Summary
The ongoing severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic and heterologous immunization approaches implemented worldwide for booster doses call for diversified vaccine portfolios. GRAd-COV2 is a gorilla adenovirus-based COVID-19 vaccine candidate encoding prefusion-stabilized spike. The safety and immunogenicity of GRAd-COV2 is evaluated in a dose- and regimen-finding phase 2 trial (COVITAR study, ClinicalTrials.gov: NCT04791423) whereby 917 eligible participants are randomized to receive a single intramuscular GRAd-COV2 administration followed by placebo, or two vaccine injections, or two doses of placebo, spaced over 3 weeks. Here, we report that GRAd-COV2 is well tolerated and induces robust immune responses after a single immunization; a second administration increases binding and neutralizing antibody titers. Potent, variant of concern (VOC) cross-reactive spike-specific T cell response peaks after the first dose and is characterized by high frequencies of CD8s. T cells maintain immediate effector functions and high proliferative potential over time. Thus, GRAd vector is a valuable platform for genetic vaccine development, especially when robust CD8 response is needed.
Keywords: COVID-19, Sars-CoV-2 vaccine, simian adenoviral vector, neutralizing antibodies, T cell response, CD8, CD4, phase 2 clinical trial, safety, immunological memory
Graphical abstract

Highlights
-
•
GRAd-COV2 vaccine based on a recently developed group C gorilla adenoviral vector
-
•
Phase 2 trial confirms favorable safety profile for single- and double-dose regimens
-
•
SARS-CoV-2 binding/neutralizing Ab boosted by homologous or heterologous vaccine
-
•
Potent, VOC cross-reactive, functional, and durable CD4 and CD8 T cell responses
Capone et al. show that GRAd-COV2 vaccine is well tolerated and immunogenic in phase 2 trial; as an alternative Ad serotype, GRAd represents a valid addition to the SARS-CoV-2 vaccine toolbox for heterologous prime/boost regimens and a promising platform for genetic vaccines, particularly when T cell response is key.
Introduction
Since mid-2022, COVID-19 vaccine supply is no longer a limiting factor in efforts to control the global pandemic.1 Over 350 COVID-19 vaccine candidates have been developed or are in development using different technology platforms.2 The need for a range of vaccines is due to the fact that multiple factors influence policy decisions and each vaccine has distinctive features, advantages, and disadvantages to be considered in different healthcare settings, economies, subpopulations, and age groups.
Moreover, the continued emergence of new severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants of concern (VOCs) is adding complexity for vaccine developers and for policy decision-makers. Omicron and its sublineages, with an unprecedented mutation burden focused in the spike protein, have rapidly displaced previous circulating variants since late 2021. Antigenic changes leading to significant evasion from humoral immunity induced either by infection with other SARS-CoV-2 variants or by vaccination, together with functional and structural modifications affecting transmissibility and pathogenicity,3,4 call for considering Omicron as a distinct SARS-CoV-2 serotype that needs vaccine adaptation.5,6 However, such an approach may be practically unfeasible given the speed at which novel variants have emerged and then disappeared and the possibility that future variants will not linearly evolve from the latest circulating variants. Preclinical and real-world data suggest that, following repeated prototype-based vaccine booster doses, the cross-reactivity of neutralizing response is widely improved, while variant-specific adapted vaccines seem to generate more restricted immunity.7,8,9,10 While initial data on bivalent vaccines are encouraging,11 the safety and immunogenicity profile associated with novel candidate vaccines based on the prototype spike are still of interest and enable crucial direct comparisons across a range of traditional and innovative vaccine platforms, an unprecedented circumstance before the COVID-19 era.
The adenoviral vaccine platform has successfully been exploited in at least 4 effective and widely approved COVID-19 vaccines: Vaxzevria, Jcovden, Sputnik V, and Convidecia. GRAd-COV2 is a candidate vaccine based on a recently developed non-replicating gorilla group C Ad encoding for a prefusion-stabilized full-length spike. The vaccine induced potent and durable humoral and Th1-skewed cellular immune response upon a single intramuscular administration in animal models12 and in healthy adult volunteers within a phase 1, dose-escalation trial.13 The vaccine was well tolerated,14 and the safety profile was similar in terms of quality and severity to that of other COVID-19 genetic vaccines.
Here, we expanded GRAd-COV2 safety and immunogenicity evaluation in a phase 2 trial, where we also compared a two-dose versus a single-dose regimen, with the aim to select the best vaccination schedule to be further progressed in efficacy studies.
These relevant clinical data in humans are of more general interest for deepening our understanding of the immunological features embedded in innovative genetic vaccine platforms that have finally demonstrated all their potential in the COVID-19 pandemic but that have much broader applications for future emerging pathogens or for the immunotherapy of cancer.
Results
Study design
Between March 18, 2021, and April 9, 2021, a total of 923 volunteers older than 18 years were screened, and 917 were randomized. A total of 917 participants were dosed: 305 were assigned to receive a single dose (SD) of GRAd-COV2 plus placebo, 308 a repeated dose (RD) of GRAd-COV2, and 304 a placebo (Figure 1). 652 participants (71.1%) belonged to the stratum of <65 years of age without risk factors, 90 participants (9.8%) to the stratum of <65 years of age with risk factors, and 175 (19.1%) participants to the stratum of ≥65 years of age. Baseline characteristics of overall population are reported in Table 1. On June 21, 2021, following the DSMB recommendation and EC approval, the randomization code was broken to allow participants assigned to the placebo group access to the public vaccination campaign that was implemented in Italy during the first quarter of 2021, hence the blinding was maintained up to day 57. Participants in the placebo group who received marketed COVID-19 vaccines withdrew from the study, while the participants assigned to the vaccine groups continued to be followed up with for 1 year. This article considers all participants dosed with GRAd-COV2 vaccine who completed 180 days of follow up for safety and assessment of immunological parameters.
Figure 1.
Trial profile
All participants were followed up with in blind condition up to day 57 after the first dose; afterward, the randomization code was opened to allow participants assigned to the placebo group to have access to the vaccination campaign. The CONSORT diagram reports the number of participants in the full analysis set (FAS) at each time point up to day 180 and the corresponding number of participants included in the immunological analysis set (IAS). FAS included all randomized participants who received the dose of the investigational medicinal product (IMP), irrespective of their protocol adherence and continued participation in the study; IAS included all participants in the safety analysis set who had immune response assessments and no protocol deviations (or up to the time point before the deviation occurred) judged to have a potential interference with the generation or interpretation of an immune response (SARS-CoV-2 infection or commercial COVID-19 vaccination).
Table 1.
Participants’ baseline characteristics
| Vaccine single dose (SD) at 2 × 1011 vp (n = 305) | Vaccine repeated dose (RD) at 1 × 1011 vp (n = 308) | Placebo (n = 304) | |
|---|---|---|---|
| Protocol strata | |||
| Age 18 to <65 years and not at risk, n (%) | 218 (71.5) | 219 (71.1) | 215 (70.7) |
| Age 18 to <65 years and at risk, n (%) | 29 (9.5) | 30 (9.7) | 31 (10.2) |
| Age ≥65 years, n (%) | 58 (19.0) | 59 (19.2) | 58 (19.1) |
| Age, years | |||
| Mean (SD) | 46.0 (16.63) | 47.2 (16.07) | 47.3 (15.54) |
| Gender, n (%) | |||
| Male | 197 (64.6) | 200 (64.9) | 184 (60.5) |
| Female | 108 (35.4) | 108 (35.1) | 120 (39.5) |
| Body mass index, kg/m2 | |||
| Mean (SD) | 25.3 (4.4) | 25.0 (4.4) | 25.6 (4.6) |
| Underlying diseases, n (%) | |||
| None | 267 (87.5) | 270 (87.7) | 261 (85.9) |
| Significant cardiac diseases | 12 (3.9) | 14 (4.5) | 17 (5.6) |
| Chronic lung diseases | 9 (3.0) | 8 (2.6) | 6 (2.0) |
| Severe obesity | 3 (1.0) | 3 (1.0) | 6 (2.0) |
| Diabetes (type 1, type 2) | 7 (2.3) | 6 (1.9) | 6 (2.0) |
| Liver diseases | 2 (0.7) | 3 (1.0) | 3 (1.0) |
| HIV infection | 9 (3.0) | 7 (2.3) | 6 (2.0) |
vp, viral particles.
A SD or RD regimen of GRAd-COV2 vaccine was well tolerated and induced mostly mild to moderate local and systemic adverse events of short duration
Overall, GRAd-COV2 recipients, both in the SD and RD groups, reported more local reactions than placebo recipients (80.7%, 75.3%, and 28%, respectively, after the first dose). Among GRAd-COV2 recipients, mild to moderate pain and tenderness at the injection site was the most commonly reported local reaction (Figures 2A and 2B). Among GRAd-COV2 recipients receiving the second dose, local reactions after the second injection were reported less frequently (68.5%; Figure 2B). Participants ≥65 years of age reported less frequent local reactions (69%, 50.8%, and 17.2%, respectively, in the three groups after the first dose) than younger participants (83.8%, 81.5%, and 30.5%, respectively; Figure 2C). A noticeably lower percentage of participants reported injection-site erythema or swelling. Grade 3 local reactions were reported in 16 (5.2%), 10 (3.2%), and 1 (0.3%) participants in the SD, RD, and placebo groups, respectively; the most frequent grade 3 local reactions were pain and tenderness. No participants reported a grade 4 local reaction (Figure 2D). In general, local reactions were mostly mild to moderate in severity and resolved within 2 days.
Figure 2.
Frequency of participants with solicited systemic and local adverse events and severity
Data are percentage of participants.
(A) Frequency of participants with solicited systemic and local adverse events (AEs) within 7 days after the first dose.
(B) Frequency of participants with solicited systemic and local AEs within 7 days after the second dose. Only events with a frequency ≥1% are reported.
(C) Frequency of participants with solicited local and systemic AEs (any) within 7 days after the first and second dose according to the age category (18–65 years; >65 years).
(D) Most frequent (>1%) AEs (solicited and unsolicited) within 28 days after any dose of vaccination. Severity was assessed for AEs according to toxicity grading scales modified and abridged from the US FDA guidance. No grade 4 AEs were observed.
Both groups of GRAd-COV2 recipients reported more solicited systemic reactions than the placebo group (87.5%, 82.1%, and 59.2%, respectively, after the first dose; Figure 2A). Systemic solicited events were reported more often by younger than by older vaccine recipients and less often after dose 2 than dose 1 (Figure 2C). The most commonly reported solicited systemic adverse events (AEs) were fatigue, headache, malaise, myalgia, and chills, although fatigue and headache were also reported by many placebo recipients. Fever, nausea, and vomiting were less reported. Among GRAd-COV2 recipients, the frequency of severe solicited systemic events was slightly higher in the group receiving SD at 2 × 1011 vp then in the group receiving RD at 1 × 1011 vp (Figure 2D). Systemic AEs were generally mild to moderate and observed within the first 2 days after vaccination and resolved shortly thereafter. Grade 3 systemic reactions were reported in 66 (21.6%), 52 (16.9%), and 9 (3%) of participants in the SD, RD, and placebo groups, respectively; the most frequent grade 3 systemic reactions were chills, headache, and fatigue. No grade 4 severity was reported.
Unsolicited AE analyses are provided for all enrolled 917 participants, with a follow-up time of 28 days after dose 2. A comparable rate of GRAd-COV2 SD, RD, and placebo recipients reported any unsolicited AE (16.7%, 13.6%, and 14.1%, respectively) or a related unsolicited AE (3.6%, 3.9%, and 3%). Very few participants in all groups had grade 3 unsolicited AEs (0, 0.3%, and 0.3%, in SD, RD, and placebo groups, respectively). Neither AEs leading to withdrawal nor related serious AEs (SAEs) or deaths were reported. A detailed description of solicited and unsolicited AEs is reported in Table S1.
Antibody response was boosted by a second GRAd-COV2 administration, with no major differences in study populations and further enhancement in spike seropositive volunteers or after SARS-CoV-2 exposure and heterologous mRNA vaccination
By 21 days after the first dose, a spike-binding immunoglobulin G (IgG) response was induced in the majority of participants (Figure 3A; Table 2); similar geometric mean titer (GMT; 40.54 and 41.13 AU/mL) and seroconversion rates (93.5% and 92.5%) were observed between SD and RD arms, despite the 2-fold difference in vaccine dosage (2 × 1011 and 1 × 1011 vp, respectively, for SD and RD arms). Spike IgG titers in the SD arm peaked at day 36 (GMT: 45.15 AU/mL) and remained quite stable until day 57, then contracted around 3-fold by 6 months (GMT: 13.40 AU/mL at day 180). The effect of a second GRAd-COV2 administration was evident at day 36 (14 days post-dose 2 in RD arm), with a significant increase of spike-binding IgG (GMT: 77.94 AU/mL, p = 0.0001) and seroconversion rate up to 99.3% in the RD arm (Table 2). Titers were diminished at day 57 (GMT: 55.74 AU/mL) but were still significantly higher than in the SD arm (p = 0.001), then declined to similar levels at day 180 (GMT: 15.50 AU/mL), a 5-fold contraction with respect to the day 36 peak.
