Significance
Coenzyme B12 [adenosylcobalamin (AdoCbl)] is an essential metallocofactor for human methylmalonyl-CoA mutase (MCM). The radical chemistry-based reaction catalyzed by MCM is, however, prone to occasional inactivation via loss of the 5′-deoxyadenosylcobalamin moiety from the active site and susceptibility of the remaining cob(II)alamin intermediate to hyperoxidation. While an elaborate system comprising two chaperones engages in the off-loading and repair of inactive cofactor, it is only effective if the cob(II)alamin oxidation state is preserved. We have discovered that bivalent molecular mimicry by the common metabolite ADP is used as a strategy for conserving the cob(II)alamin oxidation state. By sealing off solvent access via a protein conformational change, ADP promotes the repair of a high-value metallocofactor needed for mitochondrial propionate metabolism.
Keywords: cofactor, molecular mimicry, crystal structure, cobalamin, redox
Abstract
Control over transition metal redox state is essential for metalloprotein function and can be achieved via coordination chemistry and/or sequestration from bulk solvent. Human methylmalonyl-Coenzyme A (CoA) mutase (MCM) catalyzes the isomerization of methylmalonyl-CoA to succinyl-CoA using 5′-deoxyadenosylcobalamin (AdoCbl) as a metallocofactor. During catalysis, the occasional escape of the 5′-deoxyadenosine (dAdo) moiety leaves the cob(II)alamin intermediate stranded and prone to hyperoxidation to hydroxocobalamin, which is recalcitrant to repair. In this study, we have identified the use of bivalent molecular mimicry by ADP, coopting the 5′-deoxyadenosine and diphosphate moieties in the cofactor and substrate, respectively, to protect against cob(II)alamin overoxidation on MCM. Crystallographic and electron paramagnetic resonance (EPR) data reveal that ADP exerts control over the metal oxidation state by inducing a conformational change that seals off solvent access, rather than by switching five-coordinate cob(II)alamin to the more air stable four-coordinate state. Subsequent binding of methylmalonyl-CoA (or CoA) promotes cob(II)alamin off-loading from MCM to adenosyltransferase for repair. This study identifies an unconventional strategy for controlling metal redox state by an abundant metabolite to plug active site access, which is key to preserving and recycling a rare, but essential, metal cofactor.
Beyond their role as substrates and pathway intermediates, metabolites also function as signaling molecules, regulating protein structure, dynamics, and function (1). In this signaling capacity, metabolites can bind either at the active or at an allosteric site. While active site binders tend to imitate native substrates and use molecular mimicry as a strategy to control protein function, allosteric site binders trigger structural changes from a distance to transduce functional changes (2, 3). In this study, we report an uncommon example of bivalent molecular mimicry by the common metabolite ADP, which upon binding to the active site of human methylmalonyl-CoA mutase (MCM) controls the metal redox state of its inactive cobalamin cofactor and promotes its subsequent repair and recycling.
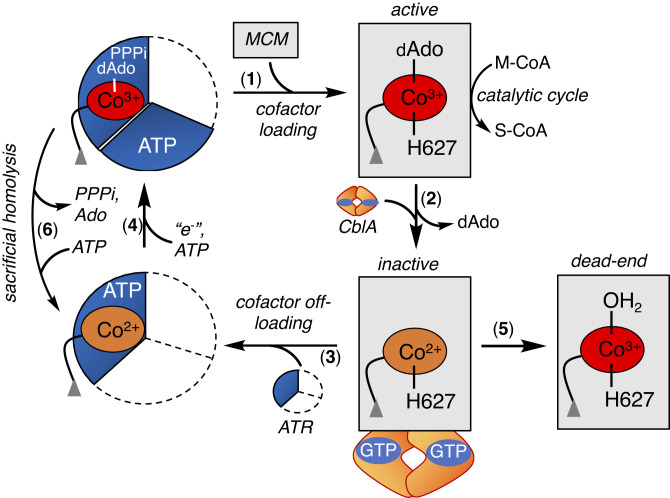
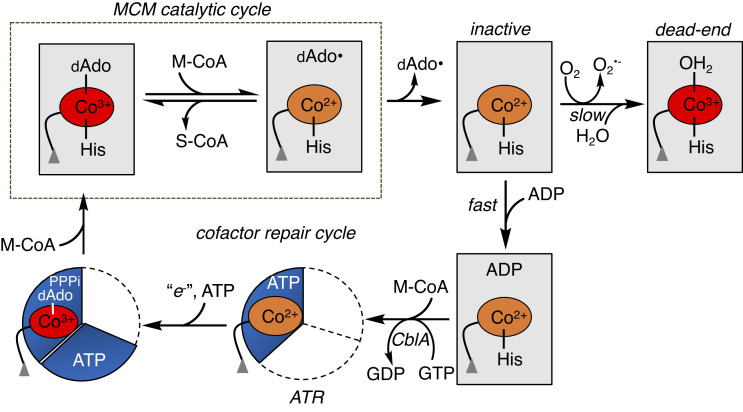
An elaborate system of trafficking proteins ensures the intracellular assimilation of the cobalamin cofactor (generically vitamin B12) into its biologically active derivatives and delivery to its client enzymes: cytoplasmic methionine synthase (MS) and mitochondrial MCM (4–7). Since cobalamin is a scarce micronutrient whose absence in mammals is incompatible with life (8), B12 chaperones also support cofactor repair when either MS or MCM is inactivated. In the mitochondrion, B12 is converted to 5′-deoxyadenosylcobalamin (AdoCbl) by ATP-dependent cob(I)alamin adenosyltransferase (ATR) (9), which doubles as an escort and loads AdoCbl directly onto MCM (Fig. 1, steps 4 and 1) (10–12). AdoCbl bound to human MCM is six-coordinate (6-c); His-627 serves as the lower axial ligand, while the dimethylbenzimidazole tail of the cofactor is tucked in a side pocket (13). If the ATR•AdoCbl•PPPi product complex is unable it off-load the cofactor onto MCM, it triggers a sacrificial cobalt–carbon bond homolysis reaction, yielding cob(II)alamin (step 6), which binds with higher affinity to ATR and averts cofactor release into solution (14, 15).
Fig. 1.
Mitochondrial chaperones support MCM function. Once loaded with AdoCbl from ATR (1), MCM catalyzes multiple rounds of isomerization of M-CoA to succinyl-CoA (S-CoA) (2). The occasional loss of dAdo from the active site leads to inactive cob(II)alamin (3), which in the presence of CblA and GTP is off-loaded onto ATR for repair (4). Formation of H2OCbl on MCM precludes cofactor off-loading (5). If the newly formed AdoCbl is not transferred to MCM, ATR catalyzes a sacrificial cobalt–carbon bond homolysis (6). ATR is a homotrimer and the occupied active sites are shown in blue.
