Abstract
DExH/D proteins are essential for all aspects of cellular RNA metabolism and processing, in the replication of many viruses and in DNA replication. DExH/D proteins are subject to current biological, biochemical and biophysical research which provides a continuous wealth of data. The DExH/D protein family database compiles this information and makes it available over the WWW (http://www.columbia.edu/~ej67/dbhome.htm ). The database can be fully searched by text based queries, facilitating fast access to specific information about this important class of enzymes.
BACKGROUND
DExH/D proteins are essential for all aspects of cellular RNA metabolism and processing, for the replication of many viruses and for DNA replication (1,2).
The DExH/D family comprises proteins from the DEAD, DEAH and DExH subgroups (3), all of which contain at least eight characteristic sequence motifs (Fig. 1) including the ATP-hydrolysis motif II from which they derive their names: DEAD, DEAH and DExH, in single-letter amino acid code (4–6). The characteristic sequence motifs are conserved from bacteria to man, underscoring the fact that DExH/D proteins represent a critical biochemical capability of living organisms (7). DExH/D proteins are a subset of the SF2 helicase family, which is related to the SF1 helicase family (5).
Figure 1.
Characteristic sequence motifs of DEAH, DEAD and DExH proteins. Within each group, letters in gray blocks represent identical amino acids, regular letters represent conservative substitutions, and x represents variable residues. The points separating the conserved motifs do not reflect spacings between the motifs, however, aligned amino acids from the different groups have comparable distance. The indicated characteristic motifs are usually surrounded by clusters of less, but still significantly conserved residues which are not indicated here. Similarity within the DEAH-group continues throughout the C-terminus. Characteristic motifs were identified from alignments of 29 DEAH, 42 DEAD and 44 DExH proteins.
All biochemically characterized DExH/D proteins possess nucleoside triphosphatase activity which, in most cases, is stimulated by RNA or DNA. Several DExH proteins are well characterized DNA helicases such as RecQ and the respective homologs in other organisms (8). However, the vast majority of DExH/D proteins are implicated in RNA-related processes. At least 26 members of the DExH/D protein family have been shown to be RNA helicases that unwind RNA duplexes in an NTP-dependent fashion in vitro. Therefore, it is hypothesized that DExH/D proteins involved in RNA metabolism play key roles in the coupling of NTP hydrolysis to RNA conformational changes in macromolecular assemblies such as the spliceosome, the degradosome or viral replication machineries (2,3,9,10).
Because DExH/D proteins are essential in numerous fundamental biological processes, these proteins are subject to intensive ongoing research in various disciplines such as biochemistry, genetics and biophysics. Therefore, available information is growing fast and, due to the quantity and variety of new data, it is difficult to maintain a comprehensive overview of the field. The purpose of the DExH/D protein family database is to compile all available information, to make it freely available over the WWW, and to facilitate convenient access to specific information by search functions.
DESCRIPTION OF THE DATABASE
Proteins of the DEAH, DEAD and DExH group have defined characteristic sequence motifs (1,2,7) that are used to identify proteins listed in the database (Fig. 1).
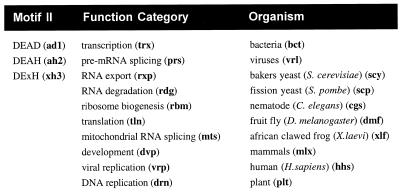
The database is searchable employing two different strategies. First, it can be searched for a protein/gene name (protein by name search). Therefore, all proteins/genes that are included in the database are compiled in one table and linked to the individual protein/gene pages where information is located. Second, a text-based string search can be performed searching any text within the database. Logical queries can be built using Boolean operators. Moreover, tailored towards the retrieval of specific information a ‘category search’ is possible. To facilitate a ‘category search’ a three-letter key has been assigned to each category (Fig. 2). This key must be entered to retrieve the protein pages in the respective category. Keys can be combined in a logical query using Boolean operators (for example: selecting translation AND yeast would return all proteins/genes that are involved in translation in yeast; more examples are given on the search page). The categories are divided into three different topics: (i) the motif II characteristics which reflect the subdivision of the DExH/D protein family into DEAD, DExH, DEAH subgroups (6); (ii) the organism from which a protein/gene is derived; (iii) a biological function category (Fig. 2). Assignment of the function categories is based on fundamental cellular processes in which DExH/D proteins are involved. However, one notable exception has been made, the development category. This category has been included because a considerable number of proteins have been found to be involved in development, implying potential interest in selectively retrieving information about this category.