Figure 3.
Spike/RBD binding and SARS-CoV2 neutralizing antibody kinetics in GRAd-COV2-vaccinated and placebo arms
(A–D) The magnitude and kinetics of antibodies binding to full-length trimeric spike (A) or RBD (B) and of live SARS-CoV-2 neutralizing antibodies, expressed as 50% (C) or 80% neutralizing titer (D), following GRAd-COV2 or placebo administration are reported over 6 months of follow up. Datapoints are the geometric mean (GM) and 95% confidence interval (CI) at each study visit for each study arm. For binding antibodies (A and B), data are expressed as arbitrary units (AU)/mL, as per assay manufacturer datasheet. For neutralizing antibody titers (C and D), data are expressed as NT50 or NT80, or the reciprocal of serum dilutions showing 50% or 80% infection reduction. Arrowheads below the x axes indicate vaccination. Statistical analysis of variance, as described in STAR Methods, is displayed only for comparison between SD and RD vaccine arms; the difference between placebo and both vaccine arms was highly significant (p ≤ 0.0001) at all post-vaccination visits.
(E and F) Neutralizing titers at day 36 visit in a subset of 100 subjects in SD or RD vaccine study arms, measured by SARS-CoV-2 pseudoparticle neutralization assay (PNA) based on VSV pseudotyped with spike from Wuhan (filled symbols) or Delta (open symbols) strains. In (E), data are expressed as 50% neutralization titer (see above for definition), while in (F) and for the Wuhan strain only as appropriate, data are converted in WHO international units (IU)/mL. Each symbol corresponds to one serum sample, and horizontal line and error bars represent GM and 95% CI, respectively. Two-tailed Mann-Whitney test was used, and the only significant difference is shown in (E). Dashed lines indicate assay LOD.
In all panels, gray symbols/lines indicate placebo arm, while red and blue symbols/lines indicate SD and RD GRAd-COV2 arms, respectively.
Table 2.
Antibody response to spike and neutralizing antibodies to live SARS-CoV-2 at baseline and post-vaccination
| Anti-S ELISA (AU/mL) |
SARS-CoV-2 neutralization (NT50) |
SARS-CoV-2 neutralization (NT80) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Visit (day) | Statistic | SD (N = 298) | RD (N = 300) | PL (N =3 01) | SD (N = 298) | RD (N = 300) | PL (N = 301) | SD (N = 298) | RD (N = 300) | PL (N = 301) |
| 1 | n | 298 | 300 | 301 | 298 | 297 | 300 | 298 | 299 | 301 |
| GM | 1.73 | 1.77 | 1.71 | 5.35 | 5.35 | 5.31 | 4.15 | 4.16 | 4.11 | |
| 95% CI for GM | 1.66,1.80 | 1.68,1.87 | 1.64,1.78 | 4.99,5.74 | 5.01,5.72 | 4.98,5.67 | 4.02,4.28 | 4.05,4.27 | 4.01,4.21 | |
| 22 | n | 291 | 295 | 288 | 291 | 294 | 288 | 291 | 295 | 288 |
| GM | 40.54 | 41.13 | 1.71 | 45.29 | 47.34 | 4.91 | 14.70 | 15.09 | 4.09 | |
| 95% CI for GM | 35.62,46.14 | 36.25,46.67 | 1.64,1.79 | 38.83,52.82 | 40.50,55.35 | 4.63,5.20 | 12.84,16.82 | 13.07,17.41 | 3.99,4.20 | |
| GMFR | 23.63 | 23.17 | 1.01 | 8.64 | 8.84 | 0.95 | 3.58 | 3.63 | 1.00 | |
| seroresponse (%) | 93.5 | 92.5 | 0 | 73.2 | 74.8 | 0 | 46.7 | 43.1 | 0 | |
| 36 | n | 287 | 293 | 279 | 286 | 292 | 278 | 287 | 292 | 278 |
| GM | 45.15 | 77.94 | 1.73 | 47.88 | 86.11 | 5.09 | 17.31 | 29.91 | 4.12 | |
| 95% CI for GM | 39.54,51.56 | 70.36,86.33 | 1.65,1.81 | 41.22,55.60 | 75.43,98.30 | 4.77,5.43 | 15.18,19.73 | 26.31,34.01 | 4.02,4.23 | |
| GMFR | 26.29 | 43.87 | 1.02 | 9.09 | 16.08 | 0.98 | 4.21 | 7.21 | 1.01 | |
| seroresponse (%) | 92 | 99.3 | 0 | 76.2 | 90.1 | 0.4 | 53 | 76 | 0 | |
| 57 | n | 279 | 288 | 232 | 279 | 288 | 232 | 279 | 288 | 232 |
| GM | 35.88 | 55.74 | 1.80 | 48.36 | 80.91 | 5.08 | 19.31 | 30.37 | 4.19 | |
| 95% CI for GM | 31.23,41.22 | 50.11,62.01 | 1.68,1.94 | 40.75,57.38 | 70.19,93.26 | 4.71,5.48 | 16.66,22.39 | 26.48,34.84 | 4.02,4.37 | |
| GMFR | 20.85 | 31.32 | 1.07 | 9.17 | 15.13 | 0.99 | 4.69 | 7.28 | 1.02 | |
| seroresponse (%) | 90.3 | 97.9 | 1.3 | 72 | 87.5 | 1.7 | 54.8 | 75.7 | 0.9 | |
| 180 | n | 193 | 238 | – | 194 | 235 | – | 194 | 235 | – |
| GM | 13.40 | 15.50 | – | 26.94 | 31.51 | – | 11.67 | 12.95 | – | |
| 95% CI for GM | 11.45,15.68 | 13.56,17.73 | – | 22.16,32.75 | 26.50,37.48 | – | 9.87,13.80 | 11.17,15.02 | – | |
| GMFR | 7.75 | 8.70 | – | 5.02 | 5.85 | – | 2.83 | 3.10 | – | |
| seroresponse (%) | 74.6 | 82.4 | – | 59.8 | 62.6 | – | 35.6 | 39.6 | – | |
The 95% CIs were calculated based on the t-distribution of the natural log-transformed values, which were then back transformed to the original scale. Data from LABCORP and VIROCLINICS-DDL Central Laboratory. Seroresponse is defined as ≥4-fold rise in titers post-baseline. N, number of volunteers in the IAS population; n, number of volunteers in the IAS population with available data at each time point; GM, geometric mean; GMFR, GM fold rise; SD, single dose; RD, repeated dose; PL, placebo.
Assessment of receptor-binding domain (RBD)-binding IgG (Figure 3B) provided substantially similar results in terms of IgG kinetics for the SD and RD vaccine regimens, with a clear effect of a GRAd-COV2 second dose (GMTs: 942 and 1,959 AU/mL at days 36 and 892 and 1,602 AU/mL at day 57 in SD and RD, respectively, p = 0.0001 at both time points). Decline of RBD IgG titers to similar levels at 6 months (day 180) was also confirmed (253 and 330 AU/mL in SD and RD, respectively).
Upon conversion into WHO binding antibody unit (BAU)/mL (Figures S1A and S1B), a single administration of GRAd-COV2 provided peak (day 36) GMTs of 212 and 134 BAU/mL on spike and RBD, respectively, in the SD arm and of 366 and 278 BAU/mL on spike and RBD, respectively, after a second GRAd-COV2 dose in the RD arm.
As measured in a live SARS-CoV-2 neutralization assay (Figure 3C; Table 2), neutralizing antibodies were induced (GM 50% neutralization titer [NT50] of 45.29 and 47.88 at days 22 and 36, respectively) and remained stable until day 57 in the SD arm (GM NT50 of 48.36 at day 57), with a peak seroconversion rate of 76.2% at day 36. A second GRAd-COV2 administration raised neutralizing titers from GM 47.34 (day 22) to 86.11 NT50 at day 36 in the RD arm, with the seroconversion rate reaching 90.1%. Titers remained stable up to day 57 and then declined at 6 months, reaching similar levels in both arms (GM around 30 NT50 at day 180), a 1.8- and 2.7-fold contraction from the peaks in the SD and RD arms, respectively. Expressed as NT80, peak neutralization GMTs of 19.31 (SD) and 30.37 (RD) were reached at day 57 in both study arms, with maximal seroconversion rates of 54.8% (day 57) for SD and 76% (day 36) for RD, again confirming the positive effect of a second GRAd-COV2 administration (Figure 3D; Table 2).
A representative sample of day 36 sera from 100 subjects per vaccine arm tested with spike pseudotyped vesicular stomatitis virus (VSV) (Figure 3E) returned GMs of 105.8 and 163.1 NT50 for the SD and RD arms, respectively, on the Wuhan strain, a difference that did not reach statistical significance with sample numerosity reduced to one-third. A 4-fold loss in neutralization potency was observed on the Delta variant (GM NT50 of 26.52 and 40.58), with the RD regimen resulting in a lower frequency of subjects with an undetectable Delta neutralizing titer (12% vs. 21% in RD and SD). When expressed in WHO international units (Figure 3F), GM neutralizing titers on the Wuhan strain were at 56.58 and 87.15 IU50/mL in the SD and RD arms. A subset of day 36 sera from 10 subjects per study arm were finally tested with a third confirmatory lentivirus-based pseudoneutralization assay, providing comparable NT50 titers to those seen with the other two neutralization assays. Reassuringly, all serology assays used to assess binding and neutralizing antibodies yielded highly correlated datasets (Figures S2A–S2C).
Only minor differences in vaccine immunogenicity were attributable to age or co-morbidities (Figures S3A, S3B, S3D, and S3E). Older volunteers in the RD arm receiving the lower vaccine dosage (1 × 1011 vp) responded less vigorously than younger, healthy volunteers throughout all time points, but GMT increased adequately after the second dose, highlighting the benefit of a second GRAd-COV2 administration in this age cohort. Conversely, immune responses were comparable in the two age cohorts at the higher vaccine dose (2 × 1011 vp, SD arm). A trend for stronger immune response to both SD and RD vaccination regimens was also noted in females (Figures S3C and S3F).
In a subset of volunteers with negative anti-N but detectable spike-binding antibodies at baseline, spike binding and neutralizing antibodies reached 10-fold higher levels at day 22 post-GRAd-COV2 administration compared with seronegative volunteers, with no appreciable boosting effect of the second vaccine dose (Figures S4A–S4D). The study also enrolled a small set of volunteers infected with HIV who responded with similar antibody titer magnitude and kinetics to both GRAd-COV2 SD and RD vaccination regimens compared with uninfected subjects (Figures S4E–S4H). A 10- to 20-fold higher level of spike binding and neutralizing antibodies were found at the day 180 visit in study participants that received approved (mostly mRNA) COVID-19 vaccines after unblinding at day 57. Such titers were clearly higher than at peak with either SD or RD primary series GRAd-COV2 vaccination (Figures S4I–S4L). Similarly, intercurrent SARS-CoV-2 infection or exposure enhanced or boosted both binding and neutralizing antibodies on top of GRAd-COV2 immunogenicity at the time when intercurrent exposure/infection was detected (Figures S5A–S5F).
A single administration of GRAd-COV2 is sufficient to induce potent, broad, Th1-skewed, cross-reactive, and polyfunctional CD4 and CD8 T cell responses to spike
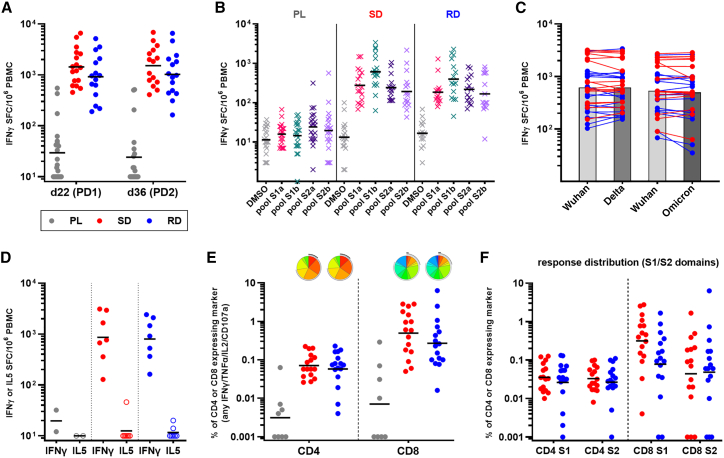
Potent spike-specific T cell response were detected by interferon γ (IFNγ) ELISpot 3 weeks after a single GRAd-COV2 vaccination (Figure 4A, day 22), with GMs of 1,438 and 920 spot-forming cells (SFCs) per million peripheral blood mononuclear cells (PBMCs) in the SD and RD arms, respectively. Administration of a second GRAd-COV2 dose did not result in an increased T cell response, which remained otherwise stable at day 36 in both SD and RD arms (GMs of 1,515 and 1,020 SFC/million PBMCs, respectively). Low to moderate levels of IFNγ secretion in response to spike stimulation was also detectable in a subset of subjects in the placebo arm. Despite the trend for slightly higher responses in the SD arm, possibly due to the 2-fold higher vaccine dose, there was not statistically significant difference between SD and RD at both day 22 and 36 visits, while the difference between each vaccine arm and the placebo arm was strongly significant (p ≤ 0.0001 by Mann-Whitney at both visits). The T cell response was broad, recognizing epitopes located throughout the whole spike protein sequence, with some preferential recognition of peptide pool S1b that included the RBD region (Figure 4B). Importantly, the vaccine-induced T cell response to spike from the Delta and Omicron variants was mostly conserved (Figure 4C). As seen in a phase 1 trial,11 the T cell response was Th1 skewed with virtually absent interleukin-5 (IL-5) secretion in response to spike antigen stimulation (Figure 4D).