Once loaded with AdoCbl, MCM catalyzes multiple rounds of a radical-based rearrangement of methylmalonyl-CoA to succinyl-CoA (S-CoA) (16, 17), which serves an anaplerotic function, linking cholesterol, odd-chain fatty acid, and branched chain amino acid catabolism to the TCA cycle. The occasional loss of the 5′-deoxyadenosine (dAdo) moiety from the active site precludes completion of the catalytic cycle and leads to cob(II)alamin accumulation (Fig. 1, step 2). Cob(II)alamin can be off-loaded to ATR for repair (step 3) or oxidized to aquo-cobalamin (H2OCbl, (step 5)), forming a dead-end MCM complex. The off-loading process engages a second chaperone, CblA, a GTPase (18), which powers the transfer of cob(II)alamin but not H2OCbl, from MCM to ATR (19, 20). Since H2OCbl bound to MCM is recalcitrant to off-loading, it raises the question as to how cob(II)alamin oxidation and formation of a repair-resistant and inactive MCM•H2OCbl complex are staved off. Mutations in ATR, MCM, or CblA result in methylmalonic aciduria, an autosomal recessive disorder that affects ~1 in 100,000 infants (21).
Proteins exist as ensembles of conformational substates, and allosteric effectors can influence this free energy landscape by altering their distribution (22, 23). While the intricacies of interprotein communication needed for cofactor off-loading from MCM await elucidation, functionally linked conformational changes have been seen in each of the component proteins. In MCM, substrate binding induces a conformational change that seals off solvent access and creates a protective cavity to house radical-based chemistry (13). In ATR on the other hand, ATP binding organizes the roof of the active site, while cob(II)alamin binding fully orders the protein and sequesters the cofactor from solvent (14, 15). The flexibility of the switch elements in CblA (18, 19, 24) is modulated by G-nucleotides, which in turn influence the formation of stable interprotein complexes with MCM as revealed by negative stain electron microscopy (20).
In this study, we report the unexpected discovery that ADP uses bivalent molecular mimicry to preserve the redox state of cob(II)alamin on inactive MCM. We furnish crystallographic evidence that ADP binds in the site vacated by dAdo and substrate and induces a conformational change that plugs solvent access. Mimicry by ADP, an abundant metabolite, is crucial for saving cob(II)alamin from further oxidation and promoting its repair. The latter is mediated by chaperones in a multiprotein complex that ensures recycling of a high-value cofactor needed for mitochondrial propionate metabolism.
Results
Adenine Nucleotides Prevent cob(II)alamin Oxidation.
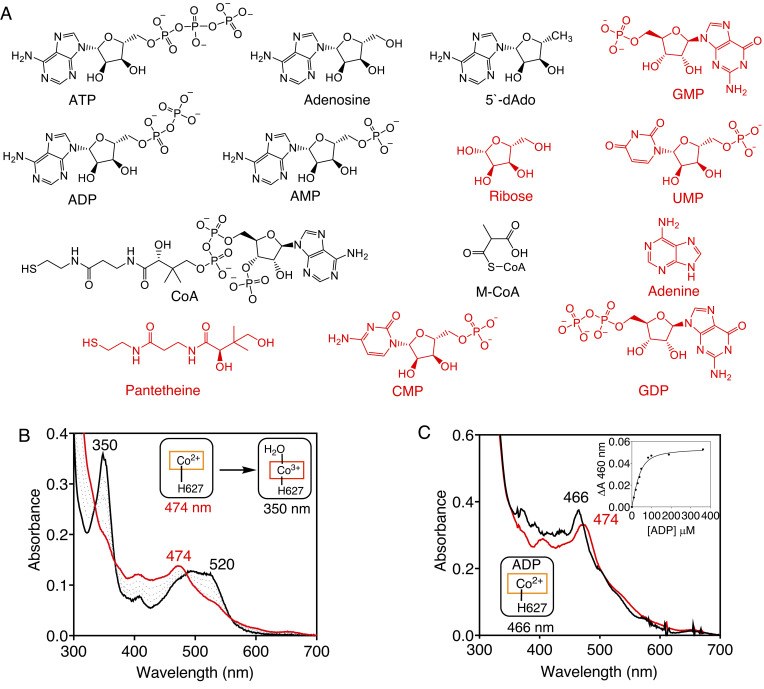
The occasional loss of dAdo during catalytic turnover leads to cob(II)alamin accumulation on MCM that is susceptible to oxidation by molecular oxygen, forming H2OCbl. To identify metabolites that protect against cob(II)alamin oxidation on MCM, we screened 14 compounds (Fig. 2A). In the absence of ligands, human MCM-bound cob(II)alamin oxidizes to H2OCbl (λmax = 350 nm) with a kobs of 0.08 min−1 at 25 ˚C (Fig. 2B). The screen identified adenosine monophosphate (AMP), adenosine diphosphate (ADP), and adenosine triphosphate (ATP) in addition to the expected binders, CoA, M-CoA, and dAdo, each of which induced a blue shift (474 to 466 nm) in the cob(II)alamin spectrum (Fig. 2C) rather than an increase at 350 nm due to H2OCbl formation.
Fig. 2.
MCM•cob(II)alamin oxidation is prevented by metabolites. (A) Structures of metabolites tested for binding to MCM•cob(II)alamin. The binders are displayed in black. (B) Human MCM•cob(II)alamin (20 µM in buffer A, red) oxidizes to OH2Cbl (black) in air. (C) ADP binding to MCM•cob(II)alamin (40 µM in buffer A) is signaled by a small blue shift from 474 to 466 nm (red to black). (Inset) The binding isotherm for ADP was monitored at 466 nm. The spectra are representative of at least three independent experiments.
Next, we determined the dissociation constants for a subset of the metabolites that bind to MCM•cob(II)alamin (Fig. 2 C, Inset and SI Appendix, Fig. S1). In comparison with the relatively high mitochondrial concentrations of CoA (25), ADP and ATP (26), the Kd values were low, supporting physiological relevance for binding to MCM (Table 1). Based on their comparable concentrations but an ~100-fold higher kon value, ADP (8.2 × 103 M−1 min−1) is expected to out compete ATP (7.6 × 101 M−1 min−1) for binding to the MCM•cob(II)alamin complex (SI Appendix, Fig. S2). Since the flux through the B12 trafficking and repair pathway is low (27), and the affinity of MCM•cob(II)alamin for ADP (and ATP) is high, these processes are unlikely to be controlled by cellular energy charge, which is tightly regulated.
Table 1.
Mitochondrial concentrations and dissociation constants of select metabolites that bind to human MCM•cob(II)alamin (25, 26)
| Metabolite | Kd (µM) | Mitochondrial concentration |
|---|---|---|
| ATP | 30 ± 5 µM | 8.0 ± 2.6 mM |
| ADP | 44 ± 4.2 µM | 6.0 ± 1.7 mM |
| 5′-dAdo | 4.2 ± 2.1 µM | NA* |
| CoA | 118 ± 13 µM | 2-5 mM |
| M-CoA | 24.2 ± 6.2 µM | NA |
*NA: not available
The generalizability of metabolite regulation of metal redox state was assessed with Mycobacterium tuberculosis MCM, which plays an important role in energy metabolism in this organism (28). Cob(II)alamin bound to M. tuberculosis MCM was oxidized to OH2Cbl with a kobs of 0.28 min−1 (SI Appendix, Fig. S3 A and B), which was prevented by ADP and M-CoA (SI Appendix, Fig. S3 C and D).
Coordination State of Metabolite-Protected MCM•cob(II)alamim.
The redox potential of the cob(III)alamin/cob(II)alamin couple in solution shifts from +200 mV to +510 mV upon loss of the lower axial nitrogen ligand (29). We therefore assessed whether metabolites modulate the redox potential of MCM-bound cob(II)alamin by inducing loss of the axial histidine ligand donated by the protein. The EPR spectra of human and M. tuberculosis MCM revealed that the axial nitrogen ligand to cob(II)alamin is retained in the presence of ADP, ATP, or CoA as signaled by triplet superhyperfine splittings (SI Appendix, Figs. S4 and S5). Hyperfine interactions between the unpaired electron and the cobalt nucleus (I = 7/2) result in an eight-line spectrum that is further split into triplets by the nitrogen (I = 1) ligand. The hyperfine lines are best resolved in the high-field region of the spectrum. These data are consistent with retention of the His-627 (human) and His-629 (M. tuberculosis) lower axial ligand to cob(II)alamin in MCM in the presence of metabolite.
Structural Basis for Metabolite Control of Metal Redox State.
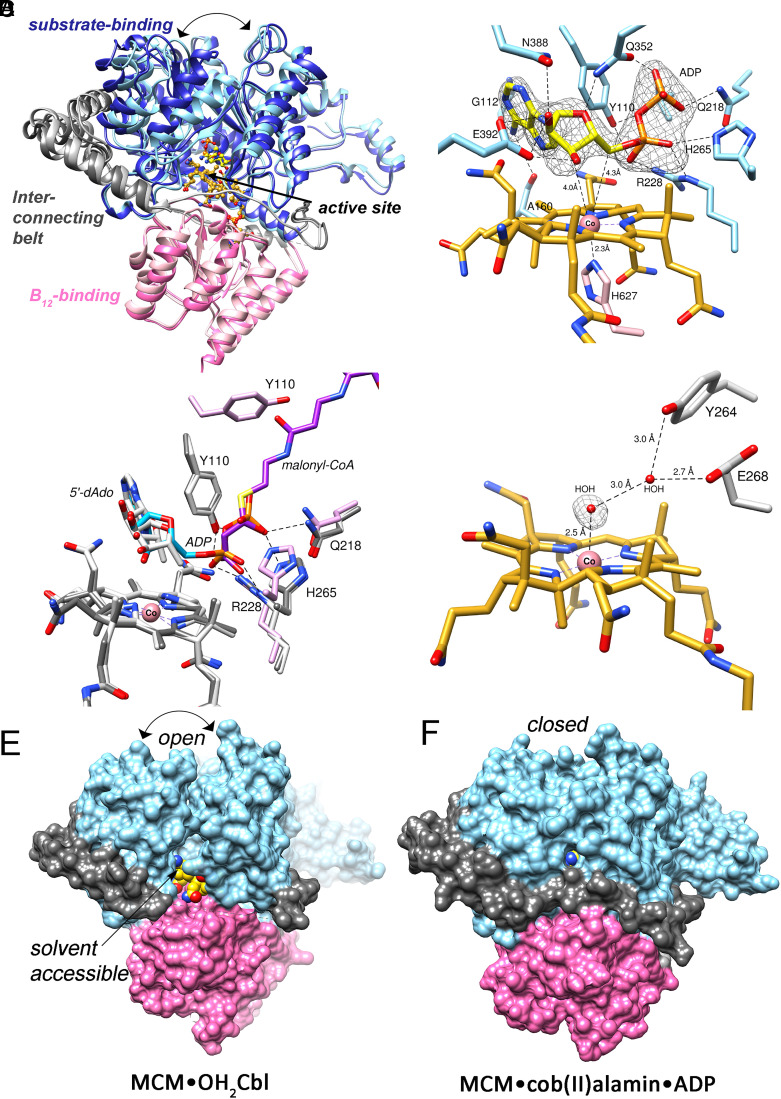
Since none of the available structures of bacterial or human MCM provide clues as to where ADP binds (13, 30, 31), we solved the structures of human MCM•cob(II)alamin in the presence and absence of ADP at 2.2 Å and 1.9 Å resolution (Fig. 3A and SI Appendix, Fig. S6), respectively, using molecular replacement (Table 2). Both crystals belonged to the C 2 2 21 space group with one molecule per asymmetric unit. ADP and cobalamin were modeled into difference density, and each ligand was refined to an occupancy of 1. Surprisingly, ADP binds with its Ado moiety occupying the same position as dAdo in AdoCbl (Fig. 3 B and C). The C3′ and C5′ of ADP are 4.0 Å and 4.3 Å from the cobalt ion, excluding water access (Fig. 3B). ADP makes hydrogen bonds with several active site residues, including the backbones of Ala-160 and Gly-112, and the side chains of Glu-392, Asn-388, Tyr-110, Gln-352, Gln-218, His-265, and Arg-228 (Fig. 3B). His-627 is 2.3 Å away from the cobalt ion, confirming the 5-c state observed by EPR spectroscopy. The diphosphate moiety of ADP overlays on the binding site of the carboxylic acid unit of CoA, revealing a high level of molecular mimicry. Thus, ADP avails some of the same binding interactions that are used by the substrate (M-CoA) and the cofactor (AdoCbl) (Fig. 3 B and C). Interactions between ADP and His-265, Arg-228 and Tyr-110 are also utilized by the substrate analog, malonyl-CoA (Fig. 3C).
Fig. 3.
Structural basis for allosteric regulation by ADP. (A) Overlay of human MCM•cob(II)alamin•ADP (light shades) and MCM•OH2Cbl (dark shades) (Cα rmsd = 1.7 Å). ADP induces closing-in of the substrate domain (light blue) and ordering of the interconnecting belt (light gray). (B) ADP (yellow) binds on the upper face of the corrin ring (gold), precluding access of water to the active site. An Fo-Fc polder omit map of ADP at 3.75 σ is shown (gray mesh). (C) Overlay of MCM•cob(II)alamin•ADP (dark gray), MCM•AdoCbl•malonyl-CoA (PDB 2XIQ, light gray) and MCM•OH2Cbl (pink). The adenine moieties in ADP (blue) and AdoCbl (gray) overlay, while the diphosphate of ADP overlays on the corresponding moiety in malonyl-CoA (purple). (D) A water molecule (red sphere) at 2.5 Å from the cobalt ion (pink sphere) in the MCM•OH2Cbl structure suggests the presence of a weak Co-O bond. A Fo-Fc omit map of water is shown at 2.75 σ (gray mesh). (E) In MCM•OH2Cbl, the substrate binding cavity (blue) is in an “open” conformation, the interconnecting belt (gray) is partially disordered, and the active site is exposed to solvent. (F) In MCM•cob(II)alamin•ADP, the substrate binding cavity is in a “closed” conformation, the interconnecting belt is ordered, and the active site is buried, explaining the role of ADP in protecting cob(II)alamin against oxidation. The B12 domain is shown in pink.
Table 2.
Crystallographic data for human MCM in the presence of cob(II)alamin and ADP
| MCM•OH2Cbl | MCM•Cob(II)•ADP | |
|---|---|---|
| Beamline |
Advanced Photon Source (APS), LS-CAT |
Advanced Photon Source (APS), LS-CAT |
| Wavelength (Å) | 1.127 | 1.127 |
| Temperature (K) | 100 | 100 |
| Space group | C 2 2 21 | C 2 2 21 |
| Cell dimension | ||
| α, β, γ (º) | 90, 90, 90 | 90, 90, 90 |
| a, b, c (Å) | 59.3, 136.1,196 | 61.0, 129.4, 188.4 |
| Resolution (Å) |
41.8-1.9 (1.94-1.9) |
47.1-2.2 (2.27-2.2) |
| Rmerge (%) | 6.6 (72) | 6.1(63) |
| Rmeas (%) | 7.4(81) | 6.9(68) |
| Rpim (%) | 2.5 (33) | 2.2 (23) |
| <I/σ> | 16.7 (2.2) | 15.8(2.7) |
| CC (½) | 1.0 (0.66) | 0.99(0.97) |
| Completeness (%) | 98.4 (82.5) | 99.2 (95.3) |
| Multiplicity | 8.3 (4.6) | 9.0 (8.0) |
| No. of reflections | 509584(14988) | 343282(24826) |
| No. of unique reflections | 61448(3258) | 37982(3114) |
|
Overall B (Å2) (Wilson plot) |
24 | 51 |
| Resolution range | 39.7-1.9 | 47.1-2.2 |
| Number of reflections (work/test) | 61384/3126 | 37922/1902 |
| Rwork /Rfree (%) | 17.5/20.9 | 19.5/22.5 |
| No. of atoms | ||
| Protein | 5315 | 5433 |
| Water | 246 | 121 |
| Ligand: B12 | 91 | 91 |
| ADP | NA | 27 |
| B-factors(Å2) | ||
| Protein | 35.3 | 66.3 |
| Ligand: B12 | 34.2 | 48.1 |
| ADP | NA | 56.8 |
| Water | 36.46 | 54.1 |
| rmsd deviations | ||
| Bond lengths (Å) | 0.007 | 0.003 |
| Bond angles (º) | 0.822 | 0.556 |
| Ramachandran plot (%) | ||
| Favored, allowed, outliers | 98.2, 1.8, 0 | 97.9, 2.1, 0 |
| MolProbity score (percentile) | 0.99 (100) | 1.1 (100) |
| PDB code | 8DYL | 8DYJ |
In the structure lacking ADP, difference density consistent with a water molecule is seen at the upper axial face of the corrin ring, which is consistent with the presence of H2OCbl (Fig. 3D). The water is, however, 2.5 Å from the cobalt ion, which is 0.6 Å longer than in the structure of free H2OCbl (1.9 Å), suggesting a weak Co-O bond (32). A hydrogen bonding network involving Tyr-264 and Glu-268 secures the position of the water ligand. A structural comparison reveals that ADP induces a conformation transition in the substrate domain from an open to a closed state and ordering of amino acids 498 to 506 in the ~100 residue long belt between the substrate- and B12-binding domains that circumscribes the active site (Fig. 3E, gray). Solvent access to the active site in the open conformation explains the oxidation propensity of cob(II)alamin. ADP binding repositions four α-helices to close off the substrate domain, precluding access to O2 for cob(II)alamin oxidation, and to H2O, the upper axial ligand in H2OCbl, which prefers a 6-c geometry. A comparable structural change is induced by the substrate analog, malonyl-CoA, which helps position the substrate for catalysis and protects radical intermediates from bulk solvent (13). We predict that ATP binds like ADP with the extra phosphate group overlapping more of the CoA tail of the substrate and induces a comparable conformational change in MCM.
Substrate Binding Promotes Cofactor Repair.
The CblA chaperone helps off-load cob(II)alamin from MCM to ATR (Fig. 1), which is signaled by an increase in the absorption intensity at 464 nm (SI Appendix, Fig. S7A) (20). Under anaerobic conditions, which preclude cob(II)alamin oxidation, ADP inhibits the rate and extent of cofactor transfer from MCM to ATR, while CoA and M-CoA enhance it (Table 3). Further, M-CoA reverses the inhibitory effect of ADP on the rate and percent transfer of cob(II)alamin (Table 3 and SI Appendix, Fig. S7B), suggesting a possible role for CoA or substrate in controlling the timing of cofactor repair.
Table 3.
CblA-dependent cob(II)alamin off-loading from human MCM to ATR*
| Metabolite | Rate of transfer (kobs) | % Completion |
|---|---|---|
| No metabolite | 0.59 ± 0.01 min−1 | 92 to 95 % |
| ADP | 0.24 ± 0.01 min−1 | 28 to 30 % |
| M-CoA | 0.77 ± 0.08 min−1 | 93 to 95 % |
| CoA | 0.92 ± 0.06 min−1 | 94 to 96 % |
| ADP and M-CoA | 0.24 ± 0.02 min−1 | 90 to 93 % |
*The data represent the mean ± SD of three independent experiments.
M-CoA modulates Cofactor Loading onto MCM.
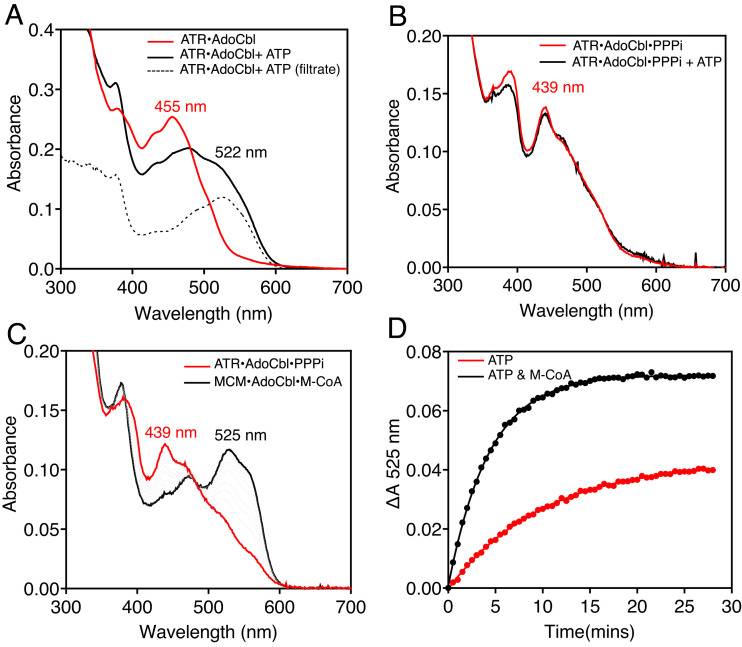
AdoCbl synthesis leads to a ternary product complex on ATR with triphosphate (PPPi) and AdoCbl (Fig. 1). PPPi, which is tightly associated (Kd < 0.4 µM), is critical for controlling the fate of the newly formed AdoCbl (14). Loss of PPPi from the ternary ATR•AdoCbl•PPPi product complex weakens the affinity for AdoCbl, and, upon binding of ATP to a vacant active site in the homotrimeric protein, the cofactor is released into solution (Fig. 4A) as described previously (14). However, in the presence of PPPi, AdoCbl is retained on ATR even in the presence of ATP (Fig. 4B). When M-CoA is added to the ternary product complex on ATR, ATP triggers AdoCbl transfer as signaled by the spectral shift from 439 nm (on ATR) to 525 nm (on MCM), reflecting the change from a 5- to 6-c environment (Fig. 4C). The rate and extent of AdoCbl transfer are promoted by M-CoA (Fig. 4D and SI Appendix, Table S1).
Fig. 4.
Metabolites regulate AdoCbl loading from human ATR onto MCM. (A) ATP triggers release of 40% AdoCbl (black) from ATR (red) into solution (dashed lines) as confirmed following separation using a 10 kDa cutoff Centricon YM10 filter. (B) The ternary ATR•AdoCbl•PPPi product complex (red) prevents cofactor release into solution in the presence of ATP (black). (C) AdoCbl loading onto MCM (black) from the ternary product complex on ATR (red) is signaled by a large spectral shift as AdoCbl converts from a 5- to 6-c state. (D) AdoCbl loading onto MCM (ΔA 525 nm) is promoted by M-CoA.
Discussion
Molecular recognition via noncovalent interactions is widely used for intra- and intermolecular signaling in biology. In metallocofactor trafficking pathways, the task of translocating reactive cargo via transient or stable protein complexes requires exquisite control of metal oxidation and coordination states. Elaborate strategies have evolved to traffic individual metals like copper ions, assemblies like iron-sulfur clusters, and organometallic complexes like cobalamin to client proteins (7, 33, 34). As the molecular details of these complex metal trafficking pathways continue to emerge, insights into the potential involvement of metabolites to control metal redox or coordination [e.g., glutathione, which serves as an iron ligand in glutaredoxin 2 (35)] remain sparse. In this study, we report that ADP functions as a bivalent mimic of the AdoCbl cofactor and the M-CoA substrate of MCM and protects cob(II)alamin from hyperoxidation to facilitate its repair.
The movement of AdoCbl to and cob(II)alamin from MCM is dependent on chaperones (Fig. 1). In the forward direction, coordination chemistry is deployed for moving AdoCbl from a low (5-c) coordination environment on ATR to a high coordination (6-c) environment on MCM with the histidine ligand playing a key role (10). While details of cofactor movement in the reverse direction await elucidation, overoxidation poses a problem since H2OCbl prefers to be 6-c [water and His-627 on human MCM serve as the axial ligands (Fig. 3D)]. A dissociative ligand exchange mechanism for off-loading H2OCbl via a 5-c cob(III)alamin species is kinetically disfavored and explains the importance of preventing formation of a dead-end MCM•H2OCbl complex (Fig. 1).
Crystallization of the MCM•cob(II)alamin complex led to the capture of two distinct conformations in the absence (“open”) and presence (“closed”) of ADP (Fig. 3), revealing the molecular mechanism for cob(II)alamin sequestration. The strategy of using ADP binding energy to elicit a protein conformational change benefits from the abundance of this metabolite in the cell and the combined use of the same binding interactions that orient the substrate and intact cofactor on MCM (Fig. 3C). This strategy is reminiscent of the rational design of multisubstrate analogs as drugs and as mechanistic probes of enzyme reactions in which at least two substrates or a substrate and a cofactor occupy the active site simultaneously (36). Binding synergy and specificity are harnessed from the combination of binding interactions resulting from the covalent tethering of a substrate–cofactor or from two or more substrates (37). Assuming a cellular ADP concentration of 6 mM (26), the rate of ADP binding is predicted to be ~600-fold faster than the oxidation of cob(II)alamin on MCM.
We can now include metabolite regulation as an important element of the mitochondrial B12 trafficking pathway (Fig. 5). Substrate binding to holo-MCM triggers a switch from the open to closed conformation, securing the active site from solvent, and supporting the radical-based transformation of M-CoA to succinyl-CoA. The occasional loss of dAdo leaves cob(II)alamin exposed in an open active site and facing one of two fates: a kinetically slow oxidation to H2OCbl and the kinetically favored protection conferred by ADP binding, which triggers the closed conformation. Cob(II)alamin off-loading to ATR from this closed state is promoted by M-CoA binding to MCM and requires guanosine triphosphate (GTP) hydrolysis by CblA (20). While the closed turnover (with AdoCbl and M-CoA) and repair (with cob(II)alamin and ADP) complexes differ in terms of their ligands, mechanistic insights into how these differences cue recruitment of the repair proteins await structural elucidation. Once off-loaded, AdoCbl is resynthesized by ATR and reloaded onto MCM, in a process that is promoted by substrates binding to ATR (ATP) (11) and MCM (M-CoA).
Fig. 5.
Model for metabolite-regulated coenzyme B12 trafficking. Loss of dAdo from MCM during the catalytic cycle leads to inactivation of MCM. The slow oxidation of the resulting MCM•cob(II)alamin is averted by the fast binding of ADP, which prevents formation of a catalytically dead MCM•OH2Cbl complex. The CblA chaperone uses GTP hydrolysis to power cofactor off-loading from MCM, which is modulated by the availability of the substrate, M-CoA. Following one-electron reduction and adenosylation, the ternary ATR•AdoCbl•PPPi complex results, which on-loads the repaired cofactor onto MCM, in a process that is triggered by ATP and M-CoA binding to ATR and MCM, respectively.
In the presence of ATP, ATR with one equivalent of AdoCbl bound ejects the cofactor into solution, which was first described with the Methylobacterium extorquens protein (11). Our study now addresses the conundrum of ATP-triggered release of AdoCbl into solution versus its transfer to MCM. Unlike the ATR•AdoCbl binary complex, the cofactor is bound tightly in the ATR•AdoCbl•PPPi ternary product complex and ATP binding promotes AdoCbl transfer to MCM but not its release into solution (Fig. 4 B and C). These data suggest that ATP binding supports the ordered release of products with AdoCbl transfer preceding PPPi release. Interestingly, loss of the C-terminal 16 residues in ATR (resulting from a Q234X truncation mutation) is associated with disease (38). While introduction of the corresponding truncation mutation in the M. extorquens ATR had only modest effects on enzyme activity, it led to the loss of ATP-dependent allosteric communication between the active sites and to a significantly weakened affinity for AdoCbl (39).
In summary, our study reveals the multiple and strategic use of protein-metabolite interactions to control metal redox state, as well as the timing and specificity of cofactor transfer in the B12 trafficking pathway. The application of global approaches for the systematic identification of metabolite–protein interactions will lead to a fuller understanding of the extent to which common metabolites are deployed as biological modifiers for pathway and network regulation (40, 41).
Materials and Methods
Materials.
adenosine triphosphate (ATP), adenosine diphosphate (ADP), adenosine monophosphate (AMP), Coenzyme A (CoA), guanosine triphosphate (GTP), guanosine diphosphate (GDP), guanosine monophosphate (GTP), uridine monophosphate (UMP), β,γ-Methyleneguanosine 5′-triphosphate (GMPPCP), sodium triphosphate pentabasic (PPPi), and AdoCbl were from Sigma-Aldrich. Isopropyl β-D-1-thiogalactopyranoside (IPTG) and Tris (2-carboxyethyl) phosphine (TCEP) were from gold biotechnology. Ni(II)-NTA resin was from Qiagen.
Expression and Purification of Human and M. tuberculosis Proteins.
The recombinant human (MCM, CblA, and ATR) and M. tuberculosis (MCM) proteins were expressed in Escherichia coli and purified as described (28, 42, 43). The proteins were stored in Buffer A (50 mM HEPES, 150 mM KCl, 2 mM MgCl2, 2 mM TCEP 5% glycerol pH 7.5), which was also used in experiments unless noted otherwise. The protein concentrations refer to those for the homodimer (human MCM and CblA), trimer (human ATR), and heterodimer (M. tuberculosis MCM).
M-CoA Synthesis.
M-CoA was synthesized as described (28). The concentration of M-CoA was estimated spectrally after resuspending the lyophilized powder in water, using ε258nm = 19.84 mM−1. Prior to use, the M-CoA stock was diluted 1:1 with 500 mM sodium phosphate, pH 7.5.
Metabolite Binding to MCM•cob(II)alamin.
Binding of metabolites to MCM•cob(II)alamin was monitored in an omni-lab anaerobic chamber (Vacuum atmospheres) at <0.2 ppm O2 levels. The stock cob(II)alamin solution was prepared by photolysis of 1.2 mM AdoCbl dissolved in anaerobic water. The concentration of cob(II)alamin was determined by using ε474nm= 9.2 cm−1 mM−1. The metabolites (5 mM each) were screened for binding following their addition to a solution containing 40 µM cob(II)alamin and 20 µM MCM (dimer concentration) in Buffer A at 20 °C, and the spectrum was recorded after 15 min. The change in cob(II)alamin absorbance from 474 nm to 466 nm was observed to detect ligand binding. Dissociation constants were determined by monitoring the change in absorbance at 460 nm, 10 min after addition of increasing concentrations of each metabolite to a solution containing 40 µM cob(II)alamin and 20 µM MCM (dimer concentration) in Buffer A at 20 °C. The data were fitted to Eq. 1 using the Prism software to obtain the respective Kd values (A = absorbance at 460 nm, [E]t= [MCM] added, [L]t= [cob(II)alamin]).
| [1] |
Stopped-Flow Spectroscopy to Determine ADP/ATP Binding to MCM•cob(II)alamin.
All stopped-flow experiments were performed under anaerobic conditions at 25 °C using an Applied Photophysics double mixing stopped-flow spectrometer equipped with a photodiode array detector (300 to 700 nm) and housed in an omni-lab anaerobic chamber (<0.2 ppm O2 levels). A stock solution of MCM•cob(II)alamin was prepared by mixing 80 µM cob(II)alamin and 40 µM MCM (dimer) in Buffer A for 10 min. MCM•cob(II)alamin was then rapidly mixed with various concentrations (0.25 to 2.5 mM) of ADP or ATP, and the change in absorbance at 460 nm was monitored and fit to single exponential (Eq. 2) to determine the observed rate constant (kobs).
| [2] |
| [3] |
The dependence of kobs on the metabolite concentration was fitted by linear regression to determine the bimolecular association rate constant (kon) for ATD/ATP binding to MCM•cob(II)alamin (Eq. 3). The koff was derived using the Kd value determined from the binding isotherm and the kon.
Kinetics of MCM•cob(II)alamin Oxidation.
Samples containing 40 µM cob(II)alamin, 20 µM MCM (dimer) 60 µM CblA, and 1 mM G-nucleotides in anaerobic Buffer A were prepared, and cob(II)alamin oxidation was initiated by diluting the sample twofold with aerobic buffer. H2OCbl formation was monitored by the increase in absorption at 350 nm and plotted against time to determine the kobs for cob(II)alamin oxidation.
Kinetics of cob(II)alamin Off-Loading from MCM.
Transfer of cob(II)alamin from MCM to ATR was performed in an anaerobic chamber by adding a solution containing 15 µM ATR, 5 mM ATP, and 1 mM GTP to 15 µM cob(II)alamin, 9 µM MCM, and 30 µM CblA in Buffer A at 25 ˚C. The role of metabolites was evaluated by adding 5 mM ADP or 0.5 mM M-CoA to the MCM solution. The change in absorption at 464 nm was monitored over 20 min and used to determine the extent of cob(II)alamin transfer, using = 5.6 cm−1 mM−1. The change in 464 nm absorbance was fitted to a single exponential equation (Eq. 2) to determine the kobs for cob(II)alamin transfer.
Kinetics of AdoCbl Loading onto MCM.
AdoCbl transfer was initiated by adding 20 µM MCM to 20 µM AdoCbl and 30 µM ATR in the presence or absence of 100 µM triphosphate in anaerobic Buffer A at 25 ˚C, and spectra were recorded every 30 s for a total of 30 min. The concentration of AdoCbl was determined by using the = 8.0 cm−1 mM−1. The amount of AdoCbl transferred to MCM was estimated using the = 7.2 mM−1 cm−1, and the kobs for AdoCbl transfer was determined by fitting the change in 525 nm absorbance to Eq. 1.
EPR Spectroscopy.
EPR spectra at 80 K were recorded on a Bruker EPR instrument model (EMX) 300 equipped with a Bruker 4201 cavity and a ColdEdge cryostat. The temperature was controlled by an Oxford Instruments MercuryiTC temperature controller. The following parameters were used: 9.38 GHz microwave frequency, power 2 mW, modulation amplitude 10 G, modulation frequency 100 kHz, 3000 G sweep width centered at 3,500 G, conversion time 164 ms, and time constant 82 ms. Samples contained 100 µM cob(II)alamin, 5 mM metabolites (ATP/ADP/CoA), and 100 µM MCM dimer (human) or 200 µM (M. tuberculosis) in anaerobic 50 mM HEPES pH 7.5, 150 mM KCl, 2 mM MgCl2, 2 mM TCEP, and 10% glycerol. Five scans were acquired for each sample. The spectrum of buffer only was subtracted from the sample spectrum.
Crystallization of MCM•cob(II)alamin and MCM•cob(II)alamin•ADP.
The MCM•cob(II)alamin complex was prepared anaerobically by incubating 0.15 mM protein and 0.30 mM cob(II)alamin in buffer containing 100 mM Tris, pH 7.5, 2 mM MgCl2, and 2 mM TCEP for 15 min. Crystals of MCM-cob(II)alamin appeared within a week in sitting drop trays. Crystals with the best diffraction were obtained from a 2 µL drop containing 1:1 protein (12 mg/mL) to well solution (0.2 M ammonium sulfate, 0.1 M bis(2-hydroxyethyl)-imino-tris(hydroxymethyl)-methane (BIS-TRIS), pH 5.5, 16% PEG 3350). Crystals were soaked in a cryoprotect solution (well solution supplemented with 25 % glycerol) for 10 min and flash-frozen in liquid nitrogen.
MCM•cob(II)alamin•ADP was prepared anaerobically by incubating 0.34 mM protein, 0.4 mM cob(II)alamin, 4 mM ADP, and 5 mM MgCl2 in buffer containing 100 mM Tris, pH 7.5, 2 mM MgCl2, and 2 mM TCEP for 1 h. Crystals of MCM•cob(II)alamin•ADP were set up aerobically using the hanging drop vapor diffusion method. The crystals with the best diffraction were obtained after 2 wk in a 1.5 µL drop containing 1:1 or 2:1 protein (15 mg/mL) to well solution (0.1 M lithium sulfate monohydrate, 0.1 M sodium acetate trihydrate, pH 4.5, 54% PEG 400). Crystals were soaked in well solution supplemented with 5 mM ADP for 10 min and flash-frozen in liquid nitrogen.
Data Collection, Processing, and Refinement.
Diffraction data were collected at Life Sciences-Collaborative Access Team (LS-CAT) (21-ID-D) at Argonne National Laboratory. The data were indexed and integrated using Xia2 DIALS and scaled in Aimless (44). Structures of MCM•OH2Cbl and MCM•cob(II)alamin•ADP were solved by molecular replacement with a previously solved structure of MCM (PDB 2XIJ or PDB 2XIQ) using Phaser (45) in the CCP4 program suit. Crystals of MCM•OH2Cbl and MCM•cob(II)alamin•ADP belonged to the space group C 2 2 21 with one chain per asymmetric unit. Iterative rounds of model building and refinement were performed with COOT (46) and Phenix (47). Ligand restraints were generated in eLBOW (48). The geometric quality of the model was assessed in MolProbity (49). Models were analyzed, and the structure figures were generated using UCSF Chimera (50).
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
This work was supported in part by grants from the NIH (K99-GM1434820 to RM and RO1-DK45776 to R.B.). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant 085P1000817).
Author contributions
H.G., R.M., M.R., and R.B. designed research; H.G., R.M., M.R., and M.Y. performed research; H.G., R.M., M.R., and R.B. analyzed data; and H.G., R.M., and R.B. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All data are available in the manuscript or SI Appendix. The structure factors and coordinates for human MCM•H2OCbl (8DYL) and MCM•Cob(II)alamin•ADP (8DYJ) have been deposited in the Protein Data Bank.
Supporting Information
References
- 1.Feng Y., et al. , Global analysis of protein structural changes in complex proteomes. Nat. Biotechnol. 32, 1036–1044 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Ribeiro A. A., Ortiz V., A chemical perspective on allostery. Chem. Rev. 116, 6488–6502 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Kern D., Zuiderweg E. R., The role of dynamics in allosteric regulation. Curr. Opin. Struct. Biol. 13, 748–757 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Banerjee R., B12 trafficking in mammals: A case for coenzyme escort service. ACS Chem. Biol. 1, 149–159 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Banerjee R., Gherasim C., Padovani D., The tinker, tailor, soldier in intracellular B12 trafficking. Cur. Op. Chem. Biol. 13, 484–491 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gherasim C., Lofgren M., Banerjee R., Navigating the B12 road: Assimilation, delivery and disorders of cobalamin. J. Biol. Chem. 288, 13186–13193 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Banerjee R., Gouda H., Pillay S., Redox-linked coordination chemistry directs vitamin B12 trafficking. Acc. Chem. Res. 54, 2003–2013 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Green R., et al. , Vitamin B12 deficiency. Nat. Rev. Dis. Primers 3, 17040 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Yamanishi M., Labunska T., Banerjee R., Mirror “base-off” conformation of coenzyme B12 in human adenosyltransferase and its downstream target, methylmalonyl-CoA mutase. J. Am. Chem. Soc. 127, 526–527 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Padovani D., Labunska T., Palfey B. A., Ballou D. P., Banerjee R., Adenosyltransferase tailors and delivers coenzyme B12. Nat. Chem. Biol. 4, 194–196 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Padovani D., Banerjee R., A rotary mechanism for coenzyme B12 Synthesis by adenosyltransferase. Biochemistry 48, 5350–5357 (2009). [DOI] [PubMed] [Google Scholar]
- 12.Yamanishi M., Vlasie M., Banerjee R., Adenosyltransferase: An enzyme and an escort for coenzyme B12? Trends Biochem. Sci. 30, 304–308 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Froese D. S., et al. , Structures of the human GTPase MMAA and vitamin B12-dependent methylmalonyl-CoA mutase and insight into their complex formation. J. Biol. Chem. 285, 38204–38213 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Campanello G. C., et al. , Sacrificial cobalt-carbon bond homolysis in coenzyme B12 as a cofactor conservation strategy. J. Am. Chem. Soc. 140, 13205–13208 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mascarenhas R., Ruetz M., McDevitt L., Koutmos M., Banerjee R., Mobile loop dynamics in adenosyltransferase control binding and reactivity of coenzyme B12. Proc. Natl. Acad. Sci. U.S.A. 117, 30412–30422 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Banerjee R., Radical peregrinations catalyzed by coenzyme B12-dependent enzymes. Biochemistry 40, 6191–6198 (2001). [DOI] [PubMed] [Google Scholar]