Figure 2.
Categories and corresponding keys to facilitate specific category search. Combination of several categories in logical queries is possible using Boolean operators (; /, / { / }).
Search results are returned as by a common web-search engine, providing links to individual protein/gene pages. On these pages, the available information about the respective protein/gene is compiled. Each protein/gene that was the subject of at least one publication is assigned a web page that provides information in several sections. The first two sections give common protein/gene names and the organism of origin. In the next section, sequence information is supplied, including the motif II characteristics and a GenBank link to retrieve the sequence. Then the function category is given (Fig. 2). In section 4 biochemical activities are summarized, such as characteristics of helicase and ATPase activities and, where available, further mechanistical information is given. In section 5 biological functions are described, including available genetic data. Results of mutational analysis are given either in the biochemical or biological section, depending on which assays were used to characterize the mutants. The next section provides links to homologs within the database. Homologies are mainly based on information given in the literature. The last two sections contain respective literature for the protein/gene and links to other databases where further and complementary information can be obtained.
For selected organisms, proteins/genes that have not yet been described in a publication are compiled providing links to other database entries. This information can be accessed from the page listing the database entries. Moreover, the database features a short introduction to the field of DExH/D proteins as well as links to other relevant databases and lab-homepages.
ACCESS
The DExH/D protein database is available on the WWW at http://www.columbia.edu/~ej67/dbhome.htm . Although the database can be navigated and all information can be accurately viewed with text-only browsers, users are encouraged to employ browsers with JavaScript capability in order to take advantage of convenient graphic navigation and clear arrangement of information. Please cite this article when the DExH/D protein family database assists in published research.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Anna M. Pyle for comments on the manuscript as well as for continuous help and invaluable discussions. E.J. was supported by the Curt-Engelhorn postdoctoral fellowship from the German Cancer Research Center.
REFERENCES
- 1.de la Cruz J., Kressler,D. and Linder,P. (1999) Trends Biochem. Sci., 24, 192–198. [DOI] [PubMed] [Google Scholar]
- 2.Kadare G. and Haenni,A.L. (1997) J. Virol., 71, 2583–2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Staley J.P. and Guthrie,C. (1998) Cell, 90, 1041–1050. [Google Scholar]
- 4.Linder P., Lasko,P.F., Ashburner,M., Leroy,P., Nielsen,P.P., Nishi,K., Schier,J. and Slonimski,P.P. (1989) Nature, 337, 121–122. [DOI] [PubMed] [Google Scholar]
- 5.Gorbalenya A.E. and Koonin,E.V. (1993) Curr. Opin. Struct. Biol., 3, 419–429. [Google Scholar]
- 6.Fuller-Pace F.V. (1994) Trends Cell Biol., 4, 271–274. [DOI] [PubMed] [Google Scholar]
- 7.Schmid S.R. and Linder,P. (1992) Mol. Microbiol., 6, 283–291. [DOI] [PubMed] [Google Scholar]
- 8.Chakraverty R.K. and Hickson,I.D. (1999) Bioassays, 21, 286–294. [DOI] [PubMed] [Google Scholar]
- 9.Anderson J.S.J. and Parker,R. (1996) Curr. Biol., 6, 780–782. [DOI] [PubMed] [Google Scholar]
- 10.Wagner J.D.O., Jankowsky,E., Company,M., Pyle,A.M. and Abelson,J.N. (1998) EMBO J., 17, 2926–2937. [DOI] [PMC free article] [PubMed] [Google Scholar]