Figure 4.
Spike-specific T cell response after GRAd-COV2 vaccination
PBMCs were isolated and cryopreserved for the analysis of T cell responses from a subset of 54 volunteers, 21 enrolled in placebo (PL), 17 in SD, and 16 in RD arms.
(A) Total T cell response to SARS-CoV-2 spike at days 22 (post-dose 1-PD1) and 36 (post-dose 2-PD2), evaluated by IFNγ ELISpot and expressed as IFNγ spot-forming cells (SFC) per million PBMCs.
(B) Breadth of response to spike: response to DMSO (negative control: gray symbols) and peptide pools covering the S1a (pink symbols), S1b (green symbols), S2a (purple symbols), and S2b (violet symbols) portions of spike, evaluated in ELISpot at day 22.
(C) Cross-reactivity of the T cell response to variants of concern: total spike response to Wuhan and Delta or Wuhan and Omicron variants, evaluated in distinct ELISpot assays using day 22 PBMCs from all GRAd-COV2-vaccinated subjects. The response on each variant in an individual volunteer is connected by a line, and bars are set at GM.
(D) IFNγ (Th1) and IL-5 (Th2) production upon spike peptide pool stimulation, evaluated at the day 36 visit in eight subjects per vaccine arm and two PL recipients by two-color ELISpot.
(E and F) Multiparametric flow cytometry analysis of CD4 and CD8 T cells responses at day 36 in all GRAd-COV2 and eight PL recipients. Total spike response (E) and breadth (F) of response on S1 and S2 spike domains. Data are expressed as the percentage of CD4 and CD8 expressing any combination of the analyzed functions (IFNγ, TNF-α, IL-2, or CD107a) within the CD69+ fraction in response to spike antigen stimulation. Pie charts (base: median) representing the functional profile of spike-specific CD4 and CD8 are shown in (E) and are better described in Figure S7.
In all panels, red circles represent SD arm subjects, blue circles represent RD arm subjects, gray circles represent PL arm subjects, and black horizontal lines indicate GM.
Multiparameter flow cytometry analysis conducted at day 36 (Figures 4E, S6A, and S6B) showed that GRAd-COV2 vaccination induced a spike-specific T cell response composed of CD4 (GMs: 0.071% and 0.058% in SD and RD arms, respectively) and an even higher frequency of CD8 (GMs: 0.5% and 0.27% in SD and RD, respectively). Polyfunctionality analysis of the CD4 compartment showed that subsets of spike-specific cells expressing all three cytokines (IFNγ/tumor necrosis factor α [TNF-α]/IL-2) as well as any combinations of two cytokines or IFNγ only were present at similar levels (Figures 4E and S7A), while in the spike-specific CD8 compartment, the most represented subsets were dual IFNγ/CD107a, dual IFNγ/TNFα, or IFNγ only, with a fair presence of triple IFNγ/TNF-α/CD107a (Figures 4E and S7B). Overall, around one-half of the spike-specific CD8 showed degranulation/cytotoxic potential (CD107a+). The distribution of the different (poly)functional cell subsets was virtually identical in the SD and RD arms. The level of spike-specific CD154+/CD69+ CD4 was superimposable to the Th1 CD4 response as detected by any combination of IFNγ/TNF-α/IL-2 secretion or to CD4 cells secreting IFNγ only; this observation confirms that the contribution of non-Th1 (i.e. Th2, Th17) to a spike-specific CD4 response to GRAd-COV2 is negligible (Figures S8A and S8B). Spike-specific CD4 and CD8 recognized both S1 and S2 spike domains, highlighting the breadth of responses in both T cell subsets (Figure 4F).
GRAd-COV2 vaccination generates spike-specific T and B cell memory 6 months after vaccination, with high proliferative capacity and further expansion by heterologous mRNA vaccination
A high frequency of circulating spike-specific T cells readily secreting IFNγ upon antigen stimulation were found in all volunteers in RD and SD arms 6 months (day 180) after the first GRAd-COV2 vaccination (Figure 5A), at levels only minimally reduced compared with those measured at peak (GM ratio days 36/180 of 1.9 in RD and 2.3 in SD subjects with response at both time points evaluable). Receipt of approved COVID-19 vaccines after day 57 stabilized T cell responses (GM ratio days 36/180 of 1.1 in SD + vaccinated [vax] subjects), but did not further amplify them above peak levels achieved with GRAd-COV2 vaccination, at variance with the potent boosting of binding and neutralizing antibodies in the same subjects (Figures S9A and S9B).
Figure 5.
Long-term T and B memory response in the PBMC substudy
(A) Kinetics of T cell response to SARS-CoV-2 spike at days 22 (post-dose 1-PD1), 36 (post-dose 2-PD2), and 180, evaluated by IFNγ ELISpot and expressed as IFNγ spot-forming cells (SFC) per million PBMCs.
(B) Proliferative CD4 and CD8 T cell responses (percentage of CD4 or CD8 CellTrace low) following 5 day incubation with spike peptide pools, assessed in PBMCs collected at day 180 (day 36 for the four PL arm subjects).
(C) Quantification of spike-specific (Spike++ as defined in STAR Methods) memory B cell (MBC) percentages in all 54 PBMC substudy subjects at day 36 and 180 visits. Data were analysed with two-tailed, paired Wilcoxon test, and only significant differences are shown.
In all panels, gray symbols/lines indicate PL arm, while red and blue symbols/lines indicate SD and RD GRAd-COV2 arms, respectively. Open red symbols and pink shaded areas indicate volunteers in the SD cohort that received an approved COVID-19 vaccine (defined SD+vax) between the day 57 and 180 visits. Horizontal black lines are set at GM.
GRAd-COV2-induced spike-specific T cells had strong proliferative capacity (Figures 5B and S10) 6 months post-vaccination, with frequencies as high as GM 1% for CD4 and 2%–3% for CD8. A booster dose of approved COVID-19 vaccines did not significantly enhance the overall proliferating memory T cells pool, in agreement with the ELISpot data. Proliferative CD4 and CD8 response were detectable on both S1 and S2 spike domains, again highlighting the breadth of the GRAd-COV2-induced memory T cell response (Figure S11).
Long-lived spike-specific memory B cells were generated by GRAd-COV2 vaccination (Figures 5C and S12) with no major differences between SD or RD regimes. If any, the higher levels seen at day 36 in the SD regimen may relate to the higher vaccine dose received by the volunteers at priming. The memory B cell frequency increased from day 36 to 180 (p = 0.035 for RD by two-tailed, paired Wilcoxon test). Coherently with the efficient boosting of antibody responses by approved COVID-19 vaccines, the pool of memory B cells was also clearly amplified in most volunteers receiving heterologous vaccination regimes (p = 0.002 for SD + vax by two-tailed, paired Wilcoxon test).
Discussion
The first randomized controlled trial of GRAd-COV2, given in a single or a two-dose regimen, has shown that the gorilla Ad-based vaccine candidate is well tolerated and immunogenic in healthy adults and at-risk individuals, with no related SAEs. The administration of a second dose was better tolerated as previously reported for two-dose adenoviral-vectored vaccine regimens.15 Humoral and cellular immune responses were induced in the majority of vaccine recipients after a single immunization, with anti-spike (S) antibodies doubling 2 weeks after the second shot. The vaccine was better tolerated and only slightly less immunogenic in older adults, the population that benefits most from a higher vaccine dose14 and second dose administration. We also report adequate immune response in a small number of persons living with HIV, similar to what was reported for Vaxzevria.16
Importantly, no thrombotic event was recorded in COVITAR participants. A recent FDA release estimates the risk of vaccine-induced thrombosis with thrombocytopenia (VITT) at 3.23 events per million of administered doses for Jcovden,17 while estimates are higher for Vaxzevria, between 1 case per 26,500 to 1 case per 127,300 first doses.18 It is worth noticing that no VITT events have been reported for widely rolled out COVID vaccines based on Ad5 (Convidecia or Sputnik); both Ad5 and GRAd are classified as group C adenoviruses, differently from Ad26 (group D) or ChAdOx1 (group E). Therefore, it is not yet clear if VITT is a class effect of all Ads or if it is restricted to specific strains or groups. Given the low frequencies reported for this rare but serious event with other adenoviral platform-based vaccines, only implementation in clinical practice may reveal if and to what extent GRAd vector is prone to induce the syndrome.
The introduction in January 2021 of the first WHO International Standard for anti-SARS-CoV-2 Ig has been instrumental for calibration of different SARS-CoV-2 serological assays and for the expression of data in a common unitage, allowing the comparison of an immunogenicity dataset associated with different COVID-19 vaccines. Reported peak IgG concentrations against the prototype Wuhan S were in the range of 60–100 BAU/mL after a single administration of Jcovden or Vaxzevria, increasing to around 200–500 depending on assay, antigen (S or RBD), and dosing interval for homologous double-dose regimens.19,20,21,22,23 Here, we show that a single administration of GRAd-COV2 induces peak binding antibody levels in the range of 150–250 BAU/mL, reaching 250–400 on RBD and S, respectively, with a second vaccine dose; similarly, in a subset of GRAd-COV2 recipients, peak neutralizing antibody titers as detected by pseudoparticle neutralization assay (PNA) was around 60 (SD) and 80 (RD) IU50/mL, well in agreement with levels reported for Jcovden or Vaxzevria; in turn, this would predict similar vaccine efficacy.24,25,26 However, this remains to be formally proven in a phase 3 study. Besides, it is well known in the literature and clinical experience that the levels of binding and neutralizing antibodies elicited by approved COVID-19 vaccines based on adenoviral vectors (and consequently by GRAd-COV2) are about 1 log lower than those elicited by mRNA or adjuvanted protein subunit vaccines after primary series.27,28,29 This apparent platform weakness is relative in a pandemic context dominated by circulating VOCs endowed with substantial immune evasion from the humoral, but not the cellular, immune response and has been counterbalanced by the adoption of highly effective heterologous booster strategies implemented in most countries.