- 17.Banerjee R., Radical carbon skeleton rearrangements: Catalysis by coenzyme B12-dependent mutases. Chem. Rev. 103, 2083–2094 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Lofgren M., Padovani D., Koutmos M., Banerjee R., A switch III motif relays signaling between a B12 enzyme and its G-protein chaperone. Nat. Chem. Biol. 9, 535–539 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Campanello G. C., M.Lofgren, A. L. Yokom, D. R. Southworth, R. Banerjee, Switch I-dependent allosteric signaling in a G-protein chaperone-B12 enzyme complex. J. Biol. Chem. 292, 17617–17625 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruetz M., et al. , Allosteric regulation of oligomerization by a B12 trafficking G-protein is corrupted in methylmalonic aciduria. Cell Chem. Biol. 26, 960–969.e964 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watkins D., Rosenblatt D. S., Inborn errors of cobalamin absorption and metabolism. Am. J. Med. Genet. C Semin. Med. Genet. 157C, 33–44 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Goodey N. M., Benkovic S. J., Allosteric regulation and catalysis emerge via a common route. Nat. Chem. Biol. 4, 474–482 (2008). [DOI] [PubMed] [Google Scholar]
- 23.Nussinov R., Wolynes P. G., A second molecular biology revolution? The energy landscapes of biomolecular function. Phys. Chem. Chem. Phys. 16, 6321–6322 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Lofgren M., Koutmos M., Banerjee R., Autoinhibition and signaling by the switch II motif in the G-protein chaperone of a radical B12 enzyme. J. Biol. Chem. 288, 30980–30989 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gout I., Coenzyme A, protein CoAlation and redox regulation in mammalian cells. Biochem Soc. Trans. 46, 721–728 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Traut T. W., Physiological concentrations of purines and pyrimidines. Mol. Cell Biochem. 140, 1–22 (1994). [DOI] [PubMed] [Google Scholar]
- 27.Hannibal L., et al. , Processing of alkylcobalamins in mammalian cells: A role for the MMACHC (cblC) gene product. Mol. Genet. Metab. 97, 260–266 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruetz M., et al. , Itaconyl-CoA forms a stable biradical in methylmalonyl-CoA mutase and derails its activity and repair. Science 366, 589–593 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lexa D., Saveant J.-M., The electrochemistry of vitamin B12. Acc. Chem. Res. 16, 235–243 (1983). [Google Scholar]
- 30.Mancia F., et al. , How coenzyme B12 radicals are generated: The crystal structure of methylmalonyl-coenzyme A mutase at 2 Å resolution. Structure 4, 339–350 (1996). [DOI] [PubMed] [Google Scholar]
- 31.Mancia F., Evans P. R., Conformational changes on substrate binding to methylmalonyl CoA mutase and new insights into the free radical mechanism. Structure 6, 711–720 (1998). [DOI] [PubMed] [Google Scholar]
- 32.Kratky C., et al. , Accurate structural data demystify B12: High-resolution solid-state structure of Aquocobalamin perchlorate and structure analysis of the aquocobalamin ion in solution. J. Am. Chem. Soc. 117, 4654–4670 (1995). [Google Scholar]
- 33.Camponeschi F., Banci L., Metal cofactors trafficking and assembly in the cell: A molecular view. Pure. App. Chem. 91, 231–245 (2019). [Google Scholar]
- 34.Lill R., Function and biogenesis of iron-sulphur proteins. Nature 460, 831–838 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Iwema T., et al. , Structural basis for delivery of the intact [Fe2S2] cluster by monothiol glutaredoxin. Biochemistry 48, 6041–6043 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Wolfenden R., Analog approaches to the structure of the transition state in enzyme reactions. Acc. Chem. Res. 5, 10–18 (1972). [Google Scholar]
- 37.Le Calvez P. B., Scott C. J., Migaud M. E., Multisubstrate adduct inhibitors: Drug design and biological tools. J. Enzyme. Inhib. Med. Chem. 24, 1291–1318 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Lerner-Ellis J. P., et al. , Mutation and biochemical analysis of patients belonging to the cblB complementation class of vitamin B12-dependent methylmalonic aciduria. Mol. Genet. Metab. 87, 219–225 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Lofgren M., Banerjee R., Loss of allostery and coenzyme B12 delivery by a pathogenic mutation in adenosyltransferase. Biochemistry 50, 5790–5798 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Orsak T., et al. , Revealing the allosterome: Systematic identification of metabolite-protein interactions. Biochemistry 51, 225–232 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Milroy L. G., Grossmann T. N., Hennig S., Brunsveld L., Ottmann C., Modulators of protein-protein interactions. Chem. Rev. 114, 4695–4748 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Li Z., et al. , The human B12 trafficking chaperones: CblA, ATR, CblC and CblD. Methods Enzymol. 668, 137–156 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mascarenhas R., Gouda H., Ruetz M., Banerjee R., Human B12-dependent enzymes: Methionine synthase and Methylmalonyl-CoA mutase. Methods Enzymol. 668, 309–326 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Winn M. D., et al. , Overview of the CCP4 suite and current developments. Acta Crystallogr. D. 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McCoy A. J., et al. , Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr D. Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liebschner D., et al. , Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. D. Struct. Biol. 75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moriarty N. W., Grosse-Kunstleve R. W., Adams P. D., electronic Ligand Builder and Optimization Workbench (eLBOW): A tool for ligand coordinate and restraint generation. Acta Crystallogr. D. Biol. Crystallogr. 65, 1074–1080 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Williams C. J., et al. , MolProbity: More and better reference data for improved all-atom structure validation. Protein. Sci. 27, 293–315 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pettersen E. F., et al. , UCSF Chimera–A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All data are available in the manuscript or SI Appendix. The structure factors and coordinates for human MCM•H2OCbl (8DYL) and MCM•Cob(II)alamin•ADP (8DYJ) have been deposited in the Protein Data Bank.