Significance
Double-click chemistry enables high-throughput synthesis of triazole compound libraries from primary amines. However, it remains challenging to exploit large-scale triazole libraries and identify bioactive compounds in an efficient and economic manner. Herein, we built a new platform for the design, synthesis, and screening of double-click triazole libraries, which was showcased on an extremely attractive drug target for the treatment of type 2 diabetes and obesity. From screening customized triazole libraries, we rapidly identified a series of new modulators that can enhance the protein target activity in the presence of its native peptide ligand, and further discovered an unexpected binding mode of the new modulator. Our approach is anticipated to expedite the discovery of new drug candidates for various therapeutic targets.
Keywords: click chemistry, affinity selection mass spectrometry, GLP-1 receptor, allosteric modulators
Abstract
The recently developed double-click reaction sequence [G. Meng et al., Nature 574, 86–89 (2019)] is expected to vastly expand the number and diversity of synthetically accessible 1,2,3-triazole derivatives. However, it remains elusive how to rapidly navigate the extensive chemical space created by double-click chemistry for bioactive compound discovery. In this study, we selected a particularly challenging drug target, the glucagon-like-peptide-1 receptor (GLP-1R), to benchmark our new platform for the design, synthesis, and screening of double-click triazole libraries. First, we achieved a streamlined synthesis of customized triazole libraries on an unprecedented scale (composed of 38,400 new compounds). By interfacing affinity-selection mass spectrometry and functional assays, we identified a series of positive allosteric modulators (PAMs) with unreported scaffolds that can selectively and robustly enhance the signaling activity of the endogenous GLP-1(9-36) peptide. Intriguingly, we further revealed an unexpected binding mode of new PAMs which likely act as a molecular glue between the receptor and the peptide agonist. We anticipate the merger of double-click library synthesis with the hybrid screening platform allows for efficient and economic discovery of drug candidates or chemical probes for various therapeutic targets.
Click chemistry, as formalized in 2001, defines a handful of powerful, highly reliable, and selective reactions to enable facile synthesis of structurally new and diverse compounds with desirable properties (1). Copper-catalyzed azide–alkyne cycloaddition (CuAAC) click reaction has become a prime example of click chemistry owning to its high reaction efficiency, remarkable chemo- and regioselectivities, and wide substrate scope (2–4). Despite the power of CuAAC reactions and the essential role of azides in bio-orthogonal chemistry (5), the availability of azide reagents is limited owing to the risk, in some cases, of explosion during their preparation. To address this issue, we recently developed another click reaction using FSO2N3 as a diazotransfer reagent to rapidly convert almost any primary amines into an azide in a convenient and safe manner (6). The new reaction was then combined with CuAAC to build a double-click reaction sequence that achieved a streamlined synthesis of a 1,224-member triazole library from primary amines in microplates (6).
Although this double-click reaction scheme is envisaged to expediate the modular synthesis of 1,2,3-triazole libraries of substantially increased diversity, its potential in generating bioactive compounds for drug discovery has never been explored. How to design a diverse library based on double-click chemistry and how to exploit the chemical space created by diversity-oriented synthesis remain largely unknown. Furthermore, an efficient and economic screening platform is required to merge with the facile synthesis of large-scale triazole libraries. Affinity selection mass spectrometry (AS-MS), which has been mainly employed to detect chemical ligands bound to a protein target from pools of synthetic compounds (7–12), could partially serve the purpose. We expect putative ligands of high affinity for the target would be captured by the AS-MS screen of crude double-click reaction product mixtures, though it had not been demonstrated before. However, relying on the AS-MS screen alone would possibly miss certain ligands of low affinity for the recombinant protein target yet still displaying considerable bioactivity in cells.
In this study, we attempted to design, synthesize, and screening double-click triazole libraries in order to discover new allosteric modulators for a highly challenging drug target, the glucagon-like-peptide-1 receptor (GLP-1R). GLP-1R is the target of several approved peptide drugs for the treatment of type 2 diabetes mellitus and obesity (13, 14). The identification of drug-like small molecules to activate the GLP-1R has aroused wide interest with a few small molecule agonists entering clinical testing (15–17). Alternatively, small-molecule positive allosteric modulators (PAMs) of the GLP-1R could serve as another pharmacological strategy for this proven therapeutic target, given that PAMs could selectively enhance the normal GLP-1 response during nutrient consumption and have the potential for producing reduced side effects (18–20). A collection of PAMs of the GLP-1R have been reported by pharmaceutical companies through high-throuput screening (HTS) of various proprietary compound libraries followed by extensive chemical optimization of the HTS hits (19–26). The exhaustive screens of synthetic libraries have rendered the discovery of potent PAMs of unreported chemotypes extremely difficult.
Herein, we conducted a double-click synthesis of triazole libraries consisting of 38,400 new compounds, which were designed based on a known ligand that antagonizes the GLP-1R activity. Then we integrated the AS-MS screen with the cell-based screen to not only improve the screening efficiency but also capture bioactive ligands to a maximal degree. Through iterative library design and synthesis, a series of potent PAMs with unreported chemical scaffolds were obtained and they exhibited strong probe dependence and signaling bias. Unexpectedly, new PAMs turned out to target a unique extracellular pocket of the receptor which is distinct from the binding site of the initial ligand, illustrating the power of double-click chemistry in creating small-molecule modulators acting by an unpredictable mechanism.
Results
Design and Synthesis of 10 Double-Click Triazole Libraries.