Indeed, it is highly improbable that substantial cross-neutralization on Omicron would be detected in serum of GRAd-COV2 vaccinated subjects, as suggested by the moderate neutralizing antibody (nAb) titers on ancestral and the clear reduction on Delta variants as detected by PNA assay. However, this is the case for most vaccines after primary series, and a multiple boosting strategy or bivalent vaccines is indeed the current standard for vaccination campaigns. Of note, extending the interval between first and second GRAd-COV2 administration beyond the 3 weeks explored in this study is expected to further increase immunogenicity, as has now been clearly established for both adenoviral vectors and mRNA-based vaccines.30,31,32,33,34,35
Vaccine-elicited antibody and T cell responses exert different and complementary roles in resistance to infection, severe disease, hospitalization, and death. Antibodies block infection by binding virus and preventing viral entry into host cells and are correlates of protection for many vaccines. Memory T cells can provide an important additional layer of protective immunity. While T cells cannot prevent host cells from initially becoming infected, they can respond rapidly once infection has occurred to limit virus replication and spread via either the secretion of antiviral cytokines or by killing infected host cells. Thus, although not formally assessed as a correlate of protection for SARS-CoV-2 infection, vaccine-induced T cell responses may ensure long-term protection from severe disease also thanks to their broad cross-reactivity against all circulating VOCs.14,36,37 Recent evidence accumulated during the SARS-CoV-2 pandemic has revealed that variants that escaped from nAbs that were induced by either natural infection or vaccination could not escape from CD8 T cell-mediated immunity.38 As the vaccine-elicited antibody response wore off, most CD4 and CD8 T cells responses were preserved.39 Accordingly, T cells have been credited with playing a key role in the sustained vaccine-mediated protection against hospitalization and death.40
Here, we show that GRAd-COV2 vaccine induces a potent and broad S-specific, Th1-skewed cellular response following the first dose. Administration of a second GRAd-COV2 dose does not increase the magnitude or alter the polyfunctionality profile of the T cell response, at least with a short 3 week interval, similar to other adeno-based vaccines.15 Polyfunctional CD4 responses induced by GRAd-COV2 are well in range of those reported for other COVID-19 vaccines, while frequencies of CD8 T cells are remarkably high when compared with published ICS datasets from Ad5, ChAdOx1, or Ad26 clinical trials,34,41,42 placing GRAd-COV2 as potential best in class for CD8 responses.29 This finding may be attributed to the slightly higher vaccine dosage than other adenoviral-based vaccines but is also well aligned with previous experience indicating group C adenoviral vectors as the most potent inducers of T cell responses.43 As expected, the T cell response cross-recognized S from both Delta and Omicron, suggesting high potential for cross-recognition of even highly divergent variants. Interestingly, GRAd-COV2 vaccination induced CD4 and CD8 responses targeted to both the S1 and S2 subunits of the S protein. The S2 subunit, which is crucial for virus and host cell membrane fusion and entry, is more conserved across human and animal coronaviruses than the S1 subunit, and cross-reactive, class-switched IgGs to S2 have been described in SARS-CoV-2 uninfected individuals, especially those of young age.44 It is tempting to speculate that potent S2-targeted T cell responses may mediate some degree of cross-recognition and cross-protection against newly emerging coronaviruses and SARS-CoV-2 variants. Importantly, vaccine-induced T cells retained immediate effector functions as well as proliferative capacity up to 6 months after immunization, demonstrating the establishment of long-lasting and proficient antigen-specific memory T cells that is a hallmark of successful vaccination. Moreover, GRAd-COV2 induced S-specific memory B cells that increase over time, as reported for other COVID-19 vaccines.29,45,46
Although the implementation of this vaccine for primary course is of limited or no value in light of Omicron prevalence, GRAd-COV2 could be deployed as a component of highly immunogenic heterologous prime-boost regimens as has been shown with other mRNA/Ad-based vaccines19,21,22,47 and, incidentally, demonstrated in the GRAd-COV2 phase 1 trial48 and in the current article. While it is now well established that an mRNA booster dose provides best peak immunogenicity in the short term, recent evidence suggests that adenoviral-vectored vaccines may be a sound option as booster doses in mRNA primary cycle recipients, in light of slower antibody decay rates and superior T cell boost, especially of (VOC cross-reactive) CD8.19,49,50 While large controlled clinical studies with all available permutations of COVID-19 vaccines are needed to make the results applicable in a broader perspective, the improved immunogenicity, effectiveness and flexibility observed so far make the heterologous prime-boost regimen approach an important strategy for policymakers globally. This is of particular relevance during challenging vaccine rollout conditions. Indeed, GRAd-COV2 is based on a different non-cross-reacting adenoviral serotype from all approved Ad-based vaccines already deployed in worldwide vaccination campaigns; this may represent an advantage for future booster doses thanks to absent or low preexisting anti-GRAd immunity in the human population. In addition, the favorable “tractability” typical of the platform, i.e., low cost of goods, reasonable thermostability, and easier manufacturing process that facilitates tech-transfer, ultimately allowing local production, makes GRAd-COV2 a valid vaccine option, especially in lower income countries.
In conclusion, we propose that the GRAd adenoviral vector is an attractive alternative vaccine platform; simply by changing the antigenic load, the platform is suitable for developing effective second-generation COVID-19 or pan-(sarbe)coronavirus vaccines or for any new emerging pathogens requiring the generation of well-coordinated antibody and T cell responses. Specifically, the induction of a potent and durable CD8 T cell response by the GRAd might be instrumental to fight SARS-CoV-2 variants and future emerging pathogens as well as diseases where this type of immune response is defective or suppressed. In addition, the adenoviral platform is suitable for mucosal administration, as shown in many preclinical and clinical settings even for SARS-CoV-2,51,52 which may be a highly desirable feature for the development of a vaccine with transmission-blocking potential.53
Limitations of the study
The study has several limitations. First, the epidemiological context and the national vaccine campaign leading to study unblinding at day 57 has made it impossible to transition to the phase 3 part of the study and to assess the vaccine efficacy, as originally planned in the study protocol. Nevertheless, the blinded phase of the study has allowed us to rigorously evaluate the safety of SD and RD regimens and has enabled the study’s steering committee jointly with DSMB to recommend the two-dose regimen for further clinical development. Second, the study was conducted in Italian clinical centers only, with population ethnicity limited to White subjects. Further studies are needed to ascertain the vaccine safety and immunogenicity in more diverse populations. Third, the durability of the immune response can only be assessed up to the presently reported 6 month follow up; in fact, during the interval of time between day 180 and 360 visits, almost all subjects enrolled in the SD and RD arms had received COVID-19 approved vaccines as booster doses. The dataset at day 360 will, anyway, provide further interesting immunogenicity data on the combination of GRAd-COV2 with existing approved (mostly mRNA-based) COVID-19 vaccines. Fourth, since Omicron was not yet circulating at the time of our study, it was not included in the preplanned serology panel.
Consortia
The members of the COVITAR study group are Luigi Ziviani, Feliciana Malescio, Irene Turrini, Rita Lawlor, Annamaria Romano, Mariagrazia Nunziata, Salvatore Armato, Nicole Mazzeo, Maria Aurora Carleo, Chiara Dell’Isola, Raffaella Pisapia, Agostina Pontarelli, Andrea Olivani, Sara Grasselli, Diletta Laccabue, Maria Cristina Leoni, Franco Paolillo, Annalisa Mancini, Barbara Ruaro, Marco Confalonieri, Francesco Salton, Giulia Mancarella, Raffaella Marocco, Margherita De Masi, Valeria Belvisi, Silvia Lamonica, Antonella Cingolani, Cristina Seguiti, Paola Brambilla, Alice Ferraresi, Matteo Lupi, Serena Ludovisi, Giulia Renisi, Roberta Massafra, Martina Pellicciotta, Luciana Armiento, Stefania Vimercati, Mariagrazia Piacenza, Paolo Bonfanti, Paola Columpsi, Marina Elena Cazzaniga, Cristina Rovelli, Mariaelena Ceresini, Letizia Previtali, Laura Trentini, Chiara Alcantarini, Walter Rugge, Stefano Biffi, Federica Poletti, Roberto Rostagno, Roberta Moglia, Ferdinando De Negri, Elisabetta Fini, Alice Cangialosi, Serena Rita Bruno, Marianna Rizzo, Mariangela Niglio, Anna Dello Stritto, Alfredo Matano, Arnolfo Petruzziello, Pietro Valsecchi, Teresa Pieri, Mauro Altamura, Angela Calamo, Anna Giannelli, Stefania Menolascina, Silvia Di Bari, Vera Mauro, Raissa Aronica, Daniela Segala, Rosario Cultrera, Laura Sighinolfi, Michelle Abbott, Andrea Gizzi, Federica Guida Marascia, Giacomo Valenti, Marcello Feasi, Nicoletta Bobbio, Filippo Del Puente, Alfredo Nicosia, Martina Frascà, Miriam Mazzoleni, Nadia Garofalo, Virginia Ammendola, Fabiana Grazioli, Federico Napolitano, Alessandra Vitelli, and Valentina Marcellini.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti human CD19 conjugated to APC-Vio770 (clone LT19) | Miltenyi Biotec | Cat# 130-113-166; RRID: AB_2725994 |
| mouse anti human CD27 conjugated to Vio Bright FITC (clone M-T271) | Miltenyi Biotec | Cat# 130-113-634; RRID: AB_2751160 |
| mouse anti human IgA conjugated to VioGreen (clone IS11-8E10) | Miltenyi Biotec | Cat# 130-113-481; RRID: AB_2734099 |
| mouse anti human IgM conjugated to APC (clone Pj2-22H3) | Miltenyi Biotec | Cat# 130-122-915; RRID: AB_2801965 |
| mouse anti human IgG conjugated to VioBlue (clone IS11-3B2.2.3) | Miltenyi Biotec | Cat# 130-119-881; RRID: AB_2751904 |
| mouse anti human CD24 conjugated to BV711 (clone ML5) | BD Biosciences | Cat# 563401; RRID: AB_2631261 |
| mouse anti human CD107a conjugated to PE-CF594 (clone H4A3) | BD Biosciences | Cat# 562628; RRID: AB_2737686 |
| mouse anti human CD3 conjugated to BUV395 (clone UCHT1) | BD Biosciences | Cat# 563546; RRID: AB_2744387 |
| mouse anti human CD4 conjugated to BUV737 clone SK3 | BD Biosciences | Cat# 612748; RRID: AB_2870079 |
| mouse anti human CD8 conjugated to BV605 (clone SK1) | BD Biosciences | Cat# 564116; RRID: AB_2869551 |
| mouse anti human CD154 conjugated to BB700 (clone TRAP1) | BD Biosciences | Cat# 745814; RRID: AB_2743265 |
| mouse anti human IFNγ conjugated to APC clone B27 | BD Biosciences | Cat# 554702; RRID: AB_398580 |
| rat anti human IL-2 conjugated to PE (clone mQ1-17H12) | BD Biosciences | Cat# 559334; RRID: AB_397231 |
| mouse anti human TNFα conjugated to BV650 (clone Mab11) | BD Biosciences | Cat# 563418; RRID: AB_2738194 |
| mouse anti human CD69 conjugated to APC-R700 (clone FN50) | BD Biosciences | Cat# 565154; RRID: AB_2744449 |
| Biological samples | ||
| Human Serum | Study Participants | N/A |
| Human PBMC | Study Participants | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| PepMix™ SARS-CoV-2 (Spike Glycoprotein) | JPT | Cat# PM-WCPV-S-1 |
| PepMix™ SARS-CoV-2 (Spike B.1.617.2/Delta) | JPT | Cat# PM-SARS2-SMUT06-1 |
| PepMix™ SARS-CoV-2 (Spike B.1.1.529/BA.1/Omicron) | JPT | Cat# PM-SARS2-SMUT08-1 |
| CEFX Ultra SuperStim Pool | JPT | Cat# PM-CEFX-1 |
| Overlapping SARS-CoV-2 Spike 15mers | custom ordered from Elabscience Biotech Inc (Lanini S. et al., SciTraslMed 2021) | N/A, |
| Staphylococcal enterotoxin B from Staphylococcus aureus | Sigma-Aldrich | Cat# S4881 |
| GolgiPlug | BD Biosciences | Cat# 555029 |
| Brilliant Stain Buffer | BD Biosciences | Cat# 563794 |
| BD Fixation/Permeabilization Kit | BD Biosciences | Cat# 554714 |
| FVS780 | BD Biosciences | Cat# 565388 |
| HyClone Defined FBS, US Origin, 500 mL | Cytiva | Cat# SH30070.03 |
| CTL-Wash™ Supplement 10x | Immunospot | Cat# CTLW-010 |
| CellTrace Violet | ThermoFisher | Cat# C34571 |
| Critical commercial assays | ||
| COVID-SeroIndex, Kantaro Quantitative SARS-CoV-2 IgG Antibody Kit (Spike) |
R&D Systems | Cat# DSR200 |
| ARCHITECT SARS-CoV-2 IgG kit (Nucleocapsid) |
Abbot | Cat# 6R86 |
| ARCHITECT SARS-CoV-2 IgG II Quant (RBD) |
Abbot | Cat# 6S60 |
| Human IFN-γ ELISpot plus kit | Mabtech | Cat# 3420-4APT |
| IFNγ-IL5 Fluorospot | Mabtech | Cat# FSP-0108 |
| SARS-CoV-2 Spike B cell analysis kit | Miltenyi Biotec | Cat# 130-128-022 |
| SARS-CoV-2 live neutralization assay | Viroclinics Biosciences-Cerba | N/A |
| Pseudo-neutralization assay (D614 and Δ strains) | Nexelis-Q2 solutions | https://nexelis.com/our-expertise/infectious-diseases/covid-19/ |
| Pseudo-neutralization assay (D614) | Monogram Biosciences-Labcorp | https://monogrambio.labcorp.com/phenosense-sars-cov-2-neutralizing-antibody |
| Software and algorithms | ||
| Graphpad PRISM 7 | GraphPad | RRID:SCR_002798 |
| FlowJo 10 | BD Biosciences | RRID:SCR_008520 |
| SAS® System software 9.4 | SAS Institute Inc. | RRID:SCR_008567 |
| CTL Immunospot suite 2.7 | Cellular Technology Limited | RRID:SCR_011082 |
| SPICE 6 | NIH/NIAID https://niaid.github.io/spice/ |
RRID:SCR_016603 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Stefania Capone (Stefania.capone@reithera.com).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
The COVITAR study protocol was approved by the Italian Regulatory Agency (AIFA), the COVID national Ethics Committee (Lazzaro Spallanzani Institute), and the local Ethics Committees of the other 23 clinical centers. Study Protocol is available for consultation (data S1). All participants received and signed a written informed consent prior to enrollment.
An independent Data Safety Monitoring Board (DSMB) was established before the start of the trial and reviewed unblinded safety data twice during the study.
Sample size and characteristics of the study groups at randomization, including total number, age, gender, body mass index (BMI), and underlying diseases are reported in Figure 1 and Table 1.
Method details
Trial design and oversight
This phase 2 randomised, observer-blind, placebo controlled, multicenter trial (COVITAR study, ClinicalTrials.gov NCT04791423) is the first part of a phase 2/3 protocol study and was conducted at 24 centers in Italy in accordance with the Declaration of Helsinki and Good Clinical Practices. Around 900 adult female and male, ≥18 years of age were planned to be included. The main exclusion criteria included: allergy to any vaccine component, Guillain-Barré syndrome, laboratory-confirmed SARS-CoV-2 infection, immunodeficiency state, any vaccination (licensed or investigational) other than for influenza within 30 days before/after administration of study intervention, and pregnancy. Full inclusion and exclusion criteria are listed in the study protocol. Mild/moderate well controlled comorbidities were allowed, including HIV infection.
Randomisation was stratified by age (< or ≥65 years); for participants <65 years, by comorbidities representing risk factors for COVID-19 severe illness (per CDC recommendation, May 2020). At least 25% of enrolled participants had to be either ≥65 years or <65 years and “at risk”.
By the use of an interactive Web-based system, participants were randomly assigned at a ratio of 1:1:1 to receive, 21 days apart, a single administration of GRAd-COV2 at a dose of 2x1011 viral particles (vp) followed by placebo, or a repeated GRAd-vaccine dose of 1x1011 vp, or two doses of placebo. Neither participants, nor investigators or Sponsor’s staff involved in clinical management or study monitoring were aware of the study intervention administered. Since GRAd-COV2 and placebo were visually distinct prior to dose preparation, in order to maintain blindness preparation of the syringes was done by an unblinded pharmacist, and then handed over to the investigator for administration to the participant.
Investigational medicinal product and placebo
GRAd-COV2 is a group C gorilla-derived adenovirus vector, GRAd-32 (ΔE1, ΔE3, ΔE4) encoding the full length, pre-fusion stabilized Spike protein (Wuhan strain). Two mutations were introduced in the Spike sequence to convert amino acid (aa) 986–987 KV into 2 prolines (2P) to stabilize the protein in its pre-fusion state.54,55 GRAd-COV2 was manufactured by ReiThera srl under good manufacturing practice conditions in the proprietary cell line ReiCell35S, a suspension adapted packaging cell line derivative of HEK293. The vaccine was purified by an extensive downstream process including host cell DNA precipitation, depth filtration, two chromatographic purification steps followed by nuclease digestion and ultrafiltration. The clinical material was finally formulated in formulation buffer (10 mM Tris, 75 mM NaCl, 1 mM MgCl2, 0.02% PS80, 5% sucrose, 0.1 mM EDTA, 10 mM Histidine, 0.5% ethanol, pH 7.4) at a concentration of 2 x 1011 viral particles/ml.
Commercially available sterile 0.9% (w/v) saline solution was administered as Placebo, and used to dilute the investigational vaccine for the 1x1011 vp dose.
Trial procedures and outcomes
Participants received the two injections (1 mL volume) in the deltoid muscle, and remained in observation for 30 min after vaccination for acute reactions.
The primary endpoints of this trial were: solicited local or systemic adverse events (AEs) within 7 days after each dose of vaccine or placebo, recorded through an electronic diary; unsolicited AEs reported by participants through 1 month after the second dose; serious adverse events (SAEs) and adverse events of special interest (AESIs) through 1 year after the second injection. AEs data up to approximately 24 weeks after the second dose are included in this report and safety results are reported for all participants who provided informed consent and received at least one dose of vaccine or placebo.
All participants had humoral immunogenicity assessment at 1 (pre-dose), 22, 36, 57, 180 and 360 days after the first dose. A subset of participants in a single clinical site (N = 54, at CRC Verona, Italy) had cellular (T and B) immune response assessment in peripheral blood mononuclear cells (PBMC) at study day 22, 36 and 180. Main serology assays were centralized.
Primary assessment of humoral response was conducted by a semi-quantitative ELISA on full length Spike, using COVID-SeroIndex, Kantaro Quantitative SARS-CoV-2 IgG Antibody IVD Kit (R&D Systems). SARS-CoV-2 infections were monitored by testing sera for seroconversion to Nucleocapsid (N) antigen using SARS-CoV-2 IgG kit, a chemiluminescence microparticle assay (CMIA) on ARCHITECT platform (Abbott Diagnostics, Chicago, IL, USA). Both Spike ELISA and N CMIA were run at centralized lab (LabCorp, Geneva). SARS-CoV-2 neutralizing antibodies were measured by a plaque reduction neutralization test at a centralized lab (Viroclinics-DDL, the Netherlands) using SARS-CoV-2 Bav/Pat1/2020 strain and determining 50% and 80% neutralization titers (NT50 and NT80). Additional exploratory serology on selected samples or study visits, as detailed in the results section, were also conducted: IgG to receptor binding domain (RBD) were measured with SARS-CoV-2 IgG II Quant, a CMIA assay on ARCHITECT platform (Abbott Diagnostics, Chicago, IL, USA); neutralization activity was assessed with two pseudovirus neutralization assays (PNA) either based on vescicular stomatitis virus (VSV) pseudotyped with SARS-CoV-2 Spike glycoproteins from Wuhan (D614) or Delta strains (Nexelis Laval, QC, Canada), or by PhenoSense Anti-SARS CoV-2 Neutralizing Antibody Assay, based on D614 Spike pseudotyped lentivirus (Monogram Biosciences Inc., CA).
Cellular immune responses to Spike from prototype Wuhan, Delta and Omicron variants were measured primarily by IFNγ enzyme-linked Immunospot (ELISpot). Additional investigation was performed by multi-parametric flow cytometry analysis for the characterization of CD4 and CD8 T cell functional profile and proliferation capacity, and for the enumeration of Spike-specific memory B cells (MBC). All cellular assays were conducted at ReiThera or Ospedale Pediatrico Bambino Gesù (Rome).
Serum and PBMC isolation and cryopreservation
Blood for serology was collected at 1 (pre-dose), 22, 36, 57, 180 and 360 days after the first dose. Yellow-Gold Top, Plastic, SST II, 6mL tubes (BD cat. N. 366444) were used for collection and care was taken to thoroughly mix the blood with the clotting activation agent by inverting the tube not less than five times. Blood was allowed to clot for 30 min (tube standing upright), and tubes centrifuged at a minimum of 1500–2000 x g for 15 min until clot and serum were well separated. Serum was collected and aliquoted into polypropylene cryovials and frozen immediately at −70°C until shipment to Central lab.
PBMC were isolated at study day 22, 36 and 180 after first vaccine dose by collecting ∼40mL of blood into 4 × 10mL Green Top Lithium-Heparin Vacutainer (BD cat. N. 367526), and immediately inverting tubes 8–10 times gently to mix blood and anticoagulant and avoid micro clotting. Blood was processed within 2 h from collection. Blood was diluted 2:1 with HBSS (20mL of blood and 10 mL of HBSS) and loaded on sterile Leucosep Bio-One 50mL tubes (2 tubes per donor) pre-filled with Leucosep separation medium (Greiner, cat#227288). Tubes were centrifuged at 800 x g for 15 min at 20°C with the centrifuge brake off. PBMC rings were collected, diluted with HBSS, pelleted and washed with HBSS +5% FBS Defined (HyClone, cat#SH30070.03 heat-inactivated for 45 min at 56°C). Pellets from the same donor were pooled before second wash. Only in case of heavy red blood cell (RBC) contamination, an RBC lysis step was performed by resuspending PBMC pellet in 3mL of ACK lysing buffer (Gibco, cat#A10492), let sit for 5 min and then diluting to 30mL with PBS and pelleted. Viable cells were counted with Muse instrument (Luminex) and immediately frozen in 1mL/vial of cold freezing medium (90% heat-inactivated FBS, 10% DMSO. 0.22μm filtered). A cell content of ideally 12–15 million cells per vial (no less than 10 million per vial and not more than 20 million cells per vial) was targeted. Vials were quickly placed in a pre-chilled StrataCooler cell freezing box (Stratagene, cat#400005) and moved to −70°C for maximum three days, before transferring to ultralow temperature in Nitrogen vapor tanks. PBMC were shipped in Dry shipper-Nitrogen Vapors containers to avoid temperature excursions to ReiThera, and were further stored at ultralow temperature in Nitrogen vapor tanks until T cell analysis.
Main and exploratory SARS-CoV-2 serology
Spike, RBD and nucleocapsid serology
The main serology assessment by validated commercially available assays was run at Central lab (LabCorp-Geneva). Determination of Spike binding antibodies in serum from all participants at all visits was performed using COVID-SeroIndex, Kantaro Quantitative SARS-CoV-2 IgG Antibody Kit (R&D Systems) following manufacturer’s instruction. Data are expressed as arbitrary units (AU)/mL; LLOQ is 3,2 AU/mL and ULOQ is 160 AU/mL. Conversion into IgG concentrations expressed as binding antibody international units (BAU)/mL was calculated by multiplying AU/mL values by the conversion factor of 0,0235 and by the final serum dilution (i.e. 200), according to manufacturer’s indication. For antibodies to Nucleocapsid and RBD, the semiquantitative SARS-CoV-2 IgG and the SARS-CoV-2 IgG II Quant chemiluminescence microparticle assays (CMIA, Abbott Diagnostics, Chicago, IL, USA) were respectively used, all run on the ARCHITECT platform and according to manufacturer’s instructions. For N, data are expressed as index, whereby an index value >1.4 is considered positive; For RBD assay, data are expressed as arbitrary units (AU)/mL, and the positivity cutoff is 50 AU/ml. For quantitative analysis, the LOD is 6.8 AU/ml, the LOQ is 21 AU/ml, and the analytical Measuring Interval is 21–40.000 (extendable to 80.000). For conversion purpose, the mathematical relationship of the Abbott AU/mL unit to the WHO BAU/mL unit follow the equation: BAU/mL = 0.142× AU/mL. For plotting and calculations, all values below LOD/LOQ were reported as 1/2 LOD/LOQ.
SARS-CoV-2 plaque reduction neutralization test (PRNT)
The SARS-CoV-2 neutralizing antibody titers of vaccinated volunteers’ serum samples were determined by means of a plaque reduction neutralization test (PRNT) developed and run at Viroclinics Biosciences. Briefly, a standard number of SARS-CoV-2 (Bav/Pat1/2020 strain) infectious units were incubated with eight 2-fold serial dilutions of heat inactivated serum, starting from 1:8 and up to 1:1024. After a 1 h pre-incubation, the virus/serum mixtures were inoculated on Vero E6 cells (ATCC) for 1 h, than washed, replaced with infection medium and the cells were left overnight. After 16 to 24 h, the cells were formalin-fixed, permeabilized with ethanol, and incubated with primary anti-SARS-CoV-2 nucleocapsid monoclonal antibody (Sino Biological 40143-MM05 clone #05) followed by a secondary anti-mouse IgG horseradish peroxidase conjugate (Life Technologies A16072) and TrueBlue substrate (KPL 50-78-02), which forms a blue precipitate on virus-positive cells. Images of all wells were acquired by an ImmunoSpot analyzer (Cellular Technology Limited, CTL), equipped with software capable to accurately count the virus positive cells. The 50 and 80% neutralization titers (PRNT50 and PRNT80, or the reciprocal serum dilutions showing 50% or 80% infection reduction) were calculated according to a method described earlier.56 During assay validation, PRNT80 met the criteria for specificity, accuracy, precision, linearity, dilutional linearity and end of run analysis. The LLOQ, ULOQ and the assay range were set at PRNT80 21–3972. For plotting and calculations, all values below LLOQ were reported as 1/2 LLOQ. The LOD is not experimentally determined during validation as for the LLOQ, but it is represented by the lowest serum dilution tested (1:8).
Pseudotyped VSV neutralization assay
A subset serum samples from 200 participants at visit 36 (peak of antibody response) from both SD and RD cohorts was tested by a validated pseudotyped virus neutralisation assay (PNA) that assessed particle entry-inhibition, developed and run at Nexelis Laval (Canada). The assay is described in detail in an earlier publication.57 Briefly, pseudotyped virus particles containing a luciferase reporter were made from a modified vesicular stomatitis virus (VSVDG) backbone expressing the full-length S protein of SARS-CoV-2 from which the last 19 amino acids of the cytoplasmic tail were removed. For the purpose of this study, pseudotyped Spike were from either Wuhan-Hu-1 (D614-validated) or from Delta variant (B.1.617.2). Seven 2-fold serial dilutions of heat-inactivated serum samples were prepared and incubated at 37°C with 5% CO2 for 1h with pseudotyped virus at a predefined target working dilution. Serum-virus complexes were then transferred onto 96 well white flat-bottom plates (Corning), previously seeded overnight with Vero E6 cells and incubated at 37°C and 5% CO2 for 20 h. Following this incubation, luciferase substrate from ONE GloTM Ex luciferase assay system (Promega) was added to the cells. Plates were then read to quantify relative luminescence units (RLU), inversely proportional to the level of neutralising antibodies present in the serum. The neutralising titer of a serum sample was calculated as the reciprocal serum dilution corresponding to the 50% neutralisation antibody titer (NT50) for that sample; for Wuhan dataset, the NT50 titers were transformed to international units per mL (IU/mL), based on the WHO international standard for anti-SARS-CoV-2 immunoglobulin, using a conversion factor determined during assay validation (1/1872). The assay’s cut-off and LLOQ were 5.3 IU/mL (10 as NT50) and 5.9 IU/mL, respectively. For plotting and calculations, all values below LLOQ were reported as 1/2 LLOQ.
