjuvanted with CF501 (n = 3) or Alum (n = 3), respectively, as previously described (Fig. 1B) (7). After two shots, the endpoint titers of IgG specific for RBD of these Omicron subvariants ranged from 512,000 to 1,792,000 in the CF501/RBD-Fc group at day 28, about 3- to 28-fold higher than that induced in the Alum/RBD-Fc group (Fig. 1 C–J). Although the endpoint titers gradually decreased at days 51, 78, and 113, the titers in the CF501/RBD-Fc group remained significantly higher than those in the Alum/RBD-Fc group. Notably, compared with the RBD-binding antibodies elicited by Alum/RBD-Fc, those induced by CF501/RBD-Fc were significantly higher with endpoint titers of 1,024,000 to 7,168,000 after three doses (Fig. 1 C–J). Moreover, the CF501/RBD-Fc group also maintained long-lasting potent RBD-binding antibodies against the Omicron subvariants, even at day 191 after the first vaccination (Fig. 1 C–J).
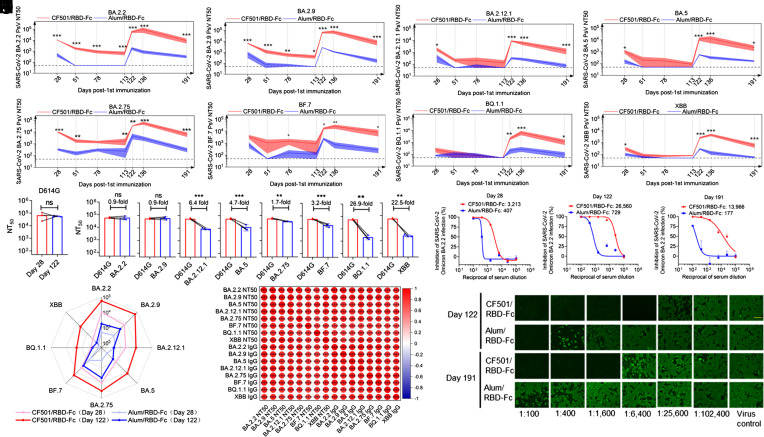
We then tested whether sera from immunized rhesus macaques could neutralize these pseudotyped Omicron subvariants. It was shown that 50% neutralization titer (NT50) of sera from CF501/RBD-Fc group against BA.2.2, BA.2.9, BA.2.12.1, BA.5, BA.2.75, BF.7, BQ.1.1 and XBB could attain 10,694, 6,364, 1,874, 1,075, 9,926, 3,624, 436 and 313 respectively, at day 28, much higher than those in Alum/RBD-Fc group (Fig. 2 A–H). After two shots, almost no detectable bnAbs were found in the Alum/RBD-Fc group against these Omicron subvariants, during days 51 to 113, while the bnAbs in the sera of CF501/RBD-Fc group were still highly effective against most of these Omicron subvariants from the same period. The NT50s against these Omicron subvariants at day 113 (92 d post the second immunization) in CF501/RBD-Fc group were 4.2- to 26.9-fold lower than that at day 28 (7 d post the second immunization). Importantly, cross-neutralizing titers in the CF501/RBD-Fc group surged to high levels after the third vaccination with NT50s of 57,126, 61,742, 8,885, 12,110, 33,393, 17,704, 2,118, and 2,526 against BA.2.2, BA.2.9, BA.2.12.1, BA.5, BA.2.75, BF.7, BQ.1.1, and XBB respectively, at day 122 post first vaccination. Serum-neutralizing activities against Omicron subvariants in the Alum/RBD-Fc group also markedly improved after three doses. However, Alum/RBD-Fc elicited only marginal level of nAbs against BA.2.12.1, BA.5, BQ.1.1, and XBB. We found that the NT50s gradually decreased in both groups. Either weak, or no, serum-neutralizing activities against Omicron subvariants were observed in the Alum/RBD-Fc group at day 191 post the first vaccination. By contrast, the macaques in CF501/RBD-Fc group maintained long-lasting bnAbs against Omicron subvariants even including the BQ.1.1 and XBB at day 191 post the first immunization, with NT50s of 9,477, 8,255, 1,354, 2,315, 6,402, 8,358, 1,153, and 596 against BA.2.2, BA.2.9, BA.2.12.1, BA.5, BA.2.75, BF.7, BQ.1.1, and XBB, respectively (Fig. 2 A–H). A decline of 1.8- to 7.4-fold at day 191 (76 d post the third immunization) relative to day 122 (7 d post the third immunization) was observed for the NT50s against these Omicron subvariants.