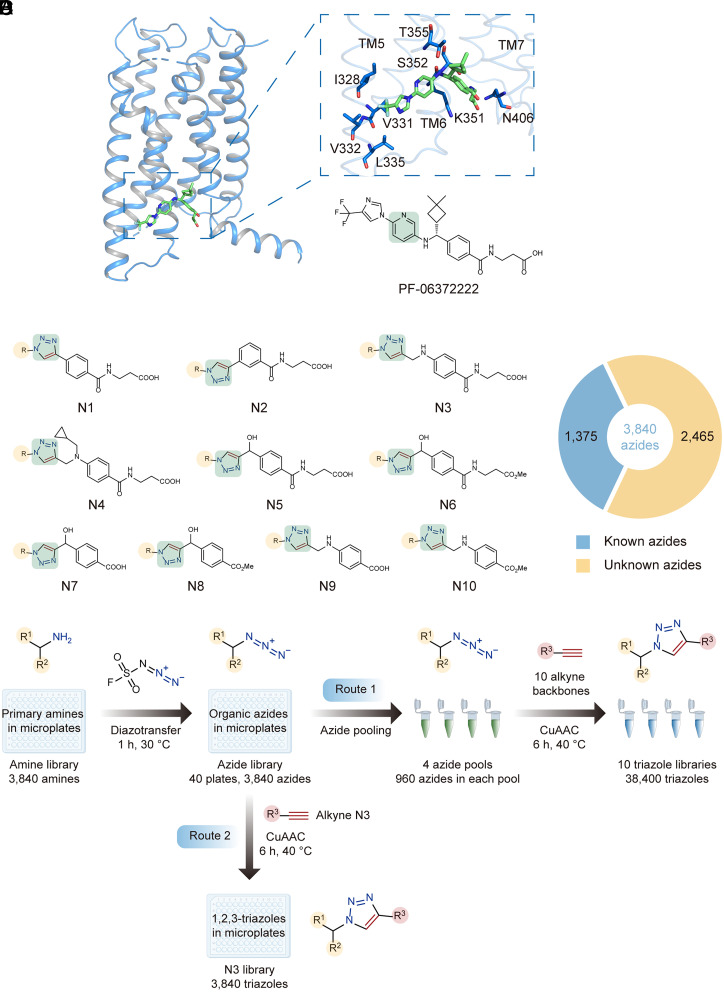
We selected a negative allosteric modulator (NAM) of the GLP-1R, PF-06372222, as an initial template for the library design. According to the crystal structure of the NAM-bound GLP-1R transmembrane domain (TMD) (27), the carboxylic acid moiety of PF-06372222 forms hydrogen bond interactions with a polar cleft between helices VI and VII, and its trifluoromethyl–pyrazole group binds a hydrophobic surface area of helix V (Fig. 1A). Notably, an agonist positive allosteric modulator (PAM) of GLP-1R is located in close proximity to this NAM-binding pockets by forming a covalent attachment to the cytoplasmic end of helix VI (28). Thus, we hypothesized that building chemical libraries based on the initial template to target this flexible pocket may allow us to discover both negative and positive allosteric modulators with unreported structures.
Fig. 1.
Design and synthesis of double-click triazole libraries. (A) The structure and binding pocket of PF-06372222 as an initial template for the library design. The functional group to be replaced with 1,2,3-triazole is highlighted in green. (B) The structural representation of 10 triazole libraries (N1-N10). (C) The number of previously unknown azides in the 3,840-azide library according to the Reaxys database. (D) Double-click reaction sequence for generation of 10 triazole libraries using multiple azide pools prepared in microtubes (Route 1) or generation of a specific library N3 using individual azides prepared in microplates (Route 2).
When designing the double-click triazole libraries, we chose to mimic the central pyridine ring with 1,2,3-triazole, replace its right portion with selected alkyne backbones, and vary its left portion with diverse R substituents derived from an azide library (Fig. 1B). Using our previously developed double-click chemistry sequence, each 1,2,3-triazole library can be readily prepared through CuAAC reactions between a given alkyne backbone and thousands of azides that were converted from primary amines in a streamlined fashion (6). Herein, we first prepared an azide library of the largest diversity which consisted of 3,840 azides via the diazotransfer reaction using FSO2N3. Within this library, 2,465 members are unknown azides not synthesized before (Fig. 1C). Unlike the previous work in which CuAAC reactions of different azide substrates were performed in individual wells, we mixed 960 azides in one pool to react with a single alkyne backbone in order to expedite the library synthesis with minimal starting materials (Route 1 in Fig. 1D). The reaction of the azide pool and a specific alkyne was carried out on a nanomole scale (150 ng of each azide consumed) and completed with high efficiency.
Each triazole library consisted of 3,840 members yielded from CuAAC reactions of four azide pools and a designed alkyne backbone. In total, we fulfilled reactions between 10 different alkyne backbones with azide pools to generate 10 triazole libraries composed of 38,400 reaction products (Fig. 1D). These products were all structurally related to PF-06372222 with extensive structural diversity.
AS-MS Screen to Select a Bioactive Triazole Library.
As our library synthesis generated highly complex reaction product mixtures that cannot be screened using conventional bioassays, we turned to an efficient and cost-effective screening approach AS-MS which enables the detection of chemical ligands specifically associated with a protein target from compound mixtures. Our laboratory has established this approach to identify new ligands modulating the activity of different G protein-coupled receptor (GPCR) targets (11, 12, 29–32).
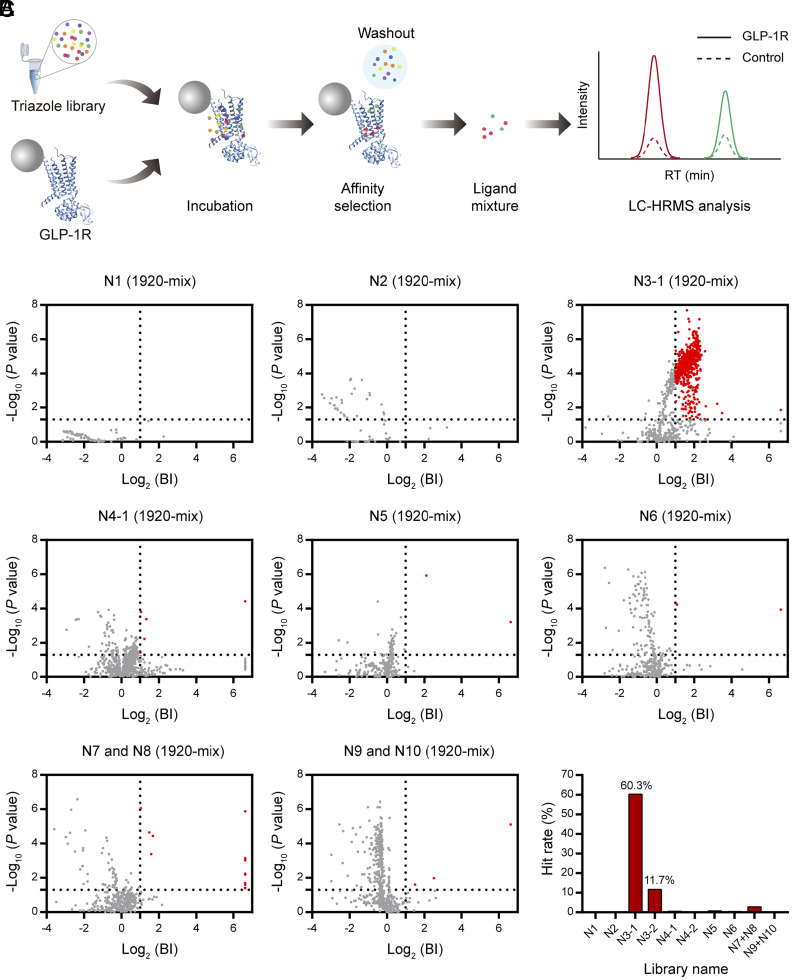
In this study, we purified the apo GLP-1R transmembrane domain in a homogeneous monomeric state (SI Appendix, Fig. S1), which was the original construct used for receptor co-crystallization with ligand PF-06372222 (27). For each triazole library, we randomly selected half of the compound pools to obtain a product mixture from 1,920 crude CuAAC reactions without purification (1,920-mix pool). The purified receptor immobilized on affinity resin was then incubated with each 1,920-mix pool. The ligand-bound GLP-1R complexes were isolated from solution before ligands were dissociated and subjected to liquid chromatography coupled to high-resolution mass spectrometry (LC-HRMS) analysis (Fig. 2A). Importantly, products in the crude CuAAC reaction mixtures can be readily differentiated from reactants, catalysts or potential intermediates based on the accurate mass measurement and retention time alignment (details in Materials and Methods). Furthermore, quantitative comparison of the MS responses of different reaction products detected in the target vs. control allowed us to distinguish receptor-binding ligands from nonspecific binders (Fig. 2A). The AS-MS assay for ligand detection was first verified with a set of known allosteric modulators of the GLP-1R (SI Appendix, Fig. S2).