PhenoSense SARS CoV-2 nAb assay
A subset of 20 d36 serum samples were tested for measurement of nAb activity using the PhenoSense SARS CoV-2 nAb Assay (Monogram Biosciences, South San Francisco, CA), based on HIV-1 pseudovirions that express the SARS-CoV-2 spike protein. The pseudovirus is prepared by co-transfecting HEK293 producer cells with an HIV-1 genomic vector that contains a firefly luciferase reporter gene together with a SARS CoV-2 spike protein expression vector. For the purpose of this study, the pseudotyped Spike was from SARS-CoV-2 Wuhan strain (D614). Neutralizing antibody activity is measured by assessing the inhibition of luciferase activity in HEK293 target cells co-trasfected to express the ACE2 receptor and TMPRSS2 protease following pre-incubation of the pseudovirions with serial dilutions of the (previously heat-inactivated) serum specimen for 1 h at 37°C. Cell suspension and virus-serum mix are incubated 3 days, then Steady Glo (Promega) is added and RLUs are measured at a luminometer. Neutralizing antibody titers are reported as the reciprocal of the serum dilution of serum samples at which RLUs were reduced by 50% (ID50) compared to pseudovirus control wells. To ensure that the measured nAb activity is SARS CoV-2 nAb specific, each test specimen is also assessed using a non-specific pseudovirus (specificity control) that expresses a nonreactive envelope protein of an unrelated virus (avian influenza virus H10N3). The LLOQ, corresponding to the reciprocal of the serum starting dilution, is set at 40, while the ULOQ is established by the final serum dilution, corresponding to 787320.
Spike-specific T and B cell response
IFNγ ELISpot assay
The frequency of IFNγ-producing T cells was assessed by enzyme-linked immunosorbent assay (Human IFN-γ ELISpot plus kit; Mabtech) after specific stimulation. PBMCs were thawed quickly in 37°C water bath and collected in thawing medium [CTL Wash supplemented medium in RPMI-1640 (Sigma-Aldrich) and supplementing with L-glutamine (Gibco) and Benzonase (Merck) 50U/ml]. After one wash, cells were suspended into 50 mL polypropylene vented cap tubes with prewarmed medium [RPMI-1640 (Sigma-Aldrich) supplemented with 10% heat-inactivated highly defined FBS (Cytiva HyClone), 2 mmol/L L-glutamine, 10 mmol/L HEPES buffer (N-2-hydroxyethylpiperazine-N-2-ethane sulfonic acid, Sigma-Aldrich), 100 U/ml penicillin, and 100 μg/mL streptomycin (Gibco)], hereafter termed R10. Cells were rested at 2x106 cells/ml density in incubator at 37°C and 5% of CO2 for at least 16 h before plating.
PBMCs were then counted using Muse instrument (Luminex), plated in ELISpot assays at 2 × 105 cells/and stimulated with specific peptides pools. For main IFNγ ELISpot analysis at all three available study visits (d22, d36 and d180), cells were stimulated with 4 peptide pools (S1a, S1b-including RBD domain, S2a and S2b) covering the full-length Spike Wuhan protein.14 Furthermore, PBMCs at day 22 were tested using commercially available S1 and S2 peptide pools (JPT, Pepmix) of the Delta (Spike B.1.617.2) and Omicron (Spike B.1.1.52) variants, in comparison with peptide pools covering Wuhan reference spike from the same commercial source. Plates with cells and stimuli were incubated for 18–20 h at 37°C and 5% of CO2. At the end of incubation, the colorimetric assay was developed according to manufacturer’s instructions. Single color spots were counted at S6 ImmunoSpot Ultimate UV image analyzer (Cellular Technology Limited, CTL). Spontaneous cytokine production (background) was assessed by incubating PBMC with DMSO, the peptides diluent (Sigma). Results are expressed as spot forming cells (SFC)/106 PBMCs in stimulating cultures after subtracting spontaneous background, and then summing the counts for the four or two peptide pools to express total Spike response. Data from four volunteers (PL: 1020036 d36; 1020049 d22 and d36; RD: 1020025 d22 and d36; SD: 1020057 d36) were excluded from IFNγ ELISpot main analyses since their spontaneous IFNγ secretion in DMSO wells was above the mean+1standard deviation of the study population (mean = 32 SFC, SD = 76.37, mean+1SD = 109 SFC/million PBMC), leading to unreliable quantitative analysis.
Human IFNγ-IL5 fluorospot
Th1/Th2 response on day 36 frozen PBMCs was assessed using human IFNγ-IL5 Fluorospot (Mabtech). Cells were thawed and plated at 2,5x105 cells/well and stimulated with 4 peptide pools covering the full-length Spike Wuhan protein (S1a, S1b, S2a and S2b). Precoated plates were activated and blocked with R10 medium, and, after 18–20 h of cells and stimuli incubation at 37°C with 5% of CO2, the ELISpot assay was developed according to manufacturer’s instructions. Spot analysis was performed with S6 ImmunoSpot Ultimate UV image analyzer (Cellular Technology Limited, CTL) ELISpot reader equipped with filters for excitation 480 nm/emission 520 nm (FITC) for IFNγ detection and excitation 570 nm/emission 600 nm (Cy3) for IL5 detection. Results are expressed as spot forming cells (SFC)/106 PBMCs in stimulating cultures after subtracting spontaneous background.
Intracellular cytokines staining
PBMC were thawed and treated as described before for ELISpot assays. After ON resting and counting, cells were aliquoted into 5mL polypropylene conical snap-cap tubes (Grenier Bio-one, AU), 1x106 cells/tubes (200μL final volume) and stimulated with either DMSO as negative control, CEFX (20 μg/mL, JPT, GE) or SEB (0,1 mg/mL Sigma-Aldrich, USA, Mo) as positive controls, S1a+b pool or S2a+b pool (0,3 mg/mL) for 5 h at 37°C 5% CO2. 1μL of GolgiPlug (BD, USA, NJ) was added after 1h of incubation for intracellular cytokine accumulation.
After stimulation, cells were transferred in clean FACS tube, washed with cold PBS to stop the stimulation, and stained with anti-CD107a PE-CF594 clone (H4A3) and FVS780 (both BD) for 20′ at RT. Cells were washed with PBS 2% FBS 2mM EDTA, then fixed and permeabilized with BD Fixation/Permeabilization Kit (BD) following manufacturer instructions and stained intracellularly with CD3 BUV395 clone UCHT1, CD4 BUV737 clone SK3, CD8 BV605 clone SK1, CD154 BB700 clone TRAP1, IFNγ APC clone B27, IL-2 PE clone mQ1-17H12, TNFa BV650 clone Mab11, CD69 APC-R700 clone FN50 in the presence of Brilliant Stain Buffer (all from BD) for 30′ at 4°C. Cells were finally acquired at Fortessa LSR (BD).
Sample analysis was performed with FlowJo software (version 10.8.1, BD) following gating strategy depicted in Figure S6. Multi-functional cells were identified by combining each single-function-positive gate with Boolean tools available in FlowJo. For each donor, the percentage of cells responding to Spike peptides-pools were subtracted of their negative control. Polyfunctional analysis of CD4 and CD8 T cells were run using SPICE software (version 6.1, NIH, USA, MD).
Proliferation assay
PBMC were thawed as described before, counted after centrifugation and stained with 1 μL/sample of CellTrace Violet (Thermofisher, USA, MA). After staining, cells were washed extensively, resuspended in R10 medium and aliquoted into a 24-well plate at 2x106 cells/well (1mL final volume). Cells were then stimulated with either DMSO as negative control, SEB (0,1 mg/mL Sigma-Aldrich, USA, Mo) as positive control, S1a+b pool or S2a+b pool (0,3 mg/mL) or CEFX (1 μg/ml) for 5 days at 37°C 5% CO2. After stimulation, cells were harvested and transferred into new FACS tubes to be stained with CD3 BUV395 clone UCHT1, CD4 BUV737 clone SK3, CD8 BV605 clone SK1 (all from BD) and CD19 APC-Vio770 clone LT19 (Miltenyi) in the presence of Brilliant Stain Buffer (BD) for 30′ at 4°C. Sample analysis was performed with FlowJo software (version 10.8.1, BD) following gating strategy depicted in Figure S10. For each donor, the percentage of cells proliferating to Spike peptides-pools were subtracted of their negative control and summed to provide total Spike proliferative response.
Detection of spike-specific memory B cells
Detection of antigen-specific memory B cells was performed with the SARS-CoV-2 Spike B cell analysis kit (Miltenyi Biotec #130-128-022) at the Ospedale Pediatrico Bambino Gesù (Rome). Briefly, the kit allows the formation of tetramers made of a recombinant SARS-CoV-2 Spike-Protein (expressed in HEK)-Biotin with either Streptavidin-PE or Streptavidin-PE-Vio770. ∼5x106 previously frozen PBMC samples were prepared and stained with antibody staining mix containing the Spike tetramers, the 7-AAD and the fluorochrome-conjugated antibodies (CD19 APC-Vio770 clone LT19, CD27 Vio Bright FITC clone M-T271, CD24 BV711 clone ML5, IgA VioGreen clone IS11-8E10, IgM APC clone Pj2-22H3, IgG VioBlue clone IS11-3B2.2.3-all from Miltenyi; CD24 BV711 clone ML5 from BD). Memory B cells were identified as CD19+CD24+CD27+ and Spike-specific memory B cells were double-positive for PE and PE-Vio770 (Spike++). Samples were acquired on Fortessa LSR (BD) (BD) and analyzed using FlowJo (version 10.7.1, BD). Limit of detection (LOD) and limit of quantification (LOQ) were calculated as previously reported.58,59,60,61 Briefly, LOD was calculated as 20x100/total no. of events and LOQ was computed as 30x100/total no. of events. Gating strategy is illustrated in Figure S12.
Quantification and statistical analysis
All statistical analyses and data processing were performed using the SAS System software (release 9.4). The safety analyses included all participants who received at least one dose of GRAd-COV2 or placebo. The findings are descriptive in nature and not based on formal statistical hypothesis testing. Safety analyses are presented as counts, percentages, and associated Clopper–Pearson 95% confidence intervals for local reactions, systemic events, and any adverse events after vaccination, according to preferred terms in the Medical Dictionary for Regulatory Activities (MedDRA), version 24.0, for each group. The immunogenicity analysis set (IAS) included all participants in the safety analysis set who had immune response assessments and no protocol deviations judged to have a potential interference with the generation or interpretation of an immune response (SARS-CoV-2 infection or commercial COVID-19 vaccination). Participants were analyzed according to the treatment they actually received. Deviations which could interfere with the generation or interpretation of an immune response were reviewed case by case by the clinical team during the blind data review meeting. Participants who were seropositive to Nucleocapsid antigen at baseline were excluded by default from this analysis set. For subjects included in the IAS set, their time points were included in the immunogenicity analysis until the last immunogenicity assessment before occurrence of the deviation (subject received national COVID-19 vaccine, subject with intercurrent laboratory confirmed SARS-CoV-2 infection or asymptomatic subject seroconverting to Anti-N positive). Number of cases and geometric mean (GM) with its 95% confidence interval were calculated for anti-S ELISA, SARS-CoV-2 neutralization NT50 and NT80. The 95% CIs for geometric means were calculated based on the t-distribution of the natural log-transformed values than back transformed to the original scales for presentation. Comparisons among treatment groups were performed by means of the analysis of variance for repeated measures on natural log-transformed values of immunogenicity outcomes where treatment group, study day, treatment group by study day interaction as fixed effects, with adjustment for protocol stratification factor.
Additional resources
The COVITAR study was registered on ClinicalTrials.gov (NCT04791423), and on EU Clinical trials Register (www.clinicaltrialsregister.eu, EudraCT 2020-005915-39).
Acknowledgments
ReiThera srl manufactured the vaccine, funded and sponsored the trial, and coordinated interactions with contract Clinical Research Organization (CRO) Exom staff and regulatory authorities. The CRO took charge of trial operation to meet the required standards of the International Council for Harmonization of Technical Requirements for Pharmaceuticals for Human Use and GCP guidelines. We are grateful to all participants who volunteered for this study and the members of the COVITAR data and safety monitoring board and steering committee for their dedication and their diligent review of the data. We also acknowledge the contribution of Antonella Bacchieri (statistician consultant) for data interpretation; the ReiThera GMP, QC, and QA staff for manufacturing and release of the GRAd-COV2 clinical lot, and all ReiThera colleagues not named here who contributed to the success of this trial. We also thank Abbott for supplying the SARS-CoV-2 IgG II Quant assay kits for the study. Pseudovirus neutralization assays performed by Nexelis was supported by the Global Health Discovery Collaboratory Platforms of the Bill & Melinda Gates Foundation. The graphical abstract was created with BioRender.com.