Fig. 2.
Potent and durable bnAb responses against Omicron subvariants induced by the pan-sarbecovirus vaccine in rhesus macaques. (A–H) Titers of bnAbs elicited by CF501/RBD-Fc (n = 3) or Alum/RBD-Fc (n = 3) vaccination in rhesus macaques against pseudotyped BA.2.2 (A), BA.2.9 (B), BA.2.12.1 (C), BA.5 (D), BA.2.75 (E), BF.7 (F), BQ.1.1 (G), and XBB (H). 1:50 was defined as the limit of detection. Data shown are means ± SEM. Statistical analyses were performed using two-way ANOVA. *P < 0.05, **P < 0.001, ***P < 0.0001. (I) Comparison of NT50s of the sera collected on day 28 and day 122 against D614G. Statistical analyses were performed using a student’s t test. ns, not significant. (J) Comparison of NT50s against D614G and Omicron subvariants at day 122 post first vaccination with CF501/RBD-Fc. Statistical analyses were performed using a student’s t test. *P < 0.05, **P < 0.001, ***P < 0.0001. (K) The radar figure showed the titer of nAbs elicited by CF501/RBD-Fc or Alum/RBD-Fc against Omicron subvariants at days 28 and 122 post the first vaccination. (L) Correlation matrices from binding IgG titers and nAb titers displaying Pearson rank order correlation values indicated by the color and size of the circle. Asterisks represent the P value. *P < 0.05, **P < 0.001, ***P < 0.0001. (M–O) Neutralizing activities of antisera at day 28 (M), 122 (N), and 191 (O) against authentic Omicron BA.2.2 infection. (P) Immunofluorescence assay showed SARS-CoV-2 N protein expression in cells treated with antisera at days 122 and 191 post the first vaccination. Scale bar, 150 μm.
Interestingly, we found that although the third immunization did not elicit increased NT50 against D614G (Fig. 2I), it did dramatically increase the titers of bnAbs against the Omicron subvariants. A decline of 0.9- to 4.7-fold was observed in the neutralizing activity of sera against BA.2.2, BA.2.9, BA.5, BA.2.75, and BF.7 relative to D614G after three doses (Fig. 2J). Although the titers of nAbs in the CF501/RBD-Fc vaccine group were reduced about 6.4-, 26.9-, and 22.5-fold against BA.2.12.1, BQ.1.1, and XBB, respectively, relative to D614G after three doses (Fig. 2J), their folds of reduction were lower than those in individuals who received mRNA vaccines (66- to 155-fold) reported before (4).
Among these Omicron subvariants, XBB and BQ.1.1 exhibited the strongest evasion against nAbs (Fig. 2K), yet the antisera of macaques in the CF501/RBD-Fc group maintained active against all of these variants after three doses (Fig. 2K). To assess relationships between neutralizing and binding antibodies specific to these Omicron subvariants, we correlated individual parameters by pairwise comparisons. As shown in Fig. 2L, strong correlations were observed between any two parameters (r: 0.74 to 0.97; P < 0.001).
Results revealed that sera in the CF501/RBD-Fc group could potently neutralize authentic BA.2.2 infection with NT50 of 3,213, 26,560, and 13,966 respectively, at day 28, 122, and 191 post the first immunization, about 7-, 35-, and 78-fold higher than those in the respective Alum/RBD-Fc group (Fig. 2 M–O). mmunofluorescence assay further demonstrated that sera from CF501/RBD-Fc could potently inhibit the replication of BA.2.2 (Fig. 2P).