Fig. 2.
AS-MS screen of double-click triazole libraries. (A) The AS-MS experimental scheme. Each pool of crude triazole products was incubated with the GLP-1R protein target before putative ligands were selected, released, and subjected to LC-HRMS analysis for structural assignment and specificity evaluation. (B) AS-MS results for screening 1,920-mix pools from different libraries. BI, binding index, a quantitative metric for determination of specific binders to the target. Hits of individual screens are indicated by red dots. BI data represent means of three independent experiments. (C) Hit rates of screening different triazole libraries. Libraries N3 and N4 were divided into two 1,920-mix pools for separate AS-MS screens.
Previous AS-MS screens of conventional synthetic libraries against various protein targets typically resulted in 0.5 to 3% hit rates, with hits almost evenly distributed among different compound pools (8–12, 30). However, the AS-MS screen of 10 triazole libraries (N1 to N10) gave rise to the largest number of primary hits solely from library N3 (Fig. 2B). Specifically, 60.3% of detectable compounds from the 1,920-mix pool of this library were regarded as putative GLP-1R ligands, respectively, whereas hit rates were below 3.0% for the other nine triazole libraries (Fig. 2C). We further screened the other half of library N3, which gave rise to a hit rate of 11.7% (Fig. 2C and SI Appendix, Fig. S3). The significant enrichment of hits in one library indicated bioactive ligands are more likely to be obtained from this particular domain of chemical space. Of note, we also assayed GLP-1-elicited cyclic adenosine monophosphate (cAMP) signaling of 10 alkyne backbones used in the triazole library synthesis, yet none of them exhibited strong activity (SI Appendix, Fig. S4), suggesting the functional assay failed to distinguish the bioactive library. In contrast, the high-throughput AS-MS screen of triazole libraries enabled rapid and economic discovery of one library enriched in bioactive compounds that can be then verified by more costly functional assays.
Identification of Allosteric Modulators of the GLP-1R.
For library N3 selected by AS-MS screen, we then resynthesized all 3,840 CuAAC reaction products in a one-well-one-reaction manner (Route 2 in Fig. 1D). Briefly, an azide library was first prepared from 3,840 different amines via diazotransfer, and it was reacted with the N3 alkyne backbone though CuAAC transformation to yield 3,840 distinct triazole products.
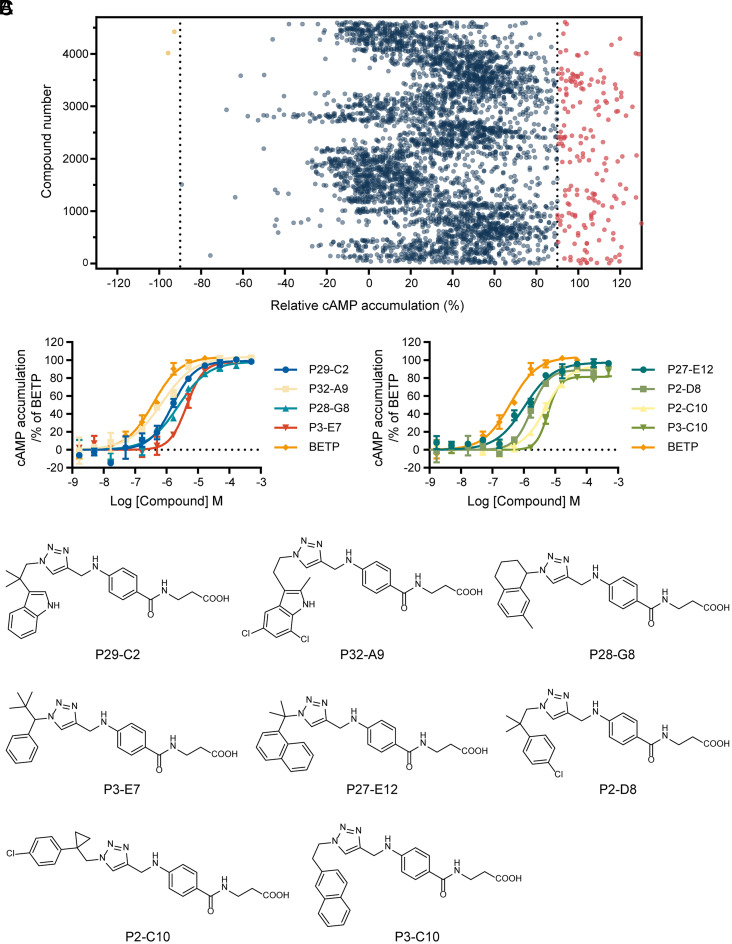
We performed a functional screen of individual crude reaction products using an intracellular cAMP accumulation assay in the presence of the endogenous peptide GLP-1(9-36). GLP-1(9-36) is a partial weak agonist for GLP-1R to stimulate Gs-coupled cAMP signaling. The initial template PF-06372222 suppresses cAMP accumulation induced by GLP-1(9-36) at a micromolar potency (SI Appendix, Fig. S5). Screening the N3 library at a single dose of each reaction product led to the identification of only two products that antagonized GLP-1(9-36)-induced cAMP signaling by over 90% efficacy (Fig. 3A). Remarkably, as many as 180 products were able to potentiate the activity of GLP-1(9-36) to over 90% efficacy (Fig. 3A). Compared to the antagonists and NAMs, discovery of agonists or PAMs for GLP-1R is of much higher value given that they are approved drugs or have the potential for the treatment of type 2 diabetes and obesity (13, 14).
Fig. 3.
Identification of GLP1-R PAMs from the functional screen of one triazole library. (A) Bioactivity screen of 3,840 crude reaction products from library N3. cAMP accumulation was measured in CHO-K1 cells expressing human GLP-1R treated by different reaction products (50 µM) in the presence of EC50 of GLP-1(9-36) (1.12 µM). Hits that potentiated or suppressed the cAMP production above 90% (relative to the full response of GLP-1(9-36) or DMSO control) were labeled in red or yellow. (B) Dose–response curves of specific new PAMs measured in CHO-K1 cells expressing human GLP-1R in the presence of EC20 of GLP-1(9-36) (0.6 µM). cAMP accumulation was normalized to the full response of BETP. Data represent means ± SEM of three independent experiments performed in triplicate. (C) The structures of new PAMs with dose–response curve data shown in B.
We then focused on these potentiating products and assayed their dose-dependent activity in comparison with a well-characterized PAM 4-(3-benzyloxyphenyl)-2-ethylsulfinyl-6-(trifluoromethyl)pyrimidine (BETP) (33). In our assay, BETP augments GLP-1(9-36)-elicited cAMP signaling to reach the full efficacy of GLP-1(7-36) at an EC50 around 500 nM (Fig. 3B). The concentration response experiment confirmed the potentiation effects of 10 crude reaction products that achieved >80% BETP efficacy with an EC50 <10 µM and thus can be regarded as PAMs of the GLP-1R (Dataset S1). After purifying the CuAAC reaction products and subjecting them to the functional assay, we verified eight new compounds with PAM activities at an EC50 of 0.6 to 6.7 µM and Emax of 77 to 102% relative to BETP (Fig. 3B and Dataset S2). Therefore, screening a selected triazole library allowed us to discover an array of new PAMs for GLP-1R all bearing a lipophilic aryl methyl/ethyl moiety which are conformationally restricted in the acyclic region (Fig. 3C).
Synthesis and Functional Screen of Focused Triazole Libraries.
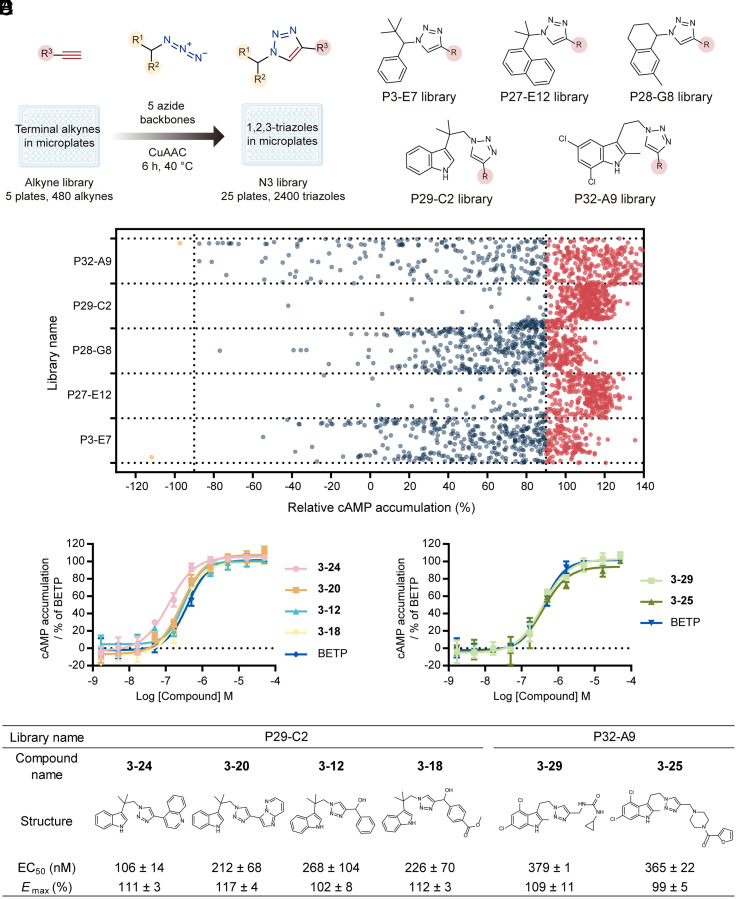
The five most potent PAMs identified from the primary functional screen then served as new templates for the second-round design of focused triazole libraries. For each template, by retaining its azide backbone and varying the alkyne module in CuAAC reactions, we prepared a new 480-member triazole library in microplates (Fig. 4A). In total, five focused libraries composed of 2,400 CuAAC reaction products were synthesized without purification (Fig. 4B). All expected products are new compounds that have never been documented.
Fig. 4.
Synthesis and functional screen of focused triazole libraries. (A) Generation of five focused libraries with the CuAAC reaction conducted in microplates. (B) The structural representation of five focused libraries. (C) Bioactivity screen of 2,400 crude reaction products from focused libraries. cAMP accumulation was measured in CHO-K1 cells expressing human GLP-1R treated by different reaction products (50 µM) in the presence of EC50 of GLP-1(9-36) (1.12 µM). Hits that potentiated or suppressed the cAMP production above 90% (relative to the full response of GLP-1(9-36) or DMSO control) were labeled in red or yellow. (D) Dose–response curves of six most potent PAMs measured in CHO-K1 cells expressing human GLP-1R in the presence of EC20 of GLP-1(9-36) (0.6 µM). cAMP accumulation was normalized to the full response of BETP. Data represent means ± SEM of three independent experiments performed in triplicate. (E) The structures and pharmacological data of these six PAMs.
Using the previously described single-dose cAMP accumulation assay, we performed another functional screen to identify 1,284 reaction products that potentiated the activity of GLP-1(9-36) by more than 90% efficacy (Fig. 4C). The concentration response experiment confirmed the potentiation effects of 55 crude reaction products that achieved >80% BETP efficacy with EC50 <2 µM (Dataset S3). Compared to the primary screen of 10 diverse triazole libraries, the secondary screen of more focused triazole libraries yielded substantially more PAM hits with at least fivefold increased potency, indicating the majority of ligands were evolving toward the PAM-enriched chemical space during our library expansion.
Re-synthesis and purification of these reaction products allowed us to verify 26 new compounds with PAM activities at an EC50 of 106 nM to 2.01 µM and Emax of 95 to 118% relative to BETP (Dataset S4). Of note, library P29-C2 and library P32-A9, which were designed based on two hits of the highest efficacy in the primary functional screen, yielded six most potent PAMs with EC50 below 500 nM and Emax above 90% relative to BETP (Fig. 4 D and E). They include four compounds from library P29-C2 and two compounds from library P32-A9, all bearing a tryptamine scaffold (Fig. 4E).
Pharmacological Characterization of New PAMs.
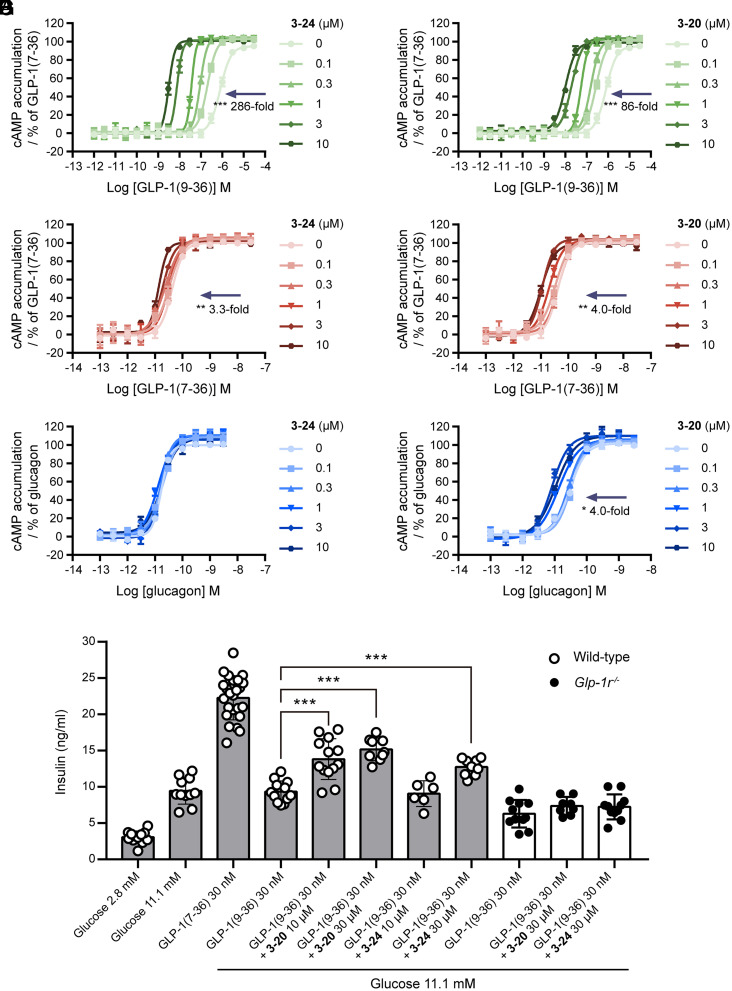
We then selected the two most potent compounds (3-24 and 3-20) to characterize the influence of allosteric modulators on the orthosteric peptide agonists. Specifically, we quantified the ability of 3-24 and 3-20 to shift the EC50 value of endogenous GLP-1(9-36) or GLP-1(7-36) at fixed compound concentrations. In the heterologous cellular system, GLP-1(7-36) is a highly potent full agonist (EC50 = 41 pM) and GLP-1(9-36) is a weak agonist (EC50 = 1.12 μM). The peptide agonist titration assays showed that both 3-24 and 3-20 significantly enhanced the potency of GLP-1(9-36) to elicit GLP-1R-dependent cAMP signaling. In the presence of a maximal concentration of compounds 3-24 and 3-20 (10 μM), the potency of GLP-1(9-36) improved to 3.1 nM and 10 nM respectively, equivalent to 286-fold and 86-fold leftward shifting in the EC50 for cAMP production (Fig. 5 A and B). However, 3-24 and 3-20 had marginal effects on the activity of GLP-1(7-36), causing only 3.3-fold and 4.0-fold left shift of the EC50 at a maximal compound concentration (Fig. 5 C and D). The selectivity of two PAMs was further tested in cells overexpressing glucagon receptor (GCGR), a class B1 receptor in close homology to GLP-1R (34). Compound 3-24 had no effect on the activity of glucagon to elicit GCGR-dependent cAMP signaling while 3-20 caused marginal potentiation of the glucagon activity (Fig. 5 E and F). These results demonstrated that 3-24 and 3-20 exhibited strong probe dependence by acting as a robust potentiator of GLP-1(9-36) but not GLP-1(7-36) or glucagon. To evaluate the effect of two PAMs on a different signaling pathway, we also characterized the activity of 3-24 and 3-20 on β-arrestin recruitment by GLP-1 peptides. Surprisingly, both PAMs produced no changes in β-arrestin recruitment by either GLP-1(9-36) or GLP-1(7-36), suggesting that the potentiation of GLP-1 peptides by these PAMs is exclusively biased toward G protein signaling (SI Appendix, Fig. S6).
Fig. 5.
The pharmacological profiles of new PAMs with different peptide agonists or receptors. (A and B) Potentiation of the cAMP accumulation produced by GLP-1(9-36) at the GLP-1R in combination with different fixed concentrations of 3-24 (A) or 3-20 (B). (C and D) Potentiation of the cAMP accumulation produced by GLP-1(7-36) at the GLP-1R in combination with fixed concentrations of 3-24 (C) or 3-20 (D). (E and F) Potentiation of the cAMP accumulation produced by glucagon at the GCGR in combination with fixed concentrations of 3-24 (E) or 3-20 (F). Data in A–D and E and F were measured in CHO-K1 cells expressing human GLP-1R and GCGR, respectively. (G) Insulin secretion in islets isolated from wild-type (gray bars) rat or GLP-1R knockout (white bars) mice treated with GLP-1(7-36), GLP-1(9-36) or GLP-1(9-36) in combination with 3-24 or 3-20 at a high-glucose condition. Data represent means ± SEM of at least three independent experiments performed in triplicate. Statistical analyses of the EC50 shift at a maximal concentration compared to the vehicle control in A–E and insulin secretion in G were performed using a two-way ANOVA test and a two-tailed t test, respectively. ***P < 0.001.
Next, to determine whether two PAMs can modulate the GLP-1R activity in more native systems, we evaluated the compounds using the rat pancreatic INS-1E β-cell line which endogenously expresses GLP-1R. In INS-1E cells, GLP-1(7-36) is still a potent full agonist (EC50 = 0.35 nM), while GLP-1(9-36) had no activity. The combination of GLP-1(9-36) with a submaximal concentration of 3-24 or 3-20 (3 μM) induced significant activity of the peptide (EC50/Emax 100 nM/60% for 3-24 and 115 nM/63% for 3-20) (SI Appendix, Fig. S7 A and B). Moreover, both PAMs potentiated the potency of GLP-1(7-36) to a greater extent than that observed in the heterologous cellular system (EC50 left-shifted by 7.7-fold for 3-24 and 27.2-fold for 3-20 at 10 μM) (SI Appendix, Fig. S7 C and D). We further investigated the ability of 3-24 and 3-20 to enhance glucose-dependent insulin secretion using ex vivo cultures of isolated islets from wild-type rat and Glp-1r knockout mice. In accordance with the PAM activity observed in cellular systems, the combination of GLP-1(9-36) and 3-24 (30 μM) or 3-20 (10 μM and 30 μM) at a high-glucose condition significantly potentiated insulin secretion from cultures of wild-type islets, though the efficacy was not as high as the full agonist GLP-1(7-36) (Fig. 5G). The enhanced responses were not observed in islets from Glp-1r knockout mice, which verified the receptor-dependent pharmacological action of these two PAMs (Fig. 5G).
Binding Mode Analysis of New PAMs.
Although we discovered a series of new PAMs from the double-click triazole libraries that were initially designed based on a negative allosteric ligand PF-06372222, the most potent PAMs obtained from focused libraries (listed in Fig. 4E) are structurally disparate from PF-06372222. Molecular docking of three representative PAMs (3-24, 3-20, and 3-29) implied plausible binding poses of three compounds within the allosteric pocket occupied by PF-06372222 (SI Appendix, Fig. S8). Binding of these PAMs to GLP-1R TMD was verified using the AS-MS assay on a ligand mixture, though their binding responses were much weaker than PF-06372222 (SI Appendix, Fig. S9A). However, site-directed mutagenesis of residues R348, S352, T355, and N407 proposed to be critical for PAM–receptor interactions did not affect their potentiation effects on GLP-1(9-36)-elicited signaling (SI Appendix, Fig. S10). Consistent with the mutagenesis experiment, AS-MS analysis demonstrated that the direct binding of three PAMs to GLP-1R TMD was not competed off in the large excess of PF-06372222. Interestingly, the binding of 3-24 and 3-20 to GLP-1R was significantly diminished in the excess of 3-29 (SI Appendix, Fig. S9B). These results indicate that the three PAMs possibly bind to the same pocket which is distinct from the PF-06372222-targeted site.
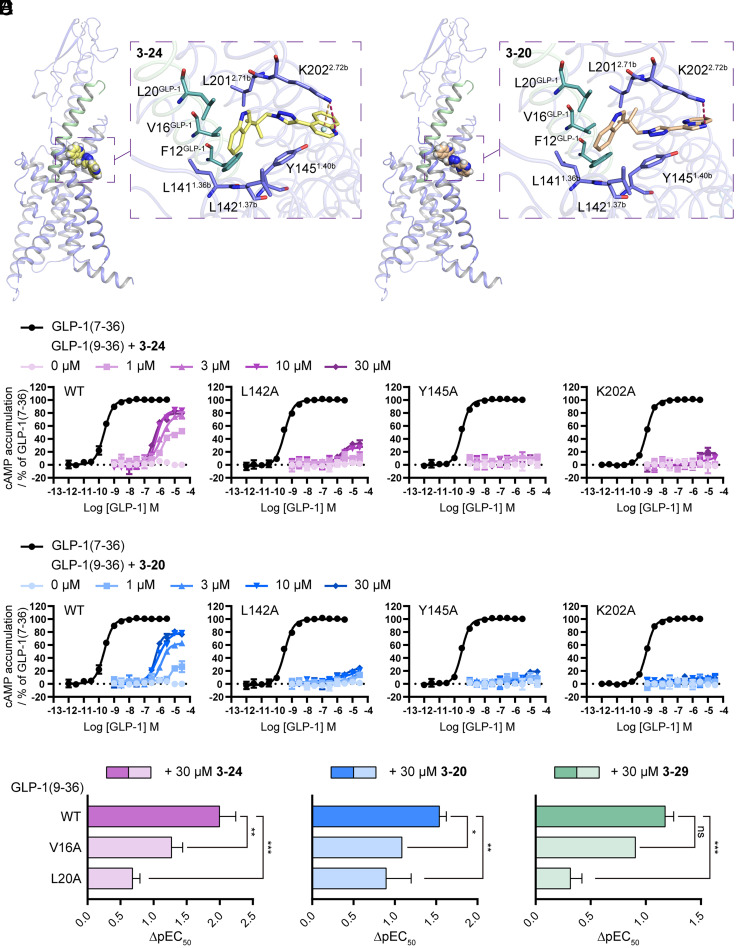
LSN3160440 is a small molecule PAM of GLP-1R which also shows strong probe-selective allosterism for GLP-1(9-36) vs. other peptide agonists (21). Structural elucidation reveals that LSN3160440 binds high in the helical bundle at an interface between helices I and II (21). To test the hypothesis that our PAMs may act at the same site as LSN3160440, we performed a docking analysis of the aforementioned three PAMs based on the structure of LSN3160440-bound GLP-1R in complex with GLP-1 and heterotrimeric Gs (PDB code: 6VCB) (21). All of them exhibited binding poses similar to LSN3160440 by occupying the extracellular pocket formed by residues in helices I and II as well as the GLP-1 peptide (Fig. 6 A and B and SI Appendix, Figs. S11 and S12A). Specifically, the tryptamine group of 3-24, 3-20, and 3-29 is proposed to insert into a hydrophobic cavity composed of L1411.36b, L1421.37b, Y1451.40b, L2012.71b, V16GLP-1, and L20GLP-1 (numbers in superscript refer to the Wootten numbering system for class B GPCRs). The dichloro-substituted tryptamine group of 3-29 also forms putative hydrogen and halogen bond with D1982.68b and K2022.72b respectively (SI Appendix, Fig. S12A). In addition, the polar group of each PAM (i.e., the heterocycle group of 3-24 and 3-20, and the urea group of 3-29) could form a putative hydrogen bond with K2022.72b (Fig. 6 A and B and SI Appendix, Fig. S12A). Binding of 3-24 and 3-29 to this pocket was further stabilized by forming cation-π or π-π interactions with K2022.72b or Y1451.40b (Fig. 6A and SI Appendix, Fig. S12A).
Fig. 6.
Characterization of the binding pocket of New PAMs. (A and B) Molecular docking models showing the binding mode of 3-24 (A) and 3-20 (B) in the extracellular allosteric pocket of the GLP-1R. Hydrogen bonding and cation–π interactions are indicated by dashed lines. (C and D) Potentiation of the cAMP accumulation produced by GLP-1(9-36) in combination with different fixed concentrations of 3-24 (C) or 3-20 (D) using wild-type (WT) or different mutant GLP-1R-transfected cells. (E) Comparison of the potentiation effects of specific PAMs on wide-type (WT) and mutant GLP1(9-36). ΔpEC50 represents the subtraction of pEC50 of GLP1(9-36) in the presence of a specific PAM with pEC50 of GLP1(9-36) alone. A largerΔpEC50 indicates a stronger potentiation effect of the test PAM. Data represent means ± SEM of three independent experiments performed in triplicate. Statistical analyses were performed using a two-way ANOVA test. *P < 0.05; **P < 0.01; ***P < 0.001; ns, no significant difference.
To verify the ligand–receptor contacts proposed by the docking model, we performed mutagenesis of three key residues in helices I and II, and compared the mutants with the wild-type receptor in the cAMP signaling assay. Single-point mutation of L142A, Y145A, and K202A either predominantly or completely abolished the ability of 3-24, 3-20, and 3-29 to potentiate GLP-1(9-36)-elicited cAMP accumulation (Fig. 6 C and D and SI Appendix, Fig. S12B). By contrast, the potentiation of GLP-1(9-36) activity by BETP which binds to helix VI was still observed in all three binding site mutants (SI Appendix, Fig. S12C). Of note, the allosteric activity of LSN3160440 was only abolished in a triple mutant, indicating the more profound effects of single-point mutations on our PAM activities may be partially interpreted by the direct hydrogen bonding between our compounds and the receptor which was absent for LSN3160440 (21). As for the ligand–peptide interactions, they are also crucial for the ligand activity as GLP-1(9-36) peptides harboring L20A or V16A mutation significantly reduced the potentiation effects of all three PAMs or two PAMs (3-24 and 3-20) (Fig. 6E and SI Appendix, Fig. S13). Taken together, these results imply that the three PAMs and possibly their analogs discovered in our study mediate their positive allosterism through engaging interactions with both the receptor and the orthosteric GLP-1 peptide.
Discussion
Using a double-click reaction scheme, we have achieved streamlined synthesis of 38,400 triazole compounds by reacting 10 designed alkyne backbones with an azide library containing 3,840 members that were readily converted from primary amines. Previously, only small libraries containing tens to hundreds of triazole derivatives have been prepared with the classical CuAAC reaction (4, 35–37). Double-click synthesis of triazole libraries in an unprecedented scale has substantially increased the structural diversity of new molecules originating from a privileged scaffold. But it has also posed a challenge in downstream bioactive screening, especially when the cell-based functional screen of a large compound library against a GPCR target is not trivial for regular research labs.
Our study has demonstrated the utility of high-throughput and high-sensitivity AS-MS as the first-line screening approach to be compatible with the facile construction of double-click triazole libraries. Notably, we reacted each alkyne backbone with an azide pool to generate mixed triazole products in the most efficient and economic manner. This crude reaction product mixture was directly subjected to the AS-MS screen which allows for simultaneous assessment of 1,920 expected products (each only consuming 5 ng) in their capability of binding to the GLP-1R transmembrane domain. Impurities such as reactants, catalysts and reaction intermediates that do not bind the protein target have almost no influence on the AS-MS screen because most of them can be washed off in the affinity selection stage. Even certain impurities remain in the final ligand elution, they are readily distinguishable from the products based on their retention time and accurate mass measurement by LC-HRMS. This in fact demonstrates a unique advantage of the AS-MS screen to identify reaction products as true binders from the crude double-click library.
The AS-MS screen quickly located a triazole library (N3) that significantly enriched a number of triazole derivatives as GLP-1R ligands. However, we realized the possibility of missing bioactive compounds of low affinity to the recombinant GLP-1R protein if only selecting binders from AS-MS for functional validation. Instead, we chose to assay the cellular activity of all 3,840 reaction products in the N3 library. In line with our published result (6), the vast majority of alkynes had estimated conversion rates above 70% in the synthesis of 10 triazole libraries. The remaining minor impurities in the crude reaction product could have certain influences on the functional screen, which may result in the small discrepancies seen in the bioactivity measurement between crude and purified compounds (Datasets S1–S4). It is noteworthy that we would have needed a functional screen for all 38,400 reaction products without leveraging the AS-MS screen for library selection. Thus, interfacing the AS-MS screen with the functional screen saved a significant amount of time and cost in the screening stage. More importantly, this hybrid screening platform facilitated the navigation of the extensive chemical space created by our double-click chemistry.
GLP-1R is a particularly challenging target for discovering new small molecule agonists or PAMs since enormous efforts have been made in the pharmaceutical industry. Our study identified a group of high-nanomolar PAMs with unreported scaffolds by iterative design and screen of double-click triazole libraries. Two new PAMs (3-24 and 3-20) were found to selectively and robustly augment GLP-1(9-36)-elicited cAMP signaling. Given that GLP-1(9-36) is the postsecretory proteolysis product of the full agonist GLP-1(7-36), it is speculated that enhancing the activity of GLP-1(9-36) could lead to a valuable therapeutic approach (18–21). Intriguingly, our study also revealed the unexpected binding mode of several new PAMs which are likely to act extracellularly as a molecular glue between the receptor and the GLP-1 peptide. Taken together, through benchmarking our strategy on a prominent GPCR target, we anticipate the merger of double-click library synthesis with the hybrid screening platform would accelerate the discovery of drug candidates or chemical probes for various therapeutic targets.
Materials and Methods
Detailed materials and methods are provided in SI Appendix, Supplementary note. These include detailed information about materials used, expression and purification of the GLP-1R TMD, synthesis of triazole libraries with azide pools for the AS-MS screen, synthesis of triazole library N3 and focused triazole library for the functional screen, AS-MS experimental procedures and data analysis, cell culture and transfections, measurement of cAMP accumulation, measurement of β-arrestin recruitment, measurement of insulin secretion in islets, molecular docking simulation, and synthesis and characterization of purified compounds.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Acknowledgments
We thank the staff members of the Assay, Cell Expression, Cloning, and Purification Core Facilities of the iHuman Institute for their support. This work was funded by National Key R&D Program of China (2022YFA1302902 to W.S. and 2018YFA0507004 to W.S.), National Natural Science Foundation of China (31971362 to W.S. and 32171439 to W.S.), Ministry of Science and Technology of China, Major State Basic Research Development Program of China (2021YFF0701704 to J.D.), Shanghai Frontiers Science Center for Biomacromolecules and Precision Medicine (to W.S.), the Science and Technology Commission of Shanghai Municipality (Grant No. 21ZR1442500 to Y.L.), and Shanghai Sailing Program (21YF1428900 to B.Z.).
Author contributions
W.S. and J.D. designed research and supervised the project; Y.X., S.L., Y.L., Z.Q., H.L., and B.Z., performed research; T.G. and G.J.T. contributed new reagents; Y.X., S.L., Y.L., Z.Q., H.L., B.Z., J.D., and W.S. analyzed data; R.C.S. and K.B.S. provided critical support; W.S., Y.X., and S.L. wrote the paper.
Competing interests
The authors have patent filings to disclose, Shanghai Institute of Organic Chemistry, CAS has filed patent applications for this technology (Chinese patent application numbers PCT/CN2018/116922, CN201810609851.4, CN201910506332.X, and CN201910605212.5). Shanghai Jiao Tong University and ShanghaiTech University have jointly filed patent applications for this technology (Chinese patent application numbers CN202210977610.1).
Footnotes
Reviewers: J.M.B., Cornell University; Xian Chen, University of North Carolina at Chapel Hill; and Xing Chen, Peking University.
Contributor Information
K. Barry Sharpless, Email: sharples@scripps.edu.
Jiajia Dong, Email: jiajiadong@sjtu.edu.cn.
Wenqing Shui, Email: shuiwq@shanghaitech.edu.cn.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Kolb H. C., Finn M. G., Sharpless K. B., Click chemistry: Diverse chemical function from a few good reactions. Angew. Chem. Int. Ed. Engl. 40, 2004–2021 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Rostovtsev V. V., Green L. G., Fokin V. V., Sharpless K. B., A stepwise Huisgen cycloaddition process: Copper(I)-catalyzed regioselective "ligation" of azides and terminal alkynes. Angew. Chem.-Int. Edit. 41, 2596-+ (2002). [DOI] [PubMed] [Google Scholar]
- 3.Tornoe C. W., Christensen C., Meldal M., Peptidotriazoles on solid phase: 1,2,3 -triazoles by regiospecific copper(I)-catalyzed 1,3-dipolar cycloadditions of terminal alkynes to azides. J. Org. Chem. 67, 3057–3064 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Kolb H. C., Sharpless K. B., The growing impact of click chemistry on drug discovery. Drug Discov. Today 8, 1128–1137 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Saxon E., Bertozzi C. R., Cell surface engineering by a modified Staudinger reaction. Science 287, 2007–2010 (2000). [DOI] [PubMed] [Google Scholar]
- 6.Meng G., et al. , Modular click chemistry libraries for functional screens using a diazotizing reagent. Nature 574, 86–89 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Muchiri R. N., van Breemen R. B., Affinity selection-mass spectrometry for the discovery of pharmacologically active compounds from combinatorial libraries and natural products. J. Mass Spectrom. 56, e4647 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Prudent R., Annis D. A., Dandliker P. J., Ortholand J. Y., Roche D., Exploring new targets and chemical space with affinity selection-mass spectrometry. Nat. Rev. Chem. 5, 62–71 (2021). [DOI] [PubMed] [Google Scholar]
- 9.O’Connell T. N., Ramsay J., Rieth S. F., Shapiro M. J., Stroh J. G., Solution-based indirect affinity selection mass spectrometry–a general tool for high-throughput screening of pharmaceutical compound libraries. Anal. Chem. 86, 7413–7420 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Deng Y., et al. , Discovery of novel, dual mechanism ERK inhibitors by affinity selection screening of an inactive kinase. J. Med. Chem. 57, 8817–8826 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Qin S., et al. , High-throughput identification of G protein-coupled receptor modulators through affinity mass spectrometry screening. Chem. Sci. 9, 3192–3199 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu Y., et al. , Accelerating the throughput of affinity mass spectrometry-based ligand screening toward a G protein-coupled receptor. Anal. Chem. 91, 8162–8169 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graaf C., et al. , Glucagon-like peptide-1 and its class B G protein-coupled receptors: A long march to therapeutic successes. Pharmacol. Rev. 68, 954–1013 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burcelin R., Gourdy P., Harnessing glucagon-like peptide-1 receptor agonists for the pharmacological treatment of overweight and obesity. Obes. Rev. 18, 86–98 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Zhao P. S., et al. , Activation of the GLP-1 receptor by a non-peptidic agonist. Nature 577, 432-+ (2020). [DOI] [PubMed] [Google Scholar]
- 16.Griffith D. A., et al. , A small-molecule oral agonist of the human glucagon-like peptide-1 receptor. J. Med. Chem. 65, 8208–8226 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawai T., et al. , Structural basis for GLP-1 receptor activation by LY3502970, an orally active nonpeptide agonist. Proc. Natl. Acad. Sci. U.S.A. 117, 29959–29967 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smelcerovic A., et al. , An overview, advantages and therapeutic potential of nonpeptide positive allosteric modulators of glucagon-like peptide-1 receptor. Chemmedchem 14, 514–521 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Willard F. S., et al. , Discovery of an orally efficacious positive allosteric modulator of the glucagon-like peptide-1 receptor. J. Med. Chem. 64, 3439–3448 (2021). [DOI] [PubMed] [Google Scholar]
- 20.Mendez M., et al. , Design, synthesis, and pharmacological evaluation of potent positive allosteric modulators of the glucagon-like peptide-1 receptor (GLP-1R). J. Med. Chem. 63, 2292–2307 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Bueno A. B., et al. , Structural insights into probe-dependent positive allosterism of the GLP-1 receptor. Nat. Chem. Biol. 16, 1105–1110 (2020). [DOI] [PubMed] [Google Scholar]
- 22.Decara J. M., et al. , Discovery of V-0219: A small-molecule positive allosteric modulator of the glucagon-like peptide-1 receptor toward oral treatment for "Diabesity". J. Med. Chem. 65, 5449–5461 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakane A., Gotoh Y., Ichihara J., Nagata H., New screening strategy and analysis for identification of allosteric modulators for glucagon-like peptide-1 receptor using GLP-1 (9-36) amide. Anal. Biochem. 491, 23–30 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Morris L. C., et al. , Discovery of (S)-2-Cyclopentyl-N-((1-isopropylpyrrolidin2-yl)-9-methyl-1-oxo-2,9-dihy dro-1H-pyrrido 3,4-b indole-4-carboxamide (VU0453379): A Novel, CNS Penetrant Glucagon-Like Peptide 1 Receptor (GLP-1R) Positive Allosteric Modulator (PAM). J. Med. Chem. 57, 10192–10197 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sloop K. W., et al. , Novel small molecule glucagon-like peptide-1 receptor agonist stimulates insulin secretion in rodents and from human islets. Diabetes 59, 3099–3107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knudsen L. B., et al. , Small-molecule agonists for the glucagon-like peptide 1 receptor. Proc. Natl. Acad. Sci. U.S.A. 104, 937–942 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song G., et al. , Human GLP-1 receptor transmembrane domain structure in complex with allosteric modulators. Nature 546, 312–315 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Cong Z. T., et al. , Molecular insights into ago-allosteric modulation of the human glucagon-like peptide-1 receptor. Nat. Commun. 12 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang B., et al. , A novel G protein-biased and subtype-selective agonist for a G protein-coupled receptor discovered from screening herbal extracts. ACS Cent Sci. 6, 213–225 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu Y., et al. , Affinity mass spectrometry-based fragment screening identified a new negative allosteric modulator of the adenosine A2A receptor targeting the sodium ion pocket. ACS Chem. Biol. 16, 991–1002 (2021). [DOI] [PubMed] [Google Scholar]
- 31.Qu X., et al. , Structural basis of tethered agonism of the adhesion GPCRs ADGRD1 and ADGRF1. Nature 604, 779–785 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma M., et al. , Targeted proteomics combined with affinity mass spectrometry analysis reveals antagonist E7 acts as an intracellular covalent ligand of orphan receptor GPR52. ACS Chem. Biol. 15, 3275–3284 (2020). [DOI] [PubMed] [Google Scholar]
- 33.Willard F. S., Ho J. D., Sloop K. W., Discovery and pharmacology of the covalent GLP-1 receptor (GLP-1R) allosteric modulator BETP: A novel tool to probe GLP-1R pharmacology. Adv. Pharmacol. 88, 173–191 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Zhang H., et al. , Structure of the glucagon receptor in complex with a glucagon analogue. Nature 553, 106–110 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Jiang X., et al. , Recent applications of click chemistry in drug discovery. Expert Opin. Drug Discov. 14, 779–789 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Tiwari V. K., et al. , Cu-catalyzed click reaction in carbohydrate chemistry. Chem. Rev. 116, 3086–3240 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Neumann S., Biewend M., Rana S., Binder W. H., The CuAAC: Principles, homogeneous and heterogeneous catalysts, and novel developments and applications. Macromol. Rapid Commun. 41, e1900359 (2020). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Data Availability Statement
All study data are included in the article and/or SI Appendix.