Author contributions
R. Camerini, S. Capone, A.F., and S. Lanini conceived and designed the trial and developed the study protocol; F.M.F., S.M., S. Borrè, S. Carbonara, S.L.C., S. Leone, G.G., P.M., A.C., M. Lichtner, R. Cauda, S.D.Z., M.V.C., A.G., S.R., P.C., S. Bonora, G.M., M.C., I.M., S. Capici, E.P., M. Libanore, A.D., and S. Lanini were study site principal investigators; S. Capone, R. Carsetti, E.P.M., S. Battella, and A.M.C. were responsible for laboratory testing and assay development; C.I. performed the statistical analyses; R.D., F.G., G.P., S. Colloca, and L.V. contributed to the implementation of the study; R. Camerini; S. Capone, R. Carsetti, L.V., and S. Lanini had full access to and verified all the data in the study and take responsibility for the integrity and accuracy of the data analysis.; R. Camerini, S. Capone, and A.F. wrote the manuscript; all authors critically reviewed and approved the final version.
Declaration of interests
S. Capone, R. Camerini, R.D., F.G., S. Battella, A.M.C., G.P., S. Colloca, and A.F. are full employees of ReiThera Srl. S. Colloca and A.F. are founders and shareholders of Keires AG. S. Colloca is named inventor of the patent application no. 20183515.4 titled “GORILLA ADENOVIRUS NUCLEIC ACID- AND AMINO ACID-SEQUENCES, VECTORS CONTAINING SAME, AND USES THEREOF.” L.V. is full employee of Exom, the CRO in charge of the COVITAR study management. S.L.C. received honoraria from Gilead, ViiV, GSK, Janssen, and MSD, has participated on the advisory boards of Gilead, ViiV, GSK, Janssen, and MSD, and has received support for attending meetings from Gilead. M. Lichtner received honoraria and support for attending meetings from Gilead, MSD, and ViiV, participated on the advisory boards of ViiV, Abbvie, and MSD, and received grants through the institution from Gilead and Abbvie. R. Carsetti was a member of the COVITAR study steering committee. C.I. received financial support from Exom for statistical analysis of the COVITAR study.
Published: May 29, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2023.101084.
Contributor Information
Stefania Capone, Email: stefania.capone@reithera.com.
COVITAR study group:
Luigi Ziviani, Feliciana Malescio, Irene Turrini, Rita Lawlor, Annamaria Romano, Mariagrazia Nunziata, Salvatore Armato, Nicole Mazzeo, Maria Aurora Carleo, Chiara Dell’Isola, Raffaella Pisapia, Agostina Pontarelli, Andrea Olivani, Sara Grasselli, Diletta Laccabue, Maria Cristina Leoni, Franco Paolillo, Annalisa Mancini, Barbara Ruaro, Marco Confalonieri, Francesco Salton, Giulia Mancarella, Raffaella Marocco, Margherita De Masi, Valeria Belvisi, Silvia Lamonica, Antonella Cingolani, Cristina Seguiti, Paola Brambilla, Alice Ferraresi, Matteo Lupi, Serena Ludovisi, Giulia Renisi, Roberta Massafra, Martina Pellicciotta, Luciana Armiento, Stefania Vimercati, Mariagrazia Piacenza, Paolo Bonfanti, Paola Columpsi, Marina Elena Cazzaniga, Cristina Rovelli, Mariaelena Ceresini, Letizia Previtali, Laura Trentini, Chiara Alcantarini, Walter Rugge, Stefano Biffi, Federica Poletti, Roberto Rostagno, Roberta Moglia, Ferdinando De Negri, Elisabetta Fini, Alice Cangialosi, Serena Rita Bruno, Marianna Rizzo, Mariangela Niglio, Anna Dello Stritto, Alfredo Matano, Arnolfo Petruzziello, Pietro Valsecchi, Teresa Pieri, Mauro Altamura, Angela Calamo, Anna Giannelli, Stefania Menolascina, Silvia Di Bari, Vera Mauro, Raissa Aronica, Daniela Segala, Rosario Cultrera, Laura Sighinolfi, Michelle Abbott, Andrea Gizzi, Federica Guida Marascia, Giacomo Valenti, Marcello Feasi, Nicoletta Bobbio, Filippo Del Puente, Alfredo Nicosia, Martina Frascà, Miriam Mazzoleni, Nadia Garofalo, Virginia Ammendola, Fabiana Grazioli, Federico Napolitano, Alessandra Vitelli, and Valentina Marcellini
Supplemental information
Data and code availability
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyse the data reported in this paper is available from the lead contact upon request.
References
- 1.WHO https://www.who.int/publications/i/item/WHO-2019-nCoV-Vaccines-SAGE-Prioritization-2022.1
- 2.Medicine L.S.o.H.T. https://vac-lshtm.shinyapps.io/ncov_vaccine_landscape/
- 3.Dejnirattisai W., Huo J., Zhou D., Zahradník J., Supasa P., Liu C., Duyvesteyn H.M.E., Ginn H.M., Mentzer A.J., Tuekprakhon A., et al. SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell. 2022;185:467–484.e15. doi: 10.1016/j.cell.2021.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Willett B.J., Grove J., MacLean O.A., Wilkie C., De Lorenzo G., Furnon W., Cantoni D., Scott S., Logan N., Ashraf S., et al. SARS-CoV-2 Omicron is an immune escape variant with an altered cell entry pathway. Nat. Microbiol. 2022;7:1161–1179. doi: 10.1038/s41564-022-01143-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kudriavtsev A.V., Vakhrusheva A.V., Novossmall ie C.V.N., Bozdaganyan M.E., Shaitan K.V., Kirpichnikov M.P., Sokolova O.S. Immune escape associated with RBD omicron mutations and SARS-CoV-2 evolution dynamics. Viruses. 2022;14:1603. doi: 10.3390/v14081603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simon-Loriere E., Schwartz O. Towards SARS-CoV-2 serotypes? Nat. Rev. Microbiol. 2022;20:187–188. doi: 10.1038/s41579-022-00708-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corbett K.S., Gagne M., Wagner D.A., O' Connell S., Narpala S.R., Flebbe D.R., Andrew S.F., Davis R.L., Flynn B., Johnston T.S., et al. Protection against SARS-CoV-2 Beta variant in mRNA-1273 vaccine-boosted nonhuman primates. Science. 2021;374:1343–1353. doi: 10.1126/science.abl8912. [DOI] [PubMed] [Google Scholar]
- 8.Gagne M., Moliva J.I., Foulds K.E., Andrew S.F., Flynn B.J., Werner A.P., Wagner D.A., Teng I.T., Lin B.C., Moore C., et al. mRNA-1273 or mRNA-Omicron boost in vaccinated macaques elicits similar B cell expansion, neutralizing responses, and protection from Omicron. Cell. 2022;185:1556–1571.e18. doi: 10.1016/j.cell.2022.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suryawanshi R.K., Chen I.P., Ma T., Syed A.M., Brazer N., Saldhi P., Simoneau C.R., Ciling A., Khalid M.M., Sreekumar B., et al. Limited cross-variant immunity from SARS-CoV-2 Omicron without vaccination. Nature. 2022;607:351–355. doi: 10.1038/s41586-022-04865-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ying B., Whitener B., VanBlargan L.A., Hassan A.O., Shrihari S., Liang C.Y., Karl C.E., Mackin S., Chen R.E., Kafai N.M., et al. Protective activity of mRNA vaccines against ancestral and variant SARS-CoV-2 strains. Sci. Transl. Med. 2022;14:eabm3302. doi: 10.1126/scitranslmed.abm3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chalkias S., Harper C., Vrbicky K., Walsh S.R., Essink B., Brosz A., McGhee N., Tomassini J.E., Chen X., Chang Y., et al. A bivalent omicron-containing booster vaccine against covid-19. N. Engl. J. Med. 2022;387:1279–1291. doi: 10.1056/NEJMoa2208343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Capone S., Raggioli A., Gentile M., Battella S., Lahm A., Sommella A., Contino A.M., Urbanowicz R.A., Scala R., Barra F., et al. Immunogenicity of a new gorilla adenovirus vaccine candidate for COVID-19. Mol. Ther. 2021;29:2412–2423. doi: 10.1016/j.ymthe.2021.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Agrati C., Castilletti C., Battella S., Cimini E., Matusali G., Sommella A., Sacchi A., Colavita F., Contino A.M., Bordoni V., et al. Safety and immune response kinetics of GRAd-COV2 vaccine: phase 1 clinical trial results. NPJ Vaccines. 2022;7:111. doi: 10.1038/s41541-022-00531-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lanini S., Capone S., Antinori A., Milleri S., Nicastri E., Camerini R., Agrati C., Castilletti C., Mori F., Sacchi A., et al. GRAd-COV2, a gorilla adenovirus-based candidate vaccine against COVID-19, is safe and immunogenic in younger and older adults. Sci. Transl. Med. 2022;14:eabj1996. doi: 10.1126/scitranslmed.abj1996. [DOI] [PubMed] [Google Scholar]
- 15.Ramasamy M.N., Minassian A.M., Ewer K.J., Flaxman A.L., Folegatti P.M., Owens D.R., Voysey M., Aley P.K., Angus B., Babbage G., et al. Safety and immunogenicity of ChAdOx1 nCoV-19 vaccine administered in a prime-boost regimen in young and old adults (COV002): a single-blind, randomised, controlled, phase 2/3 trial. Lancet. 2021;396:1979–1993. doi: 10.1016/S0140-6736(20)32466-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frater J., Ewer K.J., Ogbe A., Pace M., Adele S., Adland E., Alagaratnam J., Aley P.K., Ali M., Ansari M.A., et al. Safety and immunogenicity of the ChAdOx1 nCoV-19 (AZD1222) vaccine against SARS-CoV-2 in HIV infection: a single-arm substudy of a phase 2/3 clinical trial. Lancet. HIV. 2021;8:e474–e485. doi: 10.1016/S2352-3018(21)00103-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.FDA. https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-limits-use-janssen-covid-19-vaccine-certain-individuals#:∼:text=Today,%20the%20U.S.%20Food%20and,who%20elect%20to%20receive%20the.
- 18.Chan B., Odutayo A., Juni P., Stall N.M., Bobos P., Brown A.D., Grill A., Ivers N., Maltsev A., McGeer A., et al. Science Briefs of the Ontario COVID-19 Science Advisory Table. 2021. Risk of vaccine-induced thrombotic thrombocytopenia (VITT) following the AstraZeneca/COVISHIELD adenovirus vector COVID-19 vaccines. [DOI] [Google Scholar]
- 19.Atmar R.L., Lyke K.E., Deming M.E., Jackson L.A., Branche A.R., El Sahly H.M., Rostad C.A., Martin J.M., Johnston C., Rupp R.E., et al. Homologous and heterologous covid-19 booster vaccinations. N. Engl. J. Med. 2022;386:1046–1057. doi: 10.1056/NEJMoa2116414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Borobia A.M., Carcas A.J., Pérez-Olmeda M., Castaño L., Bertran M.J., García-Pérez J., Campins M., Portolés A., González-Pérez M., García Morales M.T., et al. Immunogenicity and reactogenicity of BNT162b2 booster in ChAdOx1-S-primed participants (CombiVacS): a multicentre, open-label, randomised, controlled, phase 2 trial. Lancet. 2021;398:121–130. doi: 10.1016/S0140-6736(21)01420-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pozzetto B., Legros V., Djebali S., Barateau V., Guibert N., Villard M., Peyrot L., Allatif O., Fassier J.B., Massardier-Pilonchéry A., et al. Immunogenicity and efficacy of heterologous ChAdOx1-BNT162b2 vaccination. Nature. 2021;600:701–706. doi: 10.1038/s41586-021-04120-y. [DOI] [PubMed] [Google Scholar]
- 22.Sablerolles R.S.G., Rietdijk W.J.R., Goorhuis A., Postma D.F., Visser L.G., Geers D., Schmitz K.S., Garcia Garrido H.M., Koopmans M.P.G., Dalm V.A.S.H., et al. Immunogenicity and reactogenicity of vaccine boosters after Ad26.COV2.S priming. N. Engl. J. Med. 2022;386:951–963. doi: 10.1056/NEJMoa2116747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmidt T., Klemis V., Schub D., Mihm J., Hielscher F., Marx S., Abu-Omar A., Ziegler L., Guckelmus C., Urschel R., et al. Immunogenicity and reactogenicity of heterologous ChAdOx1 nCoV-19/mRNA vaccination. Nat. Med. 2021;27:1530–1535. doi: 10.1038/s41591-021-01464-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng S., Phillips D.J., White T., Sayal H., Aley P.K., Bibi S., Dold C., Fuskova M., Gilbert S.C., Hirsch I., et al. Correlates of protection against symptomatic and asymptomatic SARS-CoV-2 infection. Nat. Med. 2021;27:2032–2040. doi: 10.1038/s41591-021-01540-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gilbert P.B., Montefiori D.C., McDermott A.B., Fong Y., Benkeser D., Deng W., Zhou H., Houchens C.R., Martins K., Jayashankar L., et al. Immune correlates analysis of the mRNA-1273 COVID-19 vaccine efficacy clinical trial. Science. 2022;375:43–50. doi: 10.1126/science.abm3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldblatt D., Fiore-Gartland A., Johnson M., Hunt A., Bengt C., Zavadska D., Snipe H.D., Brown J.S., Workman L., Zar H.J., et al. Towards a population-based threshold of protection for COVID-19 vaccines. Vaccine. 2022;40:306–315. doi: 10.1016/j.vaccine.2021.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chao C.H., Cheng D., Huang S.W., Chuang Y.C., Yeh T.M., Wang J.R. Serological responses triggered by different SARS-CoV-2 vaccines against SARS-CoV-2 variants in Taiwan. Front. Immunol. 2022;13:1023943. doi: 10.3389/fimmu.2022.1023943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karbiener M., Farcet M.R., Zollner A., Masuda T., Mori M., Moschen A.R., Kreil T.R. Calibrated comparison of SARS-CoV-2 neutralizing antibody levels in response to protein-mRNA-and vector-based COVID-19 vaccines. NPJ Vaccines. 2022;7:22. doi: 10.1038/s41541-022-00455-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Z., Mateus J., Coelho C.H., Dan J.M., Moderbacher C.R., Gálvez R.I., Cortes F.H., Grifoni A., Tarke A., Chang J., et al. Humoral and cellular immune memory to four COVID-19 vaccines. Cell. 2022;185:2434–2451.e17. doi: 10.1016/j.cell.2022.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flaxman A., Marchevsky N.G., Jenkin D., Aboagye J., Aley P.K., Angus B., Belij-Rammerstorfer S., Bibi S., Bittaye M., Cappuccini F., et al. Reactogenicity and immunogenicity after a late second dose or a third dose of ChAdOx1 nCoV-19 in the UK: a substudy of two randomised controlled trials (COV001 and COV002) Lancet. 2021;398:981–990. doi: 10.1016/S0140-6736(21)01699-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hall V.G., Ferreira V.H., Wood H., Ierullo M., Majchrzak-Kita B., Manguiat K., Robinson A., Kulasingam V., Humar A., Kumar D. Delayed-interval BNT162b2 mRNA COVID-19 vaccination enhances humoral immunity and induces robust T cell responses. Nat. Immunol. 2022;23:380–385. doi: 10.1038/s41590-021-01126-6. [DOI] [PubMed] [Google Scholar]
- 32.Payne R.P., Longet S., Austin J.A., Skelly D.T., Dejnirattisai W., Adele S., Meardon N., Faustini S., Al-Taei S., Moore S.C., et al. Immunogenicity of standard and extended dosing intervals of BNT162b2 mRNA vaccine. Cell. 2021;184:5699–5714.e11. doi: 10.1016/j.cell.2021.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sadoff J., Le Gars M., Brandenburg B., Cárdenas V., Shukarev G., Vaissiere N., Heerwegh D., Truyers C., de Groot A.M., Jongeneelen M., et al. Durable antibody responses elicited by 1 dose of Ad26.COV2.S and substantial increase after boosting: 2 randomized clinical trials. Vaccine. 2022;40:4403–4411. doi: 10.1016/j.vaccine.2022.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sadoff J., Le Gars M., Shukarev G., Heerwegh D., Truyers C., de Groot A.M., Stoop J., Tete S., Van Damme W., Leroux-Roels I., et al. Interim results of a phase 1-2a trial of Ad26.COV2.S covid-19 vaccine. N. Engl. J. Med. 2021;384:1824–1835. doi: 10.1056/NEJMoa2034201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Voysey M., Costa Clemens S.A., Madhi S.A., Weckx L.Y., Folegatti P.M., Aley P.K., Angus B., Baillie V.L., Barnabas S.L., Bhorat Q.E., et al. Single-dose administration and the influence of the timing of the booster dose on immunogenicity and efficacy of ChAdOx1 nCoV-19 (AZD1222) vaccine: a pooled analysis of four randomised trials. Lancet. 2021;397:881–891. doi: 10.1016/S0140-6736(21)00432-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alter G., Yu J., Liu J., Chandrashekar A., Borducchi E.N., Tostanoski L.H., McMahan K., Jacob-Dolan C., Martinez D.R., Chang A., et al. Immunogenicity of Ad26.COV2.S vaccine against SARS-CoV-2 variants in humans. Nature. 2021;596:268–272. doi: 10.1038/s41586-021-03681-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tarke A., Coelho C.H., Zhang Z., Dan J.M., Yu E.D., Methot N., Bloom N.I., Goodwin B., Phillips E., Mallal S., et al. SARS-CoV-2 vaccination induces immunological T cell memory able to cross-recognize variants from Alpha to Omicron. Cell. 2022;185:847–859.e11. doi: 10.1016/j.cell.2022.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hirai T., Yoshioka Y. Considerations of CD8+ T cells for optimized vaccine strategies against respiratory viruses. Front. Immunol. 2022;13:918611. doi: 10.3389/fimmu.2022.918611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wherry E.J., Barouch D.H. T cell immunity to COVID-19 vaccines. Science. 2022;377:821–822. doi: 10.1126/science.add2897. [DOI] [PubMed] [Google Scholar]
- 40.Barouch D.H. Covid-19 vaccines — immunity, variants, boosters. N. Engl. J. Med. 2022;387:1011–1020. doi: 10.1056/NEJMra2206573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swanson P.A., 2nd, Padilla M., Hoyland W., McGlinchey K., Fields P.A., Bibi S., Faust S.N., McDermott A.B., Lambe T., Pollard A.J., et al. AZD1222/ChAdOx1 nCoV-19 vaccination induces a polyfunctional spike protein-specific T(H)1 response with a diverse TCR repertoire. Sci. Transl. Med. 2021;13:eabj7211. doi: 10.1126/scitranslmed.abj7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu F.C., Li Y.H., Guan X.H., Hou L.H., Wang W.J., Li J.X., Wu S.P., Wang B.S., Wang Z., Wang L., et al. Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: a dose-escalation, open-label, non-randomised, first-in-human trial. Lancet. 2020;395:1845–1854. doi: 10.1016/S0140-6736(20)31208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Colloca S., Barnes E., Folgori A., Ammendola V., Capone S., Cirillo A., Siani L., Naddeo M., Grazioli F., Esposito M.L., et al. Vaccine vectors derived from a large collection of simian adenoviruses induce potent cellular immunity across multiple species. Sci. Transl. Med. 2012;4:115ra2. doi: 10.1126/scitranslmed.3002925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ng K.W., Faulkner N., Cornish G.H., Rosa A., Harvey R., Hussain S., Ulferts R., Earl C., Wrobel A.G., Benton D.J., et al. Preexisting and de novo humoral immunity to SARS-CoV-2 in humans. Science. 2020;370:1339–1343. doi: 10.1126/science.abe1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cho A., Muecksch F., Schaefer-Babajew D., Wang Z., Finkin S., Gaebler C., Ramos V., Cipolla M., Mendoza P., Agudelo M., et al. Anti-SARS-CoV-2 receptor-binding domain antibody evolution after mRNA vaccination. Nature. 2021;600:517–522. doi: 10.1038/s41586-021-04060-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Piano Mortari E., Russo C., Vinci M.R., Terreri S., Fernandez Salinas A., Piccioni L., Alteri C., Colagrossi L., Coltella L., Ranno S., et al. Highly specific memory B cells generation after the 2nd dose of BNT162b2 vaccine compensate for the decline of serum antibodies and absence of mucosal IgA. Cells. 2021;10:2541. doi: 10.3390/cells10102541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stuart A.S.V., Shaw R.H., Liu X., Greenland M., Aley P.K., Andrews N.J., Cameron J.C., Charlton S., Clutterbuck E.A., Collins A.M., et al. Immunogenicity, safety, and reactogenicity of heterologous COVID-19 primary vaccination incorporating mRNA, viral-vector, and protein-adjuvant vaccines in the UK (Com-COV2): a single-blind, randomised, phase 2, non-inferiority trial. Lancet. 2022;399:36–49. doi: 10.1016/S0140-6736(21)02718-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Agrati C., Capone S., Castilletti C., Cimini E., Matusali G., Meschi S., Tartaglia E., Camerini R., Lanini S., Milleri S., et al. Strong immunogenicity of heterologous prime-boost immunizations with the experimental vaccine GRAd-COV2 and BNT162b2 or ChAdOx1-nCOV19. NPJ Vaccines. 2021;6:131. doi: 10.1038/s41541-021-00394-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu X., Munro A.P.S., Feng S., Janani L., Aley P.K., Babbage G., Baxter D., Bula M., Cathie K., Chatterjee K., et al. Persistence of immunogenicity after seven COVID-19 vaccines given as third dose boosters following two doses of ChAdOx1 nCov-19 or BNT162b2 in the UK: three month analyses of the COV-BOOST trial. J. Infect. 2022;84:795–813. doi: 10.1016/j.jinf.2022.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tan C.S., Collier A.R.Y., Yu J., Liu J., Chandrashekar A., McMahan K., Jacob-Dolan C., He X., Roy V., Hauser B.M., et al. Durability of heterologous and homologous COVID-19 vaccine boosts. JAMA Netw. Open. 2022;5:e2226335. doi: 10.1001/jamanetworkopen.2022.26335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.King R.G., Silva-Sanchez A., Peel J.N., Botta D., Dickson A.M., Pinto A.K., Meza-Perez S., Allie S.R., Schultz M.D., Liu M., et al. Single-dose intranasal administration of AdCOVID elicits systemic and mucosal immunity against SARS-CoV-2 and fully protects mice from lethal challenge. Vaccines. 2021;9:881. doi: 10.3390/vaccines9080881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu S., Huang J., Zhang Z., Wu J., Zhang J., Hu H., Zhu T., Zhang J., Luo L., Fan P., et al. Safety, tolerability, and immunogenicity of an aerosolised adenovirus type-5 vector-based COVID-19 vaccine (Ad5-nCoV) in adults: preliminary report of an open-label and randomised phase 1 clinical trial. Lancet Infect. Dis. 2021;21:1654–1664. doi: 10.1016/S1473-3099(21)00396-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Topol E.J., Iwasaki A. Operation nasal vaccine—lightning speed to counter COVID-19. Science Immunology. 2022;7:eadd9947. doi: 10.1126/sciimmunol.add9947. [DOI] [PubMed] [Google Scholar]
- 54.Pallesen J., Wang N., Corbett K.S., Wrapp D., Kirchdoerfer R.N., Turner H.L., Cottrell C.A., Becker M.M., Wang L., Shi W., et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. USA. 2017;114:E7348–E7357. doi: 10.1073/pnas.1707304114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zielinska E., Liu D., Wu H.Y., Quiroz J., Rappaport R., Yang D.P. Development of an improved microneutralization assay for respiratory syncytial virus by automated plaque counting using imaging analysis. Virol. J. 2005;2:84. doi: 10.1186/1743-422X-2-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bewley K.R., Coombes N.S., Gagnon L., McInroy L., Baker N., Shaik I., St-Jean J.R., St-Amant N., Buttigieg K.R., Humphries H.E., et al. Quantification of SARS-CoV-2 neutralizing antibody by wild-type plaque reduction neutralization, microneutralization and pseudotyped virus neutralization assays. Nat. Protoc. 2021;16:3114–3140. doi: 10.1038/s41596-021-00536-y. [DOI] [PubMed] [Google Scholar]
- 58.Fernandez Salinas A., Piano Mortari E., Terreri S., Milito C., Zaffina S., Perno C.F., Locatelli F., Quinti I., Carsetti R. Impaired memory B-cell response to the Pfizer-BioNTech COVID-19 vaccine in patients with common variable immunodeficiency. J. Allergy Clin. Immunol. 2022;149:76–77. doi: 10.1016/j.jaci.2021.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Illingworth A., Marinov I., Sutherland D.R., Wagner-Ballon O., DelVecchio L. ICCS/ESCCA consensus guidelines to detect GPI-deficient cells in paroxysmal nocturnal hemoglobinuria (PNH) and related disorders part 3 - data analysis, reporting and case studies. Cytometry B Clin. Cytom. 2018;94:49–66. doi: 10.1002/cyto.b.21609. [DOI] [PubMed] [Google Scholar]
- 60.Palmieri R., Piciocchi A., Arena V., Maurillo L., Del Principe M.I., Paterno G., Ottone T., Divona M., Lavorgna S., Irno Consalvo M., et al. Clinical relevance of- limit of detection (LOD) - limit of quantification (LOQ) - based flow cytometry approach for measurable residual disease (MRD) assessment in acute myeloid leukemia (AML) Blood. 2020;136:37–38. doi: 10.1182/blood-2020-139557. [DOI] [Google Scholar]
- 61.Xiao W., Salem D., McCoy C.S., Lee D., Shah N.N., Stetler-Stevenson M., Yuan C.M. Early recovery of circulating immature B cells in B-lymphoblastic leukemia patients after CD19 targeted CAR T cell therapy: a pitfall for minimal residual disease detection. Cytometry B Clin. Cytom. 2018;94:434–443. doi: 10.1002/cyto.b.21591. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyse the data reported in this paper is available from the lead contact upon request.