Discussion
Our data suggest that CF501/RBD-Fc can elicit potent and durable bnAbs against original Omicron variant and its subvariants. Although the RBDs of these subvariants carry many mutations that result in the low- or noneffectiveness of the Alum/RBD-Fc and the first-generation COVID-19 vaccines, our results suggest that RBD of the original SARS-CoV-2 strain shares conserved epitopes with those of Omicron subvariants, which can elicit cross-neutralizing antibody responses that can be remarkably enhanced by the novel adjuvant CF501.
Taken collectively, our data support that the “nonchangeable against changeable” strategy is feasible for the development of pan-sarbecovirus vaccines against sarbecoviruses, including the SARS-CoV-2 Omicron subvariants, BQ.1.1 and XBB. Considering that boost immunization plays an important role in producing bnAb responses, we suggest that the adjuvant in the first-generation subunit COVID-19 vaccines be replaced with CF501 and using it for boost immunization, which may significantly enhance the immune responses against SARS-CoV-2 and its current and future variants.
Materials and Methods
In this study, the sera were collected from the rhesus macaques immunized with CF501/RBD-Fc or Alum/RBD-Fc as previously described (7). The expression plasmids of the RBDs were constructed in the plasmid of pFUSE-hIgG1-Fc2. Expi293F expression system was used to express the RBD proteins. RBD-binding IgG titers and neutralizing titers were evaluated by Enzyme-linked immunosorbent assay (ELISA) and virus neutralization assays, respectively (SI Appendix).
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank Q.W. at Fudan University, Shanghai Medical College for technical support. This work was supported by grants from the National Key R&D Program of China (2021YFC2300703 and 2022YFC2604102 to L.L.), Shanghai Municipal Science and Technology Major Project (ZD2021CY001 to L.L. and S.J.), Program of Shanghai Academic/Technology Research Leader (20XD1420300 to L.L.) and the National Natural Science Foundation of China (82202490 to Z.L.).
Author contributions
Z.L., S.J., and L.L. designed research; Z.L., J.Z., X.W., and W.X. performed research; Z.T., H.C., M.C., and J.H. contributed new reagents/analytic tools; Z.L., J.Z., G.Z., Y.W., Q.W., and S.J. analyzed data; and Z.L., S.J., and L.L. wrote the paper.
Competing interests
L.L., S.J., Z.L., J.Z., Q.W., X.W. and W.X. filed a patent application for the STING agonist. Other authors have no conflicts of interest to declare.
Contributor Information
Zezhong Liu, Email: zzliu17@fudan.edu.cn.
Shibo Jiang, Email: shibojiang@fudan.edu.cn.
Lu Lu, Email: lul@fudan.edu.cn.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Cao Y., et al. , Imprinted SARS-CoV-2 humoral immunity induces convergent Omicron RBD evolution. Nature 614, 521–529 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tuekprakhon A., et al. , Antibody escape of SARS-CoV-2 Omicron BA.4 and BA.5 from vaccine and BA.1 serum. Cell 185, 2422–2433.e13 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xie X., et al. , Neutralization of SARS-CoV-2 Omicron sublineages by 4 doses of the original mRNA vaccine. Cell Rep. 41, 111729 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Q., et al. , Alarming antibody evasion properties of rising SARS-CoV-2 BQ and XBB subvariants. Cell 186, 279–286.e8 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kurhade C., et al. , Low neutralization of SARS-CoV-2 Omicron BA.2.75.2, BQ.1.1, and XBB.1 by parental mRNA vaccine or a BA.5-bivalent booster. Nat. Med. 29, 344–347 (2023). [DOI] [PubMed] [Google Scholar]
- 6.Cox M., et al. , SARS-CoV-2 variant evasion of monoclonal antibodies based on in vitro studies. Nat. Rev. Microbiol. 21, 112–124 (2023), 10.1038/s41579-022-00809-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Z., et al. , A novel STING agonist-adjuvanted pan-sarbecovirus vaccine elicits potent and durable neutralizing antibody and T cell responses in mice, rabbits and NHPs. Cell Res. 32, 269–287 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Z., et al. , A pan-sarbecovirus vaccine induces highly potent and durable neutralizing antibody responses in non-human primates against SARS-CoV-2 Omicron variant. Cell Res. 32, 495–497 